Abstract
A site-specifically modified oligonucleotide containing a single 2′-deoxyribonolactone lesion was used as a template for primer extension reactions catalyzed by M-MuLV reverse transcriptase (RT) and by the Klenow fragments of Escherichia coli DNA polymerase proficient (KF exo+) or deficient (KF exo–) in exonuclease activity. Analysis of the extension products in the presence of the four dNTPs or of a single dNTP showed that the M-MuLV RT was completely blocked and did not incorporate any dNMP opposite 2′-deoxyribonolactone. KF exo– preferentially incorporated nucleotides opposite the lesion following the frequency order dAMP > dGMP >> dTMP ~ dCMP and thus appeared to obey the ‘A rule’ for preferential incorporation as has been shown previously for the 2′-deoxyribose abasic site. In the sequence context examined, the primer extension by KF exo– appeared to be less efficient when dAMP was positioned opposite the lesion as compared with dTMP or dGMP. These two nucleotides promoted a more efficient polymerization accompanied by nucleotide deletion through misalignment incorporations. We therefore predict that the sequence context may strongly influence the translesional synthesis by KF exo– and thus the miscoding and mutational potential of the 2′-deoxyribonolactone in E.coli.
INTRODUCTION
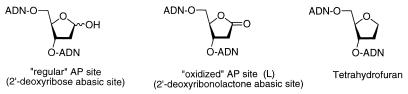
DNA is continually exposed to endogenous and exogenous factors that can damage its chemical structure. Loss of a nucleic base leaving a deoxyribose residue is probably the most frequent damaging event, and may occur spontaneously under the action of alkylating agents or enzymatically as an intermediate in the repair of abnormal or modified bases (1–3). This DNA modification has been mostly examined. A structurally related event is the formation of 2́-deoxyribonolactone (L) (Fig. 1), which corresponds to the oxidized form of the deoxyribose abasic site (or ‘regular abasic site’). It is produced in a variety of DNA oxidative processes that proceed via radical abstraction of the hydrogen at C1′(4). These notably include UV light and γ irradiation (5,6). The anticancer antibiotic neocarzinostatin that belongs to the ene-diyne family produces L damage at sequence specific sites (7–9). Chromium (V) carcinogens (10) and the cationic porphyrin Mn-TMPy P (11) lead to DNA cleavage through intermediate formation of the lactone. In model systems L formation has also been observed in the UV photoreaction of 5-halouracil incorporated in small oligonucleotides or by photosensitization of specific sequences by benzophenone (12). It has been proposed as intermediate in DNA degradation by the chemical nuclease copper phenanthroline Cu(OP)2 (13–15). The L damage has been reported to cause much greater lability of the phosphodiester linkages on both 5′ and 3′ sides than the regular abasic site and formation of the lesion has been frequently evidenced by identification of the ultimate cleavage product 5-methylenefuranone (5-MF) (16) resulting from successive β and δ elimination.
Figure 1.
Structures of the 2′-deoxyribose and 2′-L lesions and of the tetrahydrofuran stable analog.
Very little is known about the fate of L damage in the cell. It has been reported that the lesion is relatively resistant to apurinic/apyrimidinic endonucleases and might be mutagenic (17). The paucity of data is largely due to the absence of general and efficient synthesis of DNA fragments containing this highly labile lesion at predetermined positions in the sequence. Most methods described in the literature are sequence dependent or are not quantitative (5,6,9,12,18,19). We recently reported such a synthetic process (20). It was therefore of great interest to examine this damaged DNA and compare the consequences of this lesion with those reported for the regular abasic site, or for its chemically stable analog, the tetrahydrofuran. This analog has frequently been examined in enzymology studies due to the fragility of the 2′-deoxyribose lesion (21–32). This tetrahydrofuran analog could also be considered as a model for L, although reactivity and structural aspects are clearly different. Planarity of the sp2 C1 carbon atom could induce differences in terms of recognition by enzymes. Indeed the high field NMR study of an oligomer containing L in the middle of the sequence indicated absence of curvature at the site of the lesion as was observed for the tetrahydrofuranyl abasic site (33,34).
We report here our first results on the enzymology of the L lesion. We explore the miscoding properties of the lesion. Using oligonucleotides containing a L residue in the middle of the sequences as templates we show that dAMP is preferentially inserted opposite the lesion by the Klenow polymerases while reverse transcriptase (RT) is unable to bypass the lesion. We show that L constitutes a strong block for primer elongation.
MATERIALS AND METHODS
Materials
Oligonucleotides were labeled using [γ-32P]ATP (specific activity 3000 Ci/mmol) (NENTM Life Science) and T4 polynucleotide kinase purchased either from Boehringer Mannheim or MBI Fermentas. The Klenow fragments of Escherichia coli DNA polymerase I proficient (KF exo+) and deficient (KF exo–) in 3′→5′ proofreading exonuclease activity were obtained from Boehringer Mannheim and MBI Fermentas, respectively. M-MuLV RT and nucleotide triphosphates (dNTPs) were purchased from Eurogentec and MBI Fermentas. Reagents used for automated DNA synthesis were obtained from PE Biosystems. Reagents for capillary electrophoresis were from Beckman Instruments Inc.
Oligonucleotide synthesis and purification
Oligonucleotides were synthesized using phosphoramidite chemistry on an automated Perseptive Biosystem 8900 DNA synthesizer and were purified by electrophoresis on a 20% polyacrylamide gel in the presence of 7 M urea. The sequences of oligonucleotides used for these studies are shown in Table 1. The oligonucleotide primers were 5′-radiolabeled with T4 polynucleotide kinase and [γ-32P]ATP according to the manufacturer’s protocol. Primer–template substrates were prepared by hybridizing 5′-radiolabeled primer to the complementary modified (sequence 1) or non-modified (sequence 2) 34mer using a slight excess of template (20%) in 50 mM Tris–HCl pH 7.5, 10 mM MgCl2, 5 mM 2-mercaptoethanol. The oligonucleotide containing the L lesion (sequence 1) was obtained after UV-irradiation of the oligomer containing the 7-nitro-indole precursor as described previously (20). Briefly, the oligonucleotide containing the 7-nitro-indole precursor was illuminated for 1 h with a 200 W, Xe/Hg ORIEL lamp. Wavelengths >320 nm were eliminated by a 2 M KNO3 filter. The complete conversion of the precursor into 2′-L was controlled by hot alkaline treatment (100 mM NaOH, 70°C, 10 min) and the purity of the oligonucleotide was confirmed using gel capillary electrophoresis. Electropherograms were monitored on a P/ACE 5000 (Beckman) apparatus: samples were injected under 10 kV for 2–5 s in the capillary [DNA eCap, 100 µm i.d., 27 cm in length filled with single-stranded (ss)DNA 100-R gel and ssDNA 100 buffer] and separation of the products proceeded under 8.1 kV at 30°C for 40 min.
Table 1. Oligonucleotide sequences.
| Number |
Sequence (5′→3′) |
| 1 | AGCGATGAGAGGCCALGAGGAATCGCTGGTACCG |
| 2 | AGCGATGAGAGGCCATGAGGAATCGCTGGTACCG |
| 3 | CGGTACCAGCGATTCCTC |
| 4 | CGGTACCAGCGATTCCTCA |
| 5 | CGGTACCAGCGATTCCTCC |
| 6 | CGGTACCAGCGATTCCTCT |
| 7 | CGGTACCAGCGATTCCTCG |
| 8 | CGGTACCAGCGATTCCTCTG |
| 9 | CGGTACCAGCGATTCCTCTGG |
| 10 | CGGTACCAGCGATTCCTCGG |
| 11 | CGGTACCAGCGATTCCTCGGG |
| 12 | CGGTACCAGCGAT |
Insertion of a single nucleotide and DNA synthesis
Using a modified or unmodified 34mer template (0.06 µM sequence 1 or 2, respectively) annealed to a 32P-labeled 18mer primer (0.05 µM sequence 3), insertion reactions catalyzed by 0.005 U KF exo+ and 0.01 U KF exo– or 1 U RT were conducted for 20 min at 30°C in 10 µl 50 mM Tris–HCl pH 7.5, 10 mM MgCl2, 5 mM 2-mercaptoethanol and either a single dNTP (10 or 100 µM) or four dNTPs (100 µM each). Reactions were stopped by adding 20 µl formamide dye solution (95% formamide, 20 mM EDTA pH 8, 0.1% bromophenol blue, 0.1% xylene cyanol) and heated at 70°C for 5 min. Aliquots (3 µl) of these solutions were subjected to electrophoresis on a 20% denaturing PAGE gel (7 M urea, 1× TBE).
Extension and DNA synthesis
The 5′ end-labeled 19mer primers (sequences 4–7) were annealed to either the modified or unmodified template under the conditions described above. A mixture of the four dNTPs (100 µM each) was then added and the reactions were initiated by adding the DNA polymerase (0.02 or 0.2 U KF exo+; 0.01 or 0.1U KF exo– and 4 or 10 U RT). The samples were incubated at 30°C for 30 min and the reactions were stopped and analyzed as described above.
Insertion kinetics
The 5′ end-labeled 18mer (sequence 3, 0.05 µM) was annealed to the modified or unmodified template (sequence 1 or 2, 0.06 µM). Duplexes were incubated with various amounts of dNTP (10–100 µM dATP for the control and 100–500 µM dNTP for the other assays) and KF exo– (0.0005–0.05 U per sample depending on the template and dNTP) for 0.5–3 min at 30°C in order to obtain <20% elongation of the primer. Reactions were quenched by adding 20 µl formamide dye mixture. Samples were heated at 70°C for 5 min and 3 µl aliquots were subjected to electrophoresis using 20% denaturing PAGE.
Data analysis
After electrophoresis, the gels were exposed overnight at –30°C on Kodak BioMax MR-2 films. The films were scanned with an AGFA STUDIOscan IIsi scanner and bands were quantified by densitometry of the autoradiographs (using NIH Image 1.61 software). Kinetic parameters were determined from primer extension reactions. The Michaelis–Menten constant (Km) and the maximum velocity (Vmax) of the enzymatic reactions were calculated from Hanes–Woolf plots. Each kinetic data point is the average of two independent experiments with 15–25 values each. Less than 20% of the primer is extended under the steady-state conditions used, ensuring single hit kinetic conditions (35,36). The kcat was determined by dividing Vmax by enzyme concentration.
RESULTS
Oligonucleotide synthesis and stability
34mer oligonucleotide templates and primers of different lengths (13, 18 and 19mers) were prepared (Table 1). The 34mer template containing a single L residue in the middle of the sequence was synthesized as reported (20). The method is based on the photoreaction of a 7-nitro-indole nucleoside precursor site specifically incorporated in an oligonucleotide. The illumination triggers a radical process in which the nitro-indole excited state induces intramolecular H1′ abstraction, which, in turn, leads quantitatively to L. The presence of lactone in the oligonucleotide was determined using an NaOH assay comprising a 10 min incubation at 70°C with 100 mM NaOH leading to total cleavage of the oligonucleotide at the lactone site. The purity was checked by capillary electrophoresis. We verified that during the primer extension studies (i.e. hybridization with the primer and incubation with the polymerase in the enzymatic reaction buffer), <5% of the L-modified 34mer was degraded (data not shown).
Incorporation of a single dNMP opposite the L lesion
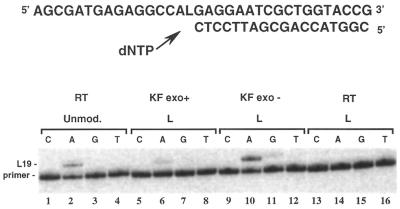
To measure deoxyribonucleotide insertion opposite the L lesion during in vitro DNA synthesis, a 5′ 32P-end-labeled 18mer primer (sequence 3) was annealed to the 34mer modified template (sequence 1). The duplex formed was subjected to primer extension by KF exo+, KF exo– and RT in the presence of a single dNTP. An unmodified template (sequence 2) was used as a control for nucleotide incorporation by RT. The products of the reactions were analyzed by denaturing PAGE (Fig. 2).
Figure 2.
Nucleotide incorporation in the presence of a single dNTP. Reaction mixtures contained the unmodified 34mer (sequence 2, 0.06 µM) or the modified 34mer L (sequence 1, 0.6 µM) as a template, primed with the 32P-labeled 18mer (sequence 3, 0.05 µM), one single dNTP (final concentration 10 µM) and either KF exo+ (0.005 U), KF exo– (0.01 U) or RT (1 U). RT was used for the control reaction containing the unmodified 34mer (lanes 1–4). Reactions were incubated for 20 min at 30°C before processing as described (Materials and Methods).
DNA synthesis by RT on the unmodified template led to the expected incorporation of dAMP opposite T at position 16 (Fig. 2, lane 2). When sequence 1 was used as a template, dAMP was preferentially incorporated opposite the L lesion in the reactions catalyzed by the KF exo+ and KF exo– (Fig. 2, lanes 6 and 10, respectively). KF exo– also catalyzed incorporation of small amounts of dGMP (Fig. 2, lane 11). Insertion of dCMP and dTMP opposite L by KF exo+ and KF exo– was not detected in the conditions used. No elongated product (19mer) was detected in the case of RT, even in the presence of a large excess of the enzyme (up to 10 U, data not shown).
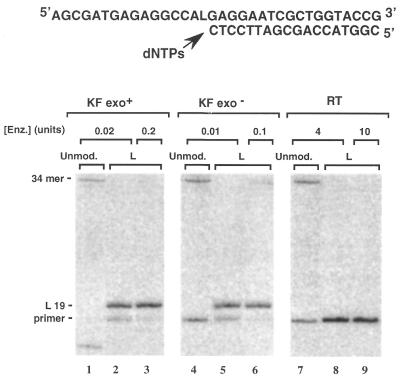
DNA synthesis on the template containing the L lesion
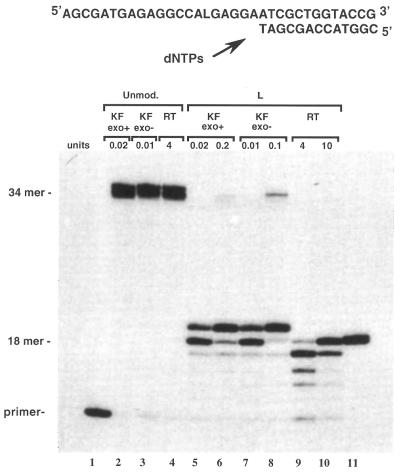
The primer sequence 3 was annealed to the modified template (sequence 1) and extended in the presence of the four dNTPs by KF exo+, KF exo– and RT. The primer extension reactions were conducted in parallel with unmodified sequence 2 (Fig. 3). As expected, all enzymes led to the fully extended 34mer product when the primer was annealed to the unmodified sequence 2 (Fig. 2, lanes 1, 4 and 7). Primer extension on the modified L template catalyzed by KF exo+ and KF exo– appeared to be blocked when the primer was elongated by one nucleotide, just opposite the L lesion (Fig. 2, lanes 2 and 5, respectively). This inhibition of the translesional synthesis could be slightly overcome in the case of KF exo– when using a 10-fold excess of the enzyme (Fig. 2, lane 6). However, the extension reaction remained strongly blocked in the case of KF exo+ even in the presence of a 10-fold excess of the enzyme (Fig. 2, lane 3).
Figure 3.
Chain extension reactions catalyzed either by KF exo+ (0.02 or 0.2 U), KF exo– (0.01 or 0.1 U) or RT (4 or 10 U). Reactions were carried out at 30°C for 30 min using the unmodified 34mer (sequence 2, 0.06 µM) or the modified 34mer L template (sequence 1, 0.06 µM) primed with the 32P-labeled 18mer (sequence 3, 0.05 µM) in the presence of the four dNTPs (at a final concentration of 100 µM each).
It is noteworthy that in the control reaction, KF exo+ caused the production of the full-length DNA (34mer; Fig. 2, lane 1). However the shorter fragment resulting from the 3′→5′ exonuclease activity was also observed. This exonuclease activity was not observed on the modified L template, even in the presence of the highest enzyme concentration (Fig. 2, lane 3). In contrast to the other enzymes, RT was unable to initiate extension even in the presence of a large enzyme excess (4 and 10 U; Fig. 2, lanes 8 and 9, respectively).
Kinetic parameters for nucleotide insertion opposite the L lesion catalyzed by KF exo–
Kinetic parameters of dNMP insertion opposite the L lesion were determined for KF exo–. These reactions measured the addition of a single dNMP to the 3′ terminus of the 18mer primer (sequence 3) when annealed to the unmodified sequence 2 or to the modified L template (sequence 1). The Michaelis–Menten constants Km and the reaction maximum rates Vmax were determined (Table 2). To allow comparison of the kinetic parameters for both substrates, the specificity constants kcat/Km were calculated. As expected, the specificity constant for nucleotide incorporation using the unmodified template (sequence 2) was highest for dAMP. kcat/Km was at least 200-fold higher for dAMP incorporation than for dCMP and dTMP and 60-fold higher than for dGMP incorporation. With the template containing the L lesion, dAMP was preferentially inserted opposite the modified site. The kcat/Km value for dAMP incorporation was two times higher than for dGMP, 20 and 40 times higher than for dTMP and dCMP, respectively. Thus the efficiency for nucleotide insertion opposite the L lesion for KF exo– follows the order: dAMP > dGMP >> dTMP ~ dCMP.
Table 2. Kinetic parameters of nucleotide insertion reactions catalyzed by KF exo–
| fig name = "gke400t01"> |
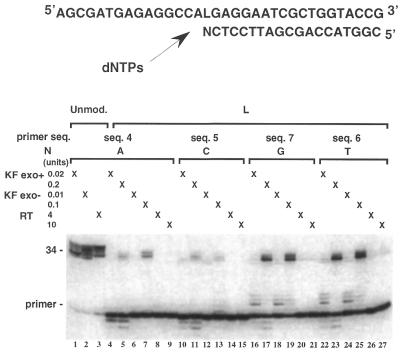
Effect of the nature of the nucleotide inserted opposite the L lesion on primer extension
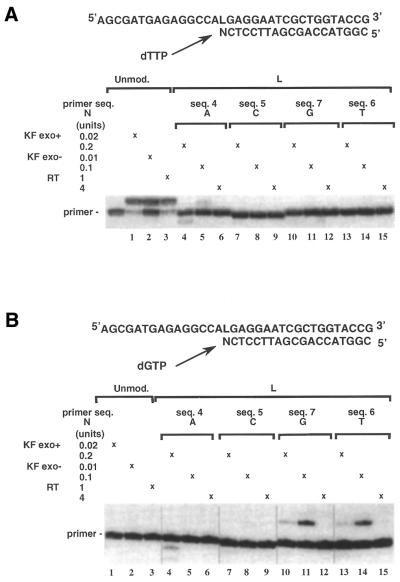
We examined the effect of changing the base at the 3′ terminus of the primer facing the L lesion in the template, on direct nucleotide extension by KF exo+, KF exo– and RT. Sequence 1 was annealed to 5′ 32P-end-labeled primers (19mers) differing only by the nature of the nucleotide (N) at their 3′ terminus (N = A, sequence 4; N = C, sequence 5; N = T, sequence 6; N = G, sequence 7). The unmodified template (sequence 2) was annealed to the 19mer primer sequence 4 (N = A) and used as a control. Primer extension experiments were first performed in the presence of the four dNTPs (Fig. 4). All three enzymes led to the expected 34mer product when the reactions were conducted on the unmodified duplex (Fig. 4, lanes 1–3) and nucleotide addition at the blunt end of the DNA duplex occurred (more slowly migrating bands). Such base addition at non-templated positions has been reported in the literature (37). Using the same enzyme concentration as in the control (0.02 U), the extension of the primers did not lead to fully extended products with the modified L template whatever the nature of N at the 3′ end. Trace amounts of elongated products were observed in the reactions catalyzed by KF exo+ or KF exo– when the primer contained a guanine or a thymine residue at the 3′ end (Fig. 4, lanes 16, 18 and 22, 24). When using higher concentrations of KF exo+ and KF exo– (10 times the concentration used for the controls) the primer containing an adenine at the 3′ terminal position was fully extended while almost no extension product was obtained for the primer ending by a cytosine (Fig. 4, lanes 5, 7 and 11, 13, respectively). The same result was obtained for primers having a thymine or a guanine at their 3′ end (Fig. 4, lanes 23, 25 and 17, 19, respectively). In fact, KF exo+ and KF exo– were even more active with these two primers. Products resulting from the proofreading activity of KF exo+ were also observed for each primer (Fig. 4, lanes 4 and 5, 10 and 11, 16 and 17, 22 and 23). The extension reaction catalyzed by RT was shown to be blocked whatever the nature of the base opposite the L lesion (Fig. 4, lanes 8, 14, 20, 26), even in the presence of an enzyme excess (Fig. 4, lanes 9, 15, 21, 27).
Figure 4.
Comparison of primer extension efficiencies past the lesion by varying the 3′ terminal base pair (N:L). The L-modified or unmodified 34mer template (sequences 1 and 2, respectively, 0.6 µM) was primed with different 32P-labeled 19mers varied by the nature of the base N at their 3′ end (N = A, sequence 4; N = C, sequence 5; N = G, sequence 7; N = T, sequence 6; 0.05 µM). Reactions were conducted for 30 min at 30°C in the presence of the four dNTPs (100 µM each) using 0.02 U KF exo+, 0.01 U KF exo– and 4 U RT for the unmodified template and using a large enzyme excess for the modified L template (0.2 U KF exo+, 0.1 U KF exo– and 10 U RT).
This experiment in the presence of the four dNTPs was repeated using a shorter primer (13mer, sequence 12) in order to see if the incorporation of several nucleotides upstream of the L position might help to bypass the lesion (Fig. 5). The primer annealed to the unmodified sequence 2 was fully extended by all three enzymes and once again, addition at the blunt end was observed (Fig. 5, lanes 2–4). On the L modified sequence 1, all three enzymes appeared to be able to elongate the 13mer primer until position 18, producing a fragment that co-migrates with the 18mer sequence 3 used as a standard (Fig. 5, lane 11). KF exo+ and KF exo– behaved as expected by producing the 19mers corresponding to the incorporation of 1 nt opposite L (Fig. 5, lanes 5–8) and the highest concentration of KF exo– (0.1 U) led to small amounts of the fully extended primer (Fig. 5, lane 8).
Figure 5.
Chain extension reaction of the 13mer (sequence 12, 0.05 µM) catalyzed either by KF exo+ (0.02 or 0.2 U), KF exo– (0.01 or 0.1 U) or RT (4 or 10 U) in the presence of the four dNTPs (100 µM each). Reactions were carried out at 30°C for 30 min using the unmodified 34mer (sequence 2, 0.06 µM) or the modified 34mer L template (sequence 1, 0.06 µM). In lane 11, the 32P-labeled 18mer (sequence 3) was used as a control.
Extension reaction of primers varying by the nature of the 3′ terminal base opposite the L lesion in the presence of dTTP or dGTP alone
This experiment was first performed in the presence of dTTP alone, i.e. the correct nucleotide to be incorporated at the 5′ side of L (Fig. 6A). As expected for the conventional duplex (sequence 2 annealed with the sequence 4 primer) all three enzymes led to the 20mer product resulting from dTMP incorporation opposite adenine (Fig. 6A, lanes 1–3). In the reactions conducted on the L-containing template none of the three enzymes led to the extension products at the concentration used for the control (0.02 U). RT remained inactive even at high concentration. The primer containing an adenine at the 3′ extremity (sequence 4) appeared to be substrate for the 3′→5′ proofreading activity of KF exo+ leading to formation of trace amounts of a shorter fragment (Fig. 6A, lane 4). Only small amounts of the primer extension product could be observed with this same sequence in the presence of the highest quantity of KF exo– (0.1 U; Fig. 6A, lane 5).
Figure 6.
Comparison of the direct extension reaction with the misalignment reaction extension at L lesion by KF exo +, KF exo– and RT. The 34mer templates containing either the L modified site (sequence 1, 0.06 µM) or a thymine at the same position (unmodified sequence 2, 0.06 µM) were primed with 19mer sequences 4, 5, 6 and 7 terminating with respectively A, C, T and G. Extension reactions catalyzed by KF exo+ and KF exo– and RT using the concentrations indicated on the left of the gels were performed at 30°C for 30 min. (A) For direct extension, dTMP was incorporated at a downstream template site adjacent to L while for misalignment extension (B), dGMP was incorporated at a template site one or two bases downstream from L. In the direct extension mode, dTMP was incorporated at a correct position by the three polymerases when the primer sequence 4 was annealed to the unmodified template sequence 2 (A).
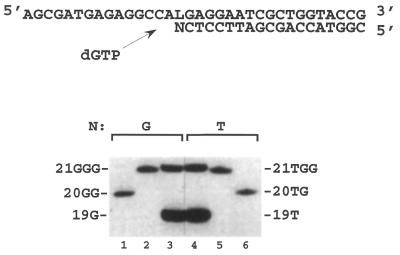
The same experiments (using the primers 4–7) were repeated in the presence of dGTP alone (Fig. 6B). As expected the primer extension performed on the unmodified template (sequence 2) did not lead to dGMP incorporation (Fig. 6B, lanes 1–3). In contrast, when the L modified template (sequence 1) was primed with the 19mers, only the sequences 6 and 7 terminated respectively by T and G (Fig. 6B, lanes 13, 14 for sequence 6 and lanes 10, 11 for sequence 7) led to extension with KF exo+ or KF exo–. No extension was observed for RT (Fig. 6B, lane 12 for sequence 7 and lane 15 for sequence 6). Identification of the products resulting from extension of sequences 6 and 7 by KF exo– was performed by comparison with synthetic reference oligonucleotides. These oligonucleotides contained one or two additional Gs at the 3′-terminus (Fig. 7), i.e. sequences 10 and 8 (Fig. 7, lanes 10 and 6) corresponding to the addition of one G to oligonucleotides 7 and 6, and sequences 11 and 9 (Fig. 7, lanes 2 and 5) corresponding to the addition of two Gs to oligonucleotides 7 and 6. The 19mers elongated by KF exo– in the presence of dGTP exhibited identical migration to the synthetic samples corresponding to the addition of two Gs.
Figure 7.
Identification of the primer extension products obtained in the reaction catalyzed by KF exo– in the presence of dGTP alone (100 µM), using the L modified template (sequence 1, 0.06 µM). Primer extension reactions were conducted for 30 min at 30°C using 0.01 U KF exo– with 32P-labeled 19mers sequence 7 and 6 terminating with G (lane 3) and T (lane 4), respectively. The mobilities of the reaction products were compared with those of authentic samples of the oligonucleotides containing either one additional dGMP (indicated as 20GG, sequence 10 or 20TG, sequence 8; lanes 1 and 6, respectively) or two (indicated as 21GGG, sequence 11 or 21TGG, sequence 9; lanes 2 and 5, respectively).
DISCUSSION
When cellular DNA is exposed to ionizing radiation, or more generally to processes involving radical formation, several types of DNA lesions may be formed among which the L is present in significant amounts. As this abasic lesion is non-informative, it is important to assess its mutagenic potential. Until now, mainly due to the absence of an efficient chemical method of incorporation, nothing has been reported on the recognition of L by DNA polymerases.
We have studied the miscoding properties of the L lesion in reactions catalyzed by the most usual polymerases, KF exo+, KF exo– and M-MuLV RT. When KF exo– or KF exo+ were used to catalyze primer extension in the presence of one single dNTP, L promoted preferential incorporation of dAMP and weaker incorporation of dGMP.
The kinetic parameters for the incorporation of one single dNMP opposite the L lesion (Km and Vmax) by KF exo– was determined. In the sequence context we used, insertion of dAMP opposite the L lesion by KF exo– was favored over dGMP > dTMP ∼ dCMP. Shibutani et al. (29) conducted similar experiments on primer–templates containing the 2′-deoxyribose abasic site and its chemically synthesized model tetrahydrofuran (Fig. 1). The relative insertion frequencies (Fins) opposite all three abasic lesions for KF exo+ and KF exo– followed the order dAMP > dGMP >> dCMP > dTMP. An A rule was proposed as a default mechanism inherent to polymerases in E.coli, favoring insertion of dAMP at non-coding abasic lesions. The results we obtain suggest that this A rule also governs nucleotide insertion opposite the L lesion by KF exo– and KF exo+.
When KF exo– and KF exo+ were used to catalyze the extension of an 18mer primer in the presence of the four dNTPs, only 1 nt was incorporated opposite the lesion, then the extension was blocked. The addition of a large excess of the enzyme was necessary to overcome the inhibition of the translesional synthesis in the case of KF exo–. A similar result was observed when a shorter primer was used (13mer, sequence 12). In this case all three enzymes behaved exactly as in the previous experiment and produced the same extension products confirming that the L lesion constitutes a strong block for primer elongation. The absence of full-length extension of the 18mer primer at the lowest enzyme concentration and the very low amount of fully extended 19mer primer ending with an adenine opposite the L lesion suggest that obeying the A rule allows elongation but at a very low rate. This was confirmed by extension experiments conducted with primers containing successively the four nucleic bases at the 3′ end opposite the L lesion. They were all elongated by addition of only 1 nt. Indeed, only small amounts of the extension product were observed for A located opposite the L lesion and, surprisingly, higher amounts of extension products were detected when a G or a T was opposite the lesion.
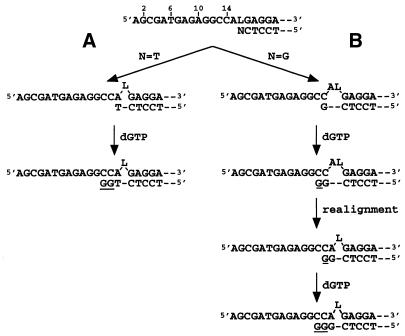
We did not investigate the effect of the sequence context on the nucleotide incorporation opposite the lesion, but our results suggest that the neighboring bases downstream to the L lesion may play an important role in the L bypass synthesis by DNA polymerases. The extension experiments of the four primers in which A, C, G or T were located successively opposite the L lesion showed that the primers terminated by T or G (sequences 6 and 7) were better substrates for KF exo+ or KF exo–. In fact, extension at the L lesion for these primers could occur by two different methods; either by direct extension of a non-distorted primer–template by addition of the next correct nucleotide or by correct extension on a DNA molecule with misaligned primer–template strands. The direct mode of extension favors nucleotide addition to a primer terminus A. The primer template misalignment mode is however strongly favored when the primer terminus is T or G. We propose that for the primer containing G at the 3′ end, this terminal G can pair with C14 located at a two base distance on the 5′ side of the L lesion in the template to form a transient two base deletion, as shown in Figure 8 path B. This leads to incorporation of a G (pairing with C15). Realignment of the extrahelical A can then occur, followed by incorporation of a new G (pairing with C13). On the other hand, the primer ending with a T (Fig. 8, path A) could pair with A13 to form a one base deletion followed by two correct incorporations of dGMP pairing with the Cs at positions 14 and 13. This scheme accounts for the structure of the elongation products that migrated like sequences 9 and 11 (Fig. 7). A comparable extension mechanism leading to base deletion has been reported by Shibutani et al. (29) for the 2′-deoxyribose abasic site and its synthetic analogs. In the sequence context examined by the authors the misalignment step could not be observed. Furthermore Kunkel (38) and Kunkel and Soni (39) have shown that some mutational hot-spots produced by DNA polymerase-β are due to an elongation process implicating correct coding on a transiently misaligned template–primer.
Figure 8.
Proposed mechanism for the misalignment extension modes in the presence of dGTP alone. (A) Extension of the primer terminating with N = T occurs by two successive dGMP incorporations opposite the adjacent 2 Cs located in the template at one and two bases downstream from the L lesion, thus promoting a one base deletion. (B) Extension of the primer terminating with G occurs by addition of one dGMP opposite the C located at two bases downstream from the L lesion (leading to transient two bases deletions). In a second step, the template realigns forming a terminal GC base pair adjacent to a GA mismatch and a second incorporation of dGMP occurs promoting here again a one base deletion. Underlined Gs indicate the dGMP incorporated by the enzyme.
We conclude from these results that the L lesion constitutes a strong block for the extension reactions catalyzed by KF exo–, KF exo+ and RT. When using a single dNTP the preferential incorporation of dAMP opposite the lesion by the two Klenow fragments shows that these enzymes obey the A rule already described for 2′-deoxyribose abasic sites and its stable analogs. Nevertheless obeying this nucleotide incorporation rule provokes a dramatic decrease in the extension efficiency at the position of the lesion. In our sequence context a by-pass mechanism was observed when the 3′ terminal base of the primer can pair with a complementary base on the template in close proximity downstream of the lesion. Most likely, the L residue is then flipped out of the realigned primer–template duplex leading to elongation products deleted by one base. Such a process should thus be strongly dependent on the sequence context of the lesion but also on the proofreading activity of the polymerases. A molecular modeling study is in progress to evaluate the structure of ‘matched’ and misaligned template–primers containing the lesion.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by Electricité de France (EDF), Service de Radioprotection and by the Association pour La Recherche sur le Cancer. Y.R. is grateful to La Ligue Contre le Cancer for a scholarship.
References
- 1.Lindahl T. and Andersson,A. (1972) Rate of chain breakage at apurinic sites in double-stranded deoxyribonucleic acid. Biochemistry, 11, 3618–3623. [DOI] [PubMed] [Google Scholar]
- 2.Singer B. and Grunberger,D. (1983) Molecular Biology of Mutagens and Carcinogens. Plenum Press, New York, NY, pp. 16–19.
- 3.Sancar A. (1996) DNA excision repair. Annu. Rev. Biochem., 65, 43–81. [DOI] [PubMed] [Google Scholar]
- 4.Pogozelski W.K. and Tullius,T.D. (1998) Oxidative strand scission of nucleic acids: routes initiated by hydrogen abstraction from the sugar moiety. Chem. Rev., 98, 1089–1107. [DOI] [PubMed] [Google Scholar]
- 5.Urata H., Yamamoto,K., Akagi,M., Hiroaki,H. and Uesugi,S. (1989) A 2′-deoxyribonolactone-containing nucleotide: isolation and characterization of the alkali-sensitive photoproduct of the trideoxyribonucleotide d(ApCpA). Biochemistry, 28, 9566–9569. [DOI] [PubMed] [Google Scholar]
- 6.Urata H. and Akagi,M. (1991) Photo-induced formation of the 2′-deoxyribonolactone-containing nucleotide for d(ApCpA); effects of neighboring bases and modification of deoxycytidine. Nucleic Acids Res., 19, 1773–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kappen L.S., Chen,C.Q. and Goldberg,I.H. (1988) Atypical abasic sites generated by neocarzinostatin at sequence-specific cytidylate residues in oligodeoxynucleotides. Biochemistry, 27, 4331–4340. [DOI] [PubMed] [Google Scholar]
- 8.Povirk L.F., Houlgrave,C.W. and Han,Y.H. (1988) Neocarzinostatin-induced DNA base release accompanied by staggered oxidative cleavage of the complementary strand. J. Biol. Chem., 263, 19263–19266. [PubMed] [Google Scholar]
- 9.Kappen L.S. and Goldberg,I.H. (1989) Identification of 2′-deoxyribonolactone at the site of neocarzinostatin-induced cytosine release in the sequence d(AGC). Biochemistry, 28, 1027–1032. [DOI] [PubMed] [Google Scholar]
- 10.Bose R.N., Fonkeng,B.S., Moghaddas,S. and Stroup,D. (1998) Mechanisms of DNA damage by chromium(V) carcinogens. Nucleic Acids Res., 27, 1588–1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pratviel G., Bernadou,J. and Meunier,B. (1998) DNA and RNA cleavage by metal complexes. Adv. Inorg. Chem., 45, 251–312. [Google Scholar]
- 12.Buchko G.W. and Cadet,J. (1992) Identification of 2-deoxy-d-ribono-1,4-lactone at the site of benzophenone photosensitized release of guanine in 2′-deoxyguanosine and thymidylyl-(3′-5′)-deoxyguanosine. Can. J. Chem., 70, 1827–1832. [Google Scholar]
- 13.Chen T. and Greenberg,M.M. (1998) Model studies indicate that copper phenanthroline induces direct strand breaks via β-elimination of the 2′-deoxyribonolactone intermediate observed in enediyne mediated DNA damage. J. Am. Chem. Soc., 120, 3815–3816. [Google Scholar]
- 14.Goyne T.E. and Sigman,D.S. (1987) Nuclease activity of 1,10-phenanthroline-copper ion. Chemistry of deoxyribose oxidation. J. Am. Chem. Soc., 109, 2846–2848. [Google Scholar]
- 15.Oyoshi T. and Sugiyama,H. (2000) Mechanism of DNA strand scission induced by (1,10-phenanthroline) copper complex: major direct DNA cleavage is not through 1′, 2′-dehydronucleotide intermediate nor β-elimination of forming ribonolactone. J. Am. Chem. Soc., 122, 6313–6314. [Google Scholar]
- 16.Hwang J.T., Tallman,K.A. and Greenberg,M.M. (1999) The reactivity of the 2′-deoxyribonolactone lesion in single-stranded DNA and its implication in reaction mechanisms of DNA damage and repair. Nucleic Acids Res., 27, 3805–3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Povirk L.F. and Goldberg,I.H. (1985) Endonuclease-resistant apyrimidinic sites formed by neocarzinostatin at cytosine residues in DNA: evidence for a possible role in mutagenesis. Proc. Natl Acad. Sci. USA, 82, 3182–3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sugiyama H., Tsutsumi,Y. and Saito,I. (1990) Highly sequence selective photoreaction of 5-bromouracil-containing deoxyhexanucleotides. J. Am. Chem. Soc., 112, 6720–6721. [Google Scholar]
- 19.Tronche C., Goodman,B.K. and Greenberg,M.M. (1998) DNA damage induced via independent generation of the radical resulting from formal hydrogen atom abstraction from the C1′ position of a nucleotide. Chem. Biol., 5, 263–271. [PubMed] [Google Scholar]
- 20.Kotera M., Bourdat,A.-G., Defrancq,E. and Lhomme,J. (1998) A highly efficient synthesis of oligodeoxyribonucleotides containing the 2′-deoxyribonolactone lesion. J. Am. Chem. Soc., 120, 11810–11811. [Google Scholar]
- 21.Takeshita M., Chang,C.N., Johnson,F., Will,S. and Grollman,A.P. (1987) Oligodeoxynucleotides containing synthetic abasic sites. Model substrates for DNA polymerases and apurinic/apyrimidinic endonucleases. J. Biol. Chem., 262, 10171–10179. [PubMed] [Google Scholar]
- 22.Randall S.K., Eritja,R., Kaplan,B.E., Petruska,J. and Goodman,M.F. (1987) Nucleotide insertion kinetics opposite abasic lesions in DNA. J. Biol. Chem., 262, 6864–6870. [PubMed] [Google Scholar]
- 23.Cai H., Bloom,L.B., Eritja,R. and Goodman,M.F. (1993) Kinetics of deoxyribonucleotide insertion and extension at abasic template lesions in different sequence contexts using HIV-1 reverse transcriptase. J. Biol. Chem., 268, 23567–23572. [PubMed] [Google Scholar]
- 24.Takeshita M. and Eisenberg,W. (1994) Mechanism of mutation on DNA templates containing synthetic abasic sites: study with a double strand vector. Nucleic Acids Res., 22, 1897–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pinz K.G., Shibutani,S. and Bogenhagen,D.F. (1995) Action of mitochondrial DNA polymerase γ at sites of base loss or oxidative damage. J. Biol. Chem., 270, 9202–9206. [DOI] [PubMed] [Google Scholar]
- 26.Ide H., Murayama,H., Sakamoto,S., Makino,K., Honda,K., Nakamuta,H., Sasaki,M. and Sugimoto,N. (1995) On the mechanism of preferential incorporation of dAMP at abasic sites in translesional DNA synthesis. Role of proofreading activity of DNA polymerase and thermodynamic characterization of model template-primers containing an abasic site. Nucleic Acids Res., 23, 123–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paz-Elizur T., Takeshita,M., Goodman,M., O’Donnell,M. and Livneh,Z. (1996) Mechanism of translesion DNA synthesis by DNA polymerase II. J. Biol. Chem., 271, 24662–24669. [DOI] [PubMed] [Google Scholar]
- 28.Paz-Elizur T., Takeshita,M. and Livneh,Z. (1997) Mechanism of bypass synthesis through an abasic site analog by DNA polymerase I. Biochemistry, 36, 1766–1773. [DOI] [PubMed] [Google Scholar]
- 29.Shibutani S., Takeshita,M. and Grollman,A.P. (1997) Translesional synthesis on DNA templates containing a single abasic site. A mechanistic study of the ‘A rule’. J. Biol. Chem., 272, 13916–13922. [DOI] [PubMed] [Google Scholar]
- 30.Efrati E., Tocco,G., Eritja,R., Wilson,S.H. and Goodman,M.F. (1997) Abasic translesion synthesis by DNA polymerase β violates the ‘A- rule’. Novel types of nucleotide incorporation by human DNA polymerase β at an abasic lesion in different sequence contexts. J. Biol. Chem., 272, 2559–2569. [DOI] [PubMed] [Google Scholar]
- 31.Hatahet Z., Zhou,M., Reha-Krantz,L.J., Ide,H., Morrical,S.W. and Wallace,S.S. (1999) In vitro selection of sequence contexts which enhance bypass of abasic sites and tetrahydrofuran by T4 DNA polymerase holoenzyme. J. Mol. Biol., 286, 1045–1057. [DOI] [PubMed] [Google Scholar]
- 32.Strauss B.S. (1991) The ‘A rule’ of mutagen specificity: a consequence of DNA polymerase bypass of non-instructional lesions? Bioessays, 13, 79–84. [DOI] [PubMed] [Google Scholar]
- 33.Jourdan M., Garcia,J., Defrancq,E., Kotera,M. and Lhomme,J. (1999) 2′-deoxyribonolactone lesion in DNA: refined solution structure determined by nuclear magnetic resonance and molecular modeling. Biochemistry, 38, 3985–3995. [DOI] [PubMed] [Google Scholar]
- 34.Coppel Y., Berthet,N., Coulombeau,C., Garcia,J. and Lhomme,J. (1997) Solution conformation of an abasic DNA undecamer duplex d(CGCACXCACGC) × d(GCGTGTGTGCG): the unpaired thymine stacks inside the helix. Biochemistry, 36, 4817–4830. [DOI] [PubMed] [Google Scholar]
- 35.Boosalis M.S., Petruska,J. and Goodman,M.F. (1987) DNA polymerase insertion fidelity. Gel assay for site-specific kinetics. J. Biol. Chem., 262, 14689–14696. [PubMed] [Google Scholar]
- 36.Goodman M.F., Creighton,S., Bloom,L.B. and Petruska,J. (1993) Biochemical basis of DNA replication fidelity. Crit. Rev. Biochem. Mol. Biol., 28, 83–126. [DOI] [PubMed] [Google Scholar]
- 37.Clark J.M., Joyce,C.M. and Beardsley,G.P. (1987) Novel blunt-end addition reactions catalyzed by DNA polymerase I of Escherichia coli. J. Mol. Biol., 198, 123–127. [DOI] [PubMed] [Google Scholar]
- 38.Kunkel T.A. (1986) Frameshift mutagenesis by eucaryotic DNA polymerases in vitro. J. Biol. Chem., 261, 13581–13587. [PubMed] [Google Scholar]
- 39.Kunkel T.A. and Soni,A. (1988) Mutagenesis by transient misalignment. J. Biol. Chem., 263, 14784–14789. [PubMed] [Google Scholar]