Abstract
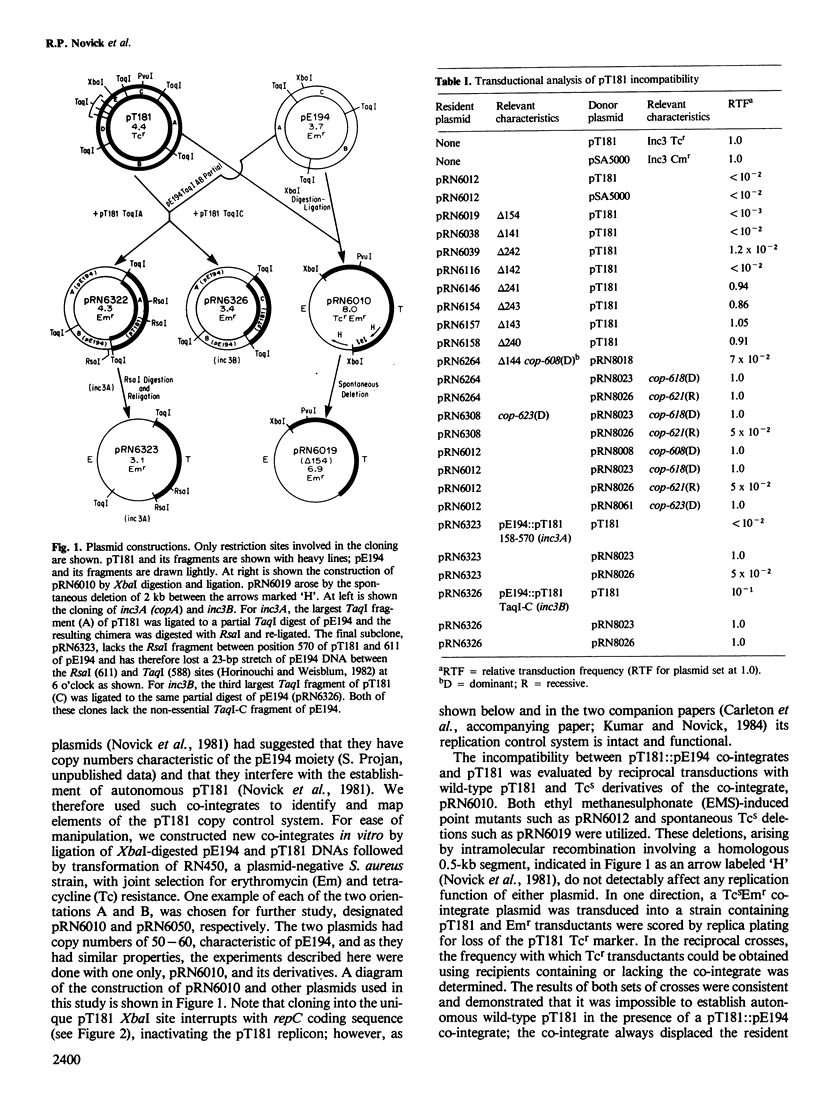
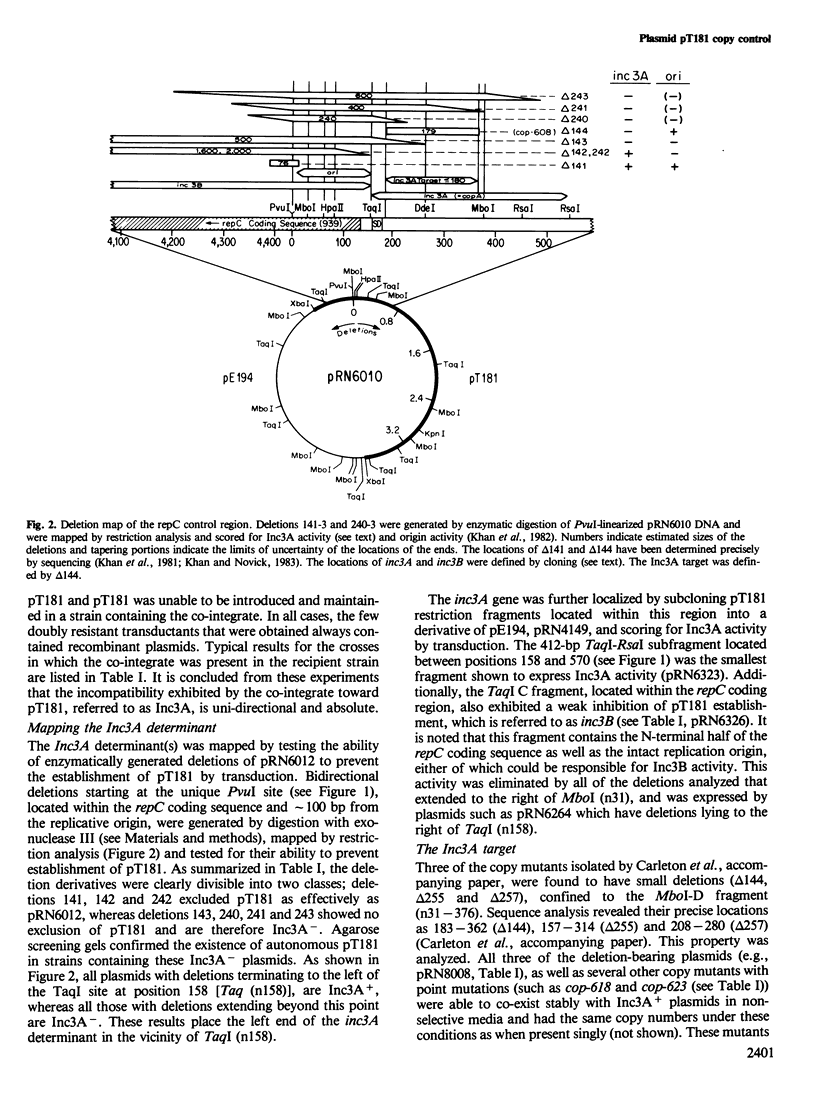
pT181 is a fully sequenced 4.4-kb 20 copy Tcr plasmid from Staphylococcus aureus. Its replication system involves a unique unidirectional origin embedded in the coding sequence for a plasmid-determined protein, RepC, that is required for initiation. When joined to a 55 copy carrier plasmid, pE194, pT181 excludes autonomous isologous replicons by inhibiting their replication. Two types of spontaneous pT181 copy mutants have been isolated, one that eliminates sensitivity to this inhibition and another that does not. A spontaneous 180-bp deletion, delta 144, eliminates both the inhibitory activity and sensitivity to it. This deletion increases copy number by 50-fold and RepC production by at least 10-fold. It is located directly upstream from the repC coding sequence and the deletion-bearing plasmid supports the replication of inhibitor-sensitive plasmids in cells containing active inhibitor. This effect is probably due to the overproduction of RepC by the delta 144 plasmid. On the basis of these results, it is suggested that RepC synthesis is negatively controlled by an inhibitor that is encoded directly upstream from the repC coding sequence and acts as a tareget set in the same region. It is likely, therefore, that pT181 replication rate is determined by the level of RepC.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Chang S., Cohen S. N. High frequency transformation of Bacillus subtilis protoplasts by plasmid DNA. Mol Gen Genet. 1979 Jan 5;168(1):111–115. doi: 10.1007/BF00267940. [DOI] [PubMed] [Google Scholar]
- Easton A. M., Rownd R. H. The incompatibility product of IncFII R plasmid NR1 controls gene expression in the plasmid replication region. J Bacteriol. 1982 Nov;152(2):829–839. doi: 10.1128/jb.152.2.829-839.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckhardt T. A rapid method for the identification of plasmid desoxyribonucleic acid in bacteria. Plasmid. 1978 Sep;1(4):584–588. doi: 10.1016/0147-619x(78)90016-1. [DOI] [PubMed] [Google Scholar]
- Heffron F., Bedinger P., Champoux J. J., Falkow S. Deletions affecting the transposition of an antibiotic resistance gene. Proc Natl Acad Sci U S A. 1977 Feb;74(2):702–706. doi: 10.1073/pnas.74.2.702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horinouchi S., Weisblum B. Nucleotide sequence and functional map of pC194, a plasmid that specifies inducible chloramphenicol resistance. J Bacteriol. 1982 May;150(2):815–825. doi: 10.1128/jb.150.2.815-825.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iordănescu S., Surdeanu M. Complementation of a plasmid replication defect by autonomous incompatible plasmids in Staphylococcus aureus. Plasmid. 1980 Jul;4(1):1–7. doi: 10.1016/0147-619x(80)90078-5. [DOI] [PubMed] [Google Scholar]
- Iordănescu S. Three distinct plasmids originating in the same Staphylococcus aureus strain. Arch Roum Pathol Exp Microbiol. 1976 Jan-Jun;35(1-2):111–118. [PubMed] [Google Scholar]
- Khan S. A., Adler G. K., Novick R. P. Functional origin of replication of pT181 plasmid DNA is contained within a 168-base-pair segment. Proc Natl Acad Sci U S A. 1982 Aug;79(15):4580–4584. doi: 10.1073/pnas.79.15.4580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan S. A., Carleton S. M., Novick R. P. Replication of plasmid pT181 DNA in vitro: requirement for a plasmid-encoded product. Proc Natl Acad Sci U S A. 1981 Aug;78(8):4902–4906. doi: 10.1073/pnas.78.8.4902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan S. A., Novick R. P. Complete nucleotide sequence of pT181, a tetracycline-resistance plasmid from Staphylococcus aureus. Plasmid. 1983 Nov;10(3):251–259. doi: 10.1016/0147-619x(83)90039-2. [DOI] [PubMed] [Google Scholar]
- LOVELESS A., HOWARTH S. Mutation of bacteria at high levels of survival by ethyl methane sulphonate. Nature. 1959 Dec 5;184:1780–1782. doi: 10.1038/1841780a0. [DOI] [PubMed] [Google Scholar]
- Lacatena R. M., Cesareni G. Base pairing of RNA I with its complementary sequence in the primer precursor inhibits ColE1 replication. Nature. 1981 Dec 17;294(5842):623–626. doi: 10.1038/294623a0. [DOI] [PubMed] [Google Scholar]
- Light J., Molin S. The sites of action of the two copy number control functions of plasmid R1. Mol Gen Genet. 1982;187(3):486–493. doi: 10.1007/BF00332633. [DOI] [PubMed] [Google Scholar]
- Masai H., Kaziro Y., Arai K. Definition of oriR, the minimum DNA segment essential for initiation of R1 plasmid replication in vitro. Proc Natl Acad Sci U S A. 1983 Nov;80(22):6814–6818. doi: 10.1073/pnas.80.22.6814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Molin S., Nordström K. Control of plasmid R1 replication: functions involved in replication, copy number control, incompatibility, and switch-off of replication. J Bacteriol. 1980 Jan;141(1):111–120. doi: 10.1128/jb.141.1.111-120.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray N. E., Bruce S. A., Murray K. Molecular cloning of the DNA ligase gene from bacteriophage T4. II. Amplification and preparation of the gene product. J Mol Biol. 1979 Aug 15;132(3):493–505. doi: 10.1016/0022-2836(79)90271-7. [DOI] [PubMed] [Google Scholar]
- Nordström K., Ingram L. C., Lundbäck A. Mutations in R factors of Escherichia coli causing an increased number of R-factor copies per chromosome. J Bacteriol. 1972 May;110(2):562–569. doi: 10.1128/jb.110.2.562-569.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick R. P., Adler G. K., Majumder S., Khan S. A., Carleton S., Rosenblum W. D., Iordanescu S. Coding sequence for the pT181 repC product: a plasmid-coded protein uniquely required for replication. Proc Natl Acad Sci U S A. 1982 Jul;79(13):4108–4112. doi: 10.1073/pnas.79.13.4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick R. P., Brodsky R. Studies on plasmid replication. I. Plasmid incompatibility and establishment in Staphylococcus aureus. J Mol Biol. 1972 Jul 21;68(2):285–302. doi: 10.1016/0022-2836(72)90214-8. [DOI] [PubMed] [Google Scholar]
- Novick R. P., Iordanescu S., Surdeanu M., Edelman I. Transduction-related cointegrate formation between Staphylococcal plasmids: a new type of site-specific recombination. Plasmid. 1981 Sep;6(2):159–172. doi: 10.1016/0147-619x(81)90064-0. [DOI] [PubMed] [Google Scholar]
- Novick R. P., Murphy E., Gryczan T. J., Baron E., Edelman I. Penicillinase plasmids of Staphylococcus aureus: restriction-deletion maps. Plasmid. 1979 Jan;2(1):109–129. doi: 10.1016/0147-619x(79)90010-6. [DOI] [PubMed] [Google Scholar]
- Novick R. P. Studies on plasmid replication. III. Isolation and characterization of replication-defective mutants. Mol Gen Genet. 1974;135(2):131–147. doi: 10.1007/BF00264781. [DOI] [PubMed] [Google Scholar]
- Novick R. Properties of a cryptic high-frequency transducing phage in Staphylococcus aureus. Virology. 1967 Sep;33(1):155–166. doi: 10.1016/0042-6822(67)90105-5. [DOI] [PubMed] [Google Scholar]
- Oertel W., Kollek R., Beck E., Goebel W. The nucleotide sequence of a DNA fragment from the replication origin of the antibiotic resistance factor R1drd19. Mol Gen Genet. 1979 Mar 27;171(3):277–285. doi: 10.1007/BF00267582. [DOI] [PubMed] [Google Scholar]
- Projan S. J., Carleton S., Novick R. P. Determination of plasmid copy number by fluorescence densitometry. Plasmid. 1983 Mar;9(2):182–190. doi: 10.1016/0147-619x(83)90019-7. [DOI] [PubMed] [Google Scholar]
- Thuring R. W., Sanders J. P., Borst P. A freeze-squeeze method for recovering long DNA from agarose gels. Anal Biochem. 1975 May 26;66(1):213–220. doi: 10.1016/0003-2697(75)90739-3. [DOI] [PubMed] [Google Scholar]
- Timmis K., Cabello F., Cohen S. N. Utilization of two distinct modes of replication by a hybrid plasmid constructed in vitro from separate replicons. Proc Natl Acad Sci U S A. 1974 Nov;71(11):4556–4560. doi: 10.1073/pnas.71.11.4556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomizawa J., Itoh T. Plasmid ColE1 incompatibility determined by interaction of RNA I with primer transcript. Proc Natl Acad Sci U S A. 1981 Oct;78(10):6096–6100. doi: 10.1073/pnas.78.10.6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomizawa J., Itoh T., Selzer G., Som T. Inhibition of ColE1 RNA primer formation by a plasmid-specified small RNA. Proc Natl Acad Sci U S A. 1981 Mar;78(3):1421–1425. doi: 10.1073/pnas.78.3.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlin B. E., Nordström K. Plasmid incompatibility and control of replication: copy mutants of the R-factor R1 in Escherichia coli K-12. J Bacteriol. 1975 Nov;124(2):641–649. doi: 10.1128/jb.124.2.641-649.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]