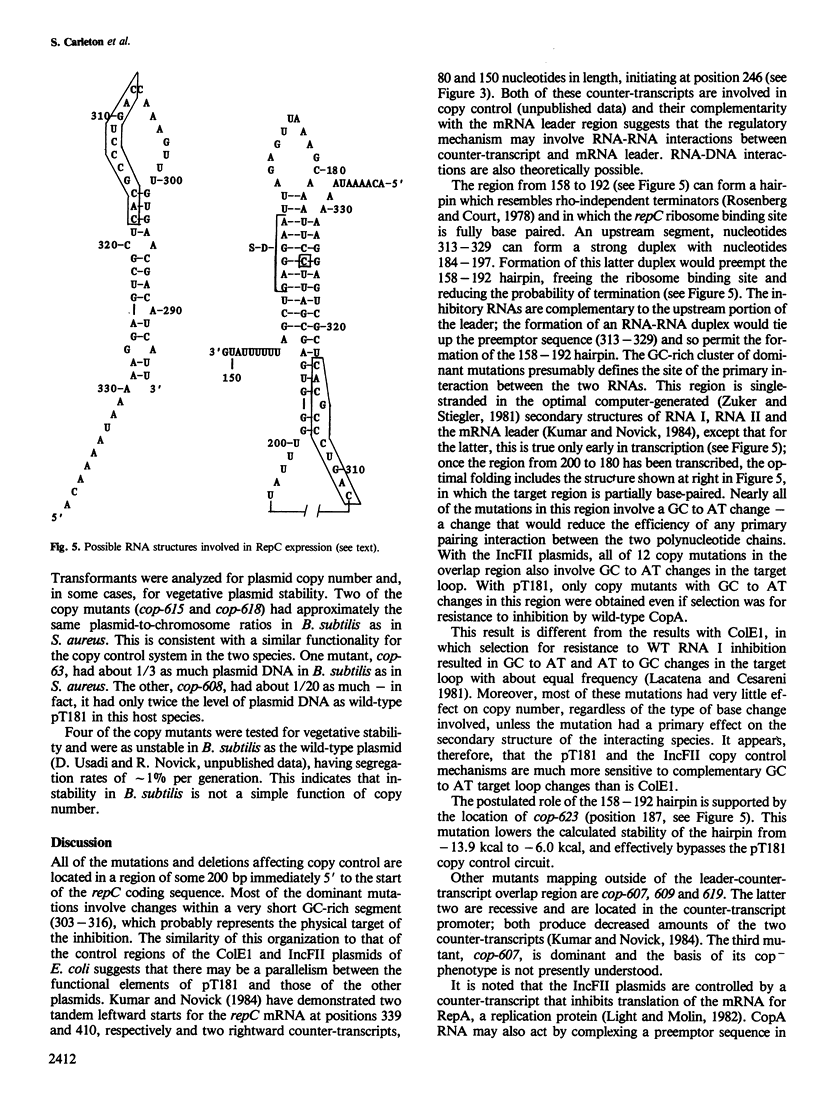
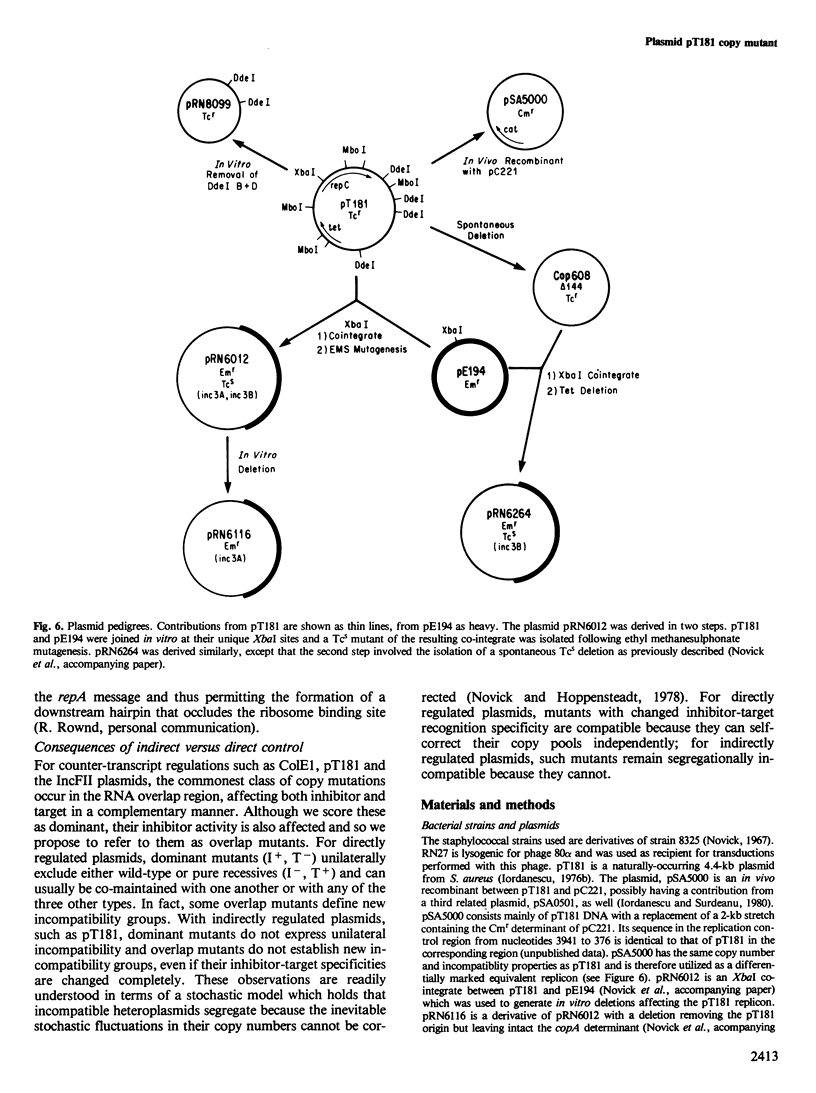
Abstract
We describe the isolation and analysis of mutations affecting the regulation of Staphylococcus aureus plasmid pT181 replication. Previous results suggested that regulation is achieved by control of the synthesis of RepC, a plasmid-coded replication protein and that the primary negative control element is CopA RNA, which consists of two transcripts that are complementary to the 5' region of the repC mRNA leader. CopA inhibition probably involves a base pairing interaction with the complementary region of the RepC mRNA leader which would facilitate the formation of a downstream stem-loop in the leader that occludes the repC ribosome binding site. RepC is freely diffusible so that regulation of pT181 replication is indirect. Both CopA RNA-sensitive (recessive) and -insensitive (dominant) mutants were isolated. The recessives have defects in CopA RNA structure or activity, the dominants have defects in the site of action (target) of the inhibitor. Some dominants were located within the copA coding sequence. These therefore affect the structure of CopA RNA as well as that of its target. Other dominant mutations mapped outside of the copA gene and therefore produced wild-type CopA RNA. In contrast to directly regulated plasmids, pT181 copy mutants producing wild-type inhibitor could be co-maintained with the wild-type plasmid and mutational changes in inhibitor-target specificity did not change incompatibility specificity.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bak A. L., Christiansen C., Stenderup A. Bacterial genome sizes determined by DNA renaturation studies. J Gen Microbiol. 1970 Dec;64(3):377–380. doi: 10.1099/00221287-64-3-377. [DOI] [PubMed] [Google Scholar]
- Chang S., Cohen S. N. High frequency transformation of Bacillus subtilis protoplasts by plasmid DNA. Mol Gen Genet. 1979 Jan 5;168(1):111–115. doi: 10.1007/BF00267940. [DOI] [PubMed] [Google Scholar]
- Easton A. M., Rownd R. H. The incompatibility product of IncFII R plasmid NR1 controls gene expression in the plasmid replication region. J Bacteriol. 1982 Nov;152(2):829–839. doi: 10.1128/jb.152.2.829-839.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GORINI L., MAAS W. K. The potential for the formation of a biosynthetic enzyme in Escherichia coli. Biochim Biophys Acta. 1957 Jul;25(1):208–209. doi: 10.1016/0006-3002(57)90450-x. [DOI] [PubMed] [Google Scholar]
- Iordănescu S., Surdeanu M. Complementation of a plasmid replication defect by autonomous incompatible plasmids in Staphylococcus aureus. Plasmid. 1980 Jul;4(1):1–7. doi: 10.1016/0147-619x(80)90078-5. [DOI] [PubMed] [Google Scholar]
- Iordănescu S. Three distinct plasmids originating in the same Staphylococcus aureus strain. Arch Roum Pathol Exp Microbiol. 1976 Jan-Jun;35(1-2):111–118. [PubMed] [Google Scholar]
- Khan S. A., Adler G. K., Novick R. P. Functional origin of replication of pT181 plasmid DNA is contained within a 168-base-pair segment. Proc Natl Acad Sci U S A. 1982 Aug;79(15):4580–4584. doi: 10.1073/pnas.79.15.4580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacatena R. M., Cesareni G. Base pairing of RNA I with its complementary sequence in the primer precursor inhibits ColE1 replication. Nature. 1981 Dec 17;294(5842):623–626. doi: 10.1038/294623a0. [DOI] [PubMed] [Google Scholar]
- Light J., Molin S. The sites of action of the two copy number control functions of plasmid R1. Mol Gen Genet. 1982;187(3):486–493. doi: 10.1007/BF00332633. [DOI] [PubMed] [Google Scholar]
- MASTER R. W. POSSIBLE SYNTHESIS OF POLYRIBONUCLEOTIDES OF KNOWN BASE-TRIPLET SEQUENCES. Nature. 1965 Apr 3;206:93–93. doi: 10.1038/206093b0. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Novick R. P., Adler G. K., Majumder S., Khan S. A., Carleton S., Rosenblum W. D., Iordanescu S. Coding sequence for the pT181 repC product: a plasmid-coded protein uniquely required for replication. Proc Natl Acad Sci U S A. 1982 Jul;79(13):4108–4112. doi: 10.1073/pnas.79.13.4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick R. P., Brodsky R. Studies on plasmid replication. I. Plasmid incompatibility and establishment in Staphylococcus aureus. J Mol Biol. 1972 Jul 21;68(2):285–302. doi: 10.1016/0022-2836(72)90214-8. [DOI] [PubMed] [Google Scholar]
- Novick R. P., Hoppensteadt F. C. On plasmid incompatibility. Plasmid. 1978 Sep;1(4):421–434. doi: 10.1016/0147-619x(78)90001-x. [DOI] [PubMed] [Google Scholar]
- Novick R. P., Murphy E., Gryczan T. J., Baron E., Edelman I. Penicillinase plasmids of Staphylococcus aureus: restriction-deletion maps. Plasmid. 1979 Jan;2(1):109–129. doi: 10.1016/0147-619x(79)90010-6. [DOI] [PubMed] [Google Scholar]
- Novick R. Properties of a cryptic high-frequency transducing phage in Staphylococcus aureus. Virology. 1967 Sep;33(1):155–166. doi: 10.1016/0042-6822(67)90105-5. [DOI] [PubMed] [Google Scholar]
- Pattee P. A., Neveln D. S. Transformation analysis of three linkage groups in Staphylococcus aureus. J Bacteriol. 1975 Oct;124(1):201–211. doi: 10.1128/jb.124.1.201-211.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Projan S. J., Carleton S., Novick R. P. Determination of plasmid copy number by fluorescence densitometry. Plasmid. 1983 Mar;9(2):182–190. doi: 10.1016/0147-619x(83)90019-7. [DOI] [PubMed] [Google Scholar]
- Rosenberg M., Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- Tomizawa J., Itoh T., Selzer G., Som T. Inhibition of ColE1 RNA primer formation by a plasmid-specified small RNA. Proc Natl Acad Sci U S A. 1981 Mar;78(3):1421–1425. doi: 10.1073/pnas.78.3.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M., Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981 Jan 10;9(1):133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]