Abstract
As typical Chinese indigenous pig breeds, Erhualian and Meishan have been widely used to produce new strain or breed in the world. However, the genetic basis of characteristics of these pig breeds is still limited. Moreover, considering cost and output of sequencing, it is necessary to further develop cost-effective method for pig genome screening. To contribute on this issue, we developed a SLAF-seq (specific-locus amplified fragment sequencing) method for pigs and applied it to analyze the genetic difference among Landrace, Erhualian, and Meishan pigs. A total of 453.75 million reads were produced by SLAF-seq. After quality-control, 165,670 SNPs (single nucleotide polymorphisms) were used in further analysis. The results showed that Landrace had distinct genetic relationship compared to Erhualian (F ST = 0.5480) and Meishan (F ST = 0.5800), respectively, while Erhualian and Meishan held the relatively close genetic relationship (F ST = 0.2335). Furthermore, a genome-wide scanning revealed 268 differentially selected regions (DSRs) with 855 genes and 256 DSRs with 347 genes between Landrace and the two Chinese indigenous pig breeds and between Erhualian and Meishan, respectively. This study provides a new cost-effective method for pig genome study and might contribute to a better understanding on the formation mechanism of genetic difference among pigs with different geographical origins.
Introduction
In 2009, the pig genome was being sequenced and annotated for public utility1, which made it possible for analysis and application of genome-wide genetic variants of pig. Shortly afterwards, Illumina developed the PorcineSNP60 Genotyping BeadChip, which included around 60,000 loci by genotyping 158 individuals across multiple pig breeds2. Due to the advantages of high density and inexpensive price, the Illumina PorcineSNP60 Genotyping BeadChip has been widely used in multiple research areas. For example, identifying candidate genes associated with economically important traits in different pig populations3–5, detecting inbreeding depression of reproductive traits in Iberian pigs6, mining genetic diversity and selection signatures in Chinese and Western pig breeds7, and discovering population structure of local pig breeds from Russia, Belorussia, Kazakhstan, and Ukraine8. These studies greatly enhance our knowledge for understanding the information implied in the pig genome.
Simultaneously, with the rapid development of the next generation sequencing technology, genome resequencing has also been introduced into the pig genome research. i.e., identifying candidate loci related to economically important traits between Korean and European origin breeds9, digging signatures of selection in European or Asian domestic pig genome10, 11, and measuring coancestry and fitness in Sus cebifrons and Pietrain pigs to design optimal breeding or conservation programs12. As a full-scanning approach for discovering all possible genome-wide variants, genome resequencing is considered as a more comprehensive solution for resolving the mystery of the genome. But so far, to some extent, the high costs of sequencing and bioinformatics analysis from genome resequencing have limited its application in population genetics of domestic animals, particularly in some research groups with insufficient funds.
Alternatively, other approaches based on reduced representation genome sequencing strategy to reduce complexity in the genome for resequencing can be applied in pig genome-wide study13–15. For example, genome reducing and sequencing (GGRS), which contains more number of loci but similar or lower costs than that of Illumina PorcineSNP60 Genotyping BeadChip, has recently been used to identify selection signatures in Yorkshire and Landrace pigs16 and discover the genetic diversity of Chinese indigenous pig breeds of the Taihu Lake region17. Meanwhile, another reduced representation genome sequencing, named specific-locus amplified fragment sequencing (SLAF-seq), was also exploited. As a reduced representation library and high-throughput sequencing based approach, SLAF-seq can pre-design the experimental project through bioinformatics and selection of fragments with a specific length in constructing library. More importantly, SLAF-seq has been considered as a high cost-effective way for large-scale genotyping due to its advantages of high genotyping accuracy, high efficiency for marker discovery, low cost, and high capacity for large populations18. Therefore, SLAF-seq is widely applied for revealing loci or genes associated with characteristic traits of multiple breeds. i.e., fruit traits in cucumber19, tolerance to low-phosphorus stress in soybean20, and disease, growth and carcass traits in chicken21–23. Obviously, these data support that reduced representation genome sequencing strategy is a convenient and effective way for genome-wide study in plants and animals.
As representatives of Chinese pigs, Erhualian and Meishan are two native pig breeds with high reproductive performance and excellent meat quality such as high litter size and intramuscular fat content. Most recently, many single nucleotide polymorphisms (SNPs) and candidate genes associated with reproductive or meat quality traits in Erhualian pigs have been identified using Illumina PorcineSNP60 Genotyping BeadChip24, 25. Additionally, due to high costs, a limited number of pigs were used (usually from 4 to 10 individuals) for genome resequencing and most of the studies classified Erhualian and Meishan as one of domesticated pig breeds rather than separate groups for further analysis10, 26, 27. Knowledge of the genetic difference between Erhualian and Meishan pig breeds is lacking. Furthermore, a genome-wide view of possible candidate loci or genes associated with the characteristics of Erhualian and Meishan pigs compared with other distant breeds such as European origin pigs has still not been well investigated. Taken together, for the first time, a SLAF-seq approach for pigs was developed in this study and then was performed to analyze the genetic difference among Landrace, Erhualian, and Meishan pigs, thus providing a cost-effective way for pig genome-wide study and new guidance for understanding the genetic basis of germplasm characteristics of these pig breeds.
Results
Development of SLAF markers in the pig genome
In order to exploit SLAF markers, in silico restriction analysis was firstly performed on the current pig reference genome (Sscrofa 10.2). As a result, two restriction enzymes, RsaI and HaeIII, were used as enzyme combination for developing around 500,000 SLAFs. Finally, a total of 516,733 SLAF tags were predicted using in silico restriction analysis, which showed similar average SLAF distance among different chromosomes (Supplementary Table S1).
Specific-locus amplified fragment sequencing (SLAF) basics
To obtain the actual SLAF markers used in this study, SLAF-seq was performed in 28 Landrace, 26 Erhualian, and 27 Meishan individuals using the same enzyme combination as in silico restriction analysis. As shown in Table 1 and Supplementary Table S2, a total of 453.75 million reads were obtained from all individuals, which showed that average Q30 and GC content were 87.67% and 44.55%, respectively. Similar to the number of expected SLAFs, 461,261 unique SLAF tags, with average SLAF numbers and sequencing depth as 398,034 (12.74×), 344,471 (6.10×), and 369,160 (6.13×) in Landrace, Erhualian, and Meishan pig populations, respectively, were identified further (Table 1 and Supplementary Table S3). Moreover, compared to the distribution of the expected 516,733 SLAF tags, similar uniformly distribution of 461,261 SLAF tags was also found on different chromosomes (Supplementary Figure S1). In addition, Oryza sativa indica was used as a control during sequencing, the results found that the percentage of digestion normally and paired-end mapped reads of control were 91.73% and 90.68%, respectively, indicating that SLAF-seq process was normal and available.
Table 1.
Characteristics of SLAF-seq among Landrace, Erhualian, and Meishan.
| Item | Landrace | Erhualian | Meishan | Average of individuals | Oryza sativa indica |
|---|---|---|---|---|---|
| Statistics of reads | |||||
| Q30 percentage (%) | 83.26–87.40 | 87.03–89.21 | 87.51–89.89 | 87.67 | 88.24 |
| GC percentage (%) | 42.93–46.89 | 44.26–46.76 | 42.74–45.32 | 44.55 | 42.48 |
| Number of reads | 7,314,304–11,830,078 | 2,572,513–5,014,955 | 3,151,950–5,353,332 | 5,601,972 | 1,452,138 |
| Average of reads | 9,038,472 | 3,601,484 | 3,964,589 | — | — |
| Statistics of SLAFs | |||||
| Number of SLAFs | 352,574–420,742 | 316,233–382,547 | 349,762–389,120 | 371,217 | — |
| Average of SLAFs | 398,034 | 344,471 | 369,160 | — | — |
| Average depth | 12.74 | 6.10 | 6.13 | — | — |
| Statistics of SNPs | |||||
| Number of SNPs | 1,615,995–2,604,078 | 1,160,477–1,663,707 | 1,322,101–1,701,153 | 1,680,561 | — |
| Heterozygosity (%) | 4.81–8.12 | 5.81–8.45 | 5.20–7.23 | 6.51 | — |
| Ratio of Integrity (%) | 30.07–48.45 | 21.59–30.95 | 24.60–31.65 | 31.26 | — |
| Average of SNPs | 2,176,131 | 1,302,927 | 1,530,285 | — | — |
Identification of SNPs in Landrace, Erhualian, and Meishan pigs
After genomic mapping and SNP calling, a total of 5,373,997 SNPs were discovered using all individuals (Supplementary Table S4). Moreover, an average of 2,176,131, 1,302,927, and 1,530,285 SNPs were discovered in Landrace, Erhualian, and Meishan pig populations, respectively (Table 1 and Supplementary Table S4). A series of quality control filtering of SNPs were performed to identify 165,670 SNPs used in the further analysis (Supplementary Table S5). Specifically, 161,377 autosomal SNPs were included in linkage disequilibrium and differentially selected region (DSR) analyses, while the 4,293 SNPs on chromosome X were used for DSRs analysis only (Supplementary Figure S2). To reduce the effect from ascertainment bias, a subset of 107,626 autosomal SNPs with minor allele frequency (MAF) ≥ 0.2 was produced and used in genetic structure analysis (Supplementary Figure S2).
Measuring of linkage disequilibrium by chromosomes
Linkage disequilibrium (r2) for pairs of loci was measured by chromosomes in each pig population. As shown in Supplementary Figure S3, the average r2 values dropped quickly when physical distances reached 40 kilo base pairs (Kb). With the increasing of physical distance, the similar decreasing trends of the average r2 values were observed among three populations. In addition, average r2 values of Landrace and Meishan were higher than that of Erhualian.
Accessing genetic structure, heterozygosity and gene diversity
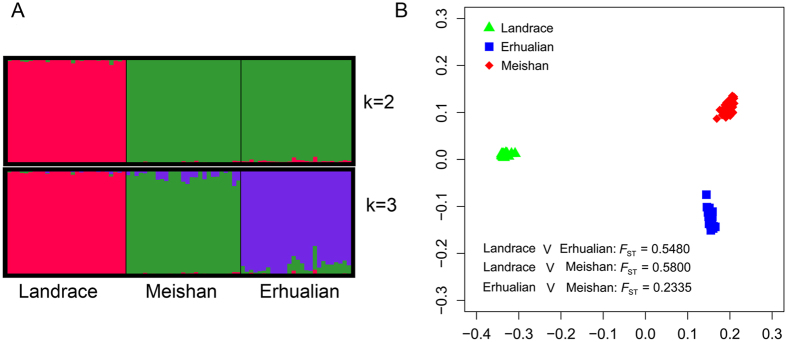
As expected, population structure analysis revealed that Landrace and Meishan formed the first two independent populations (K = 2) followed by the Erhualian (K = 3) (Fig. 1A). Moreover, a multidimensional scaling plot clearly showed that Landrace was a distant population compared with Erhualian and Meishan (Fig. 1B). Based on pair-wise estimates of F ST described by Weir and Cockerham28, the significant genetic differentiation appeared between Landrace and Erhualian (F ST = 0.5480), between Landrace and Meishan (F ST = 0.5800), and between Erhualian and Meishan (F ST = 0.2335), respectively, demonstrating that Erhualian and Meishan hold closer genetic relationship compared to Landrace. As shown in Supplementary Figure S4, observed heterozygosity of Landrace, Erhualian, and Meishan was 0.1956, 0.2410, and 0.2432, respectively, while the gene diversity of Landrace, Erhualian, and Meishan was 0.2276, 0.2952, and 0.2776, respectively. Erhualian and Meishan had the higher observed heterozygosity and gene diversity than that of Landrace.
Figure 1.
Population structure analysis of the genetic differentiation among Landrace, Erhualian, and Meishan. (A) Population structure among Landrace, Erhualian, and Meishan using STRUCTURE software. (B) Multidimensional scaling plots among Landrace, Erhualian, and Meishan using PLINK software. F ST represents the pair-wise F ST between any two pig populations.
Identification of DSRs between Landrace and the two Chinese indigenous pig breeds
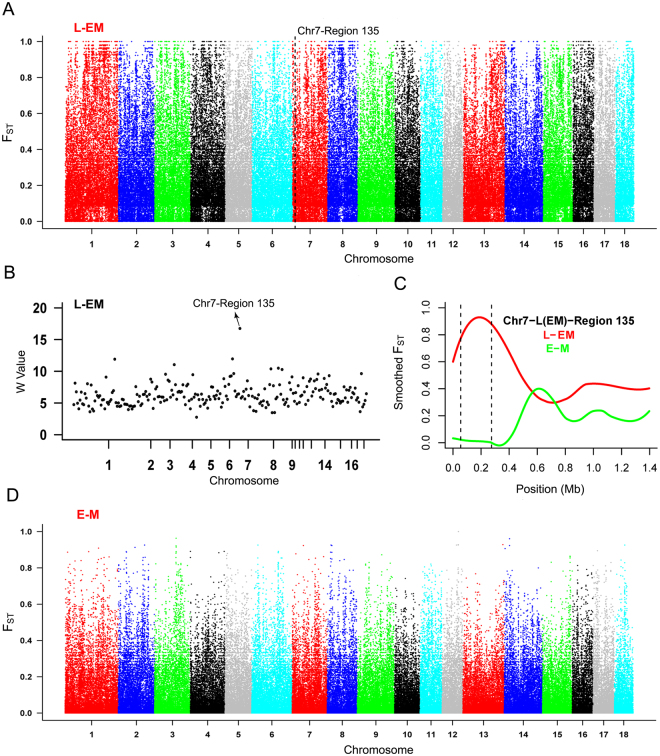
To obtain the genomic difference among different pig breeds, a SNP-based F ST estimation was performed to identify DSRs, which defined regions with shared differentially selection signals across breeds. As these two Chinese indigenous pig breeds have similar origin, Erhualian and Meishan were firstly merged as one population compared with Landrace. As a result, a total of 268 DSRs were identified between Landrace and the two Chinese indigenous pig breeds, which included 237 DSRs in autosomes and 31 DSRs in chromosome X (Supplementary Table S6). The DSR (named region 135) with both the richest top SNPs and the highest W value was located on chromosome 7 (Table 2 and Fig. 2A–C). Moreover, 855 unique known genes, including 511 genes from autosomal DSRs and 344 genes from X chromosome DSRs (Table 2 and Supplementary Table S6), were discovered further, which included many genes associated with economically important traits, i.e., ESR1 and KIT. Furthermore, the gene functional analysis showed that the biological processes between Landrace and the two Chinese indigenous pig breeds were enriched in flavonoid related metabolic process, pigment biosynthetic process, response to hormone, and regulation of developmental process (Supplementary Table S7).
Table 2.
Top 20 percent of autosomal differentially selected regions (DSRs) among Landrace, Erhualian, and Meishan.
| Region | W value | Chr | Position | Top 0.5% | Top 5% | Gene | Candidates |
|---|---|---|---|---|---|---|---|
| DSRs between Landrace and the two Chinese indigenous pig breeds | |||||||
| 2 | 8.12 | 1 | 16639564..16894373 | 0 | 16 | 1 | ESR1 |
| 12 | 8.02 | 1 | 125860631..126237234 | 1 | 13 | 5 | ALDH1A2,AQP9 |
| 15 | 7.82 | 1 | 157931791..158647002 | 2 | 16 | 8 | NA |
| 27 | 9.07 | 1 | 245581640..246493427 | 3 | 20 | 6 | SMARCA2 |
| 34 | 11.89 | 1 | 248939813..249341671 | 11 | 17 | 3 | FAM189A2,C1H9orf135 |
| 51 | 7.56 | 1 | 305534056..305560192 | 4 | 9 | 1 | RAPGEF1 |
| 63 | 8.74 | 2 | 132479091..132493356 | 4 | 7 | 1 | CSNK1G3 |
| 64 | 8.29 | 2 | 139541646..139596463 | 0 | 9 | 2 | ACSL6,MEIKIN |
| 68 | 7.56 | 2 | 153497601..153536938 | 0 | 6 | 1 | PRELID2 |
| 75 | 9.52 | 3 | 40440733..40671307 | 9 | 7 | 25 | SRRM2,TCEB2,BICDL2, FLYWCH1,CCDC64B, LOC100737121,LOC102158577 |
| 77 | 8.60 | 3 | 41967307..42842043 | 9 | 4 | 48 | KCTD5,UNKL,PDPK1,RNPS1 |
| 78 | 8.41 | 3 | 42957871..43071250 | 4 | 7 | 2 | IARS |
| 82 | 11.03 | 3 | 77090252..77374423 | 7 | 15 | 7 | ARHGAP25,BMP10, APLF |
| 83 | 7.87 | 3 | 78080175..78118235 | 2 | 6 | 0 | NA |
| 89 | 7.86 | 4 | 47500407..47599740 | 2 | 7 | 0 | NA |
| 92 | 7.69 | 4 | 53117484..53787644 | 3 | 7 | 2 | MMP16 |
| 93 | 7.95 | 4 | 55932479..56325201 | 3 | 3 | 3 | CA2,LOC100154871 |
| 97 | 9.45 | 4 | 90369121..90399119 | 0 | 17 | 2 | TBX19,SFT2D2 |
| 122 | 8.81 | 6 | 80274787..80286579 | 6 | 2 | 0 | NA |
| 123 | 7.60 | 6 | 80425263..80462131 | 4 | 3 | 0 | NA |
| 126 | 7.93 | 6 | 82230546..82355861 | 0 | 10 | 1 | NA |
| 127 | 9.18 | 6 | 82692715..82761603 | 0 | 9 | 7 | TXLNA,LOC100739580 |
| 128 | 9.25 | 6 | 82958401..83041048 | 1 | 8 | 4 | ZBTB8A,RBBP4, ZBTB8OS |
| 129 | 11.94 | 6 | 83940808..84077674 | 6 | 14 | 1 | LOC100523068 |
| 130 | 9.63 | 6 | 84350867..84495848 | 3 | 9 | 1 | C6H1orf94 |
| 135 | 16.74 | 7 | 55136..274807 | 11 | 23 | 1 | EXOC2 |
| 141 | 8.48 | 7 | 60246223..60417535 | 3 | 8 | 3 | ANPEP,ARPIN |
| 159 | 7.89 | 8 | 55208909..55760237 | 5 | 6 | 7 | LOC102159670 |
| 160 | 10.37 | 8 | 57463293..58027011 | 1 | 11 | 12 | HOPX,THEGL, REST |
| 162 | 7.70 | 8 | 60649748..62169164 | 9 | 4 | 0 | NA |
| 166 | 10.48 | 8 | 69657439..69988936 | 4 | 16 | 7 | CENPC,UBA6, LOC100512727 |
| 167 | 7.86 | 8 | 70578162..71233566 | 9 | 4 | 10 | AMTN,UGT2A1,LOC100515222 |
| 169 | 10.27 | 8 | 71666052..72013805 | 13 | 4 | 3 | SLC4A4,LOC100739482 |
| 175 | 8.18 | 8 | 148318210..148486319 | 7 | 5 | 8 | CSN1S2,ODAM, PRR27 |
| 177 | 9.10 | 9 | 44454513..44465001 | 5 | 2 | 1 | DIXDC1 |
| 182 | 7.72 | 11 | 52521631..52714330 | 3 | 6 | 2 | NA |
| 186 | 8.96 | 12 | 20061666..20149830 | 2 | 7 | 4 | BRCA1 |
| 189 | 7.58 | 13 | 42228668..42395388 | 0 | 7 | 2 | CCDC66,FAM208A |
| 191 | 8.41 | 13 | 194337969..194393374 | 0 | 8 | 0 | NA |
| 192 | 8.76 | 13 | 200757712..201353485 | 0 | 13 | 1 | NA |
| 195 | 8.46 | 13 | 210377658..210418642 | 0 | 9 | 0 | NA |
| 197 | 8.70 | 13 | 216483385..216515468 | 6 | 2 | 2 | NA |
| 198 | 9.56 | 14 | 22924788..23026160 | 0 | 9 | 0 | NA |
| 203 | 8.86 | 14 | 135519785..135551251 | 0 | 9 | 1 | NA |
| 205 | 7.91 | 14 | 137758835..137835354 | 3 | 5 | 1 | CCDC172 |
| 207 | 9.29 | 14 | 144278853..144330521 | 0 | 12 | 2 | BUB3 |
| 233 | 9.62 | 18 | 43951415..44344272 | 3 | 10 | 2 | BBS9,LOC100738822 |
| DSRs between Erhualian and Meishan | |||||||
| 7 | 9.63 | 1 | 15312287..15312564 | 4 | 2 | 0 | NA |
| 29 | 10.25 | 2 | 26179507..26497490 | 3 | 15 | 0 | NA |
| 35 | 10.89 | 2 | 122905147..123003827 | 3 | 7 | 0 | NA |
| 45 | 10.22 | 2 | 143611291..143611845 | 1 | 7 | 1 | SLC25A48 |
| 46 | 9.51 | 3 | 4355611..4371774 | 5 | 1 | 2 | RADIL,MMD2 |
| 52 | 9.87 | 3 | 90528360..90824884 | 3 | 3 | 8 | CCDC104,CLHC1, SMEK2, MTIF2 |
| 56 | 11.87 | 3 | 102297392..102494238 | 4 | 5 | 4 | PPM1B,PREPL, SLC3A1 |
| 58 | 9.91 | 3 | 102678119..102743126 | 4 | 2 | 1 | LRPPRC |
| 59 | 9.83 | 3 | 106283461..106442746 | 1 | 8 | 1 | SLC8A1 |
| 61 | 9.97 | 3 | 113055246..113120757 | 4 | 1 | 1 | LOC100626840 |
| 64 | 10.87 | 3 | 115856309..116048881 | 6 | 7 | 4 | YPEL5 |
| 67 | 8.83 | 4 | 18251390..18266000 | 0 | 6 | 0 | NA |
| 75 | 9.11 | 5 | 5199728..5220416 | 3 | 4 | 2 | SGSM3,MKL1 |
| 78 | 10.20 | 5 | 19478109..19576660 | 4 | 8 | 1 | NA |
| 98 | 10.37 | 6 | 102930166..103055274 | 5 | 1 | 2 | ZNF521 |
| 99 | 15.01 | 6 | 103154776..103528819 | 14 | 0 | 2 | NA |
| 100 | 12.80 | 6 | 103780912..104046778 | 9 | 3 | 4 | PSMA8,KCTD1,TAF4B |
| 104 | 11.46 | 6 | 124785239..124938047 | 6 | 4 | 2 | PTGFR |
| 110 | 10.20 | 7 | 34828000..34927976 | 3 | 1 | 2 | GRM4 |
| 111 | 9.08 | 7 | 39781167..39790945 | 3 | 3 | 0 | NA |
| 113 | 12.53 | 7 | 91745010..91808309 | 6 | 5 | 0 | NA |
| 117 | 10.97 | 7 | 105180004..105206797 | 3 | 3 | 2 | IFT43,TGFB3 |
| 118 | 9.18 | 8 | 10911848..10912137 | 4 | 0 | 2 | PROM1,LOC102160409 |
| 121 | 9.71 | 8 | 13697594..14004707 | 3 | 2 | 0 | NA |
| 134 | 14.91 | 9 | 40038695..40228268 | 12 | 2 | 8 | CWF19L2,ELMOD1, ALKBH8,LOC100626234 |
| 138 | 8.79 | 9 | 95612266..95753391 | 0 | 7 | 0 | NA |
| 139 | 12.75 | 9 | 105189739..105220813 | 7 | 0 | 0 | NA |
| 143 | 10.80 | 9 | 142936175..143029098 | 2 | 10 | 0 | NA |
| 146 | 10.06 | 10 | 3770676..3770960 | 1 | 5 | 0 | NA |
| 147 | 9.17 | 10 | 31350537..31364218 | 2 | 2 | 1 | C10H9orf3 |
| 164 | 8.86 | 11 | 50583012..50964515 | 2 | 17 | 2 | KLF12,LOC100622863 |
| 177 | 9.00 | 12 | 46196899..46237519 | 4 | 6 | 0 | NA |
| 180 | 14.14 | 12 | 51152923..51247795 | 8 | 0 | 2 | NA |
| 182 | 9.99 | 13 | 12498982..12503925 | 0 | 7 | 1 | THRB |
| 183 | 11.92 | 13 | 13052089..13079543 | 0 | 9 | 0 | NA |
| 185 | 15.35 | 13 | 189288665..189354590 | 1 | 10 | 1 | LIPI |
| 186 | 12.39 | 13 | 190678701..190700337 | 0 | 13 | 0 | NA |
| 188 | 10.51 | 13 | 216737510..216756384 | 2 | 3 | 0 | NA |
| 192 | 16.19 | 14 | 16460290..16486569 | 10 | 5 | 0 | NA |
| 195 | 11.06 | 14 | 21329899..21678452 | 5 | 8 | 4 | CLCN3,NEK1 |
| 201 | 10.68 | 14 | 47963670..48581378 | 1 | 11 | 5 | MN1,PITPNB,TTC28 |
| 202 | 12.12 | 14 | 48900016..49663109 | 2 | 15 | 17 | EWSR1,KREMEN1,NIPSP1, RHBDD3 |
| 218 | 10.39 | 15 | 152011389..152018640 | 2 | 2 | 1 | UBE2F |
| 223 | 8.72 | 16 | 24195627..24417936 | 2 | 15 | 1 | NA |
| 230 | 9.00 | 16 | 82918895..82946838 | 3 | 2 | 0 | NA |
| 235 | 8.67 | 17 | 11918899..11919532 | 0 | 7 | 0 | NA |
| 236 | 9.23 | 17 | 15112581..15187900 | 3 | 3 | 3 | TMEM230,CDS2 |
| 237 | 9.47 | 17 | 15425824..15480579 | 1 | 8 | 1 | GPCPD1 |
| 240 | 10.06 | 17 | 47494168..47511555 | 4 | 1 | 1 | PPP1R16B |
| 252 | 9.81 | 18 | 41156162..41171465 | 2 | 6 | 2 | AOAH |
Note: The candidate genes were given within the SNPs of top SNP (0.5% or 5%) for each region.
Figure 2.
Global distribution of F ST among Landrace, Erhualian, and Meishan. (A) Global distribution of F ST between Landrace and the two Chinese indigenous pig breeds on autosomes 1–18. A representative DSR (region 135 of SSC7) with both the richest top SNPs and the highest W value was indicated. (B) The W-statistics was performed for DSRs between Landrace and the two Chinese indigenous pig breeds. (C) Smoothed F ST showed strong selection signals in region 135 between Landrace and the two Chinese indigenous pig breeds. L-EM represents Landrace-the two Chinese indigenous pig breeds, while E-M means Erhualian-Meishan. (D) Global distribution of F ST between Erhualian and Meishan on autosomes 1–18.
DSRs associated with female characters between Landrace and the two Chinese indigenous pig breeds
In order to further investigate DSRs or genes related to female characters, a female based genome-wide screening was performed between Landrace and the two Chinese indigenous pig breeds. A dataset with MAF ≥ 0.1 containing 175,952 SNPs from autosomes and 8,320 SNPs from chromosome X was used in this analysis. Based on the SNP F ST estimates, 241 DSRs in autosomes and 60 DSRs in chromosome X were revealed. Further examination of these DSRs identified 443 unique known genes from autosomes and 327 genes from chromosome X (Supplementary Table S8). GO terms of these genes from DSRs were involved in chromatin organization, odontogenesis, animal organ development, and reproductive system development (Supplementary Table S9).
Characterization of DSRs between Erhualian and Meishan
A genome-wide distribution of F ST between Erhualian and Meishan on autosomes is shown in Fig. 2D. Between these two pig breeds, 256 DSRs, including 252 DSRs in autosomes and 4 DSRs in chromosome X, were discovered. Moreover, 341 unique known genes from autosomal DSRs and 6 genes from X chromosome DSRs were discovered further (Supplementary Table S10). The gene enrichment analysis found that DSRs between Erhualian and Meishan were enriched in sodium-independent organic anion transport, glycerophospholipid metabolic process, and phosphorus or phospholipid related metabolic process (Supplementary Table S11).
Discussion
In the present study, we used SLAF-seq to analyze the genetic structure and differentially selected regions among Landrace, Erhualian, and Meishan pigs. By using over 160,000 SNPs developed in this study, our results clearly indicated that there was a closer genetic relationship between Erhualian and Meishan compared with the genetic relationship between Landrace and Erhualian or Meishan. Moreover, 268 DSRs including 855 genes were discovered between Landrace and the two Chinese indigenous pig breeds, while 256 DSRs including 347 genes were found between Erhualian and Meishan. As such, our data provide a comprehensive view of the genome-wide basis for genetic difference among these pig breeds.
Basically, as a high-resolution method for genome-wide genotyping, SLAF-seq is considered as an enhanced reduced representation sequencing and possesses several remarkable features18, i.e., accurate genotyping by depth paired-end sequencing, reduced sequencing costs, pre-design strategy for fragmentation efficiency, and double barcode system for large populations. In this study, a total of 516,733 SLAFs from the pig reference genome (Sscrofa 10.2) were predicted using RsaI and HaeIII enzyme combination. With more than 6 × sequencing depth but below $80 cost for each individual, 5,373,997 SNPs were discovered from Landrace, Erhualian, and Meishan pig populations by SLAF-seq. After filtering, 165,670 SNPs were available for analyzing the genetic difference among these pig populations, which would have an average inter-marker distance of approximately 17 Kb and several multiples of the number of SNPs from Illumina PorcineSNP60 Genotyping BeadChip. Compared with the published pig GGRS, which contained 34,789–105,550 available SNPs for different objective analysis14, 17, our SLAF-seq method might provide more genomic variation information. More importantly, our data explored the genetic difference between Landrace and the two Chinese pig breeds using reduced representation sequencing strategy, which has not been well studied by GGRS. Considering the huge difference in genome variation between European and Chinese pig breeds, our data demonstrate that SLAF-seq is a powerful approach and has great potential for further studies in more breeds. Therefore, the SLAF-seq approach could be considered as a more competitive choice for pig genome study.
It is well known that Landrace was originally developed in Denmark, while Erhualian and Meishan were domesticated in China, implying that Landrace was as a geographically distant breed relative to Erhualian and Meishan. As expected, both population structure and cluster analysis showed that Landrace was genetically distant from two Chinese indigenous pig breeds (Fig. 1). Moreover, pair-wise F ST estimation supported Landrace had distant genetic relationship compared with two Chinese indigenous pig breeds (F ST = 0.5480 for Erhualian and F ST = 0.5800 for Meishan), demonstrating that there was a great genetic differentiation between Landrace and two Chinese indigenous pig breeds. These results were consistent with the data from other genetic markers such as mitochondrial DNA and microsatellite markers29, 30, which showed European and Chinese pig breeds exist long genetic distances. In particular, we found there was close but significant genetic differentiation (F ST = 0.2335) between Erhualian and Meishan. Actually, a significant genetic differentiation between Erhualian and Meishan had been observed using GGRS methods17. Therefore, our data provide a further evidence that Erhualian and Meishan should be two pig breeds, although they had been classified as the same pig breed named Taihu pigs in the annals of history as both pig breeds lived in Taihu Lake regions, eastern of China. Additionally, higher observed heterozygosity and gene diversity were found in these two Chinese indigenous pig breeds compared with Landrace, implying that these two Chinese indigenous pig breeds had more abundant genetic diversity. This is not surprising because Chinese indigenous breeds had more genetic diversity than that of European pig breeds30. Accordingly, this study provides a global view of the genetic structure and relationship among these three pig populations.
Generally speaking, Erhualian and Meishan are two of the most prolific pig breeds known in the world24, 31, 32 and have been mainly considered as the maternal line for synthetic strain selection. Meanwhile, these two pig breeds are used to build specific populations for identifying candidate genes or QTLs (quantitative trait loci) related to reproductive traits, i.e., Iberian x Meishan F2 population33, Large White × Meishan cross gilts34, and Duroc x Erhualian resource population35. Intuitively, compared to Landrace, a major of the maternal sources in European pig breeds, Erhualian and Meishan pigs possess the better farrowing capacity, which results in far more litter size36, 37. To further identify the candidates related to female reproductive traits, a female based dataset was used at the same time. Compared with the candidate genes from all individuals, a total of 502 genes from female based DSRs were found overlapping (Supplementary Tables S6 and S8). Many important genes related to female reproductive traits such as ESR1, RBBP4, and BRCA1, were discovered in both DSR datasets. In the previous study, as a key gene for mediating estrogen, ESR1 with its PvuII site polymorphism had been shown to be associated with litter size in different pig populations, i.e., Meishan synthetic line38, Landrace population39, a population of Erhualian and Xiang pig breeds40, and Large White x Meishan F2 crossbred gilts41. However, other studies did not find significant associations of ESR1 with litter size in a synthetic line of Duroc and Large White origin or a Meishan x Large White F2 population42, 43, demonstrating that the role of ESR1 in sow prolificacy among different pig populations was still controversial. Therefore our data might provide additional information for interpreting the difference in litter size between Landrace and the two Chinese indigenous pig breeds. RBBP4, a biomarker of oocyte competence, had different expression levels between in vivo- and in vitro-matured oocytes in bovine44 and could affect bipolar spindle assembly by regulating histone deacetylation during oocyte maturation in mouse45. However, little is known about the function of RBBP4 in pig reproductive process. Our data suggest that RBBP4 might have an important role in sow reproductive performance, but confirmation of this assumption warrants further study. Interestingly, BRCA1, a key DNA repair protein, was identified in DSRs. Previously, genetic variants of BRCA1 were mainly related to breast or ovarian related diseases in human and cattle46, 47. Besides, reducing numbers of primordial follicles and simultaneously increasing DNA double-strand break repair with age were observed in BRCA1-deficient mice compared to the wild-type mice, showing that BRCA1-deficient led to ovarian aging48, 49. In women, BRCA1 mutation produced lower numbers of eggs compared with control50. These data indicated BRCA1 might play a critical role in regulating ovarian development and the number of eggs. Hence our results might give us a novel clue for its role in pig reproductive performance.
Furthermore, a DSR with both the richest top SNPs and the highest W value in autosomes was found on Sus scrofa chromosome 7 (SSC7, 0.05 to 0.27 Mb) between Landrace and the two Chinese indigenous pig breeds (Fig. 2, Table 2 and Supplementary Table S6). Interestingly, EXOC2, a component of the exocyst complex, was found in this region. Previous studies indicated that EXOC2 played a role in filopodia formation by binding the GTPase RalA in Swiss 3T3 cells and mediated vesicle trafficking by interacting with the Rho guanine nucleotide exchange factor (GEF)-H1 in HeLa cells51, 52. Additionally, genetic variants of EXOC2 or adjacent EXOC2 had been considered as contributing to pigmentary traits such as hair color, skin pigmentation and tanning ability in Europeans as well as vitamin D status in the Caucasian population53–57. In this study, Landrace has white hair color and skin, while Erhualian and Meishan have colored hair and skin37, indicating that there is great difference in pigmentary traits between Landrace and the two Chinese indigenous pig breeds. Our data suggest that EXOC2 might be a good pigmentation trait candidate of pigs. However, the biological mechanism of EXOC2 has not been well characterized in regulating human pigmentation. Moreover, little is known about the role of EXOC2 in pigs. Therefore, confirmation and elucidation the role of EXOC2 in pigmentation trait in pigs requires further investigation. Actually, another key well-documented pigmentary gene, KIT was also discovered in DSRs (Supplementary Table S6). In the past two decades, KIT had been deeply investigated for its mutation associated with coat color of the dominant white in pigs58–60. Moreover, as a colored breed, Meishan had been found to have different KIT allele distinguish from white breed such as Landrace and Large White58, 60. Obviously, our data provide further evidence for the role of KIT in pigmentary traits. More interestingly, we discovered some genes associated with growth or meat quality traits in DSRs, i.e., ACSL6, RAPGEF1, BMPER, and KDR (Table 2 and Supplementary Table S6). ACSL6, a member of the long-chain acyl-CoA synthetase gene family for catalyzing the formation of acyl-CoA from fatty acids, ATP, and CoA, was shown to be associated with dry matter intake and metabolic mid-test body weight in cattle61. RAPGEF1, a guanine nucleotide releasing protein, was discovered to play an important role in skeletal muscle differentiation62 and was significantly associated with type 2 diabetes in the Korean population and Finns63, 64. These studies demonstrated that ACSL6 and RAPGEF1 might have important function in body growth or development of skeletal muscle and fat. Hence our results might provide new ideas for their function in affecting pig growth and development. In addition, BMPER and KDR, two key genes in the development of angiogenesis and vasculogenesis65, 66, were identified in DSRs. It is not surprising because our previous study had found genetic variants in the promoters of BMPER and KDR affected intramuscular fat deposition in Erhualian pigs67, 68. Moreover, BMPER was found to be associated with rump length and body size in Qinchuan cattle population69. Here our data suggest that BMPER and KDR might contribute to the regulation of pig growth and meat quality, but additional studies are needed to further confirm this speculation. In a word, our results provide a comprehensive clue for discovering the genetic difference between Landrace and the two Chinese indigenous pig breeds.
As described above, Erhualian and Meishan are two famous pig breeds with the typical characters of Chinese indigenous breeds such as high productive performance and good meat quality. However, as two different pig breeds, some characteristics, i.e., head wrinkles and colors of limbs and nose37, are very different between these two pig breeds. In this study, the genetic differentiation (F ST = 0.2335) between Erhualian and Meishan was observed, further confirming that there exists a significant genetic difference between these two pig breeds. But until now, only variants from single gene such as FUT1 have been used to compare the difference in gene frequencies between these two pig breeds70. In a genome-wide level, the genetic difference between these two pig breeds is still largely unknown. Our present study identified 256 DSRs containing 347 genes between Erhualian and Meishan. Importantly, GO terms associated with these genes were enriched in sodium-independent organic anion transport, glycerophospholipid metabolic process, and phospholipid related metabolic process. Some important candidates, i.e., SLCO3A1 in sodium-independent organic anion transport, CDS2, CRLS1, and MTMR7 in glycerophospholipid metabolic process, and SPTLC1, SGMS2, and CRLS1 in phospholipid related metabolic process, were discovered from enrichment analysis. The difference of the biological processes between these two pig breeds might result from a long-term artificial selection based on different breeding objectives, but further investigation of the cause of breed difference is required.
In summary, a SLAF-seq approach for pigs was developed to reveal the genetic structure and relationship among Landrace, Erhualian, and Meishan pigs. Meanwhile, DSRs including hundreds of genes between Landrace and the two Chinese indigenous pig breeds and between Erhualian and Meishan were identified further. Consequently, our study not only provides a cost-effective approach for pig genome-wide screening, but also establishes the genetic basis for further investigating important gene functions from DSRs among these pig breeds in future research.
Materials and Methods
Animal
Blood or ear tissue samples were collected from 28 Landrace (8 male and 20 female), 26 Erhualian (8 male and 18 female), and 27 Meishan (Small Meishan, 8 male and 19 female) at the Xiang Xin livestock and poultry Co., Ltd (Shanghai), Changzhou Erhualian production cooperation (Jiangsu), and Taicang pig breeding farm (Jiangsu), respectively. Total DNA was extracted by the phenol-chloroform extraction method, with concentration and purity measured using the NanodropTM 2000 spectrophotometer (Thermo Scientific, Waltham, MA, USA) and electrophoresis. All animal care and handling procedures were approved by the Animal Ethics Committee at Nanjing Agricultural University, China. All methods were performed in accordance with the guidelines and regulations of the Animal Ethics Committee at Nanjing Agricultural University.
SLAF library construction and sequencing
A simulated restriction enzyme digestion was carried out on the current pig genome (Sscrofa 10.2) to identify expected SLAF yield, avoid repetitive SLAFs, and obtain the relatively uniform distribution of restriction fragments in the genome. As a result, genomic DNA was digested with RsaI and HaeIII restriction enzyme combination. Meanwhile, in order to assess the experimental procedure, Oryza sativa indica (http://rapdb.dna.affrc.go.jp/) was used as a control for evaluating the effectiveness of enzyme digestion and paired-end mapped reads. In brief, SLAF library construction and sequencing for each individual was conducted as described previously18 with slight modifications: DNA fragments of 314–364 base pair (bp) were selected as SLAFs and used for paired-end sequencing by Illumina HiSeq 2500 system (Illumina, Inc., San Diego, CA, USA) at Beijing Biomarker Technologies Corporation.
Genome mapping, SNP calling and filtering
Raw paired-end reads were mapped to the pig reference genome (Sscrofa 10.2) using BWA software71. In general, SLAF groups produced by reads were mapped to the same position. If an accession was only partly digested by the restriction enzymes, reads mapped to the reference genome should have overlaps with two SLAF tags and were assigned to both of the SLAF tags in the same accession. SNP calling was performed by both GATK and samtools analysis72, 73, and a locus was defined as a SNP if it was simultaneously called from these two packages. Filtering high-quality SNPs was conducted by PLINK v1.0774. The analysis of Hardy-Weinberg Equilibrium (HWE) for SNPs from autosomes and chromosome X was performed by Pedstats tools and HWE package for R75, 76, respectively.
Population genetic basics analysis
Population structure analysis was performed using STRUCTURE with 10,000 iterations via the correlated allele model77, and then plotted by DISTRUCT software78. Linkage disequilibrium analysis was computed between each marker pair within each breed separately using Haploview 4.179. The population relatedness of pair-wise F ST and gene diversity were calculated using the HIERFSTAT package for R. Observed heterozygosity and IBS matrix of distance were estimated by PLINK v1.0774, and then multidimensional scaling (MDS) analysis of autosomal SNPs was determined by R 3.2.4.
Detection of differentially selected regions (DSRs)
The DSR algorithm was carried out as described previously3, 80 with slight modifications: 1) raw values were ranked; 2) Fisher’s exact test was executed in R 3.2.4 to compare the allele frequencies between Landrace and the two Chinese indigenous pig breeds and between Erhualian and Meishan, respectively. SNPs with P values < 0.05 were considered as statistically significant after Bonferroni correction; 3) F ST of SNP was estimated using the model proposed by Nicholson et al.81 and Flori et al.82, and then the significant SNPs with 0.5% or 5% highest F ST values were selected as the top significant SNPs; 4) by placing the top significant SNPs (0.5%) in the center of a DSR, adjacent SNPs were collected to determine the region boundaries until more than two consecutive SNPs were not in the top significant 5% threshold. The overlapped DSRs were combined as the same DSR. When a region contained more than five SNPs from the top significant SNPs (5%) but without top significant SNPs (0.5%), we also considered it as a DSR in our study. In addition, the W-statistics was used to identify the relative importance of DSRs83 and smoothed F ST values were obtained by a local variable bandwidth kernel estimator84.
Gene functional enrichment analysis
Genes in these DSRs were identified using the Sscrofa 10.2 assembly (www.animalgenome.org/cgi-bin/gbrowse/pig) and NCBI database. The functional enrichment analysis of target genes was performed using Panther bioinformatics resources (www.pantherdb.org). Terms with P values less than 0.01 and the enrichment value more than 1 were selected as significant or enriched terms.
Data accessibility
The datasets have been submitted to the SRA database of NCBI (accession number SRP090907).
Electronic supplementary material
Acknowledgements
This work was supported by the National Key Technology Research and Development Program of the Ministry of Science and Technology of China (2015BAD03B01), the Fundamental Research Funds for the Central Universities (KYZ201414), the National Natural Science Foundation of China (31501930), and the Natural Science Foundation of Jiangsu Province of China (BK20150656 and BK20150669).
Author Contributions
Lifan Zhang conceived and designed the experiments. Zhen Li performed the experiments. Zhen Li, Zhaowei Cai, Dongfeng Li, Wei Wei, and Shengjuan Wei analyzed the data. Hejun Li, Keliang Wu, Qifa Li, Jie Chen, and Honglin Liu participated in the collection of samples. Zhen Li, Shengjuan Wei, and Lifan Zhang wrote the manuscript. All authors read and approved the final manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-09969-6
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Archibald AL, et al. Pig genome sequence–analysis and publication strategy. BMC genomics. 2010;11 doi: 10.1186/1471-2164-11-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ramos AM, et al. Design of a high density SNP genotyping assay in the pig using SNPs identified and characterized by next generation sequencing technology. PloS one. 2009;4 doi: 10.1371/journal.pone.0006524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang L, et al. Genome wide screening of candidate genes for improving piglet birth weight using high and low estimated breeding value populations. Int J Biol Sci. 2014;10:236–244. doi: 10.7150/ijbs.7744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fontanesi L, Schiavo G, Galimberti G, Calo DG, Russo V. A genomewide association study for average daily gain in Italian Large White pigs. J Anim Sci. 2014;92:1385–1394. doi: 10.2527/jas.2013-7059. [DOI] [PubMed] [Google Scholar]
- 5.Zhang LC, et al. Genome-wide study refines the quantitative trait locus for number of ribs in a Large White x Minzhu intercross pig population and reveals a new candidate gene. Mol Genet Genomics. 2016;291:1885–1890. doi: 10.1007/s00438-016-1220-1. [DOI] [PubMed] [Google Scholar]
- 6.Saura M, et al. Detecting inbreeding depression for reproductive traits in Iberian pigs using genome-wide data. Genet Sel Evol. 2015;47 doi: 10.1186/s12711-014-0081-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ai H, Huang L, Ren J. Genetic diversity, linkage disequilibrium and selection signatures in chinese and Western pigs revealed by genome-wide SNP markers. PloS one. 2013;8 doi: 10.1371/journal.pone.0056001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Traspov A, et al. Population structure and genome characterization of local pig breeds in Russia, Belorussia, Kazakhstan and Ukraine. Genet Sel Evol. 2016;48 doi: 10.1186/s12711-016-0196-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choi JW, et al. Whole-genome resequencing analyses of five pig breeds, including Korean wild and native, and three European origin breeds. DNA Res. 2015;22:259–267. doi: 10.1093/dnares/dsv011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rubin CJ, et al. Strong signatures of selection in the domestic pig genome. Proc Natl Acad Sci USA. 2012;109:19529–19536. doi: 10.1073/pnas.1217149109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moon S, et al. A genome-wide scan for signatures of directional selection in domesticated pigs. BMC genomics. 2015;16 doi: 10.1186/s12864-015-1330-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bosse M, et al. Using genome-wide measures of coancestry to maintain diversity and fitness in endangered and domestic pig populations. Genome Res. 2015;25:970–981. doi: 10.1101/gr.187039.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Esteve-Codina A, et al. Partial short-read sequencing of a highly inbred Iberian pig and genomics inference thereof. Heredity. 2011;107:256–264. doi: 10.1038/hdy.2011.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen Q, et al. Genotyping by genome reducing and sequencing for outbred animals. PloS one. 2013;8 doi: 10.1371/journal.pone.0067500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bovo S, et al. Reduced representation libraries from DNA pools analysed with next generation semiconductor based-sequencing to identify SNPs in extreme and divergent pigs for back fat thickness. Int J Genomics. 2015;2015 doi: 10.1155/2015/950737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Z, et al. A genome-wide scan for selection signatures in Yorkshire and Landrace pigs based on sequencing data. Anim Genet. 2014;45:808–816. doi: 10.1111/age.12229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Z, et al. Genetic diversity and population structure of six Chinese indigenous pig breeds in the Taihu Lake region revealed by sequencing data. Anim Genet. 2015;46:697–701. doi: 10.1111/age.12349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun X, et al. SLAF-seq: an efficient method of large-scale de novo SNP discovery and genotyping using high-throughput sequencing. PloS one. 2013;8 doi: 10.1371/journal.pone.0058700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu WY, et al. A high-density genetic linkage map for cucumber (Cucumis sativus L.): based on specific length amplified fragment (SLAF) sequencing and QTL analysis of fruit traits in cucumber. Front Plant Sci. 2016;7 doi: 10.3389/fpls.2016.00437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang D, et al. High-density genetic mapping identifies new major loci for tolerance to low-phosphorus stress in soybean. Front Plant Sci. 2016;7 doi: 10.3389/fpls.2016.00372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang W, et al. Genome-wide association study of antibody level response to NDV and IBV in Jinghai yellow chicken based on SLAF-seq technology. J Appl Genet. 2015;56:365–373. doi: 10.1007/s13353-014-0269-y. [DOI] [PubMed] [Google Scholar]
- 22.Wang WH, et al. Genome-wide association study of growth traits in Jinghai Yellow chicken hens using SLAF-seq technology. Anim Genet. 2015 doi: 10.1111/age.12346. [DOI] [PubMed] [Google Scholar]
- 23.Wang W, et al. Genome-wide association study of 8 carcass traits in Jinghai Yellow chickens using specific-locus amplified fragment sequencing technology. Poult Sci. 2016;95:500–506. doi: 10.3382/ps/pev266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He LC, et al. Identification of new single nucleotide polymorphisms affecting total number born and candidate genes related to ovulation rate in Chinese Erhualian pigs. Anim Genet. 2016 doi: 10.1111/age.12492. [DOI] [PubMed] [Google Scholar]
- 25.Liu X, et al. Genome-wide association analyses for meat quality traits in Chinese Erhualian pigs and a Western Duroc x (Landrace x Yorkshire) commercial population. Genet Sel Evol. 2015;47 doi: 10.1186/s12711-015-0120-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ai H, et al. Adaptation and possible ancient interspecies introgression in pigs identified by whole-genome sequencing. Nat Genet. 2015;47:217–225. doi: 10.1038/ng.3199. [DOI] [PubMed] [Google Scholar]
- 27.Frantz LA, et al. Evidence of long-term gene flow and selection during domestication from analyses of Eurasian wild and domestic pig genomes. Nat Genet. 2015;47:1141–1148. doi: 10.1038/ng.3394. [DOI] [PubMed] [Google Scholar]
- 28.Weir, B. S. & Cockerham, C. C. Estimating F-statistics for the analysis of population structure. Evolution, 1358–1370 (1984). [DOI] [PubMed]
- 29.Giuffra E, et al. The origin of the domestic pig: independent domestication and subsequent introgression. Genetics. 2000;154:1785–1791. doi: 10.1093/genetics/154.4.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Megens HJ, et al. Biodiversity of pig breeds from China and Europe estimated from pooled DNA samples: differences in microsatellite variation between two areas of domestication. Genet Sel Evol. 2008;40:103–128. doi: 10.1186/1297-9686-40-1-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang, Z. Chinese Pig Breed Records. (Shanghai Science and Technology Press, Shanghai, China, 1986).
- 32.Haley CS, Lee GJ. Genetic basis of prolificacy in Meishan pigs. J Reprod Fertil Suppl. 1993;48:247–259. [PubMed] [Google Scholar]
- 33.Balcells I, et al. Analysis of porcine MUC4 gene as a candidate gene for prolificacy QTL on SSC13 in an Iberian x Meishan F2 population. BMC Genet. 2011;12 doi: 10.1186/1471-2156-12-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hernandez SC, Finlayson HA, Ashworth CJ, Haley CS, Archibald AL. A genome-wide linkage analysis for reproductive traits in F2 Large White x Meishan cross gilts. Anim Genet. 2014;45:191–197. doi: 10.1111/age.12123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li K, et al. Quantitative trait loci for litter size and prenatal loss in a White Duroc x Chinese Erhualian resource population. Anim Genet. 2009;40:963–966. doi: 10.1111/j.1365-2052.2009.01931.x. [DOI] [PubMed] [Google Scholar]
- 36.Du H, Chen J, Cui J, Wang X, Zhang X. Polymorphisms on SSC15q21-q26 Containing QTL for reproduction in Swine and its association with litter size. Genet Mol Biol. 2009;32:69–74. doi: 10.1590/S1415-47572009000100010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.China National Commission of Animal Genetic Resources. Animal Genetic Resources in China Pigs. (China Agriculture Press, Beijing, 2011).
- 38.Rothschild M, et al. The estrogen receptor locus is associated with a major gene influencing litter size in pigs. Proc Natl Acad Sci USA. 1996;93:201–205. doi: 10.1073/pnas.93.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu ZF, et al. Study on the association between estrogen receptor gene (ESR) and reproduction traits in Landrace pigs. Yi Chuan Xue Bao. 2006;33:711–716. doi: 10.1016/S0379-4172(06)60103-0. [DOI] [PubMed] [Google Scholar]
- 40.Chen KF, et al. The genetic effect of estrogen receptor (ESR) on litter size traits in pig. Yi Chuan Xue Bao. 2000;27:853–857. [PubMed] [Google Scholar]
- 41.van Rens BT, de Groot PN, van der Lende T. The effect of estrogen receptor genotype on litter size and placental traits at term in F2 crossbred gilts. Theriogenology. 2002;57:1635–1649. doi: 10.1016/S0093-691X(02)00671-4. [DOI] [PubMed] [Google Scholar]
- 42.Drogemuller C, Hamann H, Distl O. Candidate gene markers for litter size in different German pig lines. J Anim Sci. 2001;79:2565–2570. doi: 10.2527/2001.79102565x. [DOI] [PubMed] [Google Scholar]
- 43.Gibson JP, Jiang ZH, Robinson JA, Archibald AL, Haley CS. No detectable association of the ESR PvuII mutation with sow productivity in a Meishan x Large White F2 population. Anim Genet. 2002;33:448–450. doi: 10.1046/j.1365-2052.2002.00889.x. [DOI] [PubMed] [Google Scholar]
- 44.Adona PR, et al. In vitro maturation alters gene expression in bovine oocytes. Zygote. 2016;24:624–633. doi: 10.1017/S0967199415000672. [DOI] [PubMed] [Google Scholar]
- 45.Balboula AZ, Stein P, Schultz RM, Schindler K. RBBP4 regulates histone deacetylation and bipolar spindle assembly during oocyte maturation in the mouse. Biol Reprod. 2015;92 doi: 10.1095/biolreprod.115.128298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miki Y, et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science. 1994;266:66–71. doi: 10.1126/science.7545954. [DOI] [PubMed] [Google Scholar]
- 47.Yuan Z, et al. Investigation on BRCA1 SNPs and its effects on mastitis in Chinese commercial cattle. Gene. 2012;505:190–194. doi: 10.1016/j.gene.2012.05.010. [DOI] [PubMed] [Google Scholar]
- 48.Titus S, et al. Impairment of BRCA1-related DNA double-strand break repair leads to ovarian aging in mice and humans. Sci Transl Med. 2013;5 doi: 10.1126/scitranslmed.3004925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Oktay K, Turan V, Titus S, Stobezki R, Liu L. BRCA mutations, DNA repair deficiency, and ovarian aging. Biol Reprod. 2015;93 doi: 10.1095/biolreprod.115.132290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Oktay K, Kim JY, Barad D, Babayev SN. Association of BRCA1 mutations with occult primary ovarian insufficiency: a possible explanation for the link between infertility and breast/ovarian cancer risks. J Clin Oncol. 2010;28:240–244. doi: 10.1200/JCO.2009.24.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sugihara K, et al. The exocyst complex binds the small GTPase RalA to mediate filopodia formation. Nat Cell Biol. 2002;4:73–78. doi: 10.1038/ncb720. [DOI] [PubMed] [Google Scholar]
- 52.Pathak R, et al. The microtubule-associated Rho activating factor GEF-H1 interacts with exocyst complex to regulate vesicle traffic. Dev Cell. 2012;23:397–411. doi: 10.1016/j.devcel.2012.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sulem P, et al. Genetic determinants of hair, eye and skin pigmentation in Europeans. Nat Genet. 2007;39:1443–1452. doi: 10.1038/ng.2007.13. [DOI] [PubMed] [Google Scholar]
- 54.Han J, et al. A genome-wide association study identifies novel alleles associated with hair color and skin pigmentation. PLoS Genet. 2008;4 doi: 10.1371/journal.pgen.1000074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nan H, et al. Genome-wide association study of tanning phenotype in a population of European ancestry. J Invest Dermatol. 2009;129:2250–2257. doi: 10.1038/jid.2009.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Saternus R, et al. A closer look at evolution: Variants (SNPs) of genes involved in skin pigmentation, including EXOC2, TYR, TYRP1, and DCT, are associated with 25(OH)D serum concentration. Endocrinology. 2015;156:39–47. doi: 10.1210/en.2014-1238. [DOI] [PubMed] [Google Scholar]
- 57.Branicki W, et al. Model-based prediction of human hair color using DNA variants. Hum Genet. 2011;129:443–454. doi: 10.1007/s00439-010-0939-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Marklund S, et al. Molecular basis for the dominant white phenotype in the domestic pig. Genome Res. 1998;8:826–833. doi: 10.1101/gr.8.8.826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pielberg G, Olsson C, Syvanen AC, Andersson L. Unexpectedly high allelic diversity at the KIT locus causing dominant white color in the domestic pig. Genetics. 2002;160:305–311. doi: 10.1093/genetics/160.1.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fontanesi L, et al. Genetic heterogeneity and selection signature at the KIT gene in pigs showing different coat colours and patterns. Anim Genet. 2010;41:478–492. doi: 10.1111/j.1365-2052.2010.02054.x. [DOI] [PubMed] [Google Scholar]
- 61.Saatchi M, et al. QTLs associated with dry matter intake, metabolic mid-test weight, growth and feed efficiency have little overlap across 4 beef cattle studies. BMC genomics. 2014;15 doi: 10.1186/1471-2164-15-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sasi Kumar K, et al. C3G (RapGEF1), a regulator of actin dynamics promotes survival and myogenic differentiation of mouse mesenchymal cells. Biochim Biophys Acta. 2015;1853:2629–2639. doi: 10.1016/j.bbamcr.2015.06.015. [DOI] [PubMed] [Google Scholar]
- 63.Gaulton KJ, et al. Comprehensive association study of type 2 diabetes and related quantitative traits with 222 candidate genes. Diabetes. 2008;57:3136–3144. doi: 10.2337/db07-1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hong KW, et al. RAPGEF1 gene variants associated with type 2 diabetes in the Korean population. Diabetes Res Clin Pract. 2009;84:117–122. doi: 10.1016/j.diabres.2009.02.019. [DOI] [PubMed] [Google Scholar]
- 65.Holmes K, Roberts OL, Thomas AM, Cross MJ. Vascular endothelial growth factor receptor-2: structure, function, intracellular signalling and therapeutic inhibition. Cell Signal. 2007;19:2003–2012. doi: 10.1016/j.cellsig.2007.05.013. [DOI] [PubMed] [Google Scholar]
- 66.Prosdocimo DA, Jain MK. BMPing up angiogenesis via BMPER. Arterioscler Thromb Vasc Biol. 2011;31:2167–2168. doi: 10.1161/ATVBAHA.111.233890. [DOI] [PubMed] [Google Scholar]
- 67.Fu Y, et al. Genetic variants in KDR transcriptional regulatory region affect promoter activity and intramuscular fat deposition in Erhualian pigs. Anim Genet. 2014;45:373–380. doi: 10.1111/age.12148. [DOI] [PubMed] [Google Scholar]
- 68.Liu Z, et al. The effect of variants in the promoter of BMPER on the intramuscular fat deposition in longissimus dorsi muscle of pigs. Gene. 2014;542:168–172. doi: 10.1016/j.gene.2014.03.038. [DOI] [PubMed] [Google Scholar]
- 69.Zhao C, Gui L, Li Y, Plath M, Zan L. Associations between allelic polymorphism of the BMP Binding Endothelial Regulator and phenotypic variation of cattle. Mol Cell Probes. 2015;29:358–364. doi: 10.1016/j.mcp.2015.09.007. [DOI] [PubMed] [Google Scholar]
- 70.Bao WB, Wu SL, Musa HH, Zhu GQ, Chen GH. Genetic variation at the alpha-1-fucosyltransferase (FUT1) gene in Asian wild boar and Chinese and Western commercial pig breeds. J Anim Breed Genet. 2008;125:427–430. doi: 10.1111/j.1439-0388.2008.00722.x. [DOI] [PubMed] [Google Scholar]
- 71.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wigginton JE, Abecasis GR. PEDSTATS: descriptive statistics, graphics and quality assessment for gene mapping data. Bioinformatics. 2005;21:3445–3447. doi: 10.1093/bioinformatics/bti529. [DOI] [PubMed] [Google Scholar]
- 76.Graffelman J, Weir BS. Testing for Hardy-Weinberg equilibrium at biallelic genetic markers on the X chromosome. Heredity. 2016;116:558–568. doi: 10.1038/hdy.2016.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hubisz MJ, Falush D, Stephens M, Pritchard JK. Inferring weak population structure with the assistance of sample group information. Mol Ecol Resour. 2009;9:1322–1332. doi: 10.1111/j.1755-0998.2009.02591.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rosenberg NA. DISTRUCT: a program for the graphical display of population structure. Mol Ecol Notes. 2004;4:137–138. doi: 10.1046/j.1471-8286.2003.00566.x. [DOI] [Google Scholar]
- 79.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 80.Kijas JW, et al. Genome-wide analysis of the world’s sheep breeds reveals high levels of historic mixture and strong recent selection. PLoS Biol. 2012;10 doi: 10.1371/journal.pbio.1001258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nicholson G, et al. Assessing population differentiation and isolation from single-nucleotide polymorphism data. J Roy Stat Soc B. 2002;64:695–715. doi: 10.1111/1467-9868.00357. [DOI] [Google Scholar]
- 82.Flori L, et al. The genome response to artificial selection: a case study in dairy cattle. PloS one. 2009;4 doi: 10.1371/journal.pone.0006595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Beissinger TM, Rosa GJ, Kaeppler SM, Gianola D, de Leon N. Defining window-boundaries for genomic analyses using smoothing spline techniques. Genet Sel Evol. 2015;47 doi: 10.1186/s12711-015-0105-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Herrmann E. Local bandwidth choice in kernel regression estimation. J Comput Graph Stat. 1997;6:35–54. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets have been submitted to the SRA database of NCBI (accession number SRP090907).