Abstract
Myocyte enhancer binding factor 2 (MEF2) proteins belong to the MADS box family of transcription factors and four MEF2 proteins, MEF2A, MEF2B, MEF2C and MEF2D, have been found. MEF2 proteins have been shown to play critical roles in differentiation of muscles and neuronal tissues. How transactivational activity of MEF2 proteins is regulated is not fully understood. MEF2 proteins are activated by several kinases, including Erk5 and calcium/calmodulin-dependent kinase, and interact with repressors, including histone deacetylases 4 and 5 (HDAC4 and HDAC5) and Cabin1. During the effort to understand regulation of MEF2 activity, we identified 14-3-3τ as a MEF2D-interacting molecule by yeast two-hybrid screening. We found that 14-3-3τ forms a complex with MEF2D in vivo and specifically enhances MEF2 transactivational activity. The results from transient transfection and co-precipitation experiments suggest that 14-3-3τ activates MEF2D by competitively inhibiting HDAC4 from binding to MEF2D and thereby affects muscle cell differentiation.
INTRODUCTION
The 14-3-3 proteins form a family of highly conserved molecules expressed in a wide range of organisms and tissues (1,2). There are nine isotypes (α, β, γ, δ, ɛ, η, σ, τ and ζ) in vertebrates and additional isotypes from yeasts, plants, amphibians and invertebrates. All of these proteins have a molecular mass of ∼30 kDa and exist as homo- or heterodimers. The 14-3-3 proteins regulate various cellular activities by binding to phosphorylated signaling molecules, including Raf-1, Cbl, Bad, Cdc25C and the IGF-1 receptor (3–7). In addition, the 14-3-3 proteins have been shown to regulate transactivational activities of the glucocorticoid receptor and NFAT by direct interaction (8,9). Protein-binding motifs of 14-3-3 have been identified as RSXpSXP and RXY/FXpSXP and these motifs have been found in many other known 14-3-3 protein-binding molecules (10). Neverthless, other sequences containing serine or threonine have been also shown to bind 14-3-3 (11–14)
MEF2 proteins are a group of transcription factors which belong to the MADS box family. Four MEF2 members, MEF2A, MEF2B, MEF2C and MEF2D, have been found in human, mouse, Xenopus and Drosophila (15–19). They have almost identical DNA-binding domains at their N-termini, which are similar to the DNA-binding domains of other MADS box family members, including serum response factor (SRF) and MCM1 (20). The C-terminal regions are divergent among MEF2 family members and contain transactivation domains. The MEF2 proteins bind as homo- and heterodimers to a cis element with the consensus (C/T)TA(A/T)4TA(G/A) (19). In many muscle-specific gene promoters, a MEF2-binding site and a MyoD-binding site are found side by side and their interaction seems to be important during muscle cell differentiation (21,22). The MEF2C-deficient mouse was found to be embryonic lethal due to a cardiac muscle defect, demonstrating an essential role of MEF2 in muscle development (23). However, dominant negative mutants of MEF2 proteins also inhibit differentiation of neuronal and hematopoietic cells, which suggests a functional importance of MEF2 proteins in non-muscle tissues (24,25).
How transcriptional activity of MEF2 proteins is regulated is not fully understood. Calcium/calmodulin-dependent enzymes, such as the phosphatase calcineurin and kinase type IV (CaMK IV), activate transcriptional activity of MEF2A and MEF2D (26). It has recently been reported that Erk5 directly phosphorylates and activates MEF2 members except for MEF2B (27). In addition, MEF2 has been shown to interact with repressors, including Cabin1 and histone deacetylases 4 and 5 (HDAC4 and HDAC5) (28,29). Activated CaMK signaling stimulates MEF2 activity by inducing export of HDAC5 from the nucleus to the cytoplasm, which is dependent on CaMK-mediated phosphorylation of HDAC5 (30,31). However, HDAC4 and HDAC5 seem to be differentially regulated and how MEF2 is relieved from nuclear HDAC4 repression is not clear. During an effort to further understand regulation of MEF2 activity we identified 14-3-3τ as a MEF2D-interacting molecule by yeast two-hybrid screening. We found that 14-3-3τ specifically enhanced MEF2D transactivational activity and examined its mechanism.
MATERIALS AND METHODS
DNA constructs
The reporter plasmids pMEF2Fluc and pCMVβgal have been previously described (32). 14-3-3τ cDNA was cloned by PCR from reverse transcribed U937 cell RNA and subcloned into pcDNA3.1 (Invitrogen), pFlagCMV-2 (Kodak) and pGEX4T-1 (Pharmacia) to give pc14-3-3τ, pcF14-3-3τ and pGST14-3-3τ. Point mutations in pc14-3-3τ were generated via site-directed mutagenesis using the Quickchange kit (Stratagene) and the mutated 14-3-3τ coding sequence was subcloned into pFlagCMV-2 and pGEX4T-1. A MEF2D expression plasmid pcMEF2D has been described (32). pcHMEF2D was made by amplifying the MEF2D coding sequence from pcMEF2D and subcloning into pcDNA3.1His (Invitrogen). Partial cDNAs encoding MEF2D amino acids 1–86, MEF2D amino acids 86–173 and MEF2D amino acids 272–514 were amplified from pcMEF2D by PCR and subcloned into pRSET (Invitrogen). pTM173, expressing MEF2D amino acids 1–173, and a MyoD expression plasmid (pCSA-MyoD) were provided by Drs J. Martin and A. B. Lassar, respectively (17,33). HDAC4 expression plasmids pcHDAC4-Myc and pcFHDAC4 were provided by Drs T. Kouzarides and X. J. Yang, respectively (29,34).
Yeast two-hybrid screen
MEF2 amino acids 1–173 fused to the LexA DNA-binding domain was used as the bait to screen a human fetal brain cDNA library by the yeast two-hybrid system according to the manufacturer’s protocol (Clontech). Approximately 1 × 105 clones were screened.
Cell culture, transfections and reporter gene assays
U937 (human histiocytic lymphoma) and 10T1/2 cells were grown in RPMI 1640 and DMEM (Gibco) supplemented with 10% fetal bovine serum, 50 nM 2-mercaptoethanol and penicillin/streptomycin, respectively. Transfections were performed using Lipofectamine (Gibco BRL). When necessary, transfected cells were treated with 600 nM trichostatin A (Sigma) for 12 h. Preparation of cell extracts, the β-galactosidase assay and the luciferase assay using the Promega luciferase assay system were performed according to the manufacturer’s protocol and as described previously (32). All luciferase values are the averages of at least two separate experiments and were normalized to the β-galactosidase values resulting from expression of the internal control plasmid pCMVβgal.
Protein expression and interaction assay
GST–14-3-3 fusion proteins were expressed in Escherichia coli strain BL21(DE3) and purified according to the manufacturer’s protocols (Pharmacia). Truncated MEF2D proteins were in vitro translated and labeled with [35S]methionine using the Promega rabbit reticulocyte lysate system according to the manufacturer’s protocols. Aliquots of 10 µl of in vitro translated MEF2 proteins were incubated with 5 µg GST–14-3-3 fusion proteins immobilized on glutathione–Sepharose beads (Pharmacia) in 15 µl of binding buffer (25 mM HEPES, pH 7.5, 12.5 mM MgCl2, 150 mM KCl, 20% glycerol, 0.1% NP-40, 1 mM DTT). The beads were then washed six times with binding buffer. His6-tagged MEF2D, 14-3-3τ and HDAC4 fusion proteins from transfected cells were purified with nickel–agarose. The beads were then washed six times with TN-1 buffer (20 mM Tris–HCl, pH 8.0, 150 mM NaCl, 1% NP-40). Proteins retained on beads were boiled in 1× sample buffer and resolved by SDS–PAGE. The existence of labeled MEF2D proteins was detected by autoradiography.
Antibodies and western blotting
A MEF2D antiserum raised against codons 272–514 of mouse MEF2D has been described (32). Antibodies against the Flag epitope were purchased from Kodak. For immunoblots, cell lysates or nickel–agarose-bound proteins were boiled in 1× sample buffer for 3 min and resolved by SDS–PAGE. Proteins were transferred to a nitrocellulose membrane and incubated with specific antibodies. Bound antibody was visualized using an ECL western blotting kit according to the manufacturer’s protocols (Amersham).
Indirect immunofluorescence
10T1/2 cells were grown on glass coverslips and transfected with Lipofectamine. Transfected cells were fixed with 10% paraformaldehyde, permeabilized with 0.1% Triton X-100 in phosphate-buffered saline (PBS) and blocked with 2% BSA in PBS. Rabbit polyclonal antibody against MEF2D (32) and mouse monoclonal antibody against Flag (Kodak) were used at dilutions of 1:500 and 1:100, respectively. FITC–anti-mouse IgG (Vector Laboratories) and TRITC–anti-rabbit IgG (Zymax) were used at dilutions of 1:100 and 1:200, respectively. Stained cells were examined with a Zeiss Axiophot2 microscope.
Myogenic conversion assay
For myogenic differentiation of MyoD-transfected 10T1/2 cells, growth medium was replaced with DMEM containing horse serum (2%), 50 nM 2-mercaptoethanol and penicillin/streptomycin. Tranfected 10T1/2 cells were exposed to differentiation medium for 4 days. Myosin expression was assessed by immunoperoxidase staining with a Vectastain Elite ABC kit and an anti-skeletal myosin monoclonal antibody (Sigma).
RESULTS
14-3-3τ associates with MEF2D
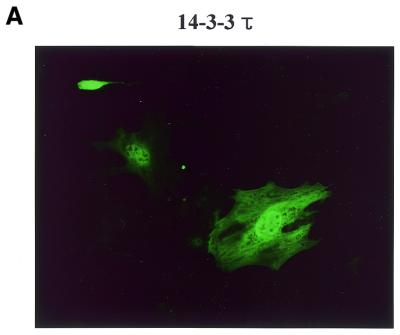
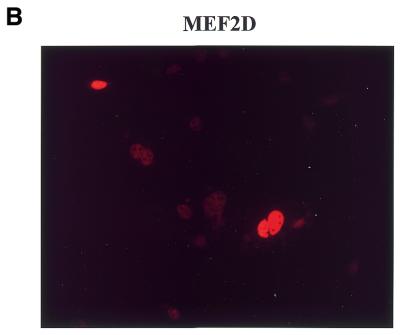
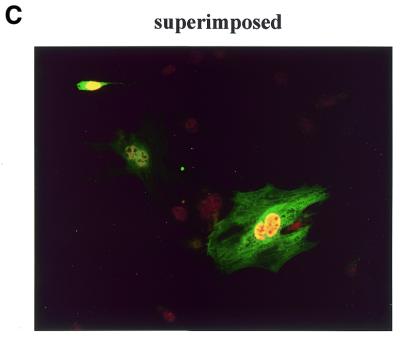
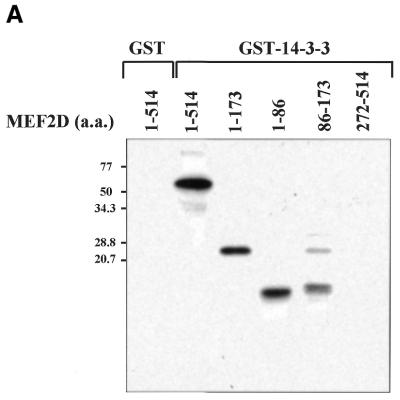
Four MEF2 proteins, MEF2A, MEF2B, MEF2C and MEF2D, have almost identical MADS box domains (amino acids 1–57) that mediate DNA binding and dimerization. An adjacent MEF2-specific domain (amino acids 58–86) influences DNA-binding specificity and interaction with regulatory molecules (35). To isolate MEF2D-interacting molecules, we performed a yeast two-hybrid screening of a human fetal brain cDNA library using a MEF2 fragment (amino acids 1–173) containing the MADS–MEF2 domain as bait. Among 19 positive clones, nine were identified as the 14-3-3τ form. First we examined whether MEF2D and 14-3-3τ co-localize in the cell by confocal immunofluorescence microscopy. Both MEF2D and Flag-tagged 14-3-3τ were co-expressed in 10T1/2 cells and labeled using specific primary antibodies and rhodamine-conjugated (MEF2D) and fluorescein-conjugated (14-3-3τ) secondary antibodies. 14-3-3τ was predominantly localized in the nucleus, although some was also detected in the cytoplasm (Fig. 1A). As expected, MEF2D was located within the nucleus (Fig. 1B). The superimposed image showed that significant amounts of 14-3-3τ co-localize with MEF2D in the nucleus (Fig. 1C). To assess the interaction between MEF2D and 14-3-3τ, a GST pull-down assay was performed with various MEF2D truncation mutants translated in vitro and bacterially expressed GST–14-3-3τ protein. Full-length MEF2D(1–514), MEF2D(1–173) and MEF2D(1–86), but not MEF2D(272–514), were retained by GST–14-3-3τ (Fig. 22). In addition, MEF2D(86–173) showed a weaker but significant interaction with GST–14-3-3τ (Fig. 2). These results show that the MADS–MEF2 domain (amino acids 1–86) of MEF2D mediates the interaction between MEF2D and 14-3-3τ in vitro and that amino acids 86–173 of MEF2D also contribute to the interaction.
Figure 1.
MEF2D and 14-3-3τ co-localize in the nucleus. 10T1/2 cells were co-transfected with plasmids expressing Flag–14-3-3τ (1.2 µg) and MEF2D (1.2 µg). Flag-14-3-3τ (A) and MEF2D (B) were detected by indirect immunofluorescence using antibodies against the Flag epitope and MEF2D and rhodamine-conjugated and fluorescein-conjugated secondary antibodies.
Figure 2.
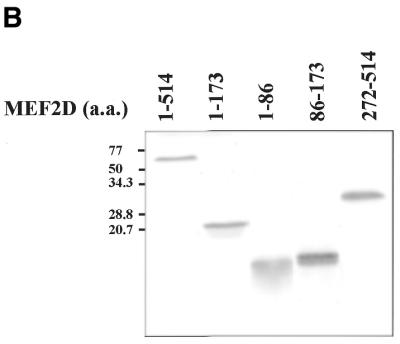
MEF2D associates with 14-3-3τ through the N-terminal region containing the MADS and MEF2 domains. The indicated MEF2D proteins were in vitro translated and [35S]methionine labeled. GST or GST–14-3-3τ proteins were conjugated to glutathione–agarose beads and incubated with 35S-labeled MEF2 proteins. 14-3-3-associated proteins were resolved by SDS–PAGE, followed by autoradiography (A). Ten percent of the input 35S-labeled MEF2 proteins were applied directly to the gel (B).
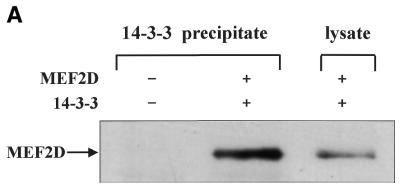
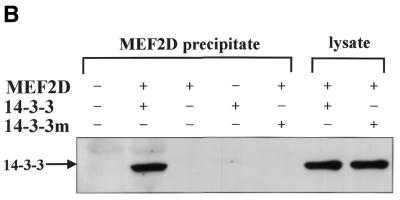
Co-precipitation assays were also performed to further examine the interaction between MEF2D and 14-3-3τ. His6-tagged 14-3-3τ (His–14-3-3τ) and MEF2D were expressed in U937 cells by transient transfection and His–14-3-3τ was precipitated with a nickel–agarose resin. Washed precipitates were separated by SDS–PAGE and blotted with anti-MEF2D antibodies. The results show that MEF2D co-precipitated with 14-3-3τ in transiently transfected cells (Fig. 3A). When His6-tagged MEF2D (His–MEF2D) and Flag-tagged 14-3-3τ (Flag–14-3-3τ) were expressed in U937 cells, Flag–14-3-3τ was co-purified with His–MEF2D on a nickel–agarose resin (Fig. 3B). The 14-3-3 dimers form a cup-like shape with an inner concave surface and a 14-3-3η protein carrying mutations in its inner concave surface failed to interact with Raf-1 (36). We made a similar 14-3-3τ mutant by changing conserved arginines at positions 56 and 60 to alanines. Although the mutant 14-3-3τ with a Flag tag (Flag–14-3-3τR56,60A) was well expressed in U937 cells, it failed to co-precipitate with MEF2D (Fig. 3B). These results showed that Arg56 and Arg60 of 14-3-3τ in its inner concave surface are critical for the interaction between MEF2D and 14-3-3τ.
Figure 3.
MEF2D forms a complex with 14-3-3τ in vivo. His–14-3-3τ and MEF2D (A) and His–MEF2D, Flag–14-3-3τ and Flag–14-3-3τR56,60A (14-3-3m) (B) were expressed as indicated in U937 cells by transient transfection. Transfected cells were lysed 48 h later in TN1 buffer and the cell lysates were incubated with a nickel–agarose resin. Eluates from the resin or whole cell lysates were analyzed by SDS–PAGE and immunoblots. Anti-MEF2D antibody (A) and anti-Flag antibody (B) were used for immunoblots.
14-3-3τ activates MEF2D by relieving repression by HDAC4
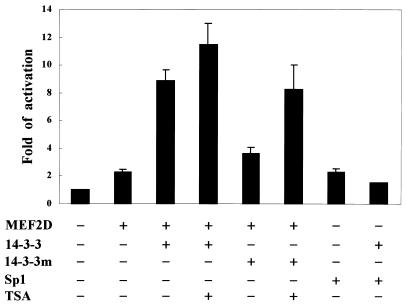
To determine whether the interaction between MEF2D and 14-3-3τ has any effect on MEF2D activity, we co-transfected a MEF2D expression plasmid (pcMEF2D) and a 14-3-3τ expression plasmid (pc14-3-3τ) with a reporter plasmid harboring two MEF2-binding sequences upstream of a minimal promoter (pMEF2FLuc) into U937 cells. As shown in Figure 4, expression of 14-3-3τ enhanced MEF2D-induced transactivation ∼4-fold. 14-3-3τ did not enhance transactivation by another transcription factor Sp1, indicating the specificity of transactivational enhancement by 14-3-3τ (Fig. 4). When the 14-3-3 mutant (14-3-3R56,60A) that could not interact with MEF2D was expressed in U937 cells, it failed to enhance MEF2D-induced transactivation, showing that enhancement of MEF2D activity depends on the interaction between MEF2D and 14-3-3τ. Because MEF2 activity is known to be repressed by HDAC4 and HDAC5 (29,30), we examined whether 14-3-3τ activates MEF2D by affecting the MEF2–HDAC interaction. When transfected cells were treated with the HDAC inhibitor trichostatin A (TSA), it did not significantly affect MEF2D activation by wild-type 14-3-3τ. In contrast, TSA treatment compensated for the inability of the 14-3-3 mutant (14-3-3R56,60A) to activate MEF2D (Fig. 4).
Figure 4.
14-3-3τ enhances the transcriptional activity of MEF2D. U937 cells were transfected with pMEF2Fluc (1 µg), pCMVβgal (0.5 µg) and plasmids expressing MEF2D (1 µg), Sp1 (1 µg), 14-3-3τ (2.5 µg) and 14-3-3τR56,60A (14-3-3m) (2.5 µg). Where indicated, transfected cells were treated with TSA for 12 h. The cells were lysed 48 h later and the lysates were analyzed by luciferase and β-galactosidase assay. All luciferase values were normalized to the β-galactosidase values.
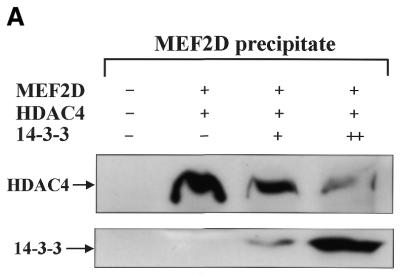
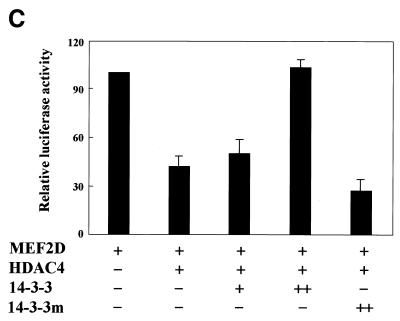
Because MEF2 has been shown to interact with HDAC4 through its MADS–MEF2 domain (30) and our experiment mapped the 14-3-3-interacting domain of MEF2D to the same region (Fig. 22A), we examined whether increasing amounts of 14-3-3τ could physically affect MEF2D–HDAC4 complex formation. Constant amounts of His–MEF2D and Flag–HDAC4 and increasing amounts of Flag–14-3-3τ were expressed in U937 cells and proteins precipitated with nickel–agarose were analyzed by immunoblotting. As increasing amounts of 14-3-3τ formed a complex with MEF2D, the amount of HDAC4 co-precipitated with MEF2D decreased (Fig. 5A). In contrast, when increasing amounts of Flag–14-3-3τ were expressed with constant amounts of His–HDAC4 and MEF2D and His–HDAC4 was precipitated, the amount of 14-3-3τ that co-precipitated with HDAC4 remained unchanged (Fig. 5B). These results strongly suggest that 14-3-3τ primarily binds to MEF2D and competes with HDAC4 for binding to MEF2D. To assess functional significance of the finding, a reporter plasmid pMEF2FLuc and the plasmids expressing MEF2D, HDAC4 and 14-3-3τ were transiently transfected into U937 cells. Figure 5C shows that activation of MEF2FLuc by MEF2D was repressed by HDAC4 and that the repression by HDAC4 was relieved by increasing expression of 14-3-3τ. However, the 14-3-3 mutant (14-3-3R56,60A) failed to relieve the repression by HDAC4 (Fig. 5C). These results strongly suggests that 14-3-3τ activates MEF2D by competitively inhibiting HDAC4 from binding to MEF2D.
Figure 5.
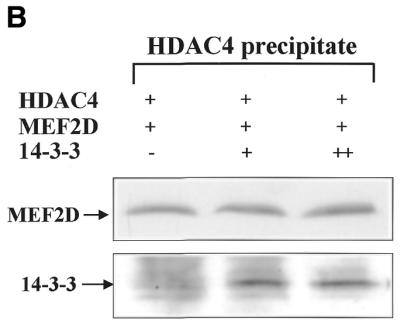
14-3-3τ relieves MEF2D from repression by HDAC4. Expression plasmids for (A) His–MEF2D (4 µg), Flag–HDAC4 (1 µg) and Flag–14-3-3τ (+, 5 µg; ++, 10 µg) and (B) His–HDAC4 (4 µg), Flag–MEF2D (1 µg) and Flag–14-3-3τ (+, 5 µg; ++, 10 µg) were transfected into U937 cells. Lysates of transfected cells were incubated with a nickel–agarose resin and eluates from the resin were analyzed on immunoblots using anti-Flag antibody. (C) U937 cells were transfected with pMEF2Fluc (1 µg), pCMVβgal (0.5 µg) and expression plasmids for MEF2D (1 µg), HDAC4 (0.2 µg), 14-3-3τ (+, 2 µg; ++, 4 µg) and 14-3-3τR56,60A (14-3-3m) (4 µg). Lysates of transfected cells were analyzed by luciferase and β-galactosidase assay. Data are presented as a percentage of the luciferase value from transfectants expressing MEF2D alone.
Control of muscle cell differentiation by 14-3-3τ
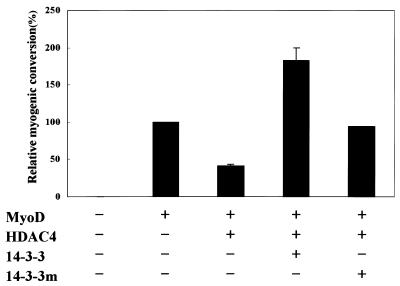
MyoD and the MEF2 proteins have been shown to induce muscle cell differentiation by cooperatively activating muscle cell-specific gene expression (22) and expression of HDAC4 or HDAC5 blocks muscle cell differentiation by repressing MEF2 activity (31). We therefore tested 14-3-3τ and its mutant for their effects on MyoD-dependent muscle cell differentiation. Transfection of 10T1/2 fibroblasts with MyoD alone resulted in efficient conversion to the muscle cell lineage and HDAC4 interfered with this process. When 14-3-3τ was co-expressed, it completely rescued the cells from the block to myogenesis imposed by HDAC4 (Fig. 6). In contrast, a 14-3-3τ mutant (14-3-3R56,60A) lacking MEF2D-interacting activity only partially released the cells from the myogenesis block by HDAC4 (Fig. 6). Taken together, these results suggest that 14-3-3τ contributes to myogenesis by releasing MEF2D from its repression by HDAC4.
Figure 6.
Control of muscle cell differentiation by 14-3-3τ. 10T1/2 fibroblasts were transiently transfected with expression plasmids for MyoD (0.4 µg), HDAC4 (0.2 µg), 14-3-3τ (1.8 µg) and 14-3-3τR56,60A (14-3-3m) (1.8 µg). Transfected cells were transferred to differentiation medium 2 days after transfection and stained with anti-myosin antibodies after an additional 3 days in culture. Myogenic conversion refers to the percentage of myosin-positive cells in transfectants relative to transfectants expressing MyoD alone (100% represents ∼600 myosin-positive cells/35 mm dish). Values represent the means ± SD from al least two experiments.
DISCUSSION
In this study we present the first experimental evidence that 14-3-3τ directly binds to MEF2D and enhances the transactivational activity of MEF2D. MEF2 proteins belong to the MADS box family, which comprises SRF, the yeast regulatory proteins MCM1 and ARG80 and several plant proteins (37). Several MADS box proteins have been shown to interact with other classes of proteins. SRF, for example, binds to its consensus sequence serum response element and forms a ternary complex with p62TCF (37). The interaction is mediated by a DNA-binding region of SRF containing a MADS domain and the ternary complex is subject to MAP kinase-dependent regulation (37). Our yeast two-hybrid screening and GST pull-down experiments also mapped the 14-3-3-interacting region of MEF2D to its N-terminal region containing a MADS domain, which suggests that interaction of MADS box proteins with other proteins through their MADS domain is a common theme. Because MEF2 proteins share almost identical MADS–MEF2 domains, we examined whether 14-3-3 could bind to other MEF2 proteins and have found that MEF2A and 14-3-3τ co-immunoprecipitate (data not shown).
Although some 14-3-3 proteins were reported to be located mostly in the cytoplasm (38,39), our experiments with a confocal microscope showed that Flag-tagged 14-3-3τ was localized in both the cytoplasm and nucleus (Fig. 1). The subcellular localization pattern of the protein, however, was not changed on treatment with the phorbol ester TPA or serum (data not shown). The 14-3-3 proteins have also been reported to interact with the transcription factors NFAT and the glucocorticoid receptor, modulating their transactivational activities (8,9). Our data further support the notion that 14-3-3 proteins have a regulatory function in transcriptional activation in the nucleus.
How 14-3-3 activates MEF2D remains to be further studied. It has been shown that 14-3-3 binds to and protects phosphorylated serine or threonine residues from phosphatase activity. MEF2 proteins have been shown to be phosphorylated and activated by several kinases, including Erk5 and CaMK (26,27,40). In our experiments MEF2D was activiated by Erk5, but co-expressed wild-type or dominant-negative 14-3-3τ did not modulate the activation by Erk5 (data not shown). While our manuscript was in preparation, McKinsey et al. reported that CaMKI and CaMKIV phosphorylate HDAC5, which then dissociates from MEF2 and translocates to the cytoplasm (31,41). In addition, binding of 14-3-3ɛ to phosphorylated HDAC5 was suggested to be important for the release of HDAC5 from MEF2 (41). However, our co-precipitation experiments showed that increasing amounts of 14-3-3τ bind to MEF2D rather than to HDAC4 (Fig. 5), suggesting that 14-3-3τ binds and activates MEF2D by competitively excluding HDAC4. This apparent discrepancy might be because our study was focused on the mechanism involving HDAC4 and 14-3-3τ. It might be possible that each 14-3-3 isotype has a different affinity for different members of the HDAC and MEF2 families. In fact, 14-3-3ɛ showed different affinities for HDAC4 and HDAC5 (41). It has also been reported that binding of calmodulin to HDAC leads to its dissociation from MEF2 (42), which suggests multiple ways of releasing HDACs from MEF2 proteins.
Our experimental results on myogenesis do not rule out the possibility that 14-3-3τ also interacts with another co-regulatory molecule(s). However, it is already known that HDAC4 and HDAC5 block muscle differentiation by binding and repressing MEF2 (31). From the results of our co-precipitation and reporter gene assay experiments showing that 14-3-3τ releases MEF2D from HDAC4, it is very likely that expression of 14-3-3τ contributes to myogenesis by releasing MEF2D from HDAC4.
Although we have shown that MEF2D forms a complex with 14-3-3τ or HDAC4, it is not clear how complex formation is regulated. The interaction between MEF2D and HDAC4 has been reported to be regulated by CaMKIV or calmodulin (30,42). In our experiments, however, treatment with phosphatase or calcium chelating agents did not affect complex formation among MEF2D, 14-3-3τ and HDAC4 (data not shown). The amino acid sequences of 14-3-3 isotypes are highly conserved and many 14-3-3 proteins show tissue-specific expression (1). These different 14-3-3 isotypes could form a complex with MEF2s or HDACs with different affinities and might contribute to tissue-specific regulation of MEF2 activity. Analysis of complex formation among MEF2, HDAC and 14-3-3 in response to different stimuli in various tissues might be important to understand the regulatory function of MEF2 proteins.
Acknowledgments
ACKNOWLEDGEMENTS
We thank T. Kouzarides, A. B. Lassar, J. Martin and X. J. Yang for plasmids and R. Prywes, J. H. Ahn, Y. J. Kim, H. W. Lee and Y. S. Seo for critical comments. This work was supported by an SRC grant (MTRC) from the Korea Science and Engineering Foundation and a Samsung Biomedical Research Institute grant (SBRI BA0001) to T.H.H.
References
- 1.Aitken A. (1995) 14-3-3 proteins on the MAP. Trends Biochem. Sci., 20, 95–97. [DOI] [PubMed] [Google Scholar]
- 2.Burbelo P.D. and Hall,A. (1995) 14-3-3 proteins: hot numbers in signal transduction. Curr. Biol., 5, 95–96. [DOI] [PubMed] [Google Scholar]
- 3.Muslin A.J., Tanner,J.W., Allen,P.M. and Shaw,A. (1996) Interaction of 14-3-3 with signaling proteins is mediated by the recognition of phosphoserine. Cell, 84, 889–897. [DOI] [PubMed] [Google Scholar]
- 4.Liu Y.C., Liu,Y., Elly,C., Yoshida,H., Lipkowitz,S. and Altman,A. (1997) Serine phosphorylation of Cbl induced by phorbol ester enhances its association with 14-3-3 proteins in T cells via a novel serine-rich 14-3-3-binding motif. J. Biol. Chem., 272, 9979–9985. [DOI] [PubMed] [Google Scholar]
- 5.Zha J., Harada,H., Yang,E., Jockel,J. and Korsmeyer,S.J. (1996) Serine phosphorylation of death against BAD in response to survival factor results in binding to 14-3-3 not BCL-X. Cell, 87, 619–628. [DOI] [PubMed] [Google Scholar]
- 6.Peng C.Y., Graver,P.R., Thomas,R.S., Wu,Z., Shaw,A.S. and Pinica-Worms,H. (1997) Mitotic and G2 check point control: regulation of 14-3-3 protein binding by phosphorylation of Cdc25C on serine-216. Science, 277, 1501–1505. [DOI] [PubMed] [Google Scholar]
- 7.Craparo A., Freund,R. and Gustafson,T.A. (1997) 14-3-3ɛ interacts with the insulin like growth factor I receptor and insulin receptor substrate I in a phosphoserine-dependent manner. J. Biol. Chem., 272, 11663–11669. [DOI] [PubMed] [Google Scholar]
- 8.Wakui H., Wright,A.P.H., Gustafsson,J.A. and Zilliacus,J. (1997) Interaction of the ligand-activated glucocorticoid receptor with the 14-3-3 η protein. J. Biol. Chem., 272, 8153–8156. [DOI] [PubMed] [Google Scholar]
- 9.Chow C.W. and Davis,R.J. (2000) Integration of calcium and cyclic AMP signaling pathways by 14-3-3. Mol. Cell. Biol., 20, 702–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yaffe M.B., Rittinger,K., Volinia,S., Caron,P.R., Aitken,A., Leffers,H., Gamblin,S.J., Smerdon,S.J. and Cantley,L.S. (1997) The structural basis for 14-3-3:phosphopeptide binding specificity. Cell, 91, 961–971. [DOI] [PubMed] [Google Scholar]
- 11.Du X., Fox,J.E. and Pei,S. (1996) Identification of a binding sequence for the 14-3-3 protein within the cytoplasmic domain of the adhesion receptor, platelet glycoprotein Ib alpha. J. Biol. Chem., 271, 7362–7367. [DOI] [PubMed] [Google Scholar]
- 12.Ku N.O., Liao,J. and Omary,M.B. (1998) Phosphorylation of human keratin 18 serine 33 regulates binding to 14-3-3 proteins. EMBO J., 17, 1892–1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Y.C., Liu,Y., Elly,C., Yishida,H., Lipkowitz,S. and Altman,A. (1997) Serine phosphorylation of Cbl induced by phorbol ester enhances its association with 14-3-3 proteins in T cells via a novel serine-rich 14-3-3 binding motif. J. Biol. Chem., 272, 9979–9985. [DOI] [PubMed] [Google Scholar]
- 14.Zhang S.H., Kobayashi,R., Graves,P.R., Piwnica-Worms,H. and Tonks,N.K. (1997) Serine phosphorylation-dependent association of the band 4.1-related protein-tyrosine phosphatase PTPH1 with 14-3-3 beta protein. J. Biol. Chem., 272, 27281–27287. [DOI] [PubMed] [Google Scholar]
- 15.Breitbart R.E., Liang,C., Smoot,L.B., Laheru,D.A., Mahdavi,V. and Nadal-Ginard,B. (1993) A fourth human MEF2 trancription factor, hMEF2D, is an early marker of the myogenic lineage. Development, 118, 1095–1106. [DOI] [PubMed] [Google Scholar]
- 16.Chambers A.E., Logan,M., Kotecha,S., Towers,N., Sparrow,D. and Mohun,T.J. (1994) The RSRF/MEF2 protein SL1 regulates cardiac muscle-specific transcription of a myosin light-chain gene in Xenopus embryos. Genes Dev., 8, 1324–1334. [DOI] [PubMed] [Google Scholar]
- 17.Martin J.F., Miano,J.M., Hustad,C.M., Copeland,N.G., Jenkins,N.A. and Olson,E.N. (1994) A mef2 gene that regulates a muscle-specific isoform via alternative mRNA splicing. Mol. Cell. Biol., 14, 1647–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McDermott J.C., Cardoso,M.C., Yu,Y., Andres,V., Leifer,D., Krainc,D., Lipton,S.A. and Nadal-Ginard,B. (1993) hMEF2C gene encodes skeletal muscle- and brain-specific transcription factor. Mol. Cell. Biol., 13, 2564–2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pollock R. and Treisman,R. (1991) Human SRF-related proteins: DNA-binding properties and potential regulatory targets. Genes Dev., 5, 2327–2341. [DOI] [PubMed] [Google Scholar]
- 20.Yu Y., Breitbart,R.B., Smoot,L.B., Lee,Y., Mahdavi,V. and Nadal-Ginard,B. (1992) Human myocyte-specific enhancer factor 2 comprises a group of tissue-restricted MADS box transcription factors. Genes Dev., 6, 1783–1798. [DOI] [PubMed] [Google Scholar]
- 21.Cserjesi P. and Olson,E.N. (1991) Myogenin induces the myocyte-specific enhancer binding factor MEF-2 independently of other muscle-specific gene products. Mol. Cell. Biol., 11, 4854–4862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Molkentin J.D., Black,B.L., Martin,J.F. and Olson,E.N. (1995) Cooperative activation of muscle gene expression by MEF2 and myogenic bHLH proteins. Cell, 83, 1125–1136. [DOI] [PubMed] [Google Scholar]
- 23.Lin Q., Schwartz,J., Bucana,C. and Olson,E.N. (1997) Control of mouse cardiac morphogenesis and myogenesis by transcription factor MEF2C. Science, 276, 1404–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mao Z., Bonni,A., Xia,F., Nadal-Vicens,M. and Greenberg,M.E. (1999) Neuronal activity-dependent cell survival mediated by transcription factor MEF2. Science, 286, 785–790. [DOI] [PubMed] [Google Scholar]
- 25.Shin H.M., Seoh,J.Y., Chung,H.Y., Choi,S.J., Hahn,M.J., Kang,J.S., Choi,M.S. and Han,T.H. (1999) Requirement of MEF2D in the induced differentiation of HL60 promyeloid cells. Mol. Immunol., 36, 1209–1214. [DOI] [PubMed] [Google Scholar]
- 26.Blaeser F., Ho,N., Prywes,R. and Chatila,T.A. (2000) Ca2+-dependent gene expression mediated by MEF2 transcription factors. J. Biol. Chem., 275, 197–209. [DOI] [PubMed] [Google Scholar]
- 27.Kato Y., Zhao,M., Morikawa,A., Sugiyama,T., Chakravortty,D., Koide,N., Yoshida,T., Tapping,R.I., Yang,Y., Yokochi,T. and Lee,J.D. (2000) Big mitogen-activated kinase regulates multiple members of the MEF2 protein family. J. Biol. Chem., 275, 18534–18540. [DOI] [PubMed] [Google Scholar]
- 28.Youn H.D., Sun,L., Prywes,R. and Liu,J.O. (1999) Apoptosis of T cells mediated by Ca2+-induced release of the transcription factor MEF2. Science, 286, 790–793. [DOI] [PubMed] [Google Scholar]
- 29.Miska E.A., Karlsson,C., Langley,M., Nielson,S.J., Pines,J. and Kouzarides,T. (1999) HDAC4 deacetylase associates with and represses the MEF2 transcription factor. EMBO J., 18, 5099–5107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu J., McKinsey,T.A., Nicol,R.L. and Olson,E.N. (2000) Signal-dependent activation of the MEF2 transcription factor by dissociation from histone deacetylase. Proc. Natl Acad. Sci. USA, 97, 4070–4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McKinsey T.A., Zhang,C.-L., Lu,J. and Olson,E.N. (2000) Signal-dependent nuclear export of a histone deacetylase regulates muscle differentiation. Nature, 408, 106–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Han T.H. and Prywes,R. (1995) Regulatory role of MEF2D in serum induction of the c-jun promoter. Mol. Cell. Biol., 15, 2907–2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Novitch B.G., Douglas,B.S., Kim,P.S., Cheung,W.L. and Lassar,A.B. (1999) pRb is required for MEF2-dependent gene expression as well as cell-cycle arrest during skeletal muscle differentiation. Curr. Biol., 9, 449–459. [DOI] [PubMed] [Google Scholar]
- 34.Wang A.H., Kruhlak,M.J., Wu,J., Bertos,N.R., Vezmar,M., Posner,B.I., Bazett-Jones,D.P. and Yang,X.J. (2000) Regulation of histone deacetylase 4 by binding of 14-3-3 proteins. Mol. Cell. Biol., 20, 6904–6912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Black B.L. and Olson,E.N. (1998) Transcriptional control of muscle development by myocyte enhancer factor-2 (MEF2) proteins. Annu. Rev. Cell. Dev. Biol., 14, 167–196. [DOI] [PubMed] [Google Scholar]
- 36.Thorson J.A., Yu,L.W.K., Hsu,A.L., Shih,N.-Y., Graves,P.R., Tanner,J.W., Allen,P.M., Piwnica-Worms,H. and Shaw,A.S. (1998) 14-3-3 proteins are required for maintenance of Raf-1 phosphorylation and kinase activity. Mol. Cell. Biol., 18, 5229–5238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Treisman R. (1994) Ternary complex factors: growth factor regulated transcriptional activators. Curr. Opin. Genet. Dev., 4, 96–101. [DOI] [PubMed] [Google Scholar]
- 38.Roth D., Morgan,A., Martin,H., Jones,D., Marten,G.J., Aitken,A. and Burgoyne,R.D. (1994) Characterization of 14-3-3 proteins in adrenal chromaffin cells and demonstration of isoform-specific phospholipid binding. Biochem. J., 301, 305–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martin H., Rostas,J., Patel,Y. and Aitken,A. (1994) Subcellular localization of 14-3-3 isoforms in rat brain using specific antibodies. J. Neurochem., 63, 2259–2265. [DOI] [PubMed] [Google Scholar]
- 40.Marinissen M.J., Chiariello,M., Plallante,M. and Gutkind,J.S. (1999) A network of mitogen-activated protein kinases links G protein-coupled receptors to the c-jun promoter: a role for c-Jun NH2-terminal kinase, p38s and extracellular signal-regulated kinase 5. Mol. Cell. Biol., 19, 4289–4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McKinsey T.A., Zhang,C.-L. and Olson,E.N. (2000) Activation of the myocyte enhancer factor-2 transcription factor by calcium/calmodulin-dependent protein kinase-stimulated binding of 14-3-3 to histone deacetylase 5. Proc. Natl Acad. Sci. USA, 97, 14400–14405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Youn H.D., Grozinger,C.M. and Liu,J.O. (2000) Calcium regulates transcriptional repression of myocyte enhancer factor 2 by histone deacetylase 4. J. Biol. Chem., 275, 22563–22567. [DOI] [PubMed] [Google Scholar]