Abstract
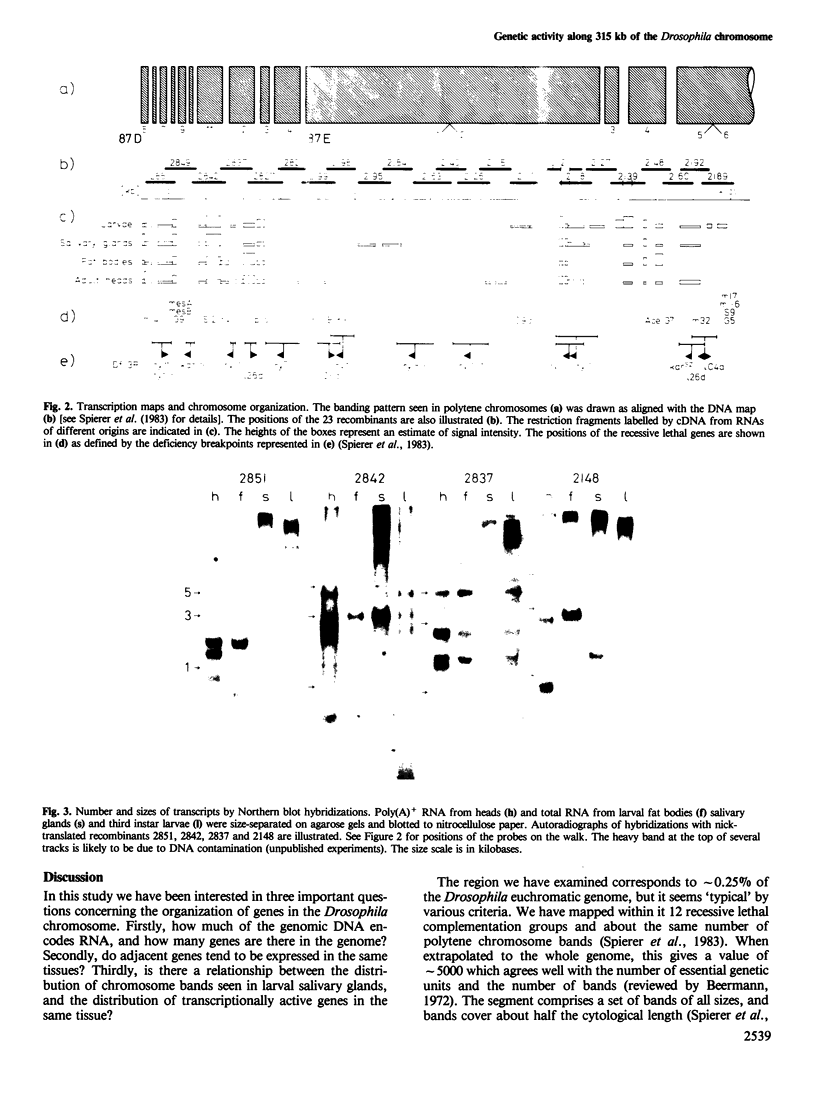
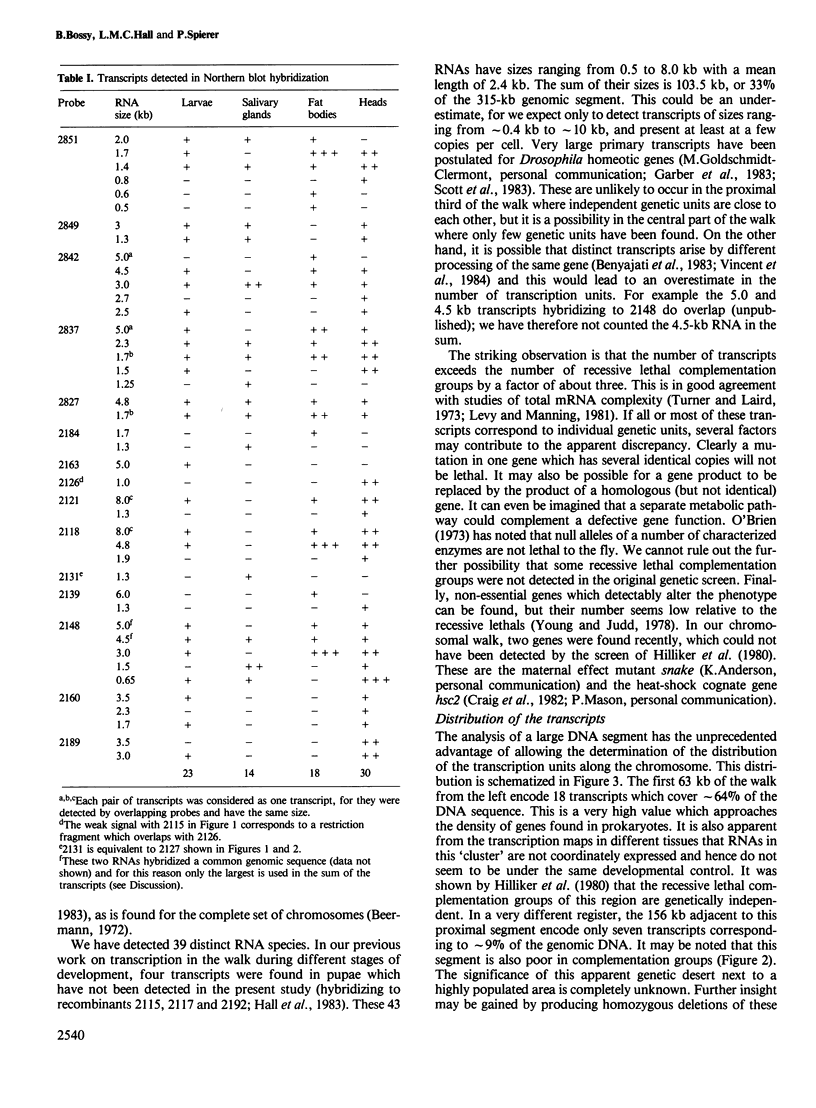
Transcripts from different tissues were mapped along a 315 kb segment of the Drosophila chromosome, a region which includes the rosy and Ace loci. Forty-three distinct RNA species were detected, though only 12 recessive lethal complementation groups had been mapped in the interval. The sum of the sizes of the transcripts covers 33% of the genomic DNA. The distribution of transcription units along the walk is very uneven. Sixty-three kb of genomic DNA at the proximal end of the walk encode 18 transcripts, while only seven are found in the next 153 kb. Each tissue exhibits a specific spectrum of transcripts. No clustering was seen among genes expressed coordinately. In salivary glands, the number of transcripts detected corresponds to the number of chromomeric units in the polytene chromosomes of this tissue. Moreover, the density distribution of transcripts along the DNA walk is parallel to the density distribution of chromomeric units.
Keywords: chromosome organization, differentiation, Drosophila melanogaster, transcription
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beermann W. Chromomeres and genes. Results Probl Cell Differ. 1972;4:1–33. doi: 10.1007/978-3-540-37164-9_1. [DOI] [PubMed] [Google Scholar]
- Bender W., Spierer P., Hogness D. S. Chromosomal walking and jumping to isolate DNA from the Ace and rosy loci and the bithorax complex in Drosophila melanogaster. J Mol Biol. 1983 Jul 25;168(1):17–33. doi: 10.1016/s0022-2836(83)80320-9. [DOI] [PubMed] [Google Scholar]
- Benyajati C., Spoerel N., Haymerle H., Ashburner M. The messenger RNA for alcohol dehydrogenase in Drosophila melanogaster differs in its 5' end in different developmental stages. Cell. 1983 May;33(1):125–133. doi: 10.1016/0092-8674(83)90341-0. [DOI] [PubMed] [Google Scholar]
- Biessmann H. Changes in the abundance of polyadenylated RNAs in development of Drosophila melanogaster analyzed with cloned DNA fragments. Dev Biol. 1981 Apr 15;83(1):62–68. doi: 10.1016/s0012-1606(81)80008-5. [DOI] [PubMed] [Google Scholar]
- Derman E., Krauter K., Walling L., Weinberger C., Ray M., Darnell J. E., Jr Transcriptional control in the production of liver-specific mRNAs. Cell. 1981 Mar;23(3):731–739. doi: 10.1016/0092-8674(81)90436-0. [DOI] [PubMed] [Google Scholar]
- Fyrberg E. A., Kindle K. L., Davidson N., Kindle K. L. The actin genes of Drosophila: a dispersed multigene family. Cell. 1980 Feb;19(2):365–378. doi: 10.1016/0092-8674(80)90511-5. [DOI] [PubMed] [Google Scholar]
- Galau G. A., Britten R. J., Davidson E. H. A measurement of the sequence complexity of polysomal messenger RNA in sea urchin embryos. Cell. 1974 May;2(1):9–20. doi: 10.1016/0092-8674(74)90003-8. [DOI] [PubMed] [Google Scholar]
- Garber R. L., Kuroiwa A., Gehring W. J. Genomic and cDNA clones of the homeotic locus Antennapedia in Drosophila. EMBO J. 1983;2(11):2027–2036. doi: 10.1002/j.1460-2075.1983.tb01696.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg D. A., Posakony J. W., Maniatis T. Correct developmental expression of a cloned alcohol dehydrogenase gene transduced into the Drosophila germ line. Cell. 1983 Aug;34(1):59–73. doi: 10.1016/0092-8674(83)90136-8. [DOI] [PubMed] [Google Scholar]
- Goldschmidt-Clermont M. Two genes for the major heat-shock protein of Drosophila melanogaster arranged as an inverted repeat. Nucleic Acids Res. 1980 Jan 25;8(2):235–252. doi: 10.1093/nar/8.2.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall L. M., Mason P. J., Spierer P. Transcripts, genes and bands in 315,000 base-pairs of Drosophila DNA. J Mol Biol. 1983 Sep 5;169(1):83–96. doi: 10.1016/s0022-2836(83)80176-4. [DOI] [PubMed] [Google Scholar]
- Hastie N. D., Bishop J. O. The expression of three abundance classes of messenger RNA in mouse tissues. Cell. 1976 Dec;9(4 Pt 2):761–774. doi: 10.1016/0092-8674(76)90139-2. [DOI] [PubMed] [Google Scholar]
- Hilliker A. J., Clark S. H., Chovnick A., Gelbart W. M. Cytogenetic analysis of the chromosomal region immediately adjacent to the rosy locus in Drosophila melanogaster. Genetics. 1980 May;95(1):95–110. doi: 10.1093/genetics/95.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefevre G., Jr The one band-one gene hypothesis: evidence from a cytogenetic analysis of mutant and nonmutant rearrangement breadpoints in Drosophila melanogaster. Cold Spring Harb Symp Quant Biol. 1974;38:591–599. doi: 10.1101/sqb.1974.038.01.063. [DOI] [PubMed] [Google Scholar]
- Levy L. S., Manning J. E. Expression of a set of head-specific genes during Drosophila development. Dev Biol. 1982 Dec;94(2):465–476. doi: 10.1016/0012-1606(82)90363-3. [DOI] [PubMed] [Google Scholar]
- Levy L. S., Manning J. E. Messenger RNA sequence complexity and homology in developmental stages of Drosophila. Dev Biol. 1981 Jul 15;85(1):141–149. doi: 10.1016/0012-1606(81)90243-8. [DOI] [PubMed] [Google Scholar]
- O'Brien S. J. On estimating functional gene number in eukaryotes. Nat New Biol. 1973 Mar 14;242(115):52–54. doi: 10.1038/newbio242052a0. [DOI] [PubMed] [Google Scholar]
- Scherer G., Telford J., Baldari C., Pirrotta V. Isolation of cloned genes differentially expressed at early and late stages of Drosophila embryonic development. Dev Biol. 1981 Sep;86(2):438–447. doi: 10.1016/0012-1606(81)90202-5. [DOI] [PubMed] [Google Scholar]
- Scholnick S. B., Morgan B. A., Hirsh J. The cloned dopa decarboxylase gene is developmentally regulated when reintegrated into the Drosophila genome. Cell. 1983 Aug;34(1):37–45. doi: 10.1016/0092-8674(83)90134-4. [DOI] [PubMed] [Google Scholar]
- Scott M. P., Weiner A. J., Hazelrigg T. I., Polisky B. A., Pirrotta V., Scalenghe F., Kaufman T. C. The molecular organization of the Antennapedia locus of Drosophila. Cell. 1983 Dec;35(3 Pt 2):763–776. doi: 10.1016/0092-8674(83)90109-5. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Spierer P., Spierer A., Bender W., Hogness D. S. Molecular mapping of genetic and chromomeric units in Drosophila melanogaster. J Mol Biol. 1983 Jul 25;168(1):35–50. doi: 10.1016/s0022-2836(83)80321-0. [DOI] [PubMed] [Google Scholar]
- Spradling A. C., Rubin G. M. The effect of chromosomal position on the expression of the Drosophila xanthine dehydrogenase gene. Cell. 1983 Aug;34(1):47–57. doi: 10.1016/0092-8674(83)90135-6. [DOI] [PubMed] [Google Scholar]
- Thomas P. S. Hybridization of denatured RNA and small DNA fragments transferred to nitrocellulose. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5201–5205. doi: 10.1073/pnas.77.9.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner S. H., Laird C. D. Diversity of RNA sequences in Drosophila melanogaster. Biochem Genet. 1973 Nov;10(3):263–274. doi: 10.1007/BF00485704. [DOI] [PubMed] [Google Scholar]
- Vincent A., O'Connell P., Gray M. R., Rosbash M. Drosophila maternal and embryo mRNAs transcribed from a single transcription unit use alternate combinations of exons. EMBO J. 1984 May;3(5):1003–1013. doi: 10.1002/j.1460-2075.1984.tb01920.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young M. W., Judd B. H. Nonessential Sequences, Genes, and the Polytene Chromosome Bands of DROSOPHILA MELANOGASTER. Genetics. 1978 Apr;88(4):723–742. doi: 10.1093/genetics/88.4.723. [DOI] [PMC free article] [PubMed] [Google Scholar]





