Abstract
The TCP transcription factors usually act as integrators of multiple growth regulatory and environmental stimuli. However, little is known about this gene family in the important tropical crop cassava (Manihot esculenta). In this study, 36 TCP genes were identified and renamed based on cassava whole-genome sequence and their sequence similarity with Arabidopsis TCPs. Typical TCP domains were detected in these proteins by multiple sequence alignment analysis. Evolutionary analysis indicated that MeTCPs could be divided into 8 subgroups, which was further supported by gene structure and conserved motif analyses. qRT-PCR analysis revealed tissue-specific and hormone-responsive expression patterns of MeTCP genes. Moreover, with global expression and promoter analysis, we found that MeTCPs showed similar or distinct expression patterns under cold and/or drought stress, suggesting that they might participate in distinct signaling pathways. Our study provides the first comprehensive analysis of TCP gene family in the cassava genome. The data will be useful for uncovering the potential functions of MeTCP genes, and their possible roles in mediating hormone and abiotic stress responses in cassava.
Introduction
As sessile organism, plant growth and yield are strongly influenced by environmental stimuli such as cold and drought1. To respond and adapt to these conditions, plants develop various mechanisms at both physiological and biochemical levels2. It has been well established that many adaptation processes are regulated by stress-responsive gene expression1, 3. Transcription factors (TFs), which are a diverse family of regulatory proteins with DNA-binding domains, play a central role in mediating various aspects of cellular processes by regulating gene expression through interacting with cis-elements in the promoter regions of various downstream genes4, 5. Series studies previously have uncovered a group of TF genes, such as AP2/ERF, MYB and bZIP, which participate in various stress-induced physiological processes and regulatory networks in higher plants6, 7.
TCP genes encode plant-specific transcription factors, which are named after the first four functionally identified members: TB1 (TEOSINTE BRANCHED 1) in Zea mays, CYC (CYCLOIDEA) in Antirrhinum majus, and PCF1 and PCF2 (PROLIFERATING CELL FACTORS 1 and 2) in Oryza sativa 8. Typically, N-terminus of this class of transcription factors exhibits a highly conserved TCP domain, which contains a non-canonical basic-Helix-Loop-Helix (bHLH) structure involved in DNA binding, protein-protein interaction and protein nuclear localization9, 10. Based on the homology of the TCP domains, TCP proteins can be further divided into two major subfamilies, Class I (represented by the rice PCF proteins) and class II (represented by CYC and TB1)8, 11. The DNA-Binding site selection assays revealed that the consensus binding sequences for these two classes are slightly different, but overlapping with GGNCCC sequences. The DNA binding sequence for class I is GGNCCCAC while class II prefer to bind the DNA motif G(T/C)GGNCCC10. Both class I and class II include TCPs that can function as transcriptional activators and repressors12.
Increasing evidences have indicated that proteins of TCP family take part in the regulation of many biological processes during plant growth and development, including plant architecture12, 13, leaf morphogenesis14–16, hormone pathways13, 17–19 and response to environmental stimuli among various species20–22. For example, studies in Arabidopsis suggest that AtTCP14 appears to function in regulating embryonic growth of seeds23. AtTCP15, along with its closest homolog AtTCP14, regulates cell proliferation in the developing leaf blade and specific floral tissues, and also modulates gibberellins and auxin responses17, 24. The expression of a repressor form of AtTCP11 caused pleiotropic developmental alterations25. AtTCP2, AtTCP3, AtTCP4, AtTCP10 and AtTCP24 have been earlier identified as targets of miRNA319, and are required for leaf development, petal growth, cell wall synthesis and JA synthesis14, 26, 27. Creeping bentgrass (Agrostis stolonifera) plants overexpressing Osa-miR319a, in which four putative target genes, AsPCF5, AsPCF6, AsPCF8, and AsTCP14 were down-regulated, displayed morphological changes and exhibited enhanced drought and salt tolerance associated with increased leaf wax content and water retention20. In rice, overexpression of OsTCP19 led to developmental abnormalities like fewer lateral root formation and contributed to better stress tolerance21.
Widely cultivated in tropical area, cassava is considered as one of the most important economic crops worldwide, providing starch for food, feed and energy production28–30. Cassava can effectively utilize light, heat and water resource; it is resistant to drought but sensitive to low temperature31. Cold and drought stress severely limit cassava plant growth and yield32. Thus, uncovering the mechanisms underlying the resistance of cassava to these stresses may provide candidate genes for genetic improvement of stress resistance for cassava. Previously, we performed strand specific RNA sequencing for cassava TMS60444 shoot apices and young leaves under cold and PEG-induced drought stress, providing an excellent resource for analysis of stress responsive genes globally.
To date, numerous TCP family members have been identified in various species33–45. However, no evidence is available regarding the TCP family in cassava. Due to the critical role of TCP transcription factors in the control of plant development and abiotic stress responses, we performed for the first time the comprehensive analysis of the MeTCP transcription factor family in cassava. In the present study, a total of 36 non-redundant MeTCP transcription factor encoding genes were identified in cassava genome and were subsequently subjected to a systematic analysis, including phylogenetic relationships, gene structure, conserved motif and expression pattern among different tissues and hormone treatments. We also analyzed the expression of the MeTCP genes under normal growth conditions and various abiotic stressors. On the basis of the expression profiles of MeTCP genes and the phylogenetic analysis among the TCP domain proteins in Arabidopsis, rice and cassava, the functions of MeTCPs were predicted. Taken together, our genome-wide analysis of MeTCP gene family will contribute to future studies on the functional characterization of MeTCP proteins in cassava, as well as the identification and comprehensive analysis of the TCP transcription factor family in other species.
Results
Identification of TCP genes in cassava
In order to extensively identify cassava TCP genes, two search strategies were used in this study: first, we used the protein sequences of all 24 TCP genes from Arabidopsis 33 as BLASTp queries to perform multiple searches against the latest whole genome of cassava (http://www.phytozome.net/cassava); then, the TCP domain (PF03634)46 was employed as query to perform a blast search against the same cassava genome database. After discarding redundant sequences, 36 candidate TCP genes were identified in cassava, and further conserved domain detection confirmed that all the identified TCPs harbor the conserved TCP domain that is the basic characteristics of this family. Due to the lack of standard annotation designated to the 36 cassava TCP genes, we named them MeTCP2 to MeTCP23 according to the Arabidopsis TCP proteins with highest sequence similarity. The length of the 36 newly identified TCP proteins varied from 73 to 562 amino acid residues and the relative molecular weight ranged from 8.3 to 58.1 kDa, with isoelectric points in the range of 5.53–10.08. The grand average of hydropathy (GRAVY) of this family genes showed that all of the proteins had a negative value, indicating that all the MeTCP proteins were hydrophilic (Table 1).
Table 1.
TCP genes identified in cassava genome.
| Gene ID | Name | Length(aa) | MW(Da) | PI | Gravy | Genomic locus |
|---|---|---|---|---|---|---|
| Manes.01G187000.1 | MeTCP15a | 392 | 42214.9 | 9.32 | −0.643 | Chromosome01:28455200..28457428 forward |
| Manes.01G263300.1 | MeTCP9a | 366 | 38916.33 | 8.94 | −0.283 | Chromosome01:33561357..33563642 forward |
| Manes.01G020700.1 | MeTCP18a | 387 | 44030.35 | 7.93 | −0.773 | Chromosome01:3502035..3504116 forward |
| Manes.01G094200.1 | MeTCP13a | 350 | 38533.91 | 8.37 | −0.667 | Chromosome01:21880606..21882675 forward |
| Manes.02G055900.1 | MeTCP13b | 361 | 39880.76 | 9.06 | −0.63 | Chromosome02:4209171..4211393 forward |
| Manes.02G066400.1 | MeTCP12 | 383 | 42814.84 | 9.78 | −0.569 | Chromosome02:4925458..4927250 reverse |
| Manes.02G194200.1 | MeTCP23a | 425 | 45005.56 | 7.98 | −0.572 | Chromosome02:15997522..15999455 forward |
| Manes.04G016700.1 | MeTCP11d | 240 | 26058.47 | 8.98 | −0.394 | Chromosome04:1959602..1961886 forward |
| Manes.04G016800.1 | MeTCP11b | 155 | 16551.84 | 9.45 | −0.415 | Chromosome04:1966346..1966813 forward |
| Manes.04G088500.1 | MeTCP20a | 312 | 33848.59 | 7.22 | −0.782 | Chromosome04:22580287..22581884 reverse |
| Manes.05G041000.1 | MeTCP9b | 346 | 36898.79 | 7.8 | −0.325 | Chromosome05:2911279..2913198 forward |
| Manes.05G100100.1 | MeTCP15b | 388 | 41706.51 | 8.23 | −0.582 | Chromosome05:8461910..8463076 reverse |
| Manes.05G123700.1 | MeTCP20c | 73 | 8314.64 | 10.08 | −1.034 | Chromosome05:14666983..14667397 forward |
| Manes.05G119300.1 | MeTCP18b | 372 | 42181.38 | 8.12 | −0.748 | Chromosome05:12207088..12208206 reverse |
| Manes.06G072800.1 | MeTCP18c | 416 | 47024.77 | 9.48 | −0.879 | Chromosome06:18738590..18740594 forward |
| Manes.06G083400.1 | MeTCP5a | 340 | 37976.22 | 8.47 | −0.611 | Chromosome06:19670814..19672682 reverse |
| Manes.06G093900.1 | MeTCP15c | 396 | 42673.9 | 7.37 | −0.73 | Chromosome06:20627278..20631189 forward |
| Manes.06G141800.1 | MeTCP19 | 358 | 37747 | 5.53 | −0.478 | Chromosome06:24669381..24671688 reverse |
| Manes.07G022400.1 | MeTCP8a | 550 | 56832.22 | 7.55 | −0.646 | Chromosome07:2093408..2096799 forward |
| Manes.08G009200.1 | MeTCP11a | 185 | 19802.37 | 6.97 | −0.497 | Chromosome08:675290..675847 reverse |
| Manes.09G051000.1 | MeTCP16 | 403 | 43569.25 | 7.58 | −0.11 | Chromosome09:6817330..6819454 forward |
| Manes.10G120400.1 | MeTCP8b | 562 | 58079.71 | 8.46 | −0.621 | Chromosome10:23214679..23218733 reverse |
| Manes.11G083000.1 | MeTCP20b | 307 | 32859.71 | 8.99 | −0.671 | Chromosome11:11404469..11406607 forward |
| Manes.11G108500.1 | MeTCP7 | 273 | 27679.86 | 9.72 | −0.271 | Chromosome11:20090517..20092799 forward |
| Manes.11G149000.1 | MeTCP11c | 198 | 21355.38 | 9.43 | −0.335 | Chromosome11:26034972..26035568 reverse |
| Manes.12G007700.1 | MeTCP20d | 299 | 32367.12 | 9.41 | −0.702 | Chromosome12:722180..724162 reverse |
| Manes.13G008300.1 | MeTCP20e | 282 | 30554.12 | 9.52 | −0.656 | Chromosome13:815757..817107 reverse |
| Manes.13G138300.1 | MeTCP3b | 343 | 37809.47 | 5.9 | −0.782 | Chromosome13:26570479..26573043 reverse |
| Manes.14G058400.1 | MeTCP3a | 336 | 36987.57 | 5.87 | −0.699 | Chromosome14:4620482..4621572 reverse |
| Manes.14G077200.1 | MeTCP15d | 398 | 42850.16 | 7.39 | −0.716 | Chromosome14:6245053..6246249 reverse |
| Manes.14G086500.1 | MeTCP5b | 387 | 43276.03 | 7.21 | −0.713 | Chromosome14:6949867..6951866 forward |
| Manes.14G097000.1 | MeTCP18d | 474 | 52697.96 | 9.31 | −0.782 | Chromosome14:7827437..7828861 reverse |
| Manes.15G091000.1 | MeTCP4 | 422 | 45753.27 | 6.17 | −0.636 | Chromosome15:6736704..6738513 reverse |
| Manes.15G123800.1 | MeTCP2a | 481 | 52167.1 | 7.86 | −0.886 | Chromosome15:9374446..9382168 reverse |
| Manes.17G072800.1 | MeTCP2b | 481 | 52472.3 | 7.06 | −0.926 | Chromosome17:21184852..21198102 reverse |
| Manes.18G103100.1 | MeTCP23b | 425 | 45159.97 | 6.81 | −0.563 | Chromosome18:9144633..9147156 forward |
Phylogenetic analysis of the MeTCP genes
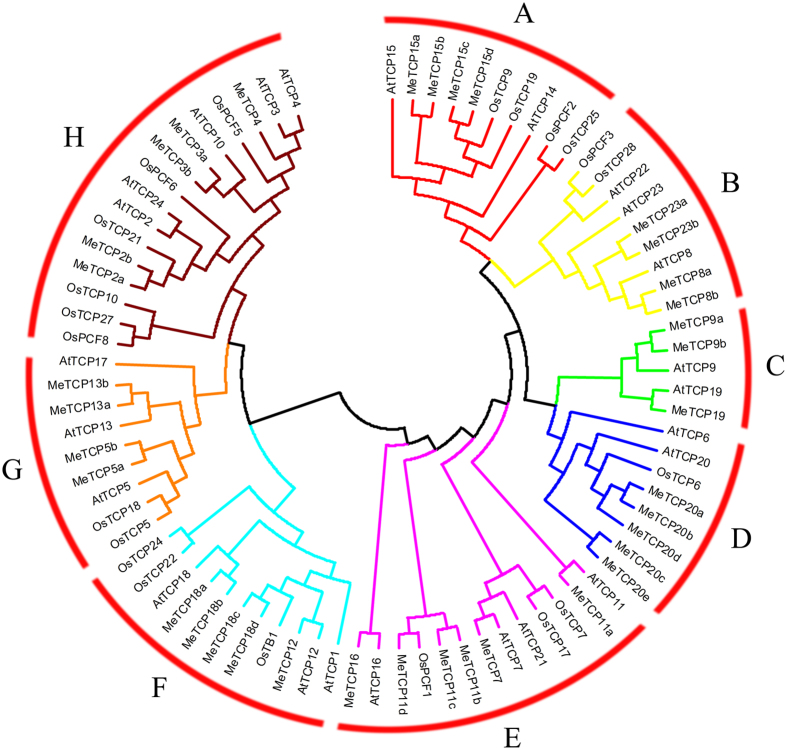
To characterize the evolutionary and phylogenetic relationships between cassava TCP genes and other known TCPs, an unrooted Neiboring-Joining (NJ) tree was constructed on the basis of multiple sequence alignment of 36 MeTCP complete protein sequences with 24 and 22 TCP protein sequences from Arabidopsis and rice, respectively (Fig. 1). At deep nodes, the phylogenetic relationship was unclear and the bootstrap values were low as a result of relatively large number of sequences. To verify the reliability of our phylogenetic tree, we also build the phylogenetic trees of TCP transcription family with Minimal Evolution methods (Fig. S1). The tree topologies were robust within different tree-building methods, except at the deep nodes. Considering these results, the NJ tree was employed for further study.
Figure 1.
Phylogenetic relationships of TCP transcription factors from cassava, Arabidopsis and rice. A total of 36 MeTCPs from cassava, 24 AtTCPs from Arabidopsis and 22 OsTCPs from rice were used to construct the Neighbor-Joining tree by MEGA 6.0 with 1000 bootstrap based on the full length sequences of TCPs. The eight subgroups are indicated with different colors.
Based on the bootstrap value of clade and topology of the tree, the MeTCP proteins could be distributed into 8 distinct groups, designated as Group A to Group H. In general, TCPs from cassava have closer relationships with the TCPs from dicot plant Arabidopsis than that from monocot plant rice, which is accord with the current understanding of plant evolutionary history. Additionally, the TCP genes showed an interspersed distribution in most clades, which is consistent with those found in previous analyses of TCP in G. raimondii and Citrullus lanatus 35, 43, indicating that the TCP family expanded before the divergence of the lineages. However, the TCP genes were not evenly distributed in some clades, such as the largest clade Group H has the maximum 7 members, whereas Group C contains only 3 MeTCP genes from cassava, suggesting the existence of a diversified MeTCP family in cassava with diverse functions (Fig. 1). Remarkably, many Arabidopsis TCP genes had more than three counterparts in cassava, such as MeTCP11, MeTCP15, MeTCP18 and MeTCP20, indicating that MeTCP genes duplicated after the divergence of cassava and Arabidopsis. It also suggests that higher number of genes in cassava as compared to Arabidopsis is the result of more gene duplication events in cassava or higher frequency of retaining copies after duplication. Group C contained three cassava TCPs, two Arabidopsis members but there were no TCP from rice, implying this group was either acquired after the divergence of monocots and dicots or lost in rice. Remarkably, 5 of the group H members (AtTCP2, AtTCP3, AtTCP4, AtTCP10, and AtTCP24) are post-transcriptionally targeted by miRNA319 in Arabidopsis. The closest homologs of these Arabidopsis genes in cassava are these five genes: MeTCP2a, MeTCP2b, MeTCP3a, MeTCP3b and MeTCP4, all containing putative target site of miR31947, 48. This suggests that regulation of leaf development by a redundant set of miRNA-regulated homologous TCP genes occurs in cassava.
Gene structure and conserved motifs of cassava TCPs
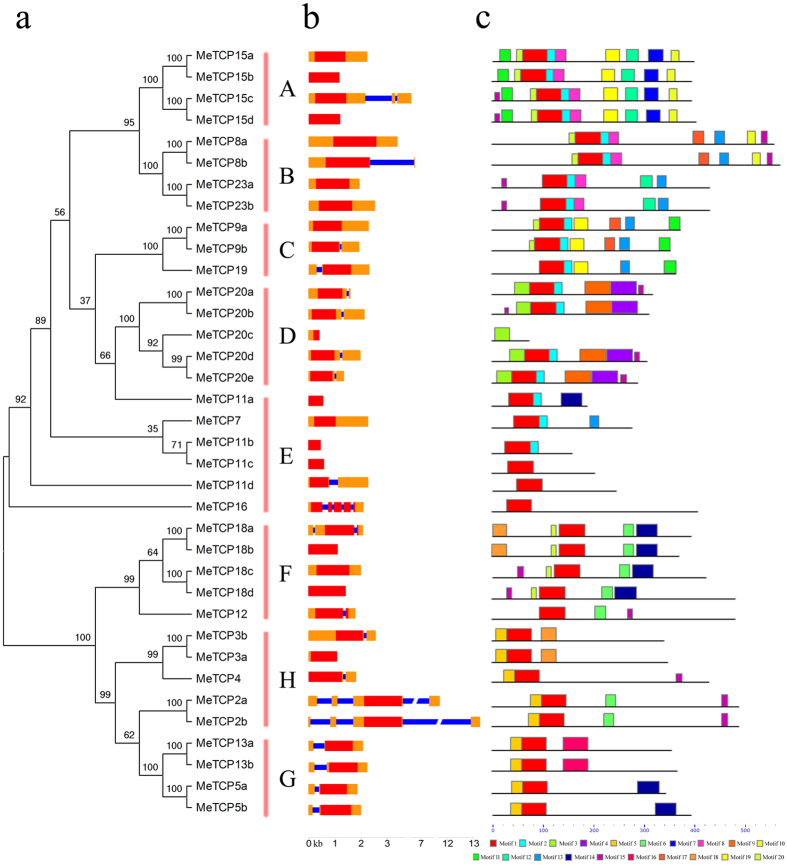
To further examine the structural features of cassava TCP genes, we investigated the exon/intron structures of individual MeTCP genes by alignment of cDNA sequences and corresponding genomic DNA sequences. Additionally, we also built an unrooted phylogenetic tree with MeTCP protein sequences (Fig. 2a), to determine whether the gene structure of MeTCPs is consistent with the phylogenetic subfamily. As illustrated in Fig. 2b, the number of introns of MeTCP genes varied from 0 to 4. For instance, 32 out of 36 MeTCP genes had no intron, while the other MeTCP genes possess 1–3 introns, with the exception of MeTCP16 containing four introns. As expected, most of MeTCP genes in the same subfamily showed similar exon-intron distribution patterns in terms of exon length and intron number, which supports their close evolutionary relationship and the classification of subgroup. However, MeTCP genes in group H showed great variability in intron number and exon length.
Figure 2.
The gene structure and conserved protein motifs of MeTCP genes according to the phylogenetic relationship. (a) The unrooted phylogenetic tree of all TCP genes in cassava was constructed using Neighbor-Joining method and the bootstrap test was performed with 1,000 iterations. (b) The gene structure with exon/intron organization of TCP genes of cassava. The orange boxes represent 5′-UTR or 3′-UTR, red boxes represent exons and blue lines indicate introns. (c) The conserved protein motifs in the TCP family were identified using MEME program. Each motif is indicated with a specific color.
To obtain more insights into the diversity of motif compositions among MeTCPs, conserved motifs were predicted by using MEME program49, and annotated by ScanProsite program50. A total of 20 conserved motifs in the MeTCP proteins, designated as motif 1 to motif 20, were captured by MEME (Fig. 2c, Fig. S2). The results showed that the only motif that hit for the database was the conserved TCP domain (motif 1). TCP domain was found in all MeTCPs, except MeTCP20c, which contained a truncated TCP domain. In general, MeTCP proteins clustered in same subgroup share similar motif composition, while high divergence was observed among different subgroups. This observation indicated that the MeTCP members within the same subgroup may have redundant functions and that some motifs may contribute to the specific function of that subfamily, which is in agreement with the previous report35, 41. According to 36 MeTCP sequence features within the TCP domain, we determined that MeTCPs from Group A, B, C, D and E belong to class I subfamily while the other MeTCPs belong to class II subfamily, as for all species so far. As reported earlier, the group members belonged to class I subfamily have extended homology C-terminal from the TCP domain, while the class II subfamily has an extended basic region, and all groups have internally conserved, but distinct loop region sequences25, 51. The motif analysis also showed that sequence conservation outside the TCP domain was low and sequence length on both sides of the TCP domain varied greatly, resulting in proteins ranging from 73 (MeTCP20c) to 563 (MeTCP8b) amino acids. For example, MeTCP20c, the smallest predicted protein, is probably truncated by a frame shift mutation that cause premature termination, since sequence homology with Arabidopsis TCP20 extends well beyond the stop codon. This result is similar to SlTCP27, which encode the smallest TCP protein with 113 amino acids in tomato36. Although MeTCP20c lacks the conserved C-terminal part of the TCP domain, which may alter its DNA binding ability, experimental evidences are required to establish the precise role of truncated TCP domain in the regulation of MeTCP20c activity.
Expression analysis of MeTCP genes in different tissues
To investigate the potential functions as well as to identify probable functional redundancy through similar expression patterns for the cassava TCP genes, the detection of their expression were carried out in different tissues including root, leaf, stem and shoot apex using qRT-PCR. As shown in Fig. 3, it is apparent that the expression levels in different tissues vary widely between the cassava TCP genes, as well as between different tissues for individual TCP genes, indicating functional specialization among TCP gene family members in cassava plant development. Of them, some genes were exclusively highly expressed in a specific tissue. For example, MeTCP20e and MeTCP11d genes exhibited higher transcriptional abundance in roots as compared to other organs; MeTCP2a, MeTCP3a, MeTCP5b, MeTCP8a, MeTCP13a and MeTCP20d showed specifically high expression in leaves, implying their specific roles in the corresponding tissues. Opposite to the tissue-specific expression pattern of MeTCP genes, many genes were more widely and less specifically expressed, such as MeTCP9b, MeTCP13b, MeTCP20a, MeTCP20b, and MeTCP23a genes, implying that these genes may play regulatory roles at multiple development stages. However, further studies are still needed to unravel the divergent roles of MeTCP genes.
Figure 3.
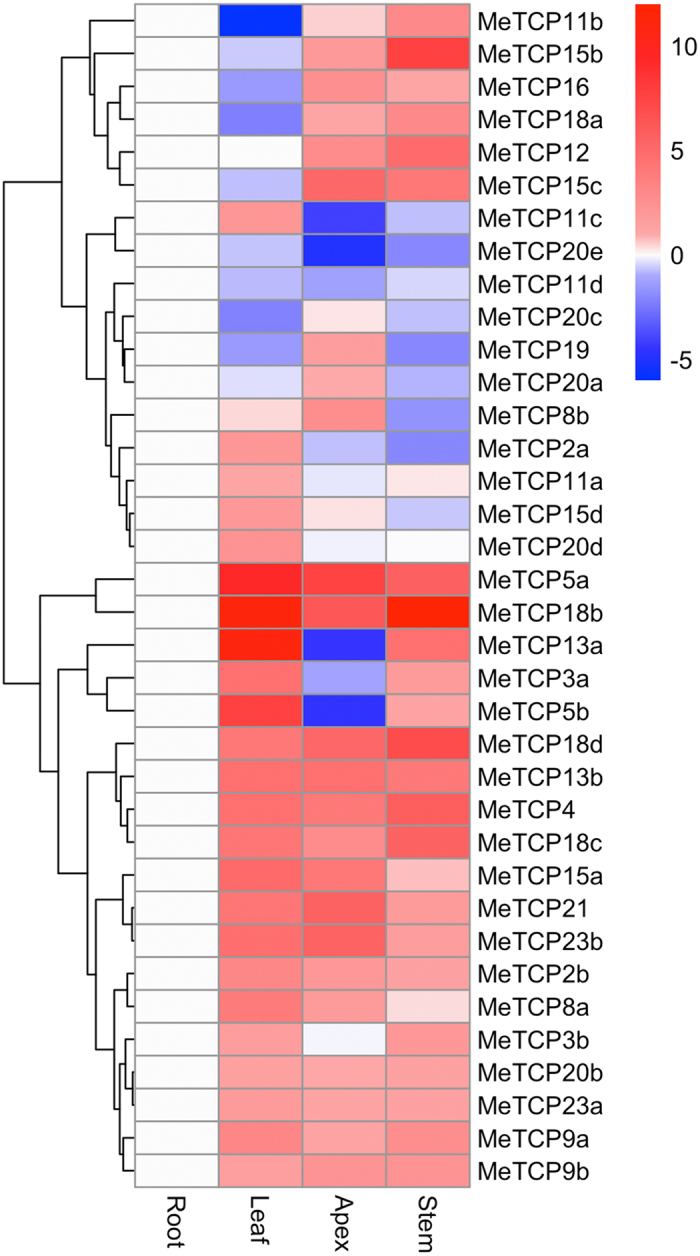
Heatmap representation for expression profiles of 36 MeTCP genes across different tissues. The expression levels of MeTCP genes were obtain through quantitative real-time PCR. MeACTIN was used as the reference gene.
To address the conservation and specificity of TCP expression pattern beyond species, we compared the expression level of homologous TCP gene pairs in these four tissues between Arabidopsis and cassava. We found a subset of TCP genes were positively correlated with pearson correlation coefficient (PCC) higher than 0.3 between Arabidopsis and cassava (Fig. S3–4, such as MeTCP2a/b and AtTCP2, MeTCP5a/b and AtTCP5, MeTCP8a/b and AtTCP8, MeTCP13a/b and AtTCP13, MeTCP15b/c and AtTCP15, MeTCP19 and AtTCP19 et al., indicating functional conserved expressional pattern of these genes. In contrast, some TCP genes show no correlation or negative correlation between Arabidopsis and cassava, such as MeTCP9a/b and AtTCP9, MeTCP11a/b/c/d and AtTCP11, suggesting their functions have been diversely changed in different species.
Expression patterns of MeTCP genes in response to hormone treatments
Multiple studies have been reported that TCP proteins regulate plant development and environmental stress adaption by mediating hormone biogenesis and response17–19, 21. To study the total effect of plant hormones on MeTCP genes, the expression levels of 36 MeTCP genes were detected in response to abscisic acid (ABA), gibberellin (GA3), indole acetic acid (IAA), jasmonate acid (JA), zeatin (ZT) and 6-benzylaminopurine (6-BA) hormone treatments by quantitative RT-PCR (Fig. 4, Supplemental Table S1). In general, hormone treatments resulted in a wide variety of MeTCP gene expression profiles. In JA treatment, 15 and 7 MeTCP genes were obviously induced and inhibited, respectively. Of them, the most up-regulated gene was MeTCP20e, and the most down-regulated gene was MeTCP3a. Similarly, 6-BA and ZT treatment led to 12 and 13 MeTCP genes were obviously induced, 11 and 5 MeTCP genes were inhibited, respectively. In GA treatment, most of MeTCPs were significantly induced and only two MeTCPs were inhibited. MeTCP7 and MeTCP23b were found to be most up-regulated. As for IAA treatment, 10 and 5 MeTCP genes showed dramatic increase and decrease, respectively. MeTCP7 and MeTCP16 went through the largest increase and decrease, respectively. ABA plays a crucial role in the adaptive response of plants to abiotic stresses52. We found 22 members showed strong sensitivity toward ABA, indicating that these genes may be regulated by ABA signal pathway. Among them, 19 genes had relative high levels of transcript abundance after ABA treatment. Notably, most of MeTCP genes responded to at least one treatment; Particularly, MeTCP11b, MeTCP12 and MeTCP21 responded to all hormone treatments, indicating these genes might play pivotal roles in the cross-talk of hormones, which would be candidates for further research in the field. However, we also found MeTCP11d, MeTCP20a and MeTCP23a were not able to respond to any treatments. Taken together, these results suggest the complicated regulatory mechanism of MeTCP genes in response to hormone treatments in cassava.
Figure 4.
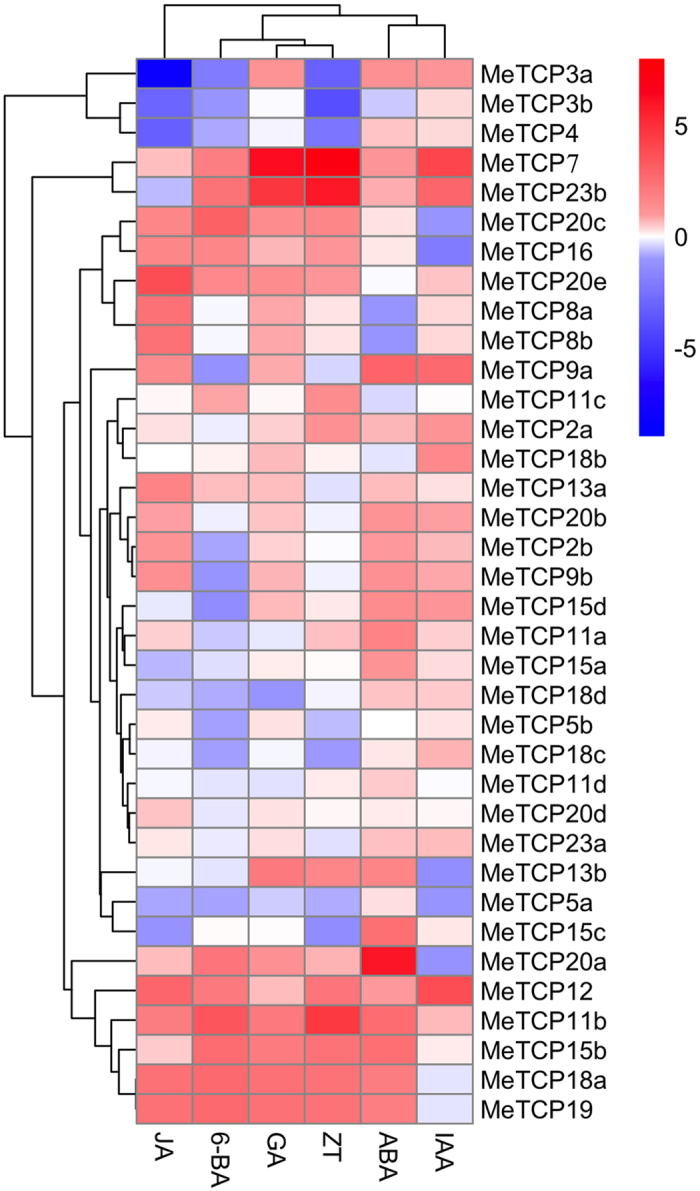
Heatmap representation for expression patterns of MeTCP genes under various hormone treatments. The expression profile data of MeTCP genes under JA, 6-BA, GA, ZT, ABA and IAA were obtain through quantitative real-time PCR. MeACTIN was used as the reference gene.
Expression profiles of MeTCP genes in response to cold and/or drought stress
Plants are frequently challenged by abiotic stressors such as cold and drought. Recent studies have suggested that TCP proteins are widely involved in signaling and response to environmental stimuli18, 22. However, information on the involvement of TCP proteins in stress responses in cassava is limited. To investigate the potential roles of MeTCP genes in response to abiotic stresses, cassava seedlings of TMS60444 genotypes were subjected to cold (4 °C) and PEG (20% PEG 6000)-induced drought stress and then the leaves tissues were sampled to extract RNA for subsequent RNA-seq analysis. According to the transcriptome data, 18 (50%) and 24 (66.7%) MeTCP genes showed significantly change (fold change >2) under cold and drought treatment, respectively. Among them, 7 (38.9%) and 11 (61.1%) MeTCP genes were up- and down-regulated by cold, respectively; 10 (41.7%) and 14 (58.3%) MeTCP genes were up- and down-regulated by drought, respectively (Fig. 5, Supplemental Table S2). These results also showed that the number of MeTCP genes down-regulated by cold and drought was greater than that were up-regulated, suggesting the comprehensive response of MeTCP genes to cold and/or drought at transcriptional levels. There were 23 MeTCP genes in total differential expressed under both cold and drought treatments, which were categorized into 4 different classes: concordant response to cold and drought, discordant response, cold-specific and drought-specific. Among concordant response, MeTCP20c/20e/11a and MeTCP18b/11c/12 were co-induced and co-repressed by two kinds of stresses, respectively. However, in discordant response class, three genes (MeTCP15d, MeTCP15b and MeTCP16) showed increased expression pattern under cold treatment, whereas down-regulated after drought treatment. By contrast, the expression levels of four genes (MeTCP2b, MeTCP19, MeTCP13a and MeTCP13b) were induced by drought treatment, but were repressed or unaltered after cold treatment. Meanwhile, MeTCP8a/5a and MeTCP20a/20b/9b/18d/3b were specifically response to cold and drought stress, respectively. Generally, the expression levels of MeTCP genes in response to cold and drought stresses were dramatically changed, implying their putative roles in stress tolerance.
Figure 5.

Expression profiles of MeTCP genes in leaves and shoot apices after cold and drought treatment. The transcript data generated from three replicates of RNA-seq data. The relative expression values were log2 transformed. The bar represents relative expression values.
Expression analysis of MeTCP genes under various abiotic stress treatments
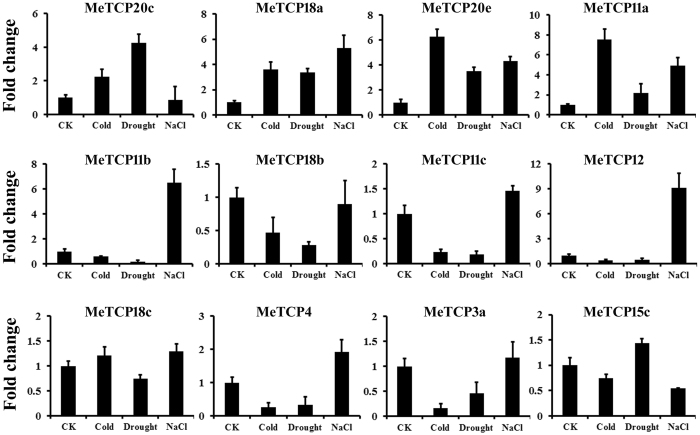
To further assess the response of MeTCP genes to various abiotic stresses and related signaling pathway at transcriptional levels, 12 MeTCP genes (MeTCP20c, MeTCP18a, MeTCP20e, MeTCP11a, MeTCP11b, MeTCP18b, MeTCP11c, MeTCP12, MeTCP18c, MeTCP4, MeTCP3a and MeTCP15c) induced or repressed by cold and drought stresses based on RNA-seq data were chosen for further examination of their expression patterns after cold, drought and salt treatments (Fig. 6). We found most of the analyzed genes exhibited differential expression in response to at least one stress treatment, implying their putative roles in these stresses tolerance. Overall, in the cold, drought and salt conditions, expression of MeTCP20c, MeTCP18a, MeTCP20e and MeTCP11a genes were significantly up-regulated, and the largest expressional change of these genes were usually observed when responding to cold and/or drought treatments. This observation is well consistent with the RNA-seq data. 6 genes (MeTCP11b, MeTCP18b, MeTCP11c, MeTCP12, MeTCP4 and MeTCP3a) were down-regulated following cold and drought treatments, among which, MeTCP12 underwent the greatest change of mRNA expression levels. Interestingly, the expression levels of all these MeTCP genes showed obviously increase under salt treatment, suggesting they may play different roles in response to the three stresses. In addition, not too many changes were observed in MeTCP18c and MeTCP15c genes when any of three stresses were carried out. These data show the potential of some MeTCP genes for enhancing adversity resistant capacity in cassava.
Figure 6.
Confirmation of the expression patterns of cold- and drought-responsive MeTCPs using qRT-PCR. The expression patterns of MeTCP under cold, drought and salt stress. The values shown are the means ± standard deviation of three replicates. MeACTIN was used as the reference gene.
Analysis of the regulatory cis-elements in the promoter of cold- and drought-responsive MeTCPs
The altered expression of 12 MeTCPs that were co-induced/repressed by cold and drought stresses indicates that they may be regulated by key stress regulatory genes. In order to elucidate the mechanism of transcriptional regulation of these genes, analysis of their promoter region was performed for the cis-elements. A 1.5 kb sequence upstream to the open reading frame of MeTCP20c, MeTCP18a, MeTCP20e, MeTCP11a, MeTCP11b, MeTCP18b, MeTCP11c, MeTCP12, MeTCP18c, MeTCP4, MeTCP3a and MeTCP15c was identified and subjected to MEME analysis. A number of common cis-acting elements were identified in the proximal promoters (Fig. 7, Fig. S5). All the identified promoters had motif 1, and most of them also contained motif 3 and 10, except for MeTCP20e, MeTCP11b, MeTCP11c, MeTCP12 and MeTCP18c, suggesting that these commonly present cis-acting elements may be involved in stress response. Notably, motifs such as 9, 13, 14 and 15 are more present in the promoters of MeTCP which are down-regulated by both cold and drought stresses, suggesting that these motifs may play key roles in the regulation of MeTCP members.
Figure 7.
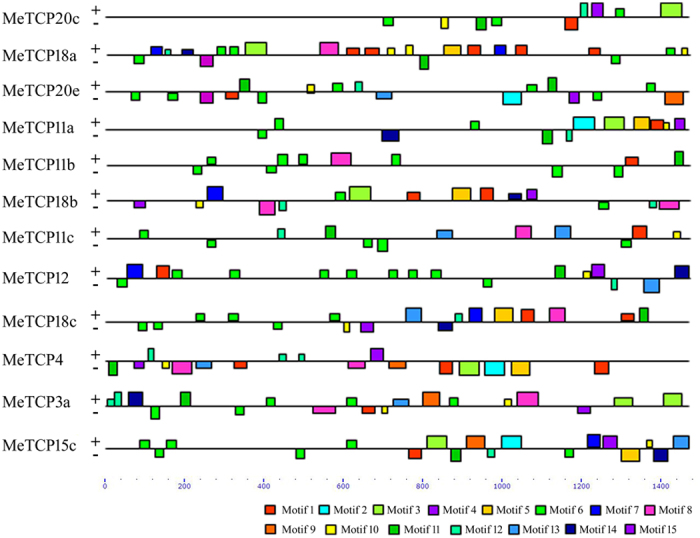
The conserved DNA sequence motifs analysis of cold- and drought-responsive MeTCP promoters. The conserved motifs in the TCP promoters were identified using MEME program. Each motif is indicated with a specific color.
Discussion
Adverse environmental conditions, such as cold and drought stress, impose severe effects on the plant growth, development and limit crop productivity and yield53. TCP transcription factors are a class of plant-specific transcription that play very important roles during plant development and abiotic stress responses22. To our knowledge, although a range of TCP family members have been described in various species35, 36, 40–45, 54, no studies have been performed on TCP genes in cassava. Additionally, the mechanisms of cassava responds to abiotic stress are poorly understood. This background knowledge prompted us to identify the full complement and expression profile of this important gene family during development and under abiotic stresses in cassava.
In the present study, a comprehensive set of 36 non-redundant TCP-encoding genes were identified and characterized from the current version of the cassava genome. Previous studies have identified 24 TCP genes in Arabidopsis 33, 22 in rice34, 38 in cotton35, 30 in tomato36, and 27 in watermelon43. The amplification of TCP gene family members in cassava can be explained by its larger genome size (~760 Mb) compared to that of Arabidopsis (~125 Mb) and gene duplication events in this family. Evolutionary analysis indicated that the cassava TCPs could be clustered into 8 subgroups, which is minimal different with previous evolutionary classification of TCPs in cotton and watermelon35, 43 (Fig. 1). The phylogenetic tree showed obvious differences in number of MeTCPs and ratio of MeTCPs/AtTCPs among subgroups. The change in the ratio of MeTCPs/AtTCPs suggested that the MeTCP family had undergone lineage-specific expansion and functional divergence during the course of evolution. Phylogenetic tree also showed that group C contained three cassava members and two Arabidopsis members, but there were no TCP from rice. On contrast, TCPs in subgroup A expanded in monocots but not in Arabidopsis, indicating that TCP genes of these subgroups expanded in a species-specific manner from common ancestral genes that were present prior to the diversification of the monocot and dicot lineages. The classification of TCP protein was further supported by gene structure analysis and conserved protein motif analysis. Gene structure analysis showed that the majority of MeTCP genes within the same subgroup exhibited very similar gene structure in terms of exon length and intron number. Furthermore, conserved protein motif analysis indicated that all the MeTCP proteins contained typical TCP domain, except MeTCP20c, which contains a truncated TCP domain. Similar to the gene structure, most subgroups of MeTCPs also exhibited conserved motif composition, with several motifs observed in some MeTCP subgroups, such as motif 4 and 17 for subgroup D, motif 7 for subgroup A (Fig. 2c). These unique motifs may contribute to the specific function of these subgroup members. In general, the majority of MeTCP genes in the same subfamilies are evolutionarily conserved, which supports their close evolutionary relationship and the classification of subgroups.
The expression pattern analysis of MeTCP genes helps us to assess their possible functions and provide a solid foundation for future functional studies. Generally, similar to the previous study, MeTCP genes exhibited greatly differential expression patterns across a variety of tissues, not only among subgroups but members within the same subgroups (MeTCP5a and MeTCP5b, MeTCP13a and MeTCP13b), suggesting that these MeTCP genes may function diversely in various tissues (Fig. 3). On the contrary, some MeTCP genes with extremely high sequence identity (MeTCP2a and MeTCP2b, MeTCP18c and MeTCP18d) showed conserved expression patterns (Fig. 3), implying they may play a redundant role in regulating plant growth.
To date, the role of plant hormones in regulating plant growth, development, and abiotic stress responses by modulating gene expression is well established52, 55. To our knowledge, although the relationship between TCP proteins and hormones has been widely known, such as cytokinins, JA and GA, the dynamic and spatially expression patterns of MeTCP genes response to various hormones was still obscure. Our current results revealed that the majority of MeTCP genes detected here displayed distinct changes under different hormone treatments. To expect, most of MeTCP were up-regulated or down-regulated by cytokinins (ZT and 6-BA). MeTCP15b and MeTCP23b, orthologs of AtTCP15 and AtTCP23, respectively, had high expression levels under ZT and 6-BA treatments. In Arabidopsis, cytokinin treatments induce TCP15 transcription and promote TCP15 (and TCP14) protein activation by post-translational modification, which in turn promote cytokinin responses56. MeTCP4, MeTCP20b, and MeTCP20e, orthologs of AtTCP4 and AtTCP20, respectively, had altered expression patterns under JA. In young leaves of Arabidopsis, AtTCP20 repressed the transcription of LIPOXYGENASE2 (AtLOX2) gene, which is involved in JA synthesis and promotes leaf senescence, while this negative control is antagonized by AtTCP4 as the leaf matures19. It is noteworthy that 22 MeTCPs were response to ABA, indicating these genes might function as key mediators of stress responses through ABA signaling pathways. Taken together, these results suggested that MeTCPs play potential regulatory roles by modulating phytohormone signaling in plant development or in the responses to stresses. Therefore, it will be particular important to further investigate the potential function of MeTCP genes in hormone signaling in the future.
Previous reports have shown that plant TCP genes are involved in plant growth and development, as well as abiotic stress responses under normal and stressed growth conditions22. In cassava, our data showed that over half of the MeTCP genes were significantly upregulated under cold and drought condition. Among them, 4 genes were up-regulated, and 8 genes were down-regulated by both cold and drought stress. Moreover, combined analysis of expression correlation and promoter content has revealed that most of these MeTCP genes exhibited differential expression in response to more than one stress treatments, suggesting the wide involvement of MeTCP genes in environmental adaptation. Previous reports have shown that knockdown of miR319-dependent TCPs (by constitutive miR319 overexpression) increases drought and salinity stress tolerance in bentgrass20. Our data showed that MeTCP3a and MeTCP4, targets of miRNA319, had altered expression patterns under cold, drought and salt stress, suggesting that these genes might play important roles under abiotic stress conditions in cassava. These data indicated that MeTCP might function in resistance to abiotic stresses in cassava.
In conclusion, we identified 36 TCP transcription factor genes from cassava. Phylogenetic analysis of cassava, Arabidopsis, and rice indicated that these MeTCP genes could be divided into 8 groups, which is supported by further conserved protein motif, and gene structure analyses. Although nearly all the MeTCP genes were expressed in the examined tissues, some genes were up-regulated in one or several specific organs. mRNA accumulation was altered by a variety of hormone treatments (ABA, IAA, GA, JA, ZT and 6-BA), environmental conditions (drought, high salinity, and low temperature). These results suggested that MeTCP family proteins play critical roles in maintaining cassava normal growth under normal or stress conditions through complicated mechanisms. Thus, additional studies on the detailed functions of each gene are warranted in cassava.
Materials and Methods
Identification and bioinformatics analysis of candidate genes
To identify potential members of the cassava TCP protein family, The Arabidopsis TCP protein sequences were used as seed queries in BLASTp searches against the cassava database (Phytozome: http://www.phytozome.net/cassava.php)57. The TCP domain (PF03634, Pfam; http://pfam.sanger.ac.uk/)46 was also employed as query to perform a blast search against the same genome database. The identified MeTCP proteins were renamed as MeTCP2 to MeTCP23 according to the Arabidopsis TCP proteins with highest sequence similarity. Information on MeTCP genes, including exons and introns number, open reading frame (ORF) and amino acid (AA) lengths, was obtained from Phytozome database. The molecular weight, theoretical isoelectric point (PI) and grand average of hydropathy (GRAVY) of the MeTCP proteins were investigated using ExPASy online tools (http://web.expasy.org/protparam/).
Analysis of phylogenetic relationships and gene structure
Multiple sequence alignments were applied to confirm the conserved domains of predicted MeTCP proteins. The Clustal × 2.058 was employed to align the full-length MeTCP proteins from cassava, Arabidopsis and rice. Then, the bootstrap neighbor-joining evolutionary tree was created by MEGA 6.0 software59 with 1000 bootstrap replicates based on the sequence alignments. The exon-intron organization of MeTCP genes was determined by comparing the coding DNA sequence (CDS) with its corresponding genomic sequences using the Gene Structure Display Server (GSDS) software (http://gsds.cbi.pku.edu.cn/)60.
Identification of conserved motif of MeTCP proteins and promoters
By using the Multiple Expectation maximization for Motif Elicitation (MEME) program (http://meme.nbcr.net/meme/cgi-bin/meme.cgi), the conserved motifs in full-length cassava MeTCP protein sequences were identified with the following parameters: maximum number of motifs was 20 and the optimum width of motifs was set between 10 and 5049. The identified protein motifs were further annotated with ScanProsite50. MeTCP promoter sequences in cassava were submitted to online MEME program for identification of conserved motifs. The optimized MEME parameters were as follows: any number of repetitions and maximum number of motifs-15.
Plant materials and hormone/stress treatment
Cassava (Manihot esculenta) cultivar (TMS60444) was used in the present study. Segments cut from cassava stems were inserted into MS plates in a greenhouse at 26 ± 2 °C, with a photoperiod of 16 h light and 8 h dark. All hormone and environmental treatments were conducted when uniform-sized seedlings developed two fully opened trifoliate leaves (approximately two weeks after sowing). For hormone treatment, 14-day-old cassava seedlings were soaked in liquid MS medium with 100 μM indole acetic acid(IAA), 100 μM gibberellin (GA3), 100 μM Methyl jasmonate (MeJA), 100 μM abscisic acid (ABA),100 μM zeatin (ZT) and 100 μM, 6-benzylaminopurine (6-BA) for 3 h, respectively, and then the young leaves and shoot apex from at least ten separate seedlings/plants were harvested. Seedlings soaked in liquid MS medium without any hormone were used as control. For cold treatment, seedlings were placed at 4 °C for 24 h, and then the young leaves and shoot apex were collected for RNA isolation. For drought and salt stress treatment, cassava seedlings were treated with 20% PEG6000 and 100 mM NaCl, and harvested at 6 h after treatment, respectively. In all cases, parallel and untreated plants at the same stage were used as controls. All samples harvested were flash-frozen in liquid nitrogen, and stored at −80 °C until RNA isolation.
RNA isolation and expression analysis
Total RNA was extracted from 0.1 g of tissue by using Plant RNA kit (OMEGA), following the manufacturer’s instructions. Reverse transcription reactions were performed using 5 μg of RNA with PrimeScript RT reagent kit with gDNA Eraser (TIANGEN, Beijing, China). Quantitative reverse transcription PCR (qRT-PCR) was performed as described elsewhere61, the PCR conditions were as follows: pre-incubation at 94 °C for 5 min, followed by 40 cycles at 94 °C for 10 s, 60 °C for 10 s, 72 °C for 30 s. After amplication was complete, a melting curve was obtained by holding at 95 °C for 5 s and then at 65 °C for 15 s, followed by heating slowly at 0.1 °C/s to 95 °C. Real-time PCR was performed with a Bio-Rad real-time thermal cycling system using SYBR® Premix Ex Taq™ II (TaKaRa, Japan) to assess gene expression levels. The relative expression levels of each gene were calculated by the 2-ΔΔCt method. The cassava actin gene was used as internal control for normalization. The primers used are listed in Supplemental Table S3. MeTCP genes that were up- or down-regulated by at least two-fold were considered as differentially expressed. The experiments were performed in triplicate.
Expression correlation analysis of level between cassava and Arabidopsis
The expression levels of AtTCPs in different tissues were extracted from the microarray data62. To compare the microarray gcRMA value of AtTCPs with qPCR relative value of MeTCPs, all values were normalized to z-score, and pearson correlation coefficient were performed for each homologous gene pairs.
Transcriptome analysis
Cassava shoot apices and youngest leaves of TMS60444 under normal conditions, cold and drought treatments were used to isolated total RNA for transcriptome analysis. As previously described61, the total RNA isolation, whole transcriptome libraries preparation and deep sequencing were performed by the Annoroad Gene Technology Corporation (Beijing, PR China). A total of 140 gigabase in-depth sequencing of library was performed initially on a HiSeq. 2500 instrument that generated paired-end reads with 125 nucleotides. Data analysis was carried out by previously described61. The generated transcriptomic data has been submitted to Sequence Read Archive (SRA) in NCBI with the accession number SRP101302.
Electronic supplementary material
Acknowledgements
This research was supported by the Program of Hainan Association for Science and Technology Plans to Youth R & D Innovation (HAST201627), the National Natural Science Foundation of China (31561143012), Central Public-interest Scientific Institution Basal Research Fund for Chinese Academy of Tropical Agricultural Sciences (1630052017022) and the Hainan Province Innovative Research Team Foundation (2016CXTD013).
Author Contributions
S.X.L. and M.P. devised the study, N.L.; X.Y; S.X.L. and C.Y.Z. conducted the experiments and analyses, all authors contributed to data interpretation and writing of the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Ning Lei, Xiang Yu and Shuxia Li contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-09398-5
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Nakashima K, Yamaguchi-Shinozaki K, Shinozaki K. The transcriptional regulatory network in the drought response and its crosstalk in abiotic stress responses including drought, cold, and heat. Frontiers in plant science. 2014;5 doi: 10.3389/fpls.2014.00170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mahajan S, Tuteja N. Cold, salinity and drought stresses: an overview. Archives of biochemistry and biophysics. 2005;444:139–158. doi: 10.1016/j.abb.2005.10.018. [DOI] [PubMed] [Google Scholar]
- 3.Nakashima K, Ito Y, Yamaguchi-Shinozaki K. Transcriptional regulatory networks in response to abiotic stresses in Arabidopsis and grasses. Plant physiology. 2009;149:88–95. doi: 10.1104/pp.108.129791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Agarwal PK, Jha B. Transcription factors in plants and ABA dependent and independent abiotic stress signalling. Biol Plantarum. 2010;54:201–212. doi: 10.1007/s10535-010-0038-7. [DOI] [Google Scholar]
- 5.Schwechheimer C, Zourelidou M, Bevan MW. Plant Transcription Factor Studies. Annual review of plant physiology and plant molecular biology. 1998;49:127–150. doi: 10.1146/annurev.arplant.49.1.127. [DOI] [PubMed] [Google Scholar]
- 6.Gahlaut V, Jaiswal V, Kumar A, Gupta PK. Transcription factors involved in drought tolerance and their possible role in developing drought tolerant cultivars with emphasis on wheat (Triticum aestivum L.) Theor Appl Genet. 2016;129:2019–2042. doi: 10.1007/s00122-016-2794-z. [DOI] [PubMed] [Google Scholar]
- 7.Wang H, Wang H, Shao H, Tang X. Recent Advances in Utilizing Transcription Factors to Improve Plant Abiotic Stress Tolerance by Transgenic Technology. Frontiers in plant science. 2016;7 doi: 10.3389/fpls.2016.00067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martin-Trillo M, Cubas P. TCP genes: a family snapshot ten years later. Trends in plant science. 2010;15:31–39. doi: 10.1016/j.tplants.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 9.Cubas P, Lauter N, Doebley J, Coen E. The TCP domain: a motif found in proteins regulating plant growth and development. The Plant journal: for cell and molecular biology. 1999;18:215–222. doi: 10.1046/j.1365-313X.1999.00444.x. [DOI] [PubMed] [Google Scholar]
- 10.Kosugi S, Ohashi Y. DNA binding and dimerization specificity and potential targets for the TCP protein family. The Plant journal: for cell and molecular biology. 2002;30:337–348. doi: 10.1046/j.1365-313X.2002.01294.x. [DOI] [PubMed] [Google Scholar]
- 11.Navaud O, Dabos P, Carnus E, Tremousaygue D, Herve C. TCP transcription factors predate the emergence of land plants. Journal of molecular evolution. 2007;65:23–33. doi: 10.1007/s00239-006-0174-z. [DOI] [PubMed] [Google Scholar]
- 12.Manassero NG, Viola IL, Welchen E, Gonzalez DH. TCP transcription factors: architectures of plant form. Biomolecular concepts. 2013;4:111–127. doi: 10.1515/bmc-2012-0051. [DOI] [PubMed] [Google Scholar]
- 13.Aguilar-Martinez JA, Poza-Carrion C, Cubas P. Arabidopsis BRANCHED1 acts as an integrator of branching signals within axillary buds. The Plant cell. 2007;19:458–472. doi: 10.1105/tpc.106.048934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Palatnik JF, et al. Control of leaf morphogenesis by microRNAs. Nature. 2003;425:257–263. doi: 10.1038/nature01958. [DOI] [PubMed] [Google Scholar]
- 15.Kieffer M, Master V, Waites R, Davies B. TCP14 and TCP15 affect internode length and leaf shape in Arabidopsis. The Plant journal: for cell and molecular biology. 2011;68:147–158. doi: 10.1111/j.1365-313X.2011.04674.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mao Y, et al. MicroRNA319a-targeted Brassica rapa ssp. pekinensis TCP genes modulate head shape in chinese cabbage by differential cell division arrest in leaf regions. Plant physiology. 2014;164:710–720. doi: 10.1104/pp.113.228007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Resentini F, et al. TCP14 and TCP15 Mediate the Promotion of Seed Germination by Gibberellins in Arabidopsis thaliana. Molecular plant. 2015;8:482–485. doi: 10.1016/j.molp.2014.11.018. [DOI] [PubMed] [Google Scholar]
- 18.Nicolas M, Cubas P. TCP factors: new kids on the signaling block. Current opinion in plant biology. 2016;33:33–41. doi: 10.1016/j.pbi.2016.05.006. [DOI] [PubMed] [Google Scholar]
- 19.Danisman S, et al. Arabidopsis class I and class II TCP transcription factors regulate jasmonic acid metabolism and leaf development antagonistically. Plant physiology. 2012;159:1511–1523. doi: 10.1104/pp.112.200303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou M, et al. Constitutive expression of a miR319 gene alters plant development and enhances salt and drought tolerance in transgenic creeping bentgrass. Plant physiology. 2013;161:1375–1391. doi: 10.1104/pp.112.208702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mukhopadhyay P, Tyagi AK. OsTCP19 influences developmental and abiotic stress signaling by modulating ABI4-mediated pathways. Scientific reports. 2015;5 doi: 10.1038/srep09998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Danisman, S. TCP Transcription Factors at the Interface between Environmental Challenges and the Plant’s Growth Responses. Frontiers in plant science7, doi:Artn 1930 10.3389/Fpls.2016.01930 (2016). [DOI] [PMC free article] [PubMed]
- 23.Tatematsu K, Nakabayashi K, Kamiya Y, Nambara E. Transcription factor AtTCP14 regulates embryonic growth potential during seed germination in Arabidopsis thaliana. The Plant journal: for cell and molecular biology. 2008;53:42–52. doi: 10.1111/j.1365-313X.2007.03308.x. [DOI] [PubMed] [Google Scholar]
- 24.Uberti-Manassero NG, Lucero LE, Viola IL, Vegetti AC, Gonzalez DH. The class I protein AtTCP15 modulates plant development through a pathway that overlaps with the one affected by CIN-like TCP proteins. Journal of experimental botany. 2012;63:809–823. doi: 10.1093/jxb/err305. [DOI] [PubMed] [Google Scholar]
- 25.Viola IL, Uberti Manassero NG, Ripoll R, Gonzalez DH. The Arabidopsis class I TCP transcription factor AtTCP11 is a developmental regulator with distinct DNA-binding properties due to the presence of a threonine residue at position 15 of the TCP domain. The Biochemical journal. 2011;435:143–155. doi: 10.1042/BJ20101019. [DOI] [PubMed] [Google Scholar]
- 26.Schommer C, et al. Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS biology. 2008;6 doi: 10.1371/journal.pbio.0060230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang H, Mao Y, Yang J, He Y. TCP24 modulates secondary cell wall thickening and anther endothecium development. Frontiers in plant science. 2015;6 doi: 10.3389/fpls.2015.00436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.E1-Sharkawy, M. A. Cassava biology and physiology. Plant molecular biology56, 481-501 (2004). [DOI] [PubMed]
- 29.Zidenga T, Leyva-Guerrero E, Moon H, Siritunga D, Sayre R. Extending cassava root shelf life via reduction of reactive oxygen species production. Plant physiology. 2012;159:1396–1407. doi: 10.1104/pp.112.200345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perera, P. I., Ordonez, C. A., Dedicova, B. & Ortega, P. E. Reprogramming of cassava (Manihot esculenta) microspores towards sporophytic development. AoB PLANTS6, 10.1093/aobpla/plu022 (2014). [DOI] [PMC free article] [PubMed]
- 31.Hu W, et al. Genome-wide characterization and analysis of bZIP transcription factor gene family related to abiotic stress in cassava. Scientific reports. 2016;6 doi: 10.1038/srep22783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Okogbenin E, et al. Phenotypic approaches to drought in cassava: review. Frontiers in physiology. 2013;4 doi: 10.3389/fphys.2013.00093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Riechmann JL, et al. Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science. 2000;290:2105–2110. doi: 10.1126/science.290.5499.2105. [DOI] [PubMed] [Google Scholar]
- 34.X Y, Ma H, Wang J, Zhang DB. Genome-Wide Comparative Analysis and Expression Pattern of TCP Gene Families in Arabidopsis thaliana and Oryza sativa. Journal of Integrative Plant Biology. 2007;49:885–897. doi: 10.1111/j.1744-7909.2007.00509.x. [DOI] [Google Scholar]
- 35.Ma, J. et al. Genome-wide identification and expression analysis of TCP transcription factors in Gossypium raimondii. Scientific reports4, doi:Artn 6645 10.1038/Srep06645 (2014). [DOI] [PMC free article] [PubMed]
- 36.Parapunova V, et al. Identification, cloning and characterization of the tomato TCP transcription factor family. BMC plant biology. 2014;14 doi: 10.1186/1471-2229-14-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Horn S, Pabon-Mora N, Theuss VS, Busch A, Zachgo S. Analysis of the CYC/TB1 class of TCP transcription factors in basal angiosperms and magnoliids. The Plant journal: for cell and molecular biology. 2015;81:559–571. doi: 10.1111/tpj.12750. [DOI] [PubMed] [Google Scholar]
- 38.Chen, L. et al. Genome-wide analysis of TCP family in tobacco. Genetics and molecular research: GMR15, 10.4238/gmr.15027728 (2016). [DOI] [PubMed]
- 39.Francis A, et al. Comparative phylogenomic analysis provides insights into TCP gene functions in Sorghum. Scientific reports. 2016;6 doi: 10.1038/srep38488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin YF, et al. Genome-wide identification and characterization of TCP genes involved in ovule development of Phalaenopsis equestris. Journal of experimental botany. 2016;67:5051–5066. doi: 10.1093/jxb/erw273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ma J, et al. Comprehensive analysis of TCP transcription factors and their expression during cotton (Gossypium arboreum) fiber early development. Scientific reports. 2016;6 doi: 10.1038/srep21535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ma X, et al. Genome-wide Identification of TCP Family Transcription Factors from Populus euphratica and Their Involvement in Leaf Shape Regulation. Scientific reports. 2016;6 doi: 10.1038/srep32795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shi P, et al. Genome-wide identification and expression analysis of the ClTCP transcription factors in Citrullus lanatus. BMC plant biology. 2016;16 doi: 10.1186/s12870-016-0765-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wei W, et al. Identification and Transcript Analysis of the TCP Transcription Factors in the Diploid Woodland Strawberry Fragaria vesca. Frontiers in plant science. 2016;7 doi: 10.3389/fpls.2016.01937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Madrigal Y, Alzate JF, Pabon-Mora N. Evolution and Expression Patterns of TCP Genes in Asparagales. Frontiers in plant science. 2017;8 doi: 10.3389/fpls.2017.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Finn RD, et al. Pfam: the protein families database. Nucleic acids research. 2014;42:D222–230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zeng C, et al. Conservation and divergence of microRNAs and their functions in Euphorbiaceous plants. Nucleic acids research. 2010;38:981–995. doi: 10.1093/nar/gkp1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen X, et al. Potential functions of microRNAs in starch metabolism and development revealed by miRNA transcriptome profiling of cassava cultivars and their wild progenitor. BMC plant biology. 2015;15 doi: 10.1186/s12870-014-0355-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brown P, et al. MEME-LaB: motif analysis in clusters. Bioinformatics. 2013;29:1696–1697. doi: 10.1093/bioinformatics/btt248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.de Castro E, et al. ScanProsite: detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic acids research. 2006;34:W362–365. doi: 10.1093/nar/gkl124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aggarwal P, et al. Identification of specific DNA binding residues in the TCP family of transcription factors in Arabidopsis. The Plant cell. 2010;22:1174–1189. doi: 10.1105/tpc.109.066647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Verma V, Ravindran P, Kumar PP. Plant hormone-mediated regulation of stress responses. BMC plant biology. 2016;16 doi: 10.1186/s12870-016-0771-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huang GT, et al. Signal transduction during cold, salt, and drought stresses in plants. Molecular biology reports. 2012;39:969–987. doi: 10.1007/s11033-011-0823-1. [DOI] [PubMed] [Google Scholar]
- 54.De Paolo S, Gaudio L, Aceto S. Analysis of the TCP genes expressed in the inflorescence of the orchid Orchis italica. Scientific reports. 2015;5 doi: 10.1038/srep16265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Larrieu A, Vernoux T. Comparison of plant hormone signalling systems. Essays in biochemistry. 2015;58:165–181. doi: 10.1042/bse0580165. [DOI] [PubMed] [Google Scholar]
- 56.Steiner E, et al. The Arabidopsis O-linked N-acetylglucosamine transferase SPINDLY interacts with class I TCPs to facilitate cytokinin responses in leaves and flowers. The Plant cell. 2012;24:96–108. doi: 10.1105/tpc.111.093518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Prochnik S, et al. The Cassava Genome: Current Progress, Future Directions. Tropical plant biology. 2012;5:88–94. doi: 10.1007/s12042-011-9088-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Larkin MA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 59.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Molecular biology and evolution. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hu B, et al. GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics. 2015;31:1296–1297. doi: 10.1093/bioinformatics/btu817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li S, et al. Genome-wide identification and functional prediction of cold and/or drought-responsive lncRNAs in cassava. Scientific reports. 2017;7 doi: 10.1038/srep45981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schmid M, et al. A gene expression map of Arabidopsis thaliana development. Nature genetics. 2005;37:501–506. doi: 10.1038/ng1543. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.