Abstract
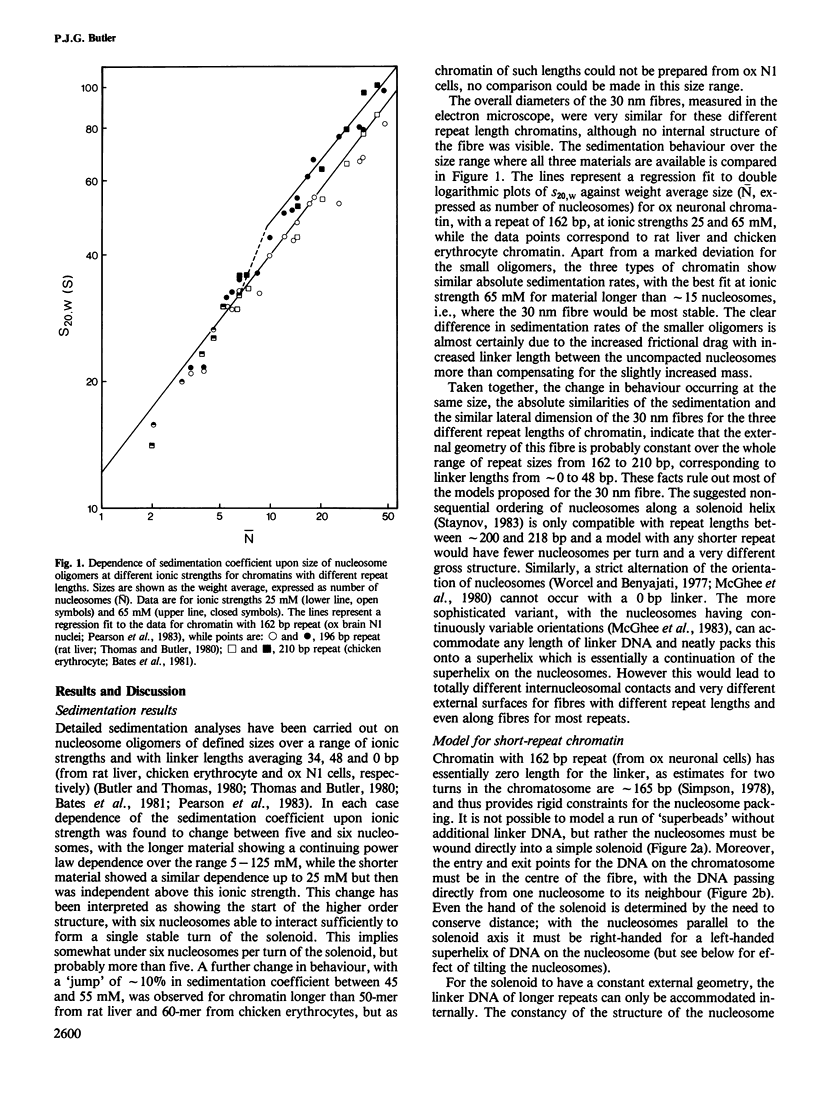
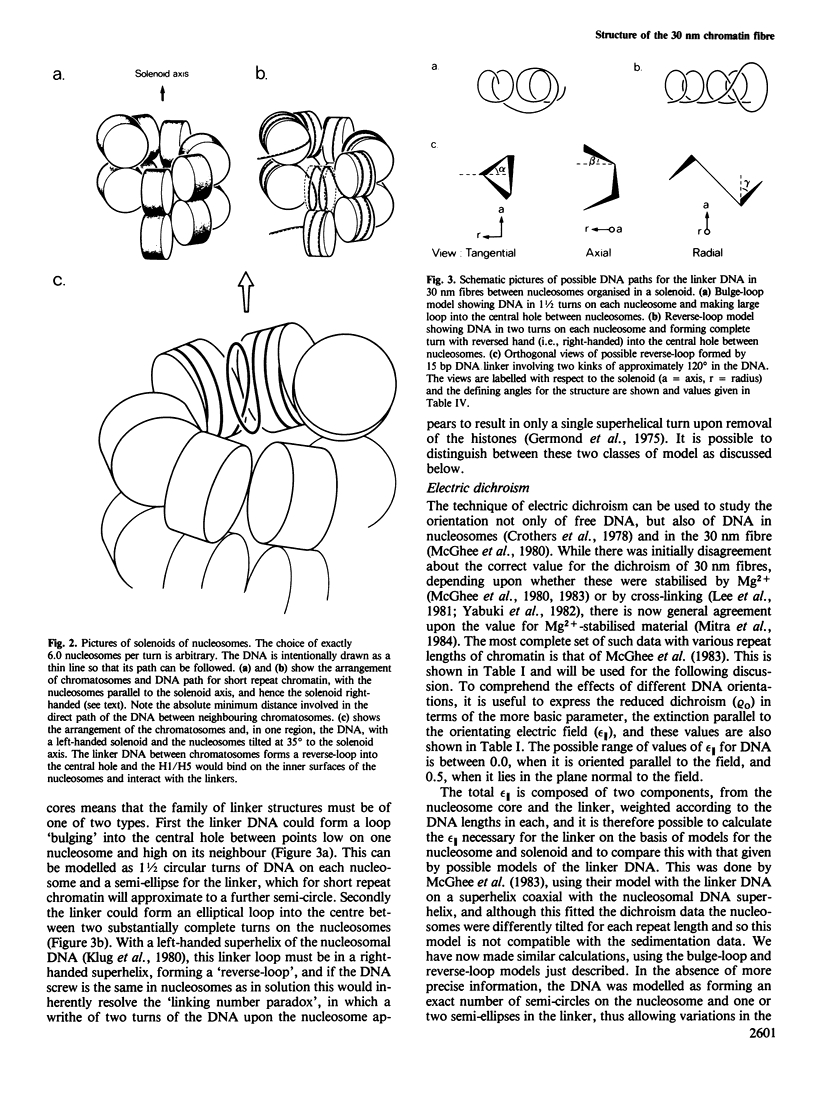
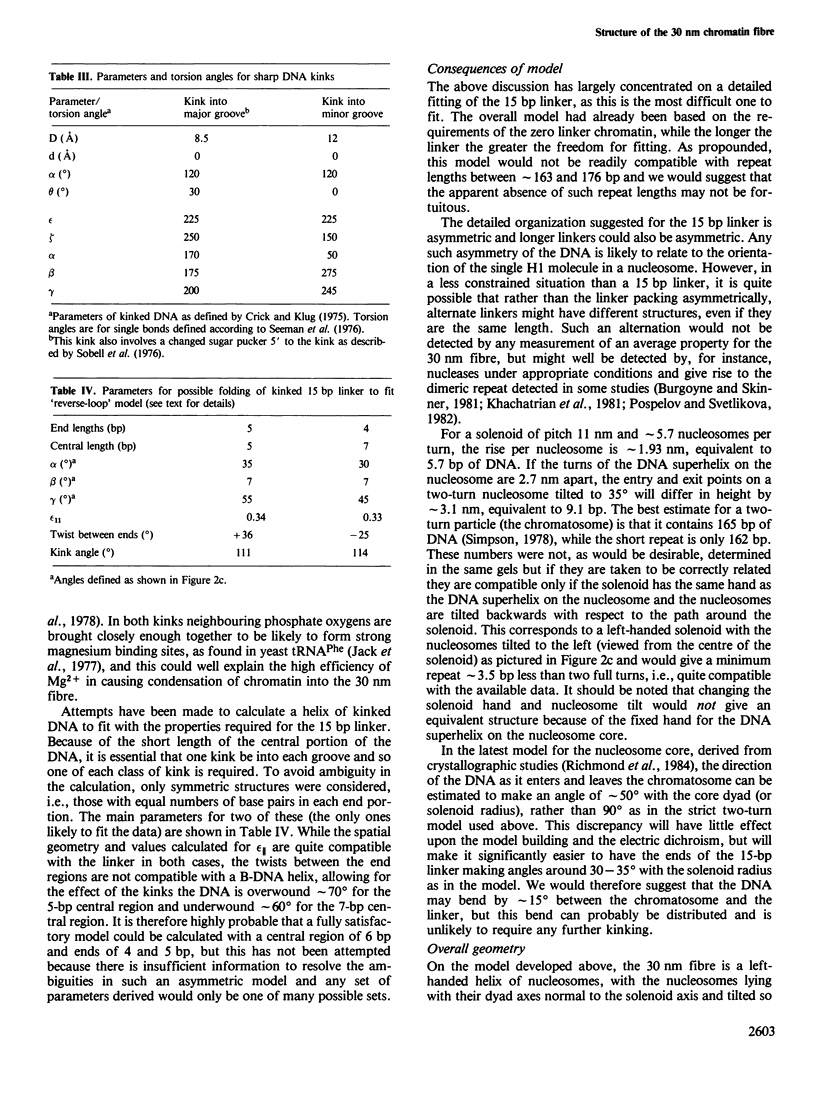
Earlier work on the condensation of chromatins of different repeat lengths into the 30 nm fibre has been surveyed and it is shown that the external geometry of the fibre must be the same for all the chromatins. This can only be fitted by a helical coiling of nucleosomes into a solenoid with the linker DNA disposed internally. On this basis, various models were calculated and compared with published electric dichroism data. The only good fit is found with a 'reverse-loop' model, where the linker DNA forms a complete turn into the hole of the solenoid, of opposite hand to the nucleosomal DNA superhelix. This gives a topological linking number of one per nucleosome and would resolve the 'linking number paradox' if the DNA screw is the same in chromatin as in solution. The feasibility of a reverse-loop for short linkers (down to 15 base pairs) was investigated by model building and kinks of approximately 120 degrees into both DNA grooves are described, which will allow such packing. There will, however, be a 'forbidden' range for the linker DNA length, between approximately 1 and 14 bp, corresponding to nucleosomal repeats of 163 and 176 bp.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allan J., Rau D. C., Harborne N., Gould H. Higher order structure in a short repeat length chromatin. J Cell Biol. 1984 Apr;98(4):1320–1327. doi: 10.1083/jcb.98.4.1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates D. L., Butler P. J., Pearson E. C., Thomas J. O. Stability of the higher-order structure of chicken-erythrocyte chromatin in solution. Eur J Biochem. 1981 Oct;119(3):469–476. doi: 10.1111/j.1432-1033.1981.tb05631.x. [DOI] [PubMed] [Google Scholar]
- Burgoyne L. A., Skinner J. D. Chromatin superstructure: the next level of structure above the nucleosome has an alternating character. A two-nucleosome based series is generated by probes armed with DNAase-I acting on isolated nuclei. Biochem Biophys Res Commun. 1981 Apr 15;99(3):893–899. doi: 10.1016/0006-291x(81)91247-x. [DOI] [PubMed] [Google Scholar]
- Butler P. J. The folding of chromatin. CRC Crit Rev Biochem. 1983;15(1):57–91. doi: 10.3109/10409238309102801. [DOI] [PubMed] [Google Scholar]
- Butler P. J., Thomas J. O. Changes in chromatin folding in solution. J Mol Biol. 1980 Jul 15;140(4):505–529. doi: 10.1016/0022-2836(80)90268-5. [DOI] [PubMed] [Google Scholar]
- Crick F. H., Klug A. Kinky helix. Nature. 1975 Jun 12;255(5509):530–533. doi: 10.1038/255530a0. [DOI] [PubMed] [Google Scholar]
- Crothers D. M., Dattagupta N., Hogan M., Klevan L., Lee K. S. Transient electric dichroism studies of nucleosomal particles. Biochemistry. 1978 Oct 17;17(21):4525–4533. doi: 10.1021/bi00614a026. [DOI] [PubMed] [Google Scholar]
- Finch J. T., Klug A. Solenoidal model for superstructure in chromatin. Proc Natl Acad Sci U S A. 1976 Jun;73(6):1897–1901. doi: 10.1073/pnas.73.6.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germond J. E., Hirt B., Oudet P., Gross-Bellark M., Chambon P. Folding of the DNA double helix in chromatin-like structures from simian virus 40. Proc Natl Acad Sci U S A. 1975 May;72(5):1843–1847. doi: 10.1073/pnas.72.5.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hingerty B., Brown R. S., Jack A. Further refinement of the structure of yeast tRNAPhe. J Mol Biol. 1978 Sep 25;124(3):523–534. doi: 10.1016/0022-2836(78)90185-7. [DOI] [PubMed] [Google Scholar]
- Jack A., Ladner J. E., Rhodes D., Brown R. S., Klug A. A crystallographic study of metal-binding to yeast phenylalanine transfer RNA. J Mol Biol. 1977 Apr 15;111(3):315–328. doi: 10.1016/s0022-2836(77)80054-5. [DOI] [PubMed] [Google Scholar]
- Khachatrian A. T., Pospelov V. A., Svetlikova S. B., Vorob'ev V. I. Nucleodisome - a new repeat unit of chromatin revealed in nuclei of pigeon erythrocytes by DNase I digestion. FEBS Lett. 1981 Jun 1;128(1):90–92. doi: 10.1016/0014-5793(81)81087-3. [DOI] [PubMed] [Google Scholar]
- Klug A., Rhodes D., Smith J., Finch J. T., Thomas J. O. A low resolution structure for the histone core of the nucleosome. Nature. 1980 Oct 9;287(5782):509–516. doi: 10.1038/287509a0. [DOI] [PubMed] [Google Scholar]
- Lee K. S., Mandelkern M., Crothers D. M. Solution structural studies of chromatin fibers. Biochemistry. 1981 Mar 17;20(6):1438–1445. doi: 10.1021/bi00509a006. [DOI] [PubMed] [Google Scholar]
- McGhee J. D., Felsenfeld G. Nucleosome structure. Annu Rev Biochem. 1980;49:1115–1156. doi: 10.1146/annurev.bi.49.070180.005343. [DOI] [PubMed] [Google Scholar]
- McGhee J. D., Nickol J. M., Felsenfeld G., Rau D. C. Higher order structure of chromatin: orientation of nucleosomes within the 30 nm chromatin solenoid is independent of species and spacer length. Cell. 1983 Jul;33(3):831–841. doi: 10.1016/0092-8674(83)90025-9. [DOI] [PubMed] [Google Scholar]
- McGhee J. D., Rau D. C., Charney E., Felsenfeld G. Orientation of the nucleosome within the higher order structure of chromatin. Cell. 1980 Nov;22(1 Pt 1):87–96. doi: 10.1016/0092-8674(80)90157-9. [DOI] [PubMed] [Google Scholar]
- Mitra S., Sen D., Crothers D. M. Orientation of nucleosomes and linker DNA in calf thymus chromatin determined by photochemical dichroism. Nature. 1984 Mar 15;308(5956):247–250. doi: 10.1038/308247a0. [DOI] [PubMed] [Google Scholar]
- Morris N. R. A comparison of the structure of chicken erythrocyte and chicken liver chromatin. Cell. 1976 Dec;9(4 Pt 1):627–632. doi: 10.1016/0092-8674(76)90045-3. [DOI] [PubMed] [Google Scholar]
- Noll M., Kornberg R. D. Action of micrococcal nuclease on chromatin and the location of histone H1. J Mol Biol. 1977 Jan 25;109(3):393–404. doi: 10.1016/s0022-2836(77)80019-3. [DOI] [PubMed] [Google Scholar]
- Olins A. L., Olins D. E. Spheroid chromatin units (v bodies). Science. 1974 Jan 25;183(4122):330–332. doi: 10.1126/science.183.4122.330. [DOI] [PubMed] [Google Scholar]
- Pearson E. C., Butler P. J., Thomas J. O. Higher-order structure of nucleosome oligomers from short-repeat chromatin. EMBO J. 1983;2(8):1367–1372. doi: 10.1002/j.1460-2075.1983.tb01593.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pospelov V. A., Svetlikova S. B. Higher order chromatin structure determines double-nucleosome periodicity of DNA fragmentation. Mol Biol Rep. 1982 Mar 31;8(2):117–122. doi: 10.1007/BF00778514. [DOI] [PubMed] [Google Scholar]
- Prunell A., Kornberg R. D. Variable center to center distance of nucleosomes in chromatin. J Mol Biol. 1982 Jan 25;154(3):515–523. doi: 10.1016/s0022-2836(82)80010-7. [DOI] [PubMed] [Google Scholar]
- Renz M., Nehls P., Hozier J. Involvement of histone H1 in the organization of the chromosome fiber. Proc Natl Acad Sci U S A. 1977 May;74(5):1879–1883. doi: 10.1073/pnas.74.5.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rill R. L. The linear dichroism of oriented helical and superhelical polymers. Biopolymers. 1972;11(9):1929–1941. doi: 10.1002/bip.1972.360110913. [DOI] [PubMed] [Google Scholar]
- Simpson R. T. Structure of the chromatosome, a chromatin particle containing 160 base pairs of DNA and all the histones. Biochemistry. 1978 Dec 12;17(25):5524–5531. doi: 10.1021/bi00618a030. [DOI] [PubMed] [Google Scholar]
- Sobell H. M., Tsai C. C., Gilbert S. G., Jain S. C., Sakore T. D. Organization of DNA in chromatin. Proc Natl Acad Sci U S A. 1976 Sep;73(9):3068–3072. doi: 10.1073/pnas.73.9.3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J. O., Butler P. J. Size-dependence of a stable higher-order structure of chromatin. J Mol Biol. 1980 Nov 25;144(1):89–93. doi: 10.1016/0022-2836(80)90215-6. [DOI] [PubMed] [Google Scholar]
- Worcel A., Benyajati C. Higher order coiling of DNA in chromatin. Cell. 1977 Sep;12(1):83–100. doi: 10.1016/0092-8674(77)90187-8. [DOI] [PubMed] [Google Scholar]
- Yabuki H., Dattagupta N., Crothers D. M. Orientation of nucleosomes in the thirty-nanometer chromatin fiber. Biochemistry. 1982 Sep 28;21(20):5015–5020. doi: 10.1021/bi00263a027. [DOI] [PubMed] [Google Scholar]



