Abstract
UK Biobank is among the world’s largest repositories for phenotypic and genotypic information in individuals of European ancestry1. We performed a genome-wide association study in UK Biobank testing ~9 million DNA sequence variants for association with coronary artery disease (4,831 cases; 115,455 controls) and carried out meta-analysis with previously published results. We identified fifteen novel loci, bringing the total number of coronary artery disease-associated loci to 95. Phenome-wide association scanning revealed that CCDC92 likely affects coronary artery disease through insulin resistance pathways whereas experimental analysis suggests that ARHGEF26 impacts the transendothelial migration of leukocytes.
Journal Subject Codes: coronary artery disease, population genetics, genome-wide association studies, gene-expression
Coronary artery disease (CAD) is a leading cause of disability and mortality worldwide2. Genome-wide association studies (GWAS) have provided new clues to the pathophysiology for this common, complex disease. Largely using a case-control design with cases ascertained based on CAD status, published studies have highlighted at least 80 loci reaching genome-wide significance3–9.
Population-based biobanks such as UK Biobank offer new potential for genetic analysis of common complex diseases. New opportunities include scale, a diverse range of traits, and the ability to explore a fuller spectrum of phenotypic consequences for identified DNA variants. Leveraging the UK Biobank resource, we sought to: 1) perform a genetic discovery analysis; 2) explore the phenotypic consequences and tissue-specific effects associated with CAD risk alleles; and 3) characterize the functional consequences of a risk mutation in a promising pathway.
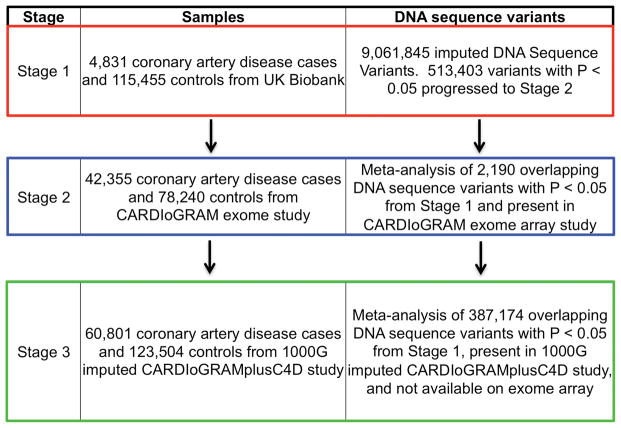
We designed a three-stage GWAS (Fig. 1). In Stage 1, we tested the association of DNA sequence variants with CAD in UK Biobank. In Stage 2 we took forward 2,190 variants that reached nominal significance in Stage 1 (P<0.05) for meta-analysis with results from an exome-focused-array analysis in 42,355 cases and 78,240 controls6. In Stage 3, we took forward 387,174 variants that reached nominal significance in Stage 1 and not tested in Stage 2 for meta-analysis with results from a genome-wide imputation study in 60,801 cases and 123,504 controls5. For each variant, we combined statistical evidence across Stages 1 and 2 (or Stages 1 and 3) and set a statistical threshold of P < 5 ×10−8 for genome-wide significance.
Figure 1. Study Design.
Stage 1 consisted of a genome-wide association study for the coronary artery disease phenotype performed in UK Biobank; variants below a threshold P value < 0.05 moving forward to meta-analysis with CARDIoGRAM Exome (Stage 2) or CARDIoGRAMplusC4D summary statistics (Stage 3).
Abbreviations: 1000G, 1000 Genomes; CARDIoGRAMplusC4D, Coronary ARtery DIsease Genome-wide Replication and Meta-analysis
Characteristics of UK Biobank participants stratified by presence of CAD are presented in Supplementary Table 1. CAD cases were more likely to be older, male, on lipid-lowering therapy, have a history of smoking, and affected with type 2 diabetes. After quality control, 9,061,845 DNA sequence variants were tested for association in 4,831 CAD patients and 115,455 controls in UK Biobank (Stage 1). A total of 269 variants at five distinct loci met the genome-wide significance threshold (P < 5×10−8) (Supplementary Fig. 1 and 2). All five have been previously reported5,10–13. In UK Biobank, the 9p21/CDKN2B-AS1 variant rs4977575 (NC_000009.12:g.22124745C>G) was the top association result (49% frequency for G allele; OR =1.24; 95%CI: 1.19–1.29; P = 5.40×10−23); the other four loci were 1p13/SORT1, PHACTR1, LPA, and KCNE2 (Supplementary Table 2). For a set of previously reported CAD loci5, we compared the effect estimates from the published literature with that from the current analysis in UK Biobank and found strong positive correlation in effect sizes (β = 0.92, 95%CI: 0.77–1.06; P = 1.8×10−17, Supplementary Fig. 3); these results validate our CAD phenotype definition in UK Biobank. A total of 513,403 variants exceeded nominal significance (P < 0.05) and were taken forward to Stages 2 or 3.
After meta-analysis, 15 new loci exceeded genome-wide significance (Tables 1–2), bringing the total number of established CAD loci to 95. Of note, while this manuscript was under review, one of the 15 loci (HNF1A) has since been reported9. Effect allele frequencies of the 15 newly identified loci ranged from 13% to 86%, with effect sizes ranging from 1.05 to 1.08. Descriptions of relevant loci appear in Supplementary Table 3, and regional association plots for novel CAD loci are shown in Supplementary Figures 4–6.
Table 1.
New loci from analysis of UK Biobank and CARDIoGRAM exome study
| UK Biobank | Stage 2 Exome Study | Combined | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lead Variant | Chr | Gene | Description | EA | EAF | OR | P | OR | P | OR | 95% CI | P |
| rs2972146 | 2 | (LOC646736) | intergenic | T | 0.65 | 1.07 | 0.0011 | 1.05 | 2.01×10−7 | 1.06 | 1.04–1.07 | 1.46×10−9 |
| rs12493885 (p.Val29Leu) | 3 | ARHGEF26 | missense | C | 0.85 | 1.07 | 0.039 | 1.09 | 8.28×10−9 | 1.08 | 1.06–1.11 | 1.02×10−9 |
| rs1800449 | 5 | LOX | missense | T | 0.17 | 1.09 | 0.0039 | 1.07 | 1.72×10−7 | 1.07 | 1.05–1.09 | 2.99×10−9 |
| rs11057401 (p.Ser70Cys) | 12 | CCDC92 | missense | T | 0.69 | 1.08 | 0.001 | 1.05 | 4.32×10−7 | 1.06 | 1.04–1.08 | 3.88×10−9 |
Genes for variants that are outside the transcript boundary of the protein-coding gene are shown in parentheses [eg, (LOC646736)].
Chr = Chromosome, CI = Confidence Interval, EA = Effect Allele, EAF = Effect Allele Frequency, OR = Odds Ratio
Table 2.
New Loci from analysis of UK Biobank and CARDIoGRAMplusC4D 1000G imputation study
| UK Biobank | Stage 3 1000G Imputed Study | Combined | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lead Variant | Chr | Gene | Description | EA | EAF | OR | P | OR | P | OR | 95% CI | P |
| rs17517928 | 2 | FN1 | intronic | C | 0.75 | 1.08 | 0.0026 | 1.06 | 5.14×10−7 | 1.06 | 1.04–1.08 | 1.06×10−8 |
| rs17843797 | 3 | UMPS-ITGB5 | intronic | G | 0.13 | 1.11 | 0.00019 | 1.07 | 2.43×10−6 | 1.07 | 1.05–1.10 | 1.52×10−8 |
| rs748431 | 3 | FGD5 | intronic | G | 0.36 | 1.04 | 0.042 | 1.05 | 2.14×10−7 | 1.05 | 1.03–1.07 | 2.63×10−8 |
| rs7623687 | 3 | RHOA | intronic | A | 0.86 | 1.09 | 0.0073 | 1.07 | 5.22×10−7 | 1.08 | 1.05–1.10 | 2.00×10−8 |
| rs10857147 | 4 | (FGF5) | regulatory region | T | 0.29 | 1.06 | 0.014 | 1.06 | 5.83×10−7 | 1.06 | 1.04–1.08 | 3.39×10−8 |
| rs7678555 | 4 | (MAD2L1) | intergenic | C | 0.29 | 1.06 | 0.027 | 1.06 | 3.26×10−7 | 1.06 | 1.04–1.08 | 2.91×10−8 |
| rs10841443 | 12 | RP11-664H17.1 | intronic | G | 0.67 | 1.06 | 0.0073 | 1.05 | 5.81×10−7 | 1.05 | 1.03–1.07 | 2.23×10−8 |
| rs2244608 | 12 | HNF1A | intronic | G | 0.32 | 1.07 | 0.003 | 1.05 | 1.02×10−6 | 1.05 | 1.03–1.07 | 2.41×10−8 |
| rs3851738 | 16 | CFDP1 | intronic | C | 0.6 | 1.07 | 0.00089 | 1.05 | 1.88×10−6 | 1.05 | 1.03–1.07 | 2.43×10−8 |
| rs7500448 | 16 | CDH13 | intronic | A | 0.75 | 1.1 | 0.00016 | 1.06 | 2.11×10−6 | 1.06 | 1.04–1.09 | 1.20×10−8 |
| rs8108632 | 19 | TGFB1 | intronic | T | 0.41 | 1.06 | 0.011 | 1.05 | 4.76×10−7 | 1.05 | 1.03–1.07 | 2.35×10−8 |
Genes for variants that are outside the transcript boundary of the protein-coding gene are shown in parentheses [eg, (FGF5)].
1000G = 1000 Genomes, Chr = Chromosome, CI = Confidence Interval, EA = Effect Allele, EAF = Effect Allele Frequency, OR = Odds Ratio
To move from these 15 DNA sequence variants to biologic insights, we took two approaches: phenome-wide association scanning and functional analysis. Understanding the full spectrum of phenotypic consequences of a given DNA sequence variant may shed light on the mechanism by which a variant/gene leads to disease. Termed a ‘phenome-wide association study’ or “PheWAS”, this approach tests the association of a mapped disease variant with a broad range of human phenotypes14. In collaboration with Genomics plc, we conducted a PheWAS combining UK Biobank data, mRNA transcript phenotypes in the Genotype-Tissue Expression Project (GTEx) dataset15, and an integrated set of GWAS results from a variety of publically available sources16–24.
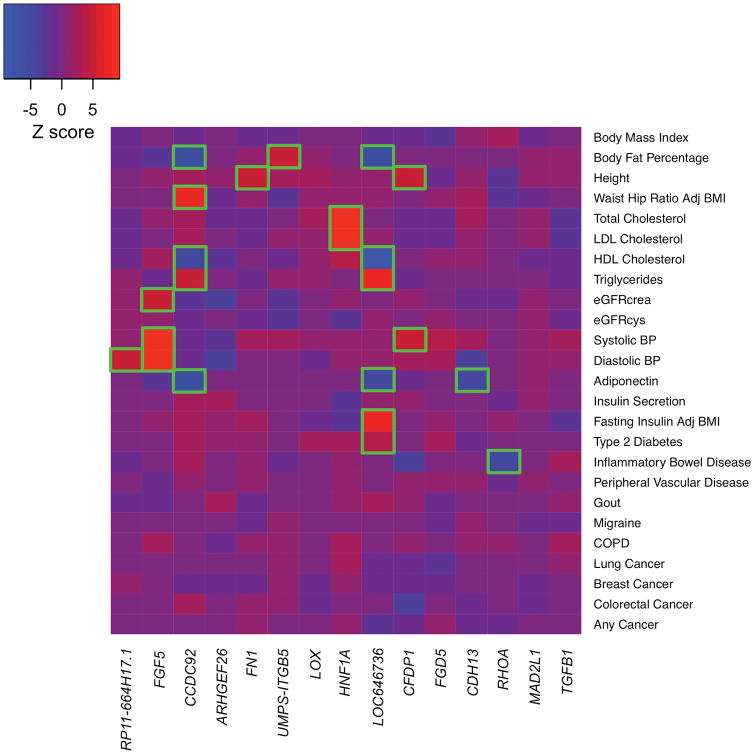
We found that several of the newly identified DNA sequence variants correlated with a range of human traits (Fig. 2, Supplementary Tables 4–5). For example, the intronic variant rs10841443 within RP11-664H17.1 is in close proximity to PDE3A, a phosphodiesterase previously implicated in an autosomal dominant form of hypertension25. PheWAS showed an association for this variant with diastolic blood pressure26, suggesting that this locus may be acting through hypertension. The variant rs2244608 within HNF1A has been previously associated with LDL cholesterol, a causal path to atherosclerosis16. The variant rs7500448 within CDH13 (encoding Cadherin 13 or T-Cadherin), a vascular adiponectin receptor implicated in hypertensive and insulin resistance biology27, associates with plasma adiponectin levels. Variant rs2972146 is downstream of IRS1 (encoding the insulin receptor substrate-1 gene24) and is a cis-eQTL for IRS1 expression in adipose tissue. rs2972146 associates with a range of phenotypes seen in the setting of insulin resistance including HDL cholesterol, triglycerides, adiponectin, fasting insulin, and type 2 diabetes.
Figure 2. Phenome-wide association results for 15 novel loci.
For the 15 novel CAD risk variants identified in our study, Z-scores (aligned to the CAD risk allele) were obtained from the Genomics plc Platform and UK Biobank. A positive Z-score (red) indicates a positive association between the CAD risk allele and the disease/trait, while a negative Z-score (blue) indicates an inverse association. Boxes are outlined in green if the variant is significantly (P < 0.00013) associated with the given trait.
Abbreviations: Adj, Adjusted; BMI, Body Mass Index; BP, Blood Pressure; crea, Creatinine; cys, cystatin-c; COPD, chronic obstructive pulmonary disease; eGFR, estimated Glomerular Filtration Rate; HDL, High Density Lipoprotein; LDL, Low Density Lipoprotein;
Compelling additional insights from the PheWAS emerged at the CCDC92 locus. Across 25 distinct traits and disorders, we observed significant associations (P < 0.00013) for CCDC92 p.Ser70Cys (rs11057401) with body fat percentage, waist-to-hip circumference ratio, as well as plasma high-density lipoprotein, triglyceride, and adiponectin levels. The directionality of these associations are hallmarks of insulin resistance and lipodystrophy17,28, and the association with plasma adiponectin levels localizes these genetic effects to adipose tissue. Recent work has highlighted two candidate genes at this locus, CCDC92 and DNAH1029, and further experimental work is necessary to define the causal gene at this locus.
However, a few of the CAD loci (FN1, LOX, ITGB5, and ARHGEF26) did not associate with any of the studied risk factor traits and thus, appear to function through pathways beyond known CAD risk factors (Fig. 2, Supplementary Tables 4–5). A common variant within an intron of FN130 (encoding Fibronectin 1) and a missense variant in LOX31 (encoding Lysyl Oxidase) suggest potential links to extracellular matrix biology. Of note, rare coding mutations in LOX were recently described to cause Mendelian forms of thoracic aortic aneurysm and dissection32,33, highlighting a potential common link between atherosclerosis and aortic disease, possibly through altered extracellular matrix biology. A variant downstream of ITGB534 (encoding Integrin Subunit Beta 5) suggests pathways underlying cell adhesion and migration.
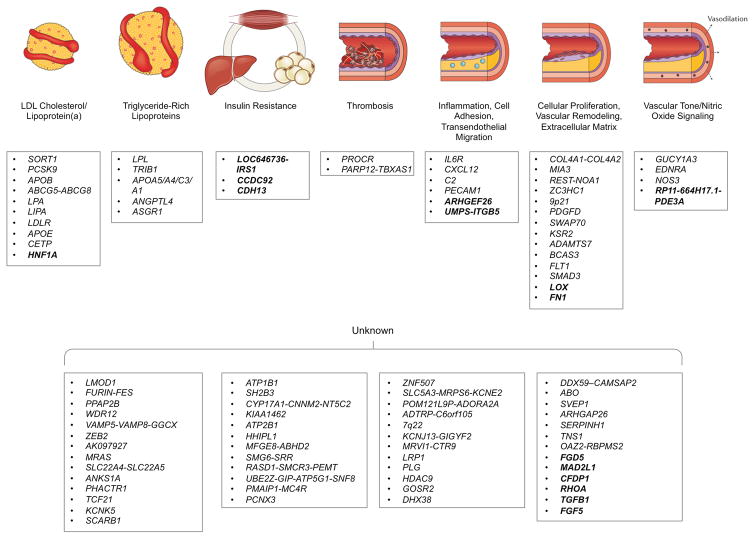
In aggregate, our analysis brings the total number of known CAD loci to 953–9, and in Figure 3, we organize these loci into plausible pathways. Of note, the causal variant, gene, cell type, and mechanism has been definitively identified at only a few of these loci and as such, additional experimental research will be required, particularly at >50% of loci without an apparent link to known risk factors.
Figure 3. Biological pathways underlying genetic loci associated with coronary artery disease.
CAD GWAS loci identified to date are depicted along with the plausible relationship to the underling biological pathway. The 15 new loci described in this paper are shown in bold. Loci names are based on the nearest genes; however, the causal gene(s) remains unclear for most associated loci and as such, the resultant annotation may prove incorrect in some cases. Adapted from Ref. 41.
At one of the new loci that did not relate to known risk factors, ARHGEF26 (encoding Rho Guanine Nucleotide Exchange Factor 26), we performed functional studies. Prior experimental work had connected this gene with murine atherosclerosis35. Earlier studies established a role for ARHGEF26 in facilitating the transendothelial migration of leukocytes, a key step in the initiation of atherosclerosis36,37. ARHGEF26 has been shown to activate RhoG GTPase by promoting the exchange of GDP by GTP and contributing to the formation of ICAM-1-induced endothelial docking structures that facilitate leukocyte transendothelial migration 36,37. In addition, Arhgef26 −/− mice, when crossed with atherosclerosis-prone Apoe null mice, displayed less aortic atherosclerosis35.
At ARHGEF26 p.Val29Leu (rs12493885), the 29Leu allele, observed in 85% of participants, is associated with increased risk for CAD. We first examined the hypothesis that a haplotype block containing this variant may alter expression of ARHGEF26 in coronary artery. While this region demonstrates eQTL effects in a variety of tissues, there is no evidence of alteration of ARHGEF26 expression in coronary artery in both eQTL and allele specific expression analyses (Supplementary Fig. 7). To further evaluate the possibility that a haplotype containing the 29Leu allele may affect gene expression, we performed a luciferase reporter assay. We cloned a 2.5 kb region immediately upstream of the ARHGEF26 start codon consisting of the promoter, 5′ untranslated region (5′ UTR), and regions with ENCODE annotations suggestive of potential cis-acting elements. We obtained the reference (in LD with Val29 G allele) and alternative (in LD with 29Leu C allele) haplotypes of this region from human rs12493885 heterozygotes. We coupled each haplotype with a luciferase reporter, and measured luciferase activity (Supplementary Fig. 8). In HEK293, human aortic endothelial cells (HAEC), and human umbilical vein endothelial cells (HUVEC), there is no significant difference in luciferase activity between reference and alternative haplotypes. These data suggest that the ARHGEF26 29Leu allele may confer CAD risk via mechanisms other than affecting ARHGEF26 transcription or promoter activity in disease-relevant tissue.
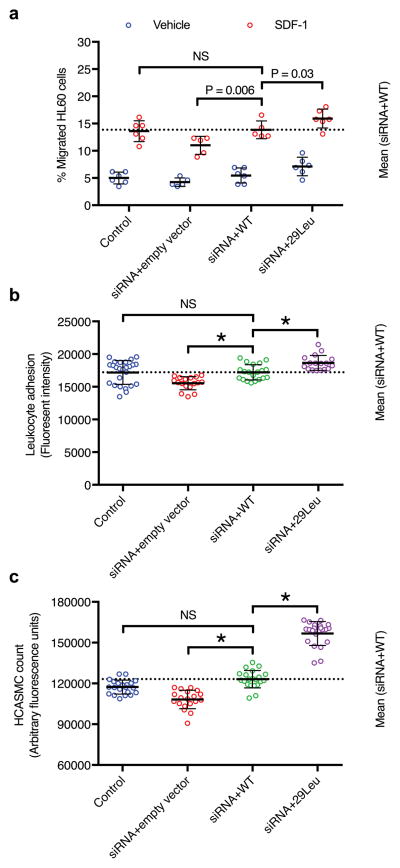
Next, we examined the hypothesis that ARHGEF26 p.Val29Leu may influence disease risk through its protein-altering consequence. We knocked down endogenous ARHGEF26 through siRNA and observed decreased leukocyte transendothelial migration, leukocyte adhesion on endothelial cells, and vascular smooth cell proliferation38 (Fig. 4, Supplementary Fig. 9). Overexpression of exogenous, wild-type ARHGEF26 rescued these phenotypes. However, ARHGEF26 29Leu mutant overexpression led to rescued phenotypes that consistently exceeded wild-type. These data support the hypothesis that the ARHGEF26 29Leu allele associated with increased CAD risk may lead to a gain-of-function ARHGEF26 protein.
Figure 4. Functional assessment of ARHGEF26 p.Val29Leu in vitro.
a) ARHGEF26-29Leu increases leukocyte transendothelial migration. HAEC were transfected with non-targeting siRNA and empty vector (control), siRNA against ARHGEF26 3′-UTR and empty vector, siRNA and ARHGEF26-WT, or siRNA and ARHGEF26-29Leu. Transfected HAEC were plated on transwell inserts and treated with 10 ng/mL TNF-α. Differentiated HL60 cells were loaded on the upper chambers of transwells and allowed to transmigrate across HAEC towards vehicle (blue) or 50 ng/mL SDF-1 (red). The migrated cells were quantified as percentage of input cells per well (n=5 or 6; mean±s.d.; F=11.89, DF=3 by two-way ANOVA within vehicle and SDF-1 subgroups with Fisher’s LSD test; variance among vehicle subgroups non-significant; NS, not significant; representative of 3 independent experiments).
b) ARHGEF26-29Leu increases leukocyte adhesion on endothelial cells. HAEC were transfected as 2a) and cultured on 96-well plates until confluent and treated with 10 ng/mL TNF-α. Calcein-AM-labeled THP-1 cells were incubated with HAEC and washed to remove non-adherent cells. The adherent cells were lysed, quantified by Calcein-AM fluorescence and compared to siRNA+WT (n=25, 17, 20, and 17; mean±s.d.; F=14.53, DF=3 by one-way ANOVA; NS, not significant; * P<0.0001 compared to siRNA+WT; representative of 3 independent experiments).
c) ARHGEF26-29Leu increases vascular smooth muscle cell proliferation. HCASMC were transfected as 2a) and made quiescent by serum starvation for 48 h, followed by 72-h proliferation in normal serum medium. Cell proliferation was quantified by a luminescent assay and compared to siRNA+WT (n=20; mean±s.d.; F=197.5, DF=3 by one-way ANOVA; NS, not significant; * P<0.0001 compared to siRNA+WT; representative of 3 independent experiments).
How could the ARHGEF26 29Leu mutation lead to a gain-of-function phenotype? We evaluated its functional impact in two ways, addressing ARHGEF26 quality and quantity, respectively. First, could the 29Leu mutation alter ARHGEF26 nucleotide exchange activity on RhoG? To answer this question, we developed a GTP-GDP nucleotide exchange assay using recombinant human full-length ARHGEF26 (wild-type or 29Leu) and RhoG proteins39. In a cell-free system, equal amount of wild-type or 29Leu ARHGEF26 protein was incubated with RhoG pre-loaded with GDP. After 60 minutes, we observed no significant difference in nucleotide exchange activity between wild-type and 29Leu mutant ARHGEF26 (Supplementary Fig. 10).
Second, could the 29Leu allele affect cellular abundance of ARHGEF26 protein? We examined this possibility by treating cells expressing wild-type or 29Leu mutant ARHGEF26 with cycloheximide, a protein synthesis inhibitor, and compared ARHGEF26 degradation over time by Western blotting. Compared to wild-type ARHGEF26, the 29Leu mutant protein displayed a longer half-life (Supplementary Fig. 11). While further work is needed to understand the mechanism in vivo, in vitro results suggest that the gain of function phenotype observed may be secondary to the 29Leu mutant protein’s resistance to degradation.
Our study should be interpreted within the context of its limitations. First, we focused on participants of European ancestry within UK Biobank and therefore results may not be generalizable to other populations. Second, our CAD phenotype definitions are based largely on interview and electronic health records and this may result in misclassification of case status. However, such misclassification should reduce statistical power for discovery and bias results toward the null. Finally, although we observed no evidence of robust changes in ARHGEF26 expression associated with the 29Leu haplotype in disease relevant tissue, it is possible that other regulatory mechanisms may potentiate the gain of function phenotypes we observed.
In summary, we performed a gene discovery study for CAD using a large population-based biobank, identified 15 new loci, and explored the phenotypic consequences of CAD risk variants through PheWAS and in vitro functional analysis. These findings permit several conclusions. First, CAD cases phenotyped via electronic health records and verbal interviews exhibit similar genetic architecture to those derived in epidemiologic cohorts and can prove useful in gene discovery efforts. Second, phenome-wide association studies with risk variants can provide initial clues on how DNA sequence variants may lead to disease. Lastly, considerable experimental evidence in cells and rodents has suggested that transendothelial migration of leukocytes is a key step in the formation of atherosclerosis40; here, we provide human genetic support for a role of this pathway in CAD.
Online Methods
Study Design and Samples
We performed a three-stage sequential analysis to identify novel genetic loci associated with CAD. In Stage 1, we first tested the association of DNA sequence variants with CAD in UK Biobank. Beginning in 2006, individuals aged 45 to 69 years old were recruited from across the United Kingdom for participation in the UK Biobank Study1. At enrollment, a trained healthcare provider ascertained participants’ medical histories through verbal interview. In addition, participants’ electronic health records (EHR) including inpatient International Classification of Disease (ICD-10) diagnosis codes and Office of Population and Censuses Surveys (OPCS-4) procedure codes, were integrated into UK Biobank. Individuals were defined as having CAD based on at least one of the following criteria:
Myocardial infarction (MI), coronary artery bypass grafting, or coronary artery angioplasty documented in medical history at time of enrollment by a trained nurse
Hospitalization for ICD-10 code for acute myocardial infarction (I21.0, I21.1, I21.2, I21.4, I21.9)
Hospitalization for OPCS-4 coded procedure: coronary artery bypass grafting (K40.1–40.4, K41.1–41.4, K45.1–45.5)
Hospitalization for OPCS-4 coded procedure: coronary angioplasty with or without stenting (K49.1–49.2, K49.8–49.9, K50.2, K75.1–75.4, K75.8–75.9)
All other individuals were defined as controls. In total, genotypes were available for 120,286 participants of European ancestry.
In Stage 2, we took forward 2,190 variants that reached nominal significance in Stage 1 for meta-analysis in the Coronary ARtery DIsease Genome wide Replication and Meta-analysis (CARDIoGRAM) Exome Consortia exome array analysis which incorporated 42,355 cases and 78,240 controls6 (Supplementary Table 6). In Stage 3, we took forward 387,174 variants that reached nominal significance in Stage 1 (and not available in Stage 2) for meta-analysis into the CARDIoGRAMplusC4D 1000 Genomes imputation study containing 60,801 cases and 123,504 controls5. Informed consent was obtained for all participants, and UK Biobank received ethical approval from the Research Ethics Committee (reference number 11/NW/0382). Our study was approved by a local Institutional Review Board at Partners Healthcare (protocol 2013P001840).
Genotyping and Quality Control
UK Biobank samples were genotyped using either the UK Bileve42 or UK Biobank Axiom Arrays having been performed in 33 separate batches of samples by Affymetrix (High Wycombe, UK). A total of 806,466 directly genotyped DNA sequence variants were available after variant quality control (QC). The UK Biobank team then performed imputation from a combined 1000 Genomes/UK10K reference panel; phasing was performed using SHAPEIT-3 and imputation carried out via IMPUTE3. Variant level QC exclusion metrics applied to imputed data for GWAS included: call rate < 95%, Hardy-Weinberg Equilibrium P-value <1×10−6, posterior call probability < 0.9, imputation quality <0.4, and minor allele frequency (MAF) < 0.005. Sex chromosome and mitochondrial genetic data were excluded from this analysis. In total, 9,061,845 imputed DNA sequence variants were included in our analysis. For sample QC, the UK Biobank analysis team removed individuals of relatedness 3rd degree or higher, and an additional 480 samples with an excess of missing genotype calls or more heterozygosity than expected were excluded. In total, genotypes were available for 120,286 participants of European ancestry.
Statistical Analysis
Stage 1 Association Analysis
The BOLT-LMM software43 was used to perform linear mixed models (LMMs) for association testing. CAD case status was analyzed while adjusting for age, gender, and chip array at run-time. This analysis was used to derive statistical significance. As effect estimates from BOLT-LMM software are unreliable due to the treatment of binary phenotype data as quantitative data, we performed logistic regression to derive effect estimates for each variant that exceeded genome-wide significance. Effect estimates of top variants were derived from logistic regression using allelic dosages adjusting for age, sex, chip at run-time, and ten principal components under the assumption of additive effects utilizing the R v3.2.0 and SNPTEST statistical software programs.
Stage 2 and 3 Meta-Analysis
In stage 2, top variants (P < 0.05) from UK Biobank were then meta-analyzed with exome chip data from the CARDIoGRAM Exome Consortium6. Tested variants in the CARDIoGRAM exome array study were analyzed through logistic regression with an additive model adjusting for study specific covariates and principal components of ancestry as appropriate. Top variants from UK Biobank that were not available for analysis in the CARDIoGRAM exome array study were then meta-analyzed with data from the 1000 Genomes imputed CARDIoGRAMplusC4D GWAS5 in Stage 3.
Given differences in effect size units between the UK Biobank Stage 1 data and the CARDIoGRAM Exome/1000 Genomes CARDIoGRAMplusC4D data, both Stage 2 and 3 meta-analyses were performed via a weighted z-score method, adjusting for an unbalanced ratio of cases to controls. To derive effect size estimates for variants exceeding genome-wide significance, we meta-analyzed logistic regression results using inverse-variance weighting with fixed effects (METAL software)44. We set a combined statistical threshold of P < 5×10−8 for genome wide significance. P values reported in analysis Stages 1, 2, and 3 are all two-sided.
Phenome-Wide Association Study
For all 15 novel DNA sequence variants associated with CAD in our study, we collaborated with Genomics plc to conduct a phenome-wide association study. This PheWAS used the Genomics plc Platform, UK Biobank, and GTEx Consortium eQTL data. The Genomics plc Platform includes PheWAS data across 545 distinct molecular and disease phenotypes, at an integrated set of over 14 million common variants, from 677 GWAS studies. UK Biobank analyses within the Genomics plc Platform were conducted under a separate research agreement. We selected 25 phenotypes across a range of relevant diseases, metabolic and anthropometric traits from either previously published GWAS datasets or UK Biobank. Complete details of phenotype definitions, sample sizes, and GWAS data sources are shown in Supplementary Tables 7 and 8. In the PheWAS, quantitative traits were standardized to have unit variance, imputation was performed to generate results for all variants within the 1000 Genomes reference panel, and P values were recalculated based on a Wald test statistic for uniformity.
Phenotypes were declared to be significantly associated with the risk variant if they met a Bonferroni corrected P value of < 0.00013 [0.05/(25 traits × 15 DNA sequence variants)]. Phenome scan results were then depicted in a heatmap based on the Z-scores for all variant-disease/trait associations aligned to the CAD risk allele as implemented by the gplots package in R v3.2.0. To identify loci that might influence gene expression, we used previously published cis-expression quantitative trait locus (eQTL) mapping data from the Genotype-Tissue Expression (GTEx) Consortium Project across 44 tissues15. We queried the 15 novel variants identified in our study for overlap with genome-wide significant variant-gene pairs from the GTEx portal.
Allele Specific Expression Analysis
Allele-specific expression (ASE) data from the GTEx project were obtained from dbGaP (accession phs000424.v6.p1). The generation of these data is summarized in Aguet et al., and relied on methods described earlier45. In brief, only uniquely mapping reads with base quality ≥ 10 at the SNP were counted, and only SNPs with coverage of at least 8 reads were reported. For ARHGEF26 p.Val29Leu, ASE counts were available for 20 heterozygous individuals. A two-sided binomial test was used to identify SNPs with significant allelic imbalance in each individual, and Benjamini-Hochberg adjusted p-values were calculated across all sites measured in an individual.
Luciferase Reporter Assay
HUVEC heterozygous for rs12493885 were identified from Caucasian donors by SNP genotyping. A 2.9 kb genomic fragment spanning from 5′ upstream of ARHGEF26 to exon 2 (rs12493885) was cloned into a pMiniT 2.0 vector (NEB) using the heterozygous HUVEC genomic DNA as a template, and sequenced for reference and alternative alleles. The −2516 to +2 reference and alternative haplotypes upstream of ARHGEF26 (NC_000003.12:154119477-154121994) were amplified from the 2.9 kb region by PCR with primers designed to create 5′ NheI and 3′ HindIII restriction sites in the PCR products. The amplified fragments were subcloned between the NheI and HindIII sites of a promoterless firefly luciferase (luc2) expression vector pGL4.10 (Promega), to create two plasmids: pGL4.10-Ref and pGL4.10-Alt. Promoterless pGL4.10-control, and pGL4.73[hRluc/SV40] vector containing the renilla luciferase hRluc reporter gene and an SV40 early enhancer/promoter, were used as negative control and co-reporter, respectively. Cells were cotransfected with equal amounts of luc2 expression plasmid (pGL4.10-control, pGL4.10-Ref and pGL4.10-Alt) and pGL4.73 vector by Lipofectamine 2000. Cells were harvested at 48 h after transfection and followed by a Dual-Glo Luciferase Assay (Promega) to measure firefly and renilla luciferase activities. The firefly luciferase activity was normalized to renilla luciferase in the same sample, and expressed as fold change relative to pGL4.10-control group.
Nucleotide Exchange Assay
Human full-length ARHGEF26 (wild-type or 29Leu) and RhoG (residues 1–188) proteins, both with N-terminal His-SUMO tags, were expressed in E. coli BL21(DE3) cells in TB media. Nucleotide exchange assay samples were prepared in buffer containing 10mM HEPES pH 7.4, 150mM NaCl, 1mM MgCl2, 0.5uM MANT-GTP, 2mM TCEP with 1μM ARHGEF26. Just prior to reading, RhoG protein, pre-loaded with GDP, was added to a final concentration of 0.4μM. MANT-GTP fluorescence was monitored for 60 minutes on a SpectraMax M2 at 37°C using an excitation wavelength of 280nm and an emissions wavelength of 440nm with a 435nm cutoff. Fluorescence data was imported into Prism GraphPad for analysis.
Functional Characterization of ARHGEF26 p.Val29Leu in Arterial Tissue
To investigate the functional effects of ARHGEF26 p.Val29Leu (rs12493885), we knocked-down the expression of endogenous ARHGEF26 in cultured human aortic endothelial cells (HAEC) and human coronary artery smooth muscle cells (HCASMC) by RNA interference. We then overexpressed wild-type or mutant ARHGEF26 (29Leu) resistant to siRNA, and measured leukocyte transendothelial migration, leukocyte adhesion on endothelial cells, and HCASMC proliferation in vitro. We also evaluated the degradation of wild-type or 29Leu mutant ARHGEF26 with a cycloheximide chase assay and Western blotting. Additional details on experimental techniques are described in the Supplementary Note.
Data Availability
Stage 2 and Stage 3 data contributed by CARDIoGRAM Exome and CARDIoGRAMplusC4D investigators is available online (see URLs). The genetic and phenotypic UK Biobank data are available upon application to the UK Biobank. Genotype-Tissue Expression Project data is available online.
Supplementary Material
Acknowledgments
This research has been conducted using the UK Biobank resource, application 7089. The PheWAS carried out by Genomics plc was also conducted using the UK Biobank resource under a separate agreement. The authors thank Dr. K. Burridge and Dr. E. Wittchen for technical advice on leukocyte transendothelial migration assay.
Funding/Support: This analysis was supported by: National Heart, Lung, and Blood Institute of the National Institutes of Health under award number T32 HL007734 (D.K.), the John S. LaDue Memorial Fellowship in Cardiology at Harvard Medical School (P.N.), the KL2/Catalyst Medical Research Investigator Training award from Harvard Catalyst funded by the National Institutes of Health (NIH) (TR001100) (A.V.K.), the Ofer and Shelly Nemirovsky Research Scholar award from the Massachusetts General Hospital (MGH), the Donovan Family Foundation and NIH R01 HL127564 (S.K.).
Footnotes
URLs: UK Biobank, https://www.ukbiobank.ac.uk/; CARDIoGRAM exome and CARDIoGRAMplusC4D data, http://www.cardiogramplusc4d.org/; R statistical software www.R-project.org; SNPTEST software, mathgen.stats.ox.ac.uk/genetics_software/snptest/snptest.html; Genotype-Tissue Expression Project Data, gtexportal.org
Role of the Funder/Sponsor: The sponsors had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
Author Contributions:
Concept and design: D.K., Q.M.Z., M.E.W., A.V.K., P.N., S.K., Acquisition, analysis, or interpretation of data: D.K., Q.M.Z., C.A.E., M.C., S.H., B.J.M., A.L., M.E.W., C.C.A.S., F.A., A.V.S., K.G.A., A.V.K., V.K.K., P.N., S.K., Drafting of the manuscript: D.K., Q.M.Z., C.A.E., M.E.W., A.V.K., P.N., S.K. Critical revision of the manuscript for important intellectual content: D.K., Q.M.Z., C.A.E., M.C., S.H., B.J.M., A.L., M.E.W, C.C.A.S., F.A., A.V.S., K.G.A., A.V.K., V.K.K., P.N., S.K., Administrative, technical, or material support: D.K., S.K.
Disclosures: Dr. Kathiresan reports grant support from Regeneron and Bayer, grant support and personal fees from Aegerion, personal fees from Regeneron Genetics Center, Merck, Celera, Novartis, Bristol-Myers Squibb, Sanofi, AstraZeneca, Alnylam, Eli Lilly, and Leerink Partners, personal fees and other support from Catabasis, and other support from San Therapeutics outside the submitted work. He is also the chair of the scientific advisory board at Genomics plc.
References
- 1.Collins R. What makes UK Biobank special? The Lancet. 2012;379:1173–1174. doi: 10.1016/S0140-6736(12)60404-8. [DOI] [PubMed] [Google Scholar]
- 2.GBD; 2015 Mortality and Causes of Death Collaborators. Global, regional and national life expectancy, all-cause mortality and cause-specific mortality for 249 causes of death, 1980–2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet. 2016;388:1459–1544. doi: 10.1016/S0140-6736(16)31012-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schunkert H, et al. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat Genet. 2011;43:333–8. doi: 10.1038/ng.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deloukas P, et al. Large-scale association analysis identifies new risk loci for coronary artery disease. Nat Genet. 2013;45:25–33. doi: 10.1038/ng.2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.CARDIoGRAMplusC4D Consortium. A comprehensive 1000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat Genet. 2015;47:1121–30. doi: 10.1038/ng.3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Myocardial Infarction Genetics and CARDIoGRAM Exome Consortia Investigators. Coding Variation in ANGPTL4, LPL, and SVEP1 and the Risk of Coronary Disease. N Engl J Med. 2016;374:1134–44. doi: 10.1056/NEJMoa1507652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nioi P, et al. Variant ASGR1 Associated with a Reduced Risk of Coronary Artery Disease. N Engl J Med. 2016;374:2131–41. doi: 10.1056/NEJMoa1508419. [DOI] [PubMed] [Google Scholar]
- 8.Webb TR, et al. Systematic Evaluation of Pleiotropy Identifies 6 Further Loci Associated With Coronary Artery Disease. J Am Coll Cardiol. 2017;69:823–836. doi: 10.1016/j.jacc.2016.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Howson JMM, et al. Fifteen new risk loci for coronary artery disease highlight arterial-wall-specific mechanisms. Nature Genetics. 2017 doi: 10.1038/ng.3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Musunuru K, et al. From noncoding variant to phenotype via SORT1 at the 1p13 cholesterol locus. Nature. 2010;466:714–9. doi: 10.1038/nature09266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Myocardial Infarction Genetics Consortium et al. Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy number variants. Nat Genet. 2009;41:334–41. doi: 10.1038/ng.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tregouet DA, et al. Genome-wide haplotype association study identifies the SLC22A3-LPAL2-LPA gene cluster as a risk locus for coronary artery disease. Nat Genet. 2009;41:283–5. doi: 10.1038/ng.314. [DOI] [PubMed] [Google Scholar]
- 13.Samani NJ, et al. Genomewide association analysis of coronary artery disease. N Engl J Med. 2007;357:443–53. doi: 10.1056/NEJMoa072366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Denny JC, et al. Systematic comparison of phenome-wide association study of electronic medical record data and genome-wide association study data. Nat Biotechnol. 2013;31:1102–10. doi: 10.1038/nbt.2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aguet F, et al. Local genetic effects on gene expression across 44 human tissues. 2016 bioRxiv. [Google Scholar]
- 16.Global Lipids Genetics Consortium et al. Discovery and refinement of loci associated with lipid levels. Nat Genet. 2013;45:1274–83. doi: 10.1038/ng.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Manning AK, et al. A genome-wide approach accounting for body mass index identifies genetic variants influencing fasting glycemic traits and insulin resistance. Nat Genet. 2012;44:659–69. doi: 10.1038/ng.2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prokopenko I, et al. A central role for GRB10 in regulation of islet function in man. PLoS Genet. 2014;10:e1004235. doi: 10.1371/journal.pgen.1004235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wood AR, et al. Defining the role of common variation in the genomic and biological architecture of adult human height. Nat Genet. 2014;46:1173–86. doi: 10.1038/ng.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berndt SI, et al. Genome-wide meta-analysis identifies 11 new loci for anthropometric traits and provides insights into genetic architecture. Nat Genet. 2013;45:501–12. doi: 10.1038/ng.2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pattaro C, et al. Genetic associations at 53 loci highlight cell types and biological pathways relevant for kidney function. Nat Commun. 2016;7:10023. doi: 10.1038/ncomms10023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu JZ, et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet. 2015;47:979–86. doi: 10.1038/ng.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dastani Z, et al. Novel loci for adiponectin levels and their influence on type 2 diabetes and metabolic traits: a multi-ethnic meta-analysis of 45,891 individuals. PLoS Genet. 2012;8:e1002607. doi: 10.1371/journal.pgen.1002607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morris AP, et al. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat Genet. 2012;44:981–90. doi: 10.1038/ng.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maass PG, et al. PDE3A mutations cause autosomal dominant hypertension with brachydactyly. Nat Genet. 2015;47:647–53. doi: 10.1038/ng.3302. [DOI] [PubMed] [Google Scholar]
- 26.Kato N, et al. Trans-ancestry genome-wide association study identifies 12 genetic loci influencing blood pressure and implicates a role for DNA methylation. Nat Genet. 2015;47:1282–93. doi: 10.1038/ng.3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chung CM, et al. A genome-wide association study reveals a quantitative trait locus of adiponectin on CDH13 that predicts cardiometabolic outcomes. Diabetes. 2011;60:2417–23. doi: 10.2337/db10-1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shungin D, et al. New genetic loci link adipose and insulin biology to body fat distribution. Nature. 2015;518:187–96. doi: 10.1038/nature14132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lotta LA, et al. Integrative genomic analysis implicates limited peripheral adipose storage capacity in the pathogenesis of human insulin resistance. Nat Genet. 2016 doi: 10.1038/ng.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sakai T, Larsen M, Yamada KM. Fibronectin requirement in branching morphogenesis. Nature. 2003;423:876–81. doi: 10.1038/nature01712. [DOI] [PubMed] [Google Scholar]
- 31.Erler JT, et al. Lysyl oxidase is essential for hypoxia-induced metastasis. Nature. 2006;440:1222–6. doi: 10.1038/nature04695. [DOI] [PubMed] [Google Scholar]
- 32.Lee VS, et al. Loss of function mutation in LOX causes thoracic aortic aneurysm and dissection in humans. Proc Natl Acad Sci U S A. 2016;113:8759–64. doi: 10.1073/pnas.1601442113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo DC, et al. LOX Mutations Predispose to Thoracic Aortic Aneurysms and Dissections. Circ Res. 2016;118:928–34. doi: 10.1161/CIRCRESAHA.115.307130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hood JD, Cheresh DA. Role of integrins in cell invasion and migration. Nat Rev Cancer. 2002;2:91–100. doi: 10.1038/nrc727. [DOI] [PubMed] [Google Scholar]
- 35.Samson T, et al. The guanine-nucleotide exchange factor SGEF plays a crucial role in the formation of atherosclerosis. PLoS One. 2013;8:e55202. doi: 10.1371/journal.pone.0055202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Rijssel J, et al. The Rho-guanine nucleotide exchange factor Trio controls leukocyte transendothelial migration by promoting docking structure formation. Mol Biol Cell. 2012;23:2831–44. doi: 10.1091/mbc.E11-11-0907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Buul JD, et al. RhoG regulates endothelial apical cup assembly downstream from ICAM1 engagement and is involved in leukocyte trans-endothelial migration. J Cell Biol. 2007;178:1279–93. doi: 10.1083/jcb.200612053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zahedi F, et al. Dicer generates a regulatory microRNA network in smooth muscle cells that limits neointima formation during vascular repair. Cell Mol Life Sci. 2016 doi: 10.1007/s00018-016-2349-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ellerbroek SM, et al. SGEF, a RhoG guanine nucleotide exchange factor that stimulates macropinocytosis. Mol Biol Cell. 2004;15:3309–19. doi: 10.1091/mbc.E04-02-0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gerhardt T, Ley K. Monocyte trafficking across the vessel wall. Cardiovasc Res. 2015;107:321–30. doi: 10.1093/cvr/cvv147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Khera AV, Kathiresan S. Genetics of coronary artery disease: discovery, biology and clinical translation. Nat Rev Genet. 2017;18:331–344. doi: 10.1038/nrg.2016.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wain LV, et al. Novel insights into the genetics of smoking behaviour, lung function, and chronic obstructive pulmonary disease (UK BiLEVE): a genetic association study in UK Biobank. Lancet Respir Med. 2015;3:769–781. doi: 10.1016/S2213-2600(15)00283-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Loh PR, et al. Efficient Bayesian mixed-model analysis increases association power in large cohorts. Nat Genet. 2015;47:284–90. doi: 10.1038/ng.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–1. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Castel SE, Levy-Moonshine A, Mohammadi P, Banks E, Lappalainen T. Tools and best practices for data processing in allelic expression analysis. Genome Biol. 2015;16:195. doi: 10.1186/s13059-015-0762-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Stage 2 and Stage 3 data contributed by CARDIoGRAM Exome and CARDIoGRAMplusC4D investigators is available online (see URLs). The genetic and phenotypic UK Biobank data are available upon application to the UK Biobank. Genotype-Tissue Expression Project data is available online.