Abstract
Although mice models rank among the most widely used tools for understanding human genetics, biology, and diseases, differences between orthologous genes among species as close as mammals are possible, particularly in orthologous gene pairs in which one or more paralogous (i.e., duplicated) genes appear in the genomes of the species. Duplicated genes can possess overlapping functions and compensate for each other. The retinoblastoma gene family demonstrates typical composite functionality in its three member genes (i.e., RB1, RB2/P130, and P107), all of which participate in controlling the cell cycle and associated phenomena, including proliferation, quiescence, apoptosis, senescence, and cell differentiation. We analyzed the role of the retinoblastoma gene family in regulating senescence in mice and humans. Silencing experiments with each member of the gene family in mesenchymal stromal cells (MSCs) and fibroblasts from mouse and human tissues demonstrated that RB1 may be indispensable for senescence in mouse cells, but not in human ones, as an example of species specificity. Furthermore, although RB2/P130 seems to be implicated in maintaining human cell senescence, the function of RB1 within any given species might differ by cell type, as an example of cell specificity. For instance, silencing RB1 in mouse fibroblasts induced a reduced senescence not observed in mouse MSCs. Our findings could be useful as a general paradigm of cautions to take when inferring the role of human genes analyzed in animal studies and when examining the role of the retinoblastoma gene family in detail.
Introduction
The mouse is the most widely used model organism for elucidating human genetics, biology, and disease. As a case in point, the function of a gene in humans can be deduced by studying the corresponding orthologous gene in mice. However, findings reveal a divergence between orthologous genes among species as close as mammals such as mice and humans [1], especially between orthologous gene pairs for which one or more paralogous genes appear in the genomes of species under investigation.
Within a genome, paralogous genes that form a gene family are due to the duplication of events involving a common ancestor gene. The duplication and divergence of a gene with more than one molecular activity can prompt the subfunctionalization of some activities or the sharing of others (i.e., overlapping), if not both. At the same time, one of the paralogous genes might acquire a new function [1]. In such cases, inferring gene function in humans by means of knockout or in situ experiments with mice can pose quite a challenge.
In response, the retinoblastoma gene family might exemplify possible orthologous gene divergence between humans and mice. The family comprises three members—RB1, RB2/P130, and P107—that regulate several aspects of cell life, including cell cycle, apoptosis, senescence, and differentiation [2], [3], [4]. Studies with mice have suggested that retinoblastoma family proteins show overlapping functions, while an initial knockout analysis suggested that Rb2/p130 and P107 played an ancillary role. Indeed, Rb1−/− mice of 129Sv or C57BL6 strains showed embryo lethality, whereas mice deficient in Rb2/p130 or P107 developed normally without any overt adult phenotype. Such genetic redundancy and functional compensation have been challenged by the observation that in mice of the BALB/c strain, the loss of Rb2/p130 induced the death of embryos, whereas P107−/− mice showed severe postnatal alterations of the phenotype [3], [5], [6].
To add to that complexity, functional compensation within a gene family can occur differently depending on whether the germline versus conditional loss-of-function mouse mutants are used. For example, embryonic fibroblasts obtained from mice with a permanent loss of the Rb1 gene maintain their capacity to arrest in a quiescent state when cultivated without growth factors. Conversely, mouse fibroblasts can lose their capacity to arrest in G0 when the Rb1 gene is acutely deleted. Indeed, the germline loss of a function of a gene with a critical function could force selection whereby another member of the gene family acquires a function to compensate for the loss of activity. In acute conditions, however, such compensation might not occur [7].
With pioneering analyses in mice, several other studies have demonstrated functional differences among proteins in the retinoblastoma family [4], [8], [9]. As a result, it is currently understood that the role of RB1, RB2/P130, and P107 depends on several parameters, including the animal species under investigation, the cell type, and the status of the cell as a stem cell, progenitor, or differentiated cell [10], [11], [12], [13].
Despite the above reported studies, following the identification of gene function in mice or in a specific cell type or tissue, if not both, researchers could perform an extrapolation to indicate the presence of a given gene's activity in humans, which could in turn negatively affect findings applicable to treating human diseases. Indeed, animal experiments often do not translate into replications in human clinical trials because they are poorly designed, conducted, or analyzed [14]. Such a concern should be taken seriously since misleading concepts can occur for well-known, thoroughly studied genes as well, including those belonging to the retinoblastoma family.
In light of that scenario, we investigated the role of retinoblastoma gene family members in regulating senescence. Initially, we focused our attention on human bone marrow mesenchymal stromal cells (MSCs). MSCs contain a subpopulation of stem cells able to differentiate in mesodermal derivatives (e.g., adipocytes, chondrocytes, and osteocytes). MSCs also contribute to the homeostasis and repair of several tissues and organs, and for that reason, MSCs continue to be scrutinized in several clinical trials [15]. The senescence of MSCs can be very deleterious since it greatly impairs tissues' renewal. At the same time, senescence promotes protective anticancer mechanisms that prompt the growth arrest of tumor cells [2], [16].
In a previous study, we demonstrated that acute silencing of the RB1 gene in human MSCs renders cells prone to DNA damage with the gradual adoption of a senescent phenotype. No cell growth deregulation or resistance to cell cycle exit was observed, despite its occurrence in other cellular systems such as in mouse embryonic fibroblasts with the inactivated Rb1 gene [3], [7]. Our finding, along with research showing that, in several cellular models, senescence-inducing signals engage either the P53 or RB1-P16 pathway, prompted us to hypothesize a complex role for retinoblastoma family members in regulating senescence. In particular, we hypothesized that the role of each member may be specific to cell type and species. To that end, we compared the biological effects of RB1, RB2/P130, and P107 silencing in human and mouse MSCs and fibroblasts, the latter chosen as a reference model given ample literature addressing the consequences of retinoblastoma gene inactivation in them.
Materials and Methods
Human MSC Cultures
After obtaining bone marrow from healthy donors who had provided their informed consent, we separated cells on a Ficoll density gradient (GE Healthcare, Milano Italy) and collected and washed the mononuclear cell fraction in phosphate-buffered saline (PBS). We seeded 1-2.5 × 105 cells/cm2 in modified Eagle's medium (alpha-MEM) containing 10% fetal bovine serum (FBS) and basic fibroblast growth factor (bFGF). After 72 hours, we discarded nonadherent cells and cultivated adherent ones to confluency. We then further propagated cells for the assays reported in what follows. Cells were used at passage 3 or 4.
Mouse MSC Cultures
We harvested MSCs from the bone marrow of femurs and tibias of adult C57BL/6J mice by inserting a 21-gauge needle into the shaft of the bone and flushing it with alpha-MEM. Cells from one animal were plated onto two 100-mm dishes with alpha-MEM containing 10% FBS and bFGF. After 24 to 48 hours, we discarded nonadherent cells and twice washed adherent cells with PBS. We then incubated cells for 7 to 10 days in proliferating medium in order to reach confluence and propagated them for additional experiments. Cells were used at passage 3 or 4.
Human and Fibroblast Cultures
As a source for fibroblast isolation, we used human dermis obtained from disposable tissues of patients undergoing surgical operations. All patients provided their informed consent. We used mice's tails to isolate murine fibroblasts.
We treated human and mouse specimens with a collagenase II solution (1 mg/ml) for several hours at 37°C and centrifuged samples at 1000g for 5 minutes at room temperature. We discarded supernatants, washed pellets several times with PBS, and filtered them through culture meshes (40 μm). We next plated cells 5000/cm2 in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum. We then expanded cells for experiments described in the results. Cells were used at passage 3 or 4.
Silencing with shRNA
We obtained shRNAs targeting the human and mouse RB1, RB2/P130, and P107 mRNA, as well as control shRNAs from Sigma-Aldrich (St. Louis, MO) that is distributor of Broad Institute RNAi consortium (Cambridge, MA). The shRNA targeting coding sequence region (CDS) of human RB1 mRNA was CCGGCCGTGGATTCTGAACGTACTTCTCGAGAAGTACGTTCAGAATCCACGGTTTTTG (code TRCN0000042543). The shRNA against CDS of human RB2/P130 was CCGGCCAGAACATCATGCGTTGTTACTCGAGTAACAACGCATGATGTTCTGGTTTTTG (code TRCN0000071277). The shRNA targeting human P107 CDS was GTACCGGATCTTTGCCAATGCTATAATGCTCGAGCATTATAGCATTGGCAAAGATTTTTTTG (code TRCN0000218550).
The shRNA against the 3′UTR region of mouse RB1 mRNA was CCGGCAGAGATCGTGTATTGAGATTCTCGAGAATCTCAATACACGATCTCTGTTTTTG (code TRCN0000010419). The 3′UTR of mouse RB2/P130 mRNA was targeted with CCGGGCTGAGAGAAATATGGAACTTCTCGAGAAGTTCCATATTTCTCTCAGCTTTTTG shRNA (code TRCN0000039923). ShRNA against 5′UTR of mouse P107 mRNA was CCGGGCACAGGCTAATGTGGAGTATCTCGAGATACTCCACATTAGCCTGTGCTTTTTG. The control shRNA was obtained from Sigma-Aldrich (code SHC001). It was a negative control containing a sequence that did not target any known mammalian genes.
The selected shRNAs were inserted into lentiviral particles. To generate knockdown cells, we produced lentiviral particles as described by the Broad Institute (http://www.broadinstitute.org/genome_bio/trc/publicProtocols.html). Briefly, we transfected 1 × 106 293FT cells (Invitrogen, Waltham, CA) with 2.25 μg of second-generation packaging plasmid, 0.75 μg of PMD2G envelop plasmid, and 3 μg of a selected pLKO.1 vector using 30 μl of Fugene HD (Roche, Mannheim, Germany) on 100-mm plates. We generated polyclonal populations of knocked down cultures by infection with 1 MOI of shRNA lentiviral particles. At 3 days postinfection, we selected cells with 2 μg/ml puromycin (Sigma-Aldrich, St. Louis, MO) for 1 week. At that time point, cells had expanded to 70% to 80%, and unless stated otherwise, we performed all experiments soon afterward.
Silencing with siRNAs
We obtained validated siRNAs targeting human or mouse RB1, RB2/P130, P16, and P27 mRNAs from Santa Cruz Biotechnology (Dallas, TX). The control siRNA had a sequence that did not target any known mammalian genes.
We incubated human MSCs with 100 pMoli of siRNA against RB1 or with control siRNA (SCR) and, at the same time, added either siRNA against RB2/P130, P27, or P16. We incubated the mix of siRNAs in Lipofectamine 2000 (Invitrogen, Milano, Italy) according to the manufacturer's instructions and collected cells for further analysis 48 hours posttreatment. We incubated mouse MSCs with 100 pMoli of siRNA against RB2/P130 or with control siRNA (scr) and, at the same time, added either siRNA against RB1, P27, or P16. We incubated the mix of siRNAs in Lipofectamine 2000 (Invitrogen, Milano, Italy) according to the manufacturer's instructions. We collected cells for further analysis at 48 hours posttreatment.
In Situ Senescence-Associated Beta-Galactosidase Assay
We calculated the percentage of senescent cells by the number of blue beta-galactosidase–positive cells out of at least 500 cells in different microscope fields, as previously reported [17].
Detection of Senescence-Associated Heterocromatic Foci (SAHFs)
Cells grown on coversplips were fixed with 4% paraformaldeyhe and then stained with DAPI for 30 minutes. Cells were observed through a fluorescence microscope (Leica Microsystem, Milano, Italy). The intensity of DAPI was acquired with a CCD camera and analyzed with Quantity One 1-D analysis software (Bio-Rad Laboratories, Hercules, CA). We calculated the sum of the fluorescent pixel values of DAPI-positive cells and then determined the average fluorescent pixel intensity, which was expressed in arbitrary units. For every experimental condition, staining intensity was determined for 200 cells.
In Vitro Follow-Up of Senescence Process
After treating cultures with reagents for senescence-associated beta-galactosidase assay, we washed samples with PBS, incubated them with blocking solution (5% BSA, 0.3% Triton X100) for 1 hour, and then incubated them overnight in PBS containing diluted primary antibodies—namely, rabbit polyclonal antibodies anti-RB1 (Santa Cruz Biotech) and anti-RB2/P130 (Abcam, Cambridge UK). We used mouse monoclonal anti-RPS6 (Cell Signaling, Danvers, MA) and goat polyclonal antibodies anti-Ki67 (Santa Cruz Biotech). Following primary antibody incubation, we washed samples with PBS and incubated them with secondary antibodies (i.e., AMCA [anti-rabbit], TRITC [antigoat], and FITC [anti-mouse]). We then analyzed samples using fluorescence or a light microscope (Leica Microsystem, Milano Italy).
RNA Extraction, RT-PCR, and Real-Time PCR
We extracted total RNA from cell cultures using Omnizol (EuroClone, Pero Italy), according to the manufacturer's protocol, and measured mRNA levels by RT-PCR amplification.
We used sequences of mRNAs from the Nucleotide Data Bank (National Center for Biotechnology Information, Bethesda, MA) to design primer pairs for real-time RT-PCRs (Primer Express; Applied Biosystems, Milano, Italy); primer sequences are available upon request. We used appropriate regions of HPRT and/or GAPDH cDNA as controls and ran real-time PCR assays on an Opticon 4 machine (MJ Research, Waltham, MA). We carried out reactions according to the manufacturer's instructions using a SYBR green PCR master mix and used the 2−ΔΔCT method as a relative quantification strategy for quantitative real-time PCR data analysis.
Western Blotting
We lysed cells in a buffer containing 0.1% Triton for 30 minutes at 4°C. A quantity of 10 to 40 μg of each lysate was electrophoresed in a polyacrylamide gel and electroblotted onto a nitrocellulose membrane. We used all primary antibodies—that is, rabbit polyclonal antibodies anti-RB1, anti-P107, and anti-P21 (Santa Cruz Biotech), anti-RB2/P130 (Abcam), and anti-P53 and anti-p27 (ProteinTech, Chicago, IL, USA)—according to the manufacturers' instructions. We also used mouse monoclonal primary antibody anti-MDM2, anti-ARF (Santa Cruz Biotech), and anti-P16 (Abcam).
We detected immunoreactive signals with a horseradish peroxidase–conjugated secondary antibody (Santa Cruz Biotech) and reacted with ECL plus reagent (GE Healthcare). We conducted a semiquantitative analysis of protein levels using a gel documentation system (Bio-Rad, Milano, Italy).
Statistical Analysis
We evaluated statistical significance using analysis of variance, followed by a Student's t and Bonferroni's tests. For data with continuous outcomes, we used mixed-model variance analysis and, in any case, analyzed all data with GraphPad Prism version 5.01 (GraphPad, La Jolla, CA).
Bioinformatic Analysis
We identified human and mouse promoters belonging to RB1, RB2/P130, and P107 according to the Transcriptional Regulatory Element Database (https://cb.utdallas.edu/cgi-bin/TRED/tred.cgi?process=searchPromForm) and its instructions. We evaluated identified promoter regions with AliBaba2 (http://www.gene-regulation.com), a pattern-based program for predicting transcription factor binding sites in DNA sequences that uses the set of binding sites from TRANSFAC Public 6.0.
Results
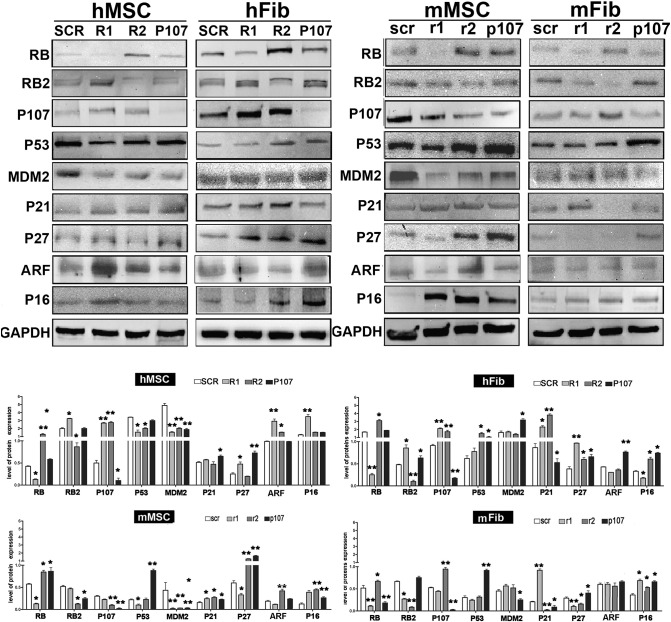
We silenced retinoblastoma genes in human and mice cells by use of shRNA technology. The selected shRNAs effectively silenced and induced a decrease of their target mRNAs in all cell types, as detected by RT-PCR (Supplementary Figure 1). We further evaluated silencing by determining the protein levels of target genes (Figure 1).
Figure 1.
Western blot analysis following silencing experiments.
RB1, RB2/P130, and P107 were silenced in human and mouse cells with specific shRNAs. The panel shows the expression levels of several proteins following treatments of cells with shRNAs. GAPDH protein was used as loading control.
R1, R2, and P107 stand for shRNAs against human RB1, RB2/P130, and P107 mRNAs, respectively. The control shRNA with a scrambled sequence was named SCR. Silencing experiments were carried out in human MSCs (hMSC) and fibroblasts (hFib).
Mouse MSCs (mMSC) and fibroblasts (mFib) were treated with shRNAs that specifically targeted mouse mRNAs of RB1, RB2/P130, and P107 (r1, r2, and p107, respectively). Control shRNAs for mouse cells was named scr.
The histogram shows the quantitative evaluation of Western blot bands. For every experimental condition, the mean expression values (±SD, n = 3) are indicated. The expression levels of the indicated proteins were evaluated in RB1- or RB2/P130- or P107-silenced cells and were compared with those of cells treated with control shRNAs (*P < .05; **P < .01).
Retinoblastoma Proteins and Senescence
The silencing of each member of the retinoblastoma family produced different effects on senescence depending on cell type and animal species.
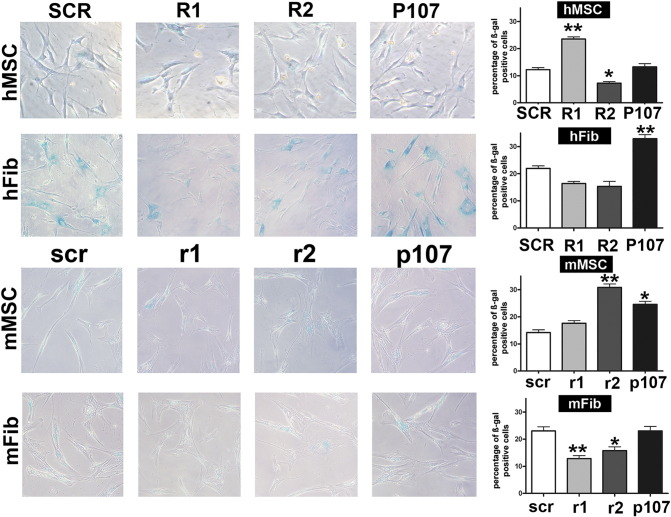
We employed in situ acid-beta-galactosidase assay to detect senescence in lentiviral transduced cells. Silencing RB1 induced a significant increase in the percentage of senescent cells in human MSC cultures yet no effect in corresponding murine cells. By contrast, in mouse fibroblasts, the downregulation of RB1 occurred with decreased senescence. Silencing RB2 triggered an increase of senescence in mouse MSCs and a diminution of the same phenomenon in human MSCs. Lastly, P107 silencing induced the upregulation of senescence in both mouse MSCs and human fibroblasts (Figure 2A).
Figure 2.
(A) Senescence levels in cells with silenced retinoblastoma proteins.
The presence of senescent cells was evaluated in human and mouse cells following the silencing of RB1, RB2/P130, and P107 with specific shRNAs. The picture shows representative microscopic fields of senescence-associated beta-galactosidase–positive cells in the different experimental conditions. The histograms show the percentage of senescent cells in MSCs and fibroblasts from both human and mouse origin (hMSC, mMSC, hFib, and mFib, respectively). R1, R2m and P107 stand for shRNAs against human RB1, RB2/P130, and P107 mRNAs, respectively. The control shRNA with a scrambled sequence was named SCR. The shRNAs against the corresponding mouse mRNAs were indicated as r1, r2, and p107, respectively. Control shRNAs for mouse cells was named scr. Data are expressed with standard deviation (n = 3, *P < .05, **P < .01).
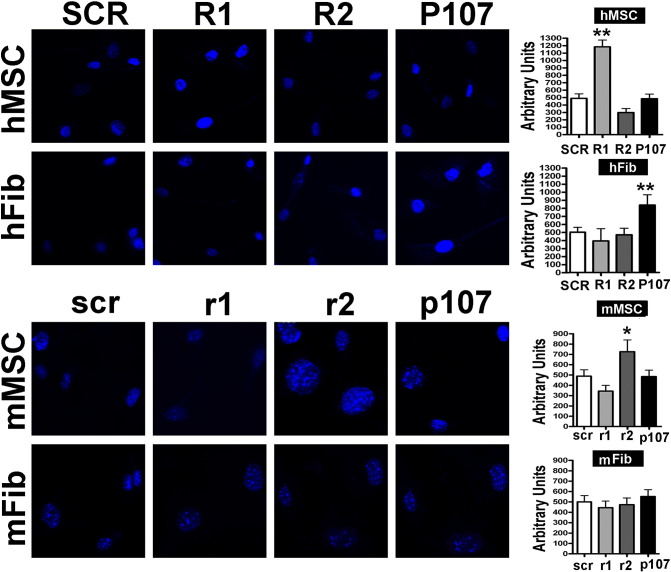
(B) DAPI staining.
Fluorescence photomicrographs show cells stained with DAPI (blue). Representative microscopic fields are shown. The graph shows the degree of DAPI staining. For each positive cell, the DAPI intensity was acquired with a CCD camera and analyzed with Quantity One 1-D analysis software (Bio-Rad Laboratories). We calculated the sum of the fluorescent pixel values of DAPI-positive cells and then determined the average fluorescent pixel intensity, which was expressed in arbitrary units. For every experimental condition, staining intensity was determined for 200 cells. Data are expressed with standard deviation (n = 3, *P < .05, **P < .01).
In many in vitro and in vivo models, senescent cells show heterochromatic nuclear foci containing silenced genes. For this reason, we evaluated these SAHFs with DAPI staining [18]. Indeed, increase in senescence, as detected with the acid-beta-galactosidase assay, was associated with an augment of SAHFs (Figure 2B).
We decided to extend our analysis to other proteins that play a key role in senescence in order to find a molecular algorithm that could be associated with different outputs induced by silencing retinoblastoma proteins in human and murine cells. In particular, we analyzed the expression of P53, MDM2, and three cyclin kinase inhibitors (CKIs): P21, P27, P16, ARF. In human cells, increased senescence seemed to be associated with the upregulation of P16 and P27 in the presence of RB2/P130 (Table 1). Indeed, in human MSCs with silenced RB1 and in human fibroblasts with silenced P107, we detected an increase of senescence related to augmented P27 and P16 levels. The increase of the two CKIs was not enough to sustain senescence; in fact, their upregulation in the absence of RB2/P130 induced either a decrease or no change in senescence, as observed in human MSCs and fibroblasts, respectively (Table 1).
Table 1.
Changes in Protein Expression Following Silencing Experiments
| hMSC | R1 | R2 | P107 | hFIB | R1 | R2 | P107 | ||
| Senescent | ↑ | ↓ | ≈ | Senescent | ↓ | ≈ | ↑↑ | ||
| RB | ⊥ | ↑↑ | ≈ | RB | ⊥ | ↑ | ≈ | ||
| RB2 | ↑ | ⊥ | ≈ | RB2 | ↑ | ⊥ | ↑ | ||
| P107 | ↑↑ | ↑↑ | ⊥ | P107 | ↑↑ | ↑↑ | ⊥ | ||
| P53 | ↓ | ↓ | ≈ | P53 | ≈ | ↑ | ↑ | ||
| MDM2 | ↓↓ | ↓↓ | ↓↓ | MDM2 | ≈ | ≈ | ↑ | ||
| P21 | ≈ | ≈ | ↑ | P21 | ↑ | ↑↑ | ↓ | ||
| P27 | ↑ | ≈ | ↑↑ | P27 | ↑↑ | ↑ | ↑ | ||
| ARF | ↑↑ | ↑ | ≈ | ARF | ≈ | ≈ | ↑ | ||
| P16 | ↑↑ | ≈ | ≈ | P16 | ↓ | ↑ | ↑ | ||
| mMSC | R1 | R2 | P107 | mFIB | R1 | R2 | P107 | ||
| Senescent | ≈ | ↑↑ | ↑ | Senescent | ↓ | ≈ | ≈ | ||
| RB | ⊥ | ↑ | ↑ | RB | ⊥ | ↑ | ↓ | ||
| RB2 | ≈ | ⊥ | ↓ | RB2 | ↓ | ⊥ | ≈ | ||
| P107 | ≈ | ↓↓ | ⊥ | P107 | ≈ | ↑↑ | ⊥ | ||
| P53 | ↓ | ≈ | ↑↑ | P53 | ≈ | ≈ | ↑↑ | ||
| MDM2 | ↓↓ | ↓↓ | ↓↓ | MDM2 | ≈ | ≈ | ↓ | ||
| P21 | ↑ | ↑ | ↑ | P21 | ↑↑ | ↓↓ | ↓ | ||
| P27 | ↓ | ↑↑ | ↑↑ | P27 | ↓↓ | ↓ | ↑ | ||
| ARF | ↓ | ↑ | ≈ | ARF | ≈ | ≈ | ≈ | ||
| P16 | ↑↑ | ↑↑ | ↑↑ | P16 | ↑ | ↑ | ↑ |
RB1, RB2/P130, and P107 were silenced in human and mouse cells with specific shRNAs.
In the different experimental conditions, the variation in the percentage of senescence is indicated. Variations are compared with controls. The table also shows changes in protein levels as determined by quantitative analysis (see also Supplementary Figure 2).
In the different experimental conditions, the variation in the percentage of senescence is indicated. Variations are compared with controls. The table also shows changes in protein levels as determined by quantitative analysis (see also Supplementary Figure 2).
The protein levels of RB1- or RB2/P130- or P107-silenced cells were compared with those of cells treated with control shRNAs. ↑ or ↑↑indicates significant or highly significant upregulation compared to control. ↓ or ↓↓ represents downregulation. ≈ means not significant changes. ⊥ means that, in a given experimental condition, the indicated gene was silenced.
R1, R2, and P107 stand for shRNAs against human RB1, RB2/P130, and P107 mRNAs, respectively. Silencing experiments were carried out in human MSCs (hMSC) and fibroblasts (hFib).
Mouse MSCs (mMSC) and fibroblasts (mFib) were treated with shRNAs that specifically targeted mouse mRNAs of RB1, RB2/P130, and P107 (r1, r2, and p107, respectively).
In murine cells, increased senescence was associated with the upregulation of P16 in the presence of RB1 (Table 1). As matter of fact, in mouse MSCs with silenced RB2/P130 or P107, we observed a significant increase in the percentage of senescent cells coupled with the upregulation of P16 and the presence of RB1. The sole increase in P16 is inadequate to induce senescence. Indeed, in murine fibroblasts with silenced RB1, we detected a decrease in senescence even in the presence of enhanced P16 expression (Table 1).
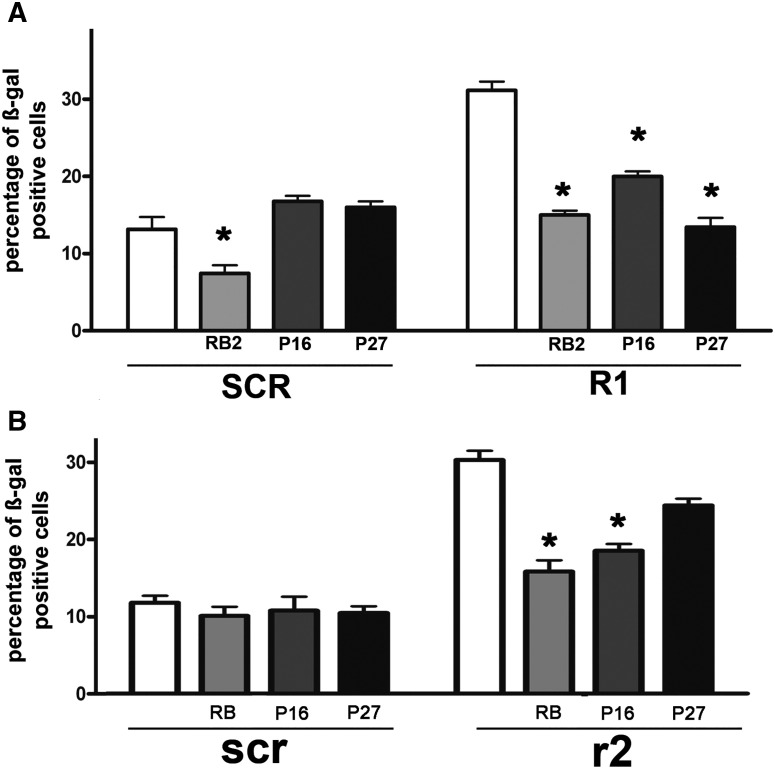
We next tested the accuracy of the identified molecular algorithms associated with senescence: RB2/P130-P27-P16 for human cells and RB1-P16 for murine ones. To that end, we focused on human MSCs with silenced RB1 and mouse MSCs with silenced RB2/P130 since we had detected increased senescence in both conditions.
Using siRNA technology, we downregulated in human MSCs the expression of RB1 and, at the same time, of RB2/P130 or P27 or P16. Silencing each CKI or RB2/P130 abrogated the increase in senescence (Figure 3), and the use of siRNAs instead of shRNAs effectively excluded any overlooked side effects associated with lentivirus vectors.
Figure 3.
Identification of senescence-associated pathways in human and mouse cells.
(A) The histogram shows the percentage of senescent cells following treatment with siRNAs against RB2/P130 or P27 or P16 in human MSCs having silenced RB1 (R1) or in control sample (SCR).
(B) The histogram shows the percentage of senescent cells following treatment with siRNAs against RB1 or P27 or P16 in mouse MSCs having silenced RB2/P130 (r2) or in control sample (scr). Data are expressed with standard deviation (n = 3, *P < .05).
In the same manner, we used siRNAs to shut down RB2 and RB1, P16, or P27 in mouse MSCs, and in like fashion, the inhibition of either RB1 or P16 expression negatively affected the onset of senescence (Figure 3).
RB1’s Different Regulation During Acute Senescence in Human and Mouse Cells
The identified pathways clearly indicate that RB1 plays a role in senescence depending on cell type and animal species. In particular, its expression is indispensable for senescence in murine cells but not human ones. Paradoxically, its silencing in human MSCs induced senescence. The phenomena may relate to compensation events that occurred with silencing experiments and may not reflect the role played by RB1. To confirm that RB1 plays a different role in senescence in human and murine cells, we analyzed its expression during acute senescence induced by peroxide hydrogen in cells with the potential to express all retinoblastoma proteins.
To that end, we selected mouse fibroblasts as a paradigmatic cell type that has RB1 associated with senescence. We also chose human MSCs given evidence that senescence can occur in the absence of RB1. We induced senescence in both cell types by incubation with peroxide hydrogen for 30 minutes. After 3 and 24 hours, we collected cells and performed immunocytochemistry to evaluate RB1 expressions and its cellular localization. We also determined RB2/P130 expression since silencing experiments in human cells proved that the protein might play a role in senescence. We selected an early and a late phase of senescence to discriminate events in cycling and permanently arrested cells. Indeed, early-senescent cells are still cycling (i.e., positive for the expression of Ki67 antigen), and the irreversible exit from the cell cycle with the onset of the frank senescent phenotype occurs later.
In mouse fibroblasts, RB1 expression was evident in the nuclei of early senescent cells 3 hours after peroxide treatment and persisted in late phase 24 hours following incubation with the stressor (Figure 4). By contrast, we hardly detected any expression of RB2/P130 in either early or late senescent cells (Figure 4). In human MSCs, RB1 expression was present in early senescent cells but completely disappeared later. Conversely, RB2/P130 was expressed only in the nuclei of late-senescent cells (Figure 4).
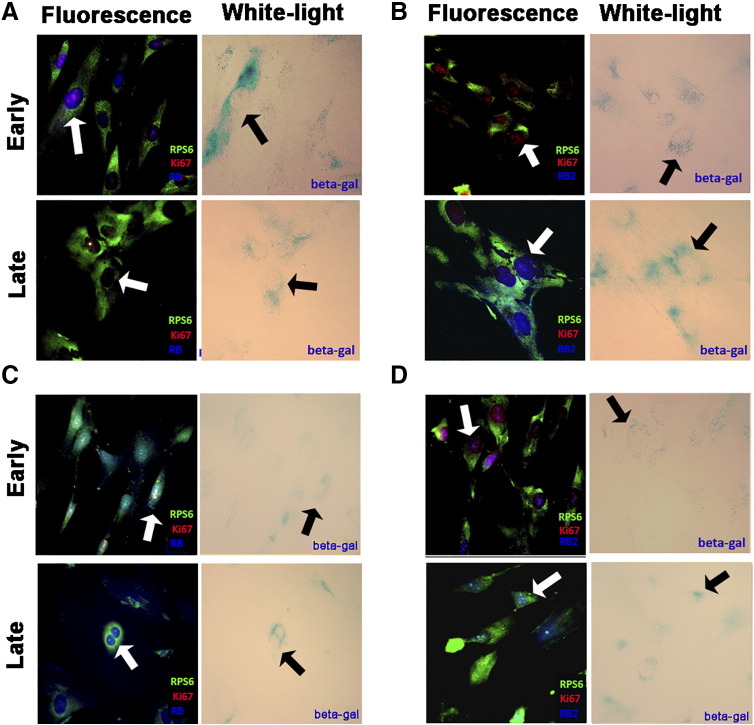
Figure 4.
Follow-up of acute senescence in human and mouse cells.
Human MSCs and mouse fibroblasts were treated with peroxide hydrogen to induce senescence. We collected cells and performed immunocytochemistry to evaluate RB1 and RB2/P130 expression 3 and 24 hours following stressor treatment. The two time points were indicated as early and late phase of senescence.
(A) Fluorescence photomicrographs show the merging of cells stained with anti-RPS6 (green), anti–Ki-67 (red), and RB1 (blue) in human MSCs during early and late senescence. Light microscopy pictures show the same fields stained to detect senescence-associated beta-galactosidase. In early senescence, the arrow indicates a senescent cell that is Ki67(+) and RB1(+). In late senescence, the arrows indicate a senescent cell that is negative for Ki67 and RB1. RPS6 stands for 40S ribosomal protein S6 and was used to detect cells.
(B) Photomicrographs show the merging of cells stained with anti-RPS6 (green), anti–Ki-67 (red), and RB2 (blue) in human MSCs during early and late senescence. Light microscopy pictures show the same fields stained to detect senescence-associated beta-galactosidase. In early phase, the arrow indicates a senescent cell that is Ki67(+) and RB2(−). In late senescence, the arrows indicate a senescent cell that is Ki67(−) and RB2 (+).
(C and D) Experiments carried out in mouse fibroblasts. The panels show the same staining we reported in panels A and B, respectively. In panel C (early), the arrow indicates a senescent cell that is positive for Ki67 and RB1. In late phase, the cells become Ki67(−) but remained RB1 positive (see arrow).
In panel D (early), the cells are Ki67 (+) and RB2 (−). In the late phase, cells were Ki67(−) and became RB2 (+) (See the arrows).
We hypothesized a different role for RB1 in human and mouse cells by globally analyzing RB1 and RB2/P130 during acute senescence induced by peroxide treatment and by evaluating senescence in cells with silenced components of the retinoblastoma family. In mouse cells, RB1 seems to play a role both in cell cycle exit events and in maintaining the senescence phenotype. In human cells, RB1 might play a role in cell cycle exit but is clearly dispensable for senescence. The senescence process appears to also be supported by RB2/P130.
Transcriptional and Posttranscriptional Events Regulating Activities of Retinoblastoma Genes
Our experiments showed that in two mammalian species (i.e., mice and humans), an orthologous gene (i.e., RB1) can execute different tasks in the same cell type. One possible explanation involves factors that might regulate gene expression at the transcriptional level and determinants that might act posttranscriptionally. For example, the upstream promoter region of human and mouse RB1 genes differs in the number and type of transcription factor binding sites. We used the Transcriptional Regulatory Element Database to identify the human and mouse RB1 promoter and selected a 1000-bp region spanning from −700 to +299 of the RB1 gene and considered it to be the 5′-flanking region, which we analyzed using AliBaba2 in search of transcription factors that might participate in regulating its expression (Supplementary Figure 2). Our analysis found 40 segments in the human sequence identified as potential binding sites. In the mouse, by contrast, we identified only 16 potential sites in the same region (Supplementary Figure 2). We used the same approach to compare promoters of human and mouse RB2/P130 and P107 genes and identified differences in the number and type of transcription factor binding sites (Supplementary Figure 2). Differences in transcription factors that could drive the expression of an orthologous gene in two species might account for different responses to environmental and internal cues and hence the involvement of the gene in different duties in the two organisms.
Posttranscription factors might also contribute to differences between roles played by an orthologous gene in several species. For example, phosphorylation events of the RB1 protein are key determinants of its functions. We used PhosphoSitePlus software (http://www.phosphosite.org/homeAction.action) to compare potential phosphorylation sites in human and mouse RB1 protein (Supplementary Figure 3), and our analysis revealed six amino acid residues that can be phosphorylated only in human protein. Conversely, in mouse RB1, there are two potentially phosphorylated residues not present in human protein (Supplementary Figure 3). Differences in phosphorylation could account for the diversity in the performed functions. The analysis of RB2/P130 and P107 proteins with PhosphoSitePlus also revealed divergences in amino acid phosphorylation between human and mouse orthologous proteins (Supplementary Figure 3).
We also observed variation in the functions of the RB1 gene in different cell types of the same organism. In that same case, factors regulating gene expression at the transcriptional and posttranscriptional levels could also play a role. For example, a cell type might exhibit a group of transcription factors that allows the expression of RB1 protein in response to a given cue. In another cell type, those factors are not present, and accordingly, the RB1 protein cannot be expressed as consequences of that cue. That dynamic might also apply to different kinase proteins that regulate the several phosphorylation sites on retinoblastoma proteins.
Discussion
The retinoblastoma gene family represents a typical example of composite functionality. Its three members—namely, RB1, RB2/P130, and P107—play a major role in controlling the cell cycle and its associated phenomena, including proliferation, quiescence, apoptosis, senescence, and cell differentiation. Accordingly, we examined the role of the retinoblastoma gene family in regulating senescence in mice and humans.
Silencing experiments of each member of the family in MSCs and fibroblasts from mouse and human tissues showed that RB1 may be indispensable for senescence in mouse cells, albeit not in human ones, as an example of species specificity (Figures 2 and 3). That hypothesis took further support from the evaluation of RB1 expressions during acute senescence induced by peroxide hydrogen treatment in mouse fibroblasts and human MSCs (Figure 4). By contrast, RB2/P130 appears to be involved in maintaining senescence in human cells (Figure 2, Figure 3, Figure 4). Our finding is in agreement with other investigations showing that, in human diploid fibroblasts, RB2/P130 is the dominant protein of the retinoblastoma family leading to replicative and accelerated senescence [19], [20]. The observation that, in human cells, RB2/P130 was highly expressed in late senescence (Figure 4) is in accordance with studies of Jackson and Pereira-Smith [21]. They evidenced that, in human breast cancer cells, the doxorubicin-induced senescence is associated with a persistent permanence (up to 8 days) of RB2/P130 on the promoters of genes involved in cell cycle regulation. They concluded that their results showed a mechanistic function of RB2/P130 in long-term maintaining of senescence.
In general, we found that RB1-P16 pathways in mouse cells are associated with senescence, whereas the RB2/P130-P27-P16 may regulate that process in human cells. It should be underlined that, besides the well-known role of P16 in senescence, there are several reports evidencing a fundamental contribute of P27 in regulation of senescence process [22], [23], [24].
Beside interspecies variations in the functions played by retinoblastoma proteins, a further complexity arose from the circumstance that their role within a given animal species can differ in distinct cell types as an example of cell specificity. For instance, silencing RB1 in mouse fibroblasts reduced senescence, an effect that was not detected in mouse MSCs.
All these circumstances are not limited to role exerted by retinoblastoma proteins in senescence, but all their functions may be cell specific and species specific. For example, cardiac myocytes of RB1-deficient mice did not show significant alteration in cell cycle distribution and apoptosis [25]. At the opposite, Rb1 silencing in mouse neuroblasts induced massive apoptosis [26]. Moreover, in another species (rat), the survival of neurons was mainly regulated by Rb2/p130 [27].
Conclusions
The assumption that orthologous genes might play the same role in different species is the basis of comparative genomics and the use of model organisms to study human biology and diseases. However, the validity of that hypothesis needs to derive from systemic examination, not only from anecdotal investigation, which is particularly true for family genes, in which more paralogous genes are present in the genome. We demonstrated that RB1 might play a different role in senescence depending on cell type and species, which could be useful as a general paradigm for cautions to take when inferring the role of human genes in animal studies. Furthermore, our finding might be interesting to clinical practitioners since we identified drivers of senescence in human MSCs under scrutiny in several clinical trials. For their safe and effective use, the senescence process needs to be under strict control.
The following are the supplementary data related to this article.
RT-PCR analysis following silencing experiments. RB1, RB2/P130, and P107 were silenced in human and mouse cells with specific shRNAs The histograms shows the expression levels of silenced genes following treatments of cells with shRNAs. The mRNA levels of RB, RB2/P130, and P107 are expressed as arbitrary units. Significant differences with respect to control in relative gene expression are indicated with *P < .05 or **P < .01. R1, R2, and P107 stand for shRNAs against human RB1, RB2/P130, and P107 mRNAs, respectively. The control shRNA with a scrambled sequence was named SCR. Silencing experiments were carried out in human MSCs (hMSC) and fibroblasts (hFib). Mouse MSCs (mMSC) and fibroblasts (mFib) were treated with shRNAs that specifically targeted mouse mRNAs of RB1, RB2/P130, and P107 (r1, r2, and p107, respectively). Control shRNAs for mouse cells was named scr.
Putative transcription factors binding sites on promoters of human and mouse retinoblastoma proteins. One thousand–base pair regions of human and mouse promoters (−700+299) belonging to RB1, RB2/P130, and P107 were identified by Transcriptional Regulatory Element Database (https://cb.utdallas.edu/cgi-bin/TRED/tred.cgi?process=searchPromForm). The retrieved accession numbers are shown in the picture. Identified promoter regions were evaluated with ALIBABA2 (http://www.gene-regulation.com), which is a pattern-based program for predicting transcription factor binding sites (TFBS) in DNA sequences. It uses the set of binding sites from TRANSFAC Public 6.0. In the pictures are shown the putative binding sites found on the genes under analysis.
Putative and validated phosphorylation sites on human and mouse retinoblastoma proteins. PhosphoSitePlus is an online system that provides comprehensive information and tools for the study of protein posttranslational modifications. We retrieved amino acid residues that can be phosphorylated in human and mouse RB1, RB2/P130, and P107. The picture shows protein multiple sequence alignment (human, mouse, and rat) with putative and validated phosphorylation sites on the proteins under analysis.
Acknowledgments
Acknowledgements
We thank Drs. Vincenzo Condè, Daniela Di Pinto, and Martina Di Martino (Servizio di Oncologia Pediatrica, AOU, Seconda Università di Napoli) for bone marrow harvests and technical assistance. We also thank Mrs. Maria Rosaria Cipollaro (Department of Experimental Medicine, Second University of Naples) for technical assistance. This work was partially supported by the AIRC Associazione Italiana Ricerca sul Cancro, IG 2014-15690 to A.G. and from Department of Experimental Medicine, University of Campania "Luigi Vanvitelli", Naples, Italy to U.G.
Footnotes
Author contributions: Nicola Alessio, Stefania Capasso, Angela Ferone, and Giovanni Di Bernardo performed data collection and assembly and contributed to data analysis and interpretation. Marilena Cipollaro, Fiorina Casale, and Gianfranco Peluso conducted data analysis and interpretation.
Umberto Galderisi and Antonio Giordano were responsible for the study conception and design, contributed to data analysis, and wrote the manuscript.
Conflict of interests: none to disclose.
References
- 1.Gharib WH, Robinson-Rechavi M. When orthologs diverge between human and mouse. Brief Bioinform. 2011;12:436–441. doi: 10.1093/bib/bbr031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Campisi J, d'Adda di Fagagna F. Cellular senescence: when bad things happen to good cells. Nat Rev Mol Cell Biol. 2007;8:729–740. doi: 10.1038/nrm2233. [DOI] [PubMed] [Google Scholar]
- 3.Classon M, Dyson N. p107 and p130: versatile proteins with interesting pockets. Exp Cell Res. 2001;264:135–147. doi: 10.1006/excr.2000.5135. [DOI] [PubMed] [Google Scholar]
- 4.Galderisi U, Cipollaro M, Giordano A. The retinoblastoma gene is involved in multiple aspects of stem cell biology. Oncogene. 2006;25:5250–5256. doi: 10.1038/sj.onc.1209736. [DOI] [PubMed] [Google Scholar]
- 5.LeCouter JE, Kablar B, Hardy WR, Ying C, Megeney LA, May LL, Rudnicki MA. Strain-dependent myeloid hyperplasia, growth deficiency, and accelerated cell cycle in mice lacking the Rb-related p107 gene. Mol Cell Biol. 1998;18:7455–7465. doi: 10.1128/mcb.18.12.7455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.LeCouter JE, Kablar B, Whyte PF, Ying C, Rudnicki MA. Strain-dependent embryonic lethality in mice lacking the retinoblastoma-related p130 gene. Development. 1998;125:4669–4679. doi: 10.1242/dev.125.23.4669. [DOI] [PubMed] [Google Scholar]
- 7.Sage J, Miller AL, Perez-Mancera PA, Wysocki JM, Jacks T. Acute mutation of retinoblastoma gene function is sufficient for cell cycle re-entry. Nature. 2003;424:223–228. doi: 10.1038/nature01764. [DOI] [PubMed] [Google Scholar]
- 8.Claudio PP, Howard CM, Baldi A, De Luca A, Fu Y, Condorelli G, Sun Y, Colburn N, Calabretta B, Giordano A. p130/pRb2 has growth suppressive properties similar to yet distinctive from those of retinoblastoma family members pRb and p107. Cancer Res. 1994;54:5556–5560. [PubMed] [Google Scholar]
- 9.Galderisi U, Jori FP, Giordano A. Cell cycle regulation and neural differentiation. Oncogene. 2003;22:5208–5219. doi: 10.1038/sj.onc.1206558. [DOI] [PubMed] [Google Scholar]
- 10.Haigis K, Sage J, Glickman J, Shafer S, Jacks T. The related retinoblastoma (pRb) and p130 proteins cooperate to regulate homeostasis in the intestinal epithelium. J Biol Chem. 2006;281:638–647. doi: 10.1074/jbc.M509053200. [DOI] [PubMed] [Google Scholar]
- 11.Herrera RE, Sah VP, Williams BO, Makela TP, Weinberg RA, Jacks T. Altered cell cycle kinetics, gene expression, and G1 restriction point regulation in Rb-deficient fibroblasts. Mol Cell Biol. 1996;16:2402–2407. doi: 10.1128/mcb.16.5.2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.MacPherson D, Sage J, Crowley D, Trumpp A, Bronson RT, Jacks T. Conditional mutation of Rb causes cell cycle defects without apoptosis in the central nervous system. Mol Cell Biol. 2003;23:1044–1053. doi: 10.1128/MCB.23.3.1044-1053.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.MacPherson D, Sage J, Kim T, Ho D, McLaughlin ME, Jacks T. Cell type–specific effects of Rb deletion in the murine retina. Genes Dev. 2004;18:1681–1694. doi: 10.1101/gad.1203304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bracken MB. Why animal studies are often poor predictors of human reactions to exposure. J R Soc Med. 2009;102:120–122. doi: 10.1258/jrsm.2008.08k033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Galderisi U, Giordano A. The gap between the physiological and therapeutic roles of mesenchymal stem cells. Med Res Rev. 2014;34:1100–1126. doi: 10.1002/med.21322. [DOI] [PubMed] [Google Scholar]
- 16.Capasso S, Alessio N, Squillaro T, Di Bernardo G, Melone MA, Cipollaro M, Peluso G, Galderisi U. Changes in autophagy, proteasome activity and metabolism to determine a specific signature for acute and chronic senescent mesenchymal stromal cells. Oncotarget. 2015;6:39457–39468. doi: 10.18632/oncotarget.6277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Debacq-Chainiaux F, Erusalimsky JD, Campisi J, Toussaint O. Protocols to detect senescence-associated beta-galactosidase (SA-betagal) activity, a biomarker of senescent cells in culture and in vivo. Nat Protoc. 2009;4:1798–1806. doi: 10.1038/nprot.2009.191. [DOI] [PubMed] [Google Scholar]
- 18.Aird KM, Zhang R. Detection of senescence-associated heterochromatin foci (SAHF) Methods Mol Biol. 2013;965:185–196. doi: 10.1007/978-1-62703-239-1_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Helmbold H, Galderisi U, Bohn W. The switch from pRb/p105 to Rb2/p130 in DNA damage and cellular senescence. J Cell Physiol. 2012;227:508–513. doi: 10.1002/jcp.22786. [DOI] [PubMed] [Google Scholar]
- 20.Helmbold H, Komm N, Deppert W, Bohn W. Rb2/p130 is the dominating pocket protein in the p53-p21 DNA damage response pathway leading to senescence. Oncogene. 2009;28:3456–3467. doi: 10.1038/onc.2009.222. [DOI] [PubMed] [Google Scholar]
- 21.Jackson JG, Pereira-Smith OM. Primary and compensatory roles for RB family members at cell cycle gene promoters that are deacetylated and downregulated in doxorubicin-induced senescence of breast cancer cells. Mol Cell Biol. 2006;26:2501–2510. doi: 10.1128/MCB.26.7.2501-2510.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bringold F, Serrano M. Tumor suppressors and oncogenes in cellular senescence. Exp Gerontol. 2000;35:317–329. doi: 10.1016/s0531-5565(00)00083-8. [DOI] [PubMed] [Google Scholar]
- 23.McConnell BB, Starborg M, Brookes S, Peters G. Inhibitors of cyclin-dependent kinases induce features of replicative senescence in early passage human diploid fibroblasts. Curr Biol. 1998;8:351–354. doi: 10.1016/s0960-9822(98)70137-x. [DOI] [PubMed] [Google Scholar]
- 24.Revandkar A, Perciato ML, Toso A, Alajati A, Chen J, Gerber H, Dimitrov M, Rinaldi A, Delaleu N, Pasquini E. Inhibition of Notch pathway arrests PTEN-deficient advanced prostate cancer by triggering p27-driven cellular senescence. Nat Commun. 2016;7:13719. doi: 10.1038/ncomms13719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.MacLellan WR, Garcia A, Oh H, Frenkel P, Jordan MC, Roos KP, Schneider MD. Overlapping roles of pocket proteins in the myocardium are unmasked by germ line deletion of p130 plus heart-specific deletion of Rb. Mol Cell Biol. 2005;25:2486–2497. doi: 10.1128/MCB.25.6.2486-2497.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Macleod KF, Hu Y, Jacks T. Loss of Rb activates both p53-dependent and independent cell death pathways in the developing mouse nervous system. EMBO J. 1996;15:6178–6188. [PMC free article] [PubMed] [Google Scholar]
- 27.Liu DX, Nath N, Chellappan SP, Greene LA. Regulation of neuron survival and death by p130 and associated chromatin modifiers. Genes Dev. 2005;19:719–732. doi: 10.1101/gad.1296405. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
RT-PCR analysis following silencing experiments. RB1, RB2/P130, and P107 were silenced in human and mouse cells with specific shRNAs The histograms shows the expression levels of silenced genes following treatments of cells with shRNAs. The mRNA levels of RB, RB2/P130, and P107 are expressed as arbitrary units. Significant differences with respect to control in relative gene expression are indicated with *P < .05 or **P < .01. R1, R2, and P107 stand for shRNAs against human RB1, RB2/P130, and P107 mRNAs, respectively. The control shRNA with a scrambled sequence was named SCR. Silencing experiments were carried out in human MSCs (hMSC) and fibroblasts (hFib). Mouse MSCs (mMSC) and fibroblasts (mFib) were treated with shRNAs that specifically targeted mouse mRNAs of RB1, RB2/P130, and P107 (r1, r2, and p107, respectively). Control shRNAs for mouse cells was named scr.
Putative transcription factors binding sites on promoters of human and mouse retinoblastoma proteins. One thousand–base pair regions of human and mouse promoters (−700+299) belonging to RB1, RB2/P130, and P107 were identified by Transcriptional Regulatory Element Database (https://cb.utdallas.edu/cgi-bin/TRED/tred.cgi?process=searchPromForm). The retrieved accession numbers are shown in the picture. Identified promoter regions were evaluated with ALIBABA2 (http://www.gene-regulation.com), which is a pattern-based program for predicting transcription factor binding sites (TFBS) in DNA sequences. It uses the set of binding sites from TRANSFAC Public 6.0. In the pictures are shown the putative binding sites found on the genes under analysis.
Putative and validated phosphorylation sites on human and mouse retinoblastoma proteins. PhosphoSitePlus is an online system that provides comprehensive information and tools for the study of protein posttranslational modifications. We retrieved amino acid residues that can be phosphorylated in human and mouse RB1, RB2/P130, and P107. The picture shows protein multiple sequence alignment (human, mouse, and rat) with putative and validated phosphorylation sites on the proteins under analysis.