Abstract
The interaction of hexamminecobalt(III), Co(NH3)63+, with 160 and 3000–8000 bp length calf thymus DNA has been investigated by circular dichroism, acoustic and densimetric techniques. The acoustic titration curves of 160 bp DNA revealed three stages of interaction: (i) Co(NH3)63+ binding up to the molar ratio [Co(NH3)63+]/[P] = 0.25, prior to DNA condensation; (ii) a condensation process between [Co(NH3)63+]/[P] = 0.25 and 0.30; and (iii) precipitation after [Co(NH3)63+]/[P] = 0.3. In the case of 3000–8000 bp DNA only two processes were observed: (i) binding up to [Co(NH3)63+]/[P] = 0.3; and (ii) precipitation after this point. In agreement with earlier observations, long DNA aggregates without changes in its B-form circular dichroism spectrum, while short DNA demonstrates a positive B→Ψ transition after [Co(NH3)63+]/[P] = 0.25. From ultrasonic and densimetric measurements the effects of Co(NH3)63+ binding on volume and compressibility have been obtained. The binding of Co(NH3)63+ to both short and long DNA is characterized by similar changes in volume and compressibility calculated per mole Co(NH3)63+: ΔV = 9 cm3 mol–1 and Δκ = 33 × 10–4 cm3 mol–1 bar–1. The positive sign of the parameters indicates dehydration, i.e. water release from Co(NH3)63+ and the atomic groups of DNA. This extent of water displacement would be consistent with the formation of two direct, hydrogen bonded contacts between the cation and the phosphates of DNA.
INTRODUCTION
DNA in viruses and cells exists in a highly condensed, tightly packed state. For instance, the concentration of DNA in the head of bacteriophages is ∼800 mg/ml and in metaphase chromosomes ∼170 mg/ml (1). In contrast, DNA concentration in spectroscopic studies is usually ∼10–2 mg/ml. Knowledge of the condensation process is essential for understanding the molecular mechanisms of biological processes such as DNA transcription and replication. Studies on DNA condensation have received additional impetus in recent years from an interest in gene therapy, which is based on delivery of foreign DNA molecules into cells (2 and references therein).
Condensation of DNA can be induced by different agents: polyamines (3,4), multivalent cations (5–7), histones (8–10), positively charged polypeptides (11–14), alcohols (15–18) and neutral polymers in conjunction with salts (19–21). Trivalent hexamminecobalt, Co(NH3)63, is able to condense DNA from very dilute aqueous solutions (6). This agent is substitutionally inert due to tightly bound amino groups and does not bind specifically to DNA (6). These properties make Co(NH3)63 a convenient agent to study changes in hydration during DNA condensation in aqueous solutions.
Hydration is an important aspect of DNA condensation (22–24). Hydration forces play a crucial role in the condensation process by reorganizing the water molecules between DNA duplexes (22,25,26). The reorganization of water molecules or changes in hydration can be accurately followed by ultrasonic and densimetric methods. These methods have been used to study the hydration of nucleic acids (27–30), hydration effects of metal binding to nucleic acids (28,30–34) and the Ni2+-induced B→Z transition of poly[d(G-C]·poly[d(G-C)] (35).
In the present work we used ultrasonic and densimetric techniques in conjunction with circular dichroism (CD) spectroscopy to study the interaction of Co(NH3)63+ with 160 and 3000–8000 bp calf thymus (CT) DNA. We investigated Co(NH3)63+ binding prior to condensation and during the condensation process itself. The binding process is accompanied by significant dehydration or release of water molecules from the hydration shells of DNA and Co(NH3)63+, in an amount that corresponds to two direct contacts, probably with negatively charged phosphates. These dehydration effects are similar for both short and long DNA. CD spectroscopy demonstrated that the condensation process is length dependent: short DNA shows a B→Ψ transition, while no significant CD changes are observed for 3000–8000 bp DNA.
MATERIALS AND METHODS
Materials
Mononucleosomal (160 bp) CT DNA was prepared according to Wang et al. (36) as adapted from Strzelecka and Rill (37). The sodium salt of CT DNA was obtained from Merck. Agarose gel electrophoresis showed that the length of this DNA was in the range 3000–8000 bp. Both DNA samples were dissolved in 0.1 M CsCl, 2 mM HEPES, 2 mM EDTA, pH 7.5, and exhaustively dialyzed against final buffer (5 mM CsCl, 0.2 mM Cs HEPES, pH 7.5) at 2–4°C for 3–4 days. The solutions of Co(NH3)6Cl3 were prepared in the final buffer and the pH was adjusted to 7.5. After binding experiments no significant changes in pH were observed. Small (20–200 bp) CT DNA was prepared by sonication of 3000–8000 bp DNA in 0.2 M NaCl, 2 mM HEPES, 2 mM EDTA, pH 7.5, buffer at 2–4°C under a nitrogen atmosphere with a Branson sonifier. The concentration of DNA was ∼1.5 mg/ml in a volume of 5 ml and sonication was performed at the lowest power output for 10–12 h. After sonication the DNA solution was filtered through a 0.45 µm filter. The UV melting curves of the duplexes before and after ultrasound treatment showed no significant changes in melting behavior. This DNA was then dialyzed against the final buffer. The concentrations of all DNA samples per mole phosphate were determined optically using a molar extinction coefficient of 6550 M–1 cm–1. The concentration of Co(NH3)6Cl3 (Aldrich) solution was determined by weighing the dry samples and the needed amount of buffer. The concentration was further checked by optical density and was in good agreement with the literature (6,38): at λmax = 476 nm, ɛ = 56.5 M–1 cm–1 and at λ max = 340 nm, ɛ = 46.1 M–1 cm–1. All measurements were performed in 5 mM CsCl, 0.2 mM Cs HEPES, pH 7.3, at 20°C.
Absorption and CD measurements
Absorption spectra were obtained with a Hewlett Packard 8452A diode array spectrophotometer and CD spectra with a Jasco J-600 spectropolarimeter using a quartz cuvette with a 0.05 cm path length. Both devices were equipped with water-jacketed cuvette holders. The titration experiments were performed by adding Co(NH3)63+ solution to DNA solution in the cuvette using Hamilton syringes. Stirring was carried out directly in the cuvette using a vibrating bar.
Ultrasound velocity measurements
Relative ultrasound velocity was measured by the differential resonator method (39–41) in a stainless steel cell of 0.8 cm3 volume with built-in stirrers (40) and a quartz cell of 0.25 cm3 volume (32). The molar increment of ultrasonic velocity (A) was calculated using the equation:
A = (U – Uo)/(UoC) 1
where U and Uo are the ultrasound velocities in the solution and solvent, respectively, and C is the molar concentration of DNA calculated per mole nucleotide. The relative experimental error in (U – Uo)/Uo was 2 × 10–5%.
Acoustic titration experiments were performed by adding Co(NH3)63+ solution to the DNA solution in a sample cell. In the case of the quartz cells stirring was performed in the sample cell using a vibrating bar. To estimate the net effects of the interactions of Co(NH3)63+ with DNA, the contribution of the salt to ultrasound velocity was eliminated by carrying out a titration of buffer in the same sample cell.
Density measurements
The densities of solutions were measured with a DMA-602 densimeter (Anton Paar, Graz, Austria) using a 0.2 ml cell. As in the case of the acoustic measurements, a differential system consisting of two cells was employed. The mixtures of DNA with Co(NH3)6Cl3 were prepared by weight in microcentrifuge tubes.
Calculation of parameters
The apparent molar volume (ΦV) was calculated using the equation (42,43):
ΦV = M/ρo – (ρ – ρo)/(ρoC) 2
where ρo and ρ are the density of the solvent and solution, respectively, and M is the molecular mass of DNA per nucleotide unit.
The apparent molar adiabatic compressibility (ΦκS) was determined as a function of ΦV and the apparent molar volume (44,45):
ΦκS = 2βo(ΦV – A – M/2ρo) 3
where βo is the adiabatic compressibility coefficient of the solvent. The value of βo was calculated from our measurements of density, ρo, and the ultrasonic velocity, Uo, in the solvent using the equation βo = (ρo Uo2)–1.
The changes in A, ΦV, and ΦκS due to the interaction of Co(NH3)63+ ions with DNA were calculated using the equations
ΔA = A – Ao 4
ΔV = ΦV – ΦVo 5
Δκ = 2βo (ΔV – ΔA) 6
where A and ΦV are, respectively, the molar increment of ultrasound velocity and apparent molar volume of DNA + Co(NH3)63+ solution relative to buffer + Co(NH3)63+ and Ao and ΦVo are, respectively, the molar increment of ultrasound velocity and apparent molar volume of a DNA solution relative to buffer.
RESULTS AND DISCUSSION
Hexamminecobalt(III) induces the Ψ conformation of short DNA molecules
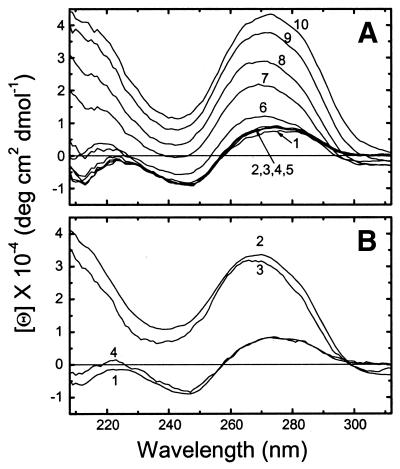
CD spectra of 160 bp DNA were recorded over the range of concentration ratios Co(NH3)63+/DNA-P = 0–0.353, as indicated in Figure 1A. The CD spectrum undergoes a sharp change, characteristic of the B→Ψ transition, at a ratio of 0.275 and above. The B→Ψ transition is associated with condensed DNA in a cholesteric liquid crystalline phase (46,47). Centrifugation of Co(NH3)63+–DNA complexes at 3000 r.p.m. for 15 min removed ∼75% of the DNA from the solution at a [Co(NH3)63+]/[P] ratio of 0.32, but the CD spectrum stayed essentially the same (Fig. 1B), indicating that small Ψ-DNA aggregates remained suspended in solution. The transition was fully reversed by adding a monovalent cation (in our case CsCl) (line 4 in Fig. 1B), as is characteristic for Co(NH3)63+-condensed DNA (6).
Figure 1.
(A) CD spectra of 160 bp CT DNA at various concentrations of hexamminecobalt(III): (1) [Co(NH3)63+]/[P] = 0; (2) 0.098; (3) 0.197; (4) 0.216; (5) 0.256; (6) 0.275; (7) 0.295; (8) 0.314; (9) 0.334; (10) 0.353. The solution initially contained DNA at 3 mM (P) concentration in 5 mM CsCl, 0.2 mM Cs HEPES, pH 7.3. (B) CD spectra of the same DNA at various conditions: (1) [Co(NH3)63+]/[P] = 0, [P] = 2.42 mM; (2) [Co(NH3)63+]/[P] = 0.32, [P] = 2.38 mM; (3) solution (2) after centrifuging, [P] = 0.63 mM; (4) solution (3) after adding CsCl solution, [Cs+]/[P] = 25.8, [P] = 0.62 mM.
Our findings appear somewhat at odds with those of Widom and Baldwin, who showed that Co(NH3)63+ efficiently aggregates random (irregular) sequence natural DNA without any CD changes (6). This likely reflects differences in experimental conditions. The present measurements were done on 160 bp CT DNA at a 2–3 mM DNA phosphate concentration, pH 7.5 and 20°C, while Widom and Baldwin (6) used 49 000 bp long λ DNA at 1.6 µM concentration in 1 mM NaOAc, pH 5.0, presumably at 25°C or ambient temperature.
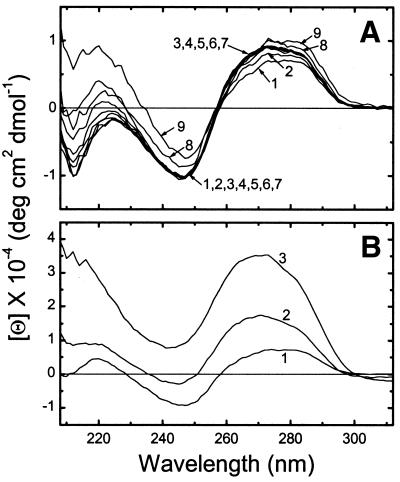
One can hypothesize at least five reasons that explain the discrepancy between the present and earlier (6) CD results: pH of the solutions, DNA concentration, DNA source, DNA preparation method and DNA length. A difference in pH should not be the reason, since lower pH is more favorable for the B→Ψ transition, due to better neutralization of the DNA negative charge. Measurements at a lower concentration of DNA (0.03 mM) also showed a B→Ψ transition (data not shown), indicating that DNA concentration is not the source of the discrepancy. Our DNA came from CT, while Widom and Baldwin used λ DNA. However, like λ DNA, the CD spectra of high molecular weight CT DNA, 3000–8000 bp in length, show no evidence of Ψ conformation regardless of Co(NH3)63+ concentration (Fig. 2A).
Figure 2.
(A) CD spectra of 3000–8000 bp CT DNA in 5 mM CsCl, 0.2 mM Cs HEPES, pH 7.3, at 20°C and at various concentrations of hexamminecobalt(III). The concentration ratio increases from 0 to 0.37 with increasing line number. For the values see Figure 3G. (B) Results on the same DNA after reducing the size to 20–200 bp: (1) the CD spectrum before adding Co(NH3)63+; (2) [Co(NH3)63+]/[P] = 0.28; (3) [Co(NH3)63+]/[P] = 0.33.
Thus we are left with the length of DNA as the most likely reason for the different CD behavior. Since our 160 and 3000–8000 bp DNA molecules were obtained from different CT sources, we sonicated the long CT DNA to 20–200 bp (see Materials and Methods) and then observed a B→Ψ transition at sufficiently high [Co(NH3)63+]/[P] (Fig. 2B). This demonstrates that DNA length, rather than source or method of preparation, is the determining factor in formation of Ψ-DNA.
This conclusion is consistent with several related investigations. It explains the discrepancy between two earlier studies on spermidine-induced condensation of DNA: Damaschun et al. (48) demonstrated a B→Ψ transition for sonicated DNA (750 bp length), while Gosule and Schellman (4) saw no changes in CD spectrum of unsonicated phage T7 DNA. Another example of length dependence is found in the work of Shin et al. (49). They demonstrated that adding Co(NH3)63+ to 250–1500 bp poly[d(A-T)]·poly[d(A-T)] converted it to the Ψ conformation, while 10 000 bp poly[d(A-T)]·poly[d(A-T)] condensation was accompanied by a B→X transition (49), which results from structural changes in the secondary structure of DNA (50,51). Maniatis et al. (52) also suggested that sonicated DNA (∼150 bp) is condensed into a more orderly structure than high molecular weight DNA. The influence of DNA length on spermine-induced condensation was investigated by Marquet et al. (53) by electric dichroism measurements. They demonstrated that 258 and 436 bp DNA condensed into rod-like particles, while 748 bp or larger DNA condensed into torus-shaped particles (53).
The development of Ψ-type spectra in aggregates of DNA and other biopolymers is thought to be due to the formation of extended chiral arrays, similar to cholesteric liquid crystals, with a pitch comparable to the wavelength of light (46). We may speculate that relatively short DNA molecules can fairly readily adjust their relative orientations to form these chiral arrays, while longer molecules are constrained by curvature and entanglements from adopting well-ordered arrangements.
Titration curves
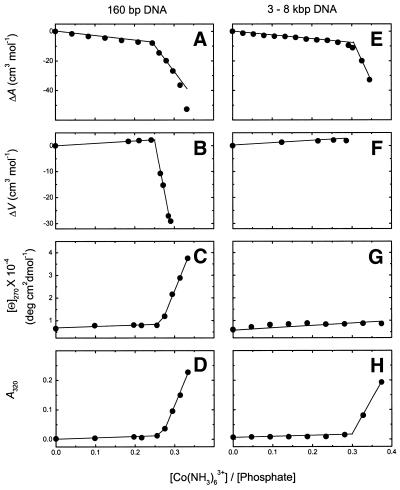
Both 160 and 3000–8000 bp DNA were titrated with Co(NH3)63+ and the interaction process was followed by ultrasound (ΔA), volume (ΔV), CD ([Θ]270) and absorbance (A320) as a function of the concentration ratio, [Co(NH3)63+]/[P] (Fig. 3).
Figure 3.
Titration of 160 bp (left column) and 3000–8000 bp DNA (right column) with Co(NH3)63+. Ultrasonic (A and E), densimetric (B and F), CD (C and G) and absorbance/turbidity (D and H) measurements.
For 160 bp DNA (left column of Fig. 3) all four parameters revealed that a dramatic change occurs at [Co(NH3)63+]/[P] = 0.25: the initial decrease in the increment of ultrasound velocity gets stronger (Fig. 3A); the initial increase in volume is followed by a strong decrease (Fig. 3B); the CD signal increases sharply after an initial moderate increase (Fig. 3C); the absorbance at 320 nm starts to rise (Fig. 3D), indicating the onset of turbidity, which is characteristic of the condensation process.
One can attribute the first, slowly changing, stage of the titration curves (0–0.25) to Co(NH3)63+ binding to DNA preceding aggregation. To be sure that aggregation does not participate in this binding event, additional centrifuge experiments at 3000 r.p.m. were performed. For instance, at the end of the binding process, [Co(NH3)63+]/[P] = 0.242, even after 140 min no visual precipitation was observed; at [Co(NH3)63+]/[P] = 0.273 some precipitate appeared, but only after 30 min centrifuging; at [Co(NH3)63+]/[P] = 0.292 precipitate appeared after 10 min centrifuging; at [Co(NH3)63+]/[P] = 0.316 the solution was already opaque and the precipitate appeared immediately upon centrifuging. Thus, one can reasonably assume that before [Co(NH3)63+]/[P] = 0.25 the ultrasound and density changes mainly reflect hydration changes, which we interpret as release of water upon overlapping of the hydration shells of the cation and DNA.
Titration experiments on long DNA are shown in the right panel of Figure 3. They are similar to those of 160 bp DNA, but there are two major differences. First, as discussed in the previous section, long DNA does not exhibit a B→Ψ transition. Second, the rapid changes in acoustic (Fig. 3E) and absorbance (Fig. 3H) properties start at [Co(NH3)63+]/[P] = 0.3 and are accompanied by immediate precipitation of DNA.
These titration results are in striking agreement with isothermal titration calorimetry measurements of the interaction of Co(NH3)63+ with plasmid DNA (54). Two stages of heat evolution were observed: the first (up to [Co(NH3)63+]/[P] = 0.25 at low salt) corresponding to binding and the second to DNA condensation. The distinction between the two stages was very sharp, just as it is in the current study. The small positive enthalpy of binding was attributed to direct hydrogen bonding between Co(NH3)63+ and DNA phosphates, consistent with the water displacement observed here.
Molecular interpretation of volume and compressibility effects
The changes in volume and compressibility due to Co(NH3)63+ binding to DNA, or to DNA condensation, can be expressed as the sum of intrinsic contributions (ΔVm and Δκm), due to DNA structural changes, and hydration contributions (ΔΔVh and ΔΔκh), due to changes in hydration (55):
ΔV = ΔVm + ΔΔVh 7
Δκ = Δκm + ΔΔκh 8
A variety of evidence shows that structural changes in DNA, and therefore contributions from ΔVm and Δκm, are negligible during the experiments performed here. The interaction of Co(NH3)63+ with long CT DNA does not significantly change the CD profile over the entire concentration range [Co(NH3)63+]/[P] = 0–0.3 (Fig. 2A). For 160 bp DNA we observed a B→Ψ transition after [Co(NH3)63+]/[P] = 0.25 (Fig. 1), but this transition is not accompanied by changes in secondary structure (51). As shown earlier (28,34), even structural changes within right-handed nucleic acid duplexes (B→C and B→A) do not influence the intrinsic volume and compressibility of the duplexes. Therefore, in the following sections we will attribute observed volume and compressibility changes solely to hydration effects.
Hydration effects of Co(NH3)63+ binding to DNA prior to condensation
The volume and compressibility changes upon Co(NH3)63+ binding to DNA molecules are positive (Table 1), which indicates a release of water molecules from the hydration shells of interacting molecules (a dehydration process). In the hydration shells the water molecules occupy less space and are less compressible than in the bulk state and therefore release of water molecules from hydration shells is accompanied by an increase in volume and compressibility. Table 1 shows no significant differences between hydration effects of Co(NH3)63+ binding to long or short DNA molecules, and CD changes before condensation are also similar for both DNAs. Therefore, one can reasonably assume that, prior to condensation, Co(NH3)63+ binds in the same manner to 160 and 3000–8000 bp DNA molecules.
Table 1. Hydration effects of Co(NH3)63+ binding to CT DNA in 5 mM CsCl, 0.2 mM Cs HEPES, pH 7.3, at 20°C.
| DNA |
ΔA (cm3 mol–1) |
ΔV (cm3 mol–1) |
Δκ × 104 (cm3 mol–1 bar–1) |
| 160 bp | 30.6 ± 2 | 9.3 ± 1.5 | 35.5 ± 3.5 |
| 3–8 kb | 26.5 ± 2 | 8.4 ± 1.5 | 31.1 ± 3.5 |
The ratio k = ΔV/Δκ = ΔΔVh/ΔΔκh is sensitive to the nature of the solute molecules (30,56–60). In the case of absolute measurements of the apparent molar volume and the adiabatic compressibility the k value is characteristic of transfer of water molecules from the bulk state to hydration shells due to dissolving a solute molecule (56,57). In the case of some association (binding) or dissociation processes the k value is also characteristic of exchange of water molecules between hydration shells and the bulk state due to overlapping of the hydration shells during binding and emergence of new surfaces for hydration during a dissociation process. Therefore, the k values of dissolution and some association/dissociation processes can be validly compared to each other. Furthermore, k values for dissolution are in very good agreement with those for interaction processes. For instance, dissolution of charged molecules (57) and interaction between them are characterized by the same value of k, 0.3–0.4 × 104 bar (58,60). The k value increases with an increase in polar or non-polar atomic groups in the solute molecules: k = 0.6 × 104 bar for DNA duplexes (27,31); k = 0.75 × 104 bar for nucleic acid bases (59). The process of nucleic acid duplex formation, which is accompanied by uptake of water molecules bridging bases in the DNA grooves, also has k = 0.75 × 104 bar (30). The k value for binding of Co(NH3)63+ to CT DNA is ∼0.3 × 104 bar, similar to k values for the hydration effects of EDTA4– + cation (60) and ion pair formation (58). This is indicative of dehydration of charged atoms: phosphates of DNA and Co(NH3)63+ itself. Although the cobalt cation is coordinated to six NH3 groups, the influence of the complex on surrounding water molecules is still very strong. The direct indication of this is its apparent molar adiabatic compressibility, which is strongly negative, ΦκS = –109.3 × 10–4 cm3 mol–1 bar–1. Similar negative values are observed for simple electrolytes, for instance, ΦκS of different divalent salts range between –81.5 × 10–4 and –109.9 × 10–4 cm3 mol–1 bar–1 (58).
One can estimate the number N of water molecules released during Co(NH3)63+ binding to DNA from the changes in volume (Table 1) according to the equation
N = ΔV/( VW – Vh) 9
where Vw is the molar volume of bulk water, equal to 18 cm3 mol–1, and Vh is the molar volume of electrostricted water, equal to 15.5 cm3 mol–1 (57,61). The data in Table 1 thus indicate that around four water molecules are released per mole Co(NH3)63+ bound. Keeping in mind that the volume and compressibility measurements mainly detect water molecules in direct contact with solute molecules (first hydration shell) (62,63) and the hydration number of cations (58) is usually equal to the total coordination number in cation–chelator complexes (64,65), one can assume that each direct contact displaces approximately two water molecules. Thus, it appears that each bound Co(NH3)63+ can create two direct contacts mainly with negatively charged DNA groups. This is the first study of volume and compressibility effects upon Co(NH3)63+ binding to an organic molecule. Therefore, interpretation of volume and compressibility effects by comparison with those in other systems is admittedly uncertain. However, to estimate the magnitude of the dehydration effects due to Co(NH3)63+ binding, one can compare volume and compressibility changes with similar data on the binding of other cations to DNA molecules. The binding of alkaline earth metal ions (Mg2+, Ca2+, Sr2+ and Ba2+) to CT DNA is also characterized by positive changes in volume and compressibility, averaging 23 cm3 mol–1 and 39 × 10–4 cm3 mol–1 bar–1, respectively (33). It was suggested that these changes correspond to two direct contacts between interacting molecules. The binding of Ni2+ ions to synthetic poly[d(G-C)]·poly[d(G-C)] is accompanied by similar increases in volume and compressibility, ΔV = 23.9 cm3 mol–1 and Δκ = 43.1 × 10–4 cm3 mol–1 bar–1, which also corresponds to two direct contacts (34). Ni2+ binding to poly[d(A-T)]·poly[d(A-T)] is accompanied by ΔV = 11.7 cm3 mol–1 and Δκ = 19.3 × 10–4 cm3 mol–1 bar–1, which corresponds to one direct or two indirect contacts (34). Thus, Co(NH3)63+ binding to DNA is characterized by significant increases in volume and compressibility which correspond to one or two direct contacts between divalent cations and DNA.
Direct contacts between Co(NH3)63+ and nucleic acids have been observed by X-ray and NMR (66–69). X-ray studies on d(CGCGCG) (66) and d(CGTACGTACG) (67) oligomers in the Z-conformation revealed that Co(NH3)63+ binds directly to O6 and N7 of a guanine and to the phosphate group of a neighboring cytosine. In the case of B-DNA decamer d(AGGCATGCCT) (68) two different binding sites have been found: one Co(NH3)63+ binds in the major groove to 5′-AGG bases, while a second binds with oxygen atoms of phosphate groups. The solution structure of RNA hairpins (69) also reveals that Co(NH3)63+ binds in the major groove of the GAAA tetraloop to N7 of guanine and to phosphate oxygen atoms of the tetraloop.
Acknowledgments
ACKNOWLEDGEMENTS
The experimental part of this work was conducted in the Department of Experimental Methods headed by L. De Maeyer at the Max-Planck Institute of Biophysical Chemistry (Göttingen, Germany). We are grateful to L. De Maeyer, D. Pörshke and Th. Funck from the same institution, L. A. Marky from the University of Nebraska Medical Center and C. Baumann from the University of Minnesota for helpful discussions.
References
- 1.Daban J.-R. (2000) Physical constraints in the condensation of eukaryotic chromosomes. Local concentration of DNA versus linear ratio in higher order chromatin structures. Biochemistry, 39, 3861–3866. [DOI] [PubMed] [Google Scholar]
- 2.Luo D. and Saltzman,W.M.(1999) Synthetic DNA delivery systems. Nat. Biotechnol., 18, 33–37. [DOI] [PubMed] [Google Scholar]
- 3.Gosule L.C. and Schellman,J.A. (1976) Compact form of DNA induced by spermidine. Nature, 259, 333–335. [DOI] [PubMed] [Google Scholar]
- 4.Gosule L.C. and Schellman,J.A. (1978) DNA condensation with polyamines. I. Spectroscopic studies. J. Mol. Biol., 121, 311–326. [DOI] [PubMed] [Google Scholar]
- 5.Wilson R.W. and Bloomfield,V.A. (1979) Counterion-induced condensation of deoxyribonucleic acid. A light-scattering study. Biochemistry, 18, 2192–2196. [DOI] [PubMed] [Google Scholar]
- 6.Widom J. and Baldwin,R.L. (1980) Cation-induced toroidal condensation of DNA. Studies with Co3+(NH3)6. J. Mol. Biol., 144, 431–453. [DOI] [PubMed] [Google Scholar]
- 7.Ma C. and Bloomfield,V.A. (1994) Condensation of supercoiled DNA induced by MnCl2. Biophys. J., 67, 1678–1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fasman G.D., Schaffhausen,B., Goldsmith,L. and Adler,A. (1970) Conformational changes associated with f-1 histone–deoxyribonucleic acid complexes. Circular dichroism studies. Biochemistry, 9, 2814–2822. [DOI] [PubMed] [Google Scholar]
- 9.Olins D.E. and Olins,A.L. (1971) Model nucleohistones: the interaction of F1 and F2al histones with native T7 DNA. J. Mol. Biol., 57, 437–455. [DOI] [PubMed] [Google Scholar]
- 10.Adler A.J., Moran,E.C. and Fasman,G.D. (1975) Complexes of DNA with histones f2a2 and f3. Circular dichroism studies. Biochemistry, 14, 4179–4185. [DOI] [PubMed] [Google Scholar]
- 11.Cohen P. and Kidson,C. (1968) Conformational analysis of DNA–poly-l-lysine complexes by optical rotatory dispersion. J. Mol. Biol., 35, 241–245. [DOI] [PubMed] [Google Scholar]
- 12.Shapiro J.T., Leng,M. and Felsenfeld,G. (1969) Deoxyribonucleic acid–polylysine complexes. Structure and nucleotide specificity. Biochemistry, 8, 3219–3232. [DOI] [PubMed] [Google Scholar]
- 13.Haynes M., Garrett,R.A. and Gratzer,W.B. (1970) Structure of nucleic acid–poly base complexes. Biochemistry, 9, 4410–4416. [DOI] [PubMed] [Google Scholar]
- 14.Suwalsky M. and Traub,W. (1972) A comparative X-ray study of a nucleoprotamine and DNA complexes with polylysine and polyarginine. Biopolymers, 11, 2223–2231. [DOI] [PubMed] [Google Scholar]
- 15.Brahms J. and Mommaerts,W.F.H.M. (1964) A study of conformation of nucleic acids in solution by means of circular dichroism. J. Mol. Biol., 10, 73–88. [DOI] [PubMed] [Google Scholar]
- 16.Girod J.C., Johnson,W.C.J., Huntington,S.K. and Maestre,M.F. (1973) Conformation of deoxyribonucleic acid in alcohol solutions. Biochemistry, 12, 5092–5096. [DOI] [PubMed] [Google Scholar]
- 17.Huey R. and Mohr,S.C. (1981) Condensed states of nucleic acids. III. Ψ(+) and Ψ(–) conformational transitions of DNA induced by ethanol and salt. Biopolymers, 20, 2533–2552. [DOI] [PubMed] [Google Scholar]
- 18.Arscott P.G., Ma,C., Wenner,J.R. and Bloomfield,V.A. (1995) DNA condensation by cobalt hexaammine (III) in alcohol–water mixtures: dielectric constant and other solvent effects. Biopolymers, 36, 345–364. [DOI] [PubMed] [Google Scholar]
- 19.Lerman L.S. (1971) A transition to a compact form of DNA in polymer solutions. Proc. Natl Acad. Sci. USA, 68, 1886–1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheng S.-M. and Mohr,S.C. (1975) Condensed states of nucleic acids. II. Effects of molecular size, base composition and presence of intercalating agents on the Ψ transition of DNA. Biopolymers, 14, 663–674. [DOI] [PubMed] [Google Scholar]
- 21.Evdokimov Y.M., Platonov,A.L., Tikhonenko,A.S. and Varshavsky,Y.M. (1972) A compact form of double-stranded DNA in solution. FEBS Lett., 23, 180–184. [DOI] [PubMed] [Google Scholar]
- 22.Rau D.C., Lee,B. and Parsegian,V.A. (1984) Measurement of the repulsive force between polyelectrolyte molecules in ionic solution: hydration forces between parallel DNA double helices. Proc. Natl Acad. Sci. USA, 81, 2621–2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bloomfield V.A. (1991) Condensation of DNA by multivalent cations: considerations on mechanism. Biopolymers, 31, 1471–1481. [DOI] [PubMed] [Google Scholar]
- 24.Bloomfield V.A. (1997) DNA condensation by multivalent cations. Biopolymers, 44, 269–282. [DOI] [PubMed] [Google Scholar]
- 25.Rau D.C. and Parsegian,V.A. (1992) Direct measurement of the intermolecular forces between counterion-condensed DNA double helices. Evidence for long range attractive hydration forces. Biophys. J., 61, 246–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rau D.C. and Parsegian,V.A. (1992) Direct measurement of temperature-dependent solvation forces between DNA double helices. Biophys. J., 61, 260–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Buckin V.A., Kankiya,B.I., Bulichov,N.V., Lebedev,A.V., Gukovsky,V.P., Sarvazyan,A.P. and Williams,A.R. (1989) Measurements of anomalously high hydration of (dA)n·(dT)n double helices in dilute solution. Nature, 340, 321–322. [DOI] [PubMed] [Google Scholar]
- 28.Buckin V.A., Kankiya,B.I., Sarvazyan,A.P. and Uedaira,H. (1989) Acoustical investigation of poly(dA)·poly(dT), poly[d(A-T)]·poly[d(A-T)], poly(A)·poly(U) and DNA hydration in dilute aqueous solutions. Nucleic Acids Res., 17, 4189–4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chalikian T.V., Sarvazyan,A.P., Plum,G.E. and Breslauer,K.J. (1994) Influence of base composition, base sequence and duplex structure on DNA hydration: apparent molar volumes and apparent molar adiabatic compressibilities of synthetic and natural DNA duplexes at 25°C. Biochemistry, 33, 2394–2401. [DOI] [PubMed] [Google Scholar]
- 30.Kankia B.I. and Marky,L.A. (1999) DNA, RNA and DNA/RNA oligomer duplexes: a comparative study of their stability, heat, hydration and Mg2+ binding properties. J. Phys. Chem., 103, 8759–8767. [Google Scholar]
- 31.Buckin V.A., Kankiya,B.I., Rentzeperis,D. and Marky,L.A. (1994) Mg2+ recognizes the sequence of DNA through its hydration shell. J. Am. Chem. Soc., 116, 9423–9429. [Google Scholar]
- 32.Buckin V.A., Tran,H., Morozov,V. and Marky,L.A. (1996) Hydration effects accompanying the substitution of counterions in the ionic atmosphere of poly(rA)·poly(rU) and poly(rA)·2poly(rU) helices. J. Am. Chem. Soc., 118, 7033–7039. [Google Scholar]
- 33.Kankia B.I. (2000) Interaction of alkaline-earth metal ions with calf thymus DNA. Volume and compressibility effects in diluted aqueous solutions. Biophys. Chem., 84, 227–237. [DOI] [PubMed] [Google Scholar]
- 34.Kankia B.I. (2000) Hydration effects of Ni2+ binding to synthetic polynucleotides with regularly alternating purine–pyrimidine sequences. Nucleic Acids Res., 28, 911–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kankia B.I. and Jovin,T.M. (1997) Hydration changes accompanying the Ni2+-induced B–Z transition of poly{d(G-C)}·poly{d(G-C)}: ultrasonic velocity and density measurements. In Proceedings of the 11th Annual Gibbs Conference on Biothermodynamics; Carbondale, IL, p. 83.
- 36.Wang L., Ferrari,M. and Bloomfield,V.A. (1990) Large-scale preparation of mononucleosomal DNA from calf thymus for biophysical studies. Biotechniques, 9, 24–27. [PubMed] [Google Scholar]
- 37.Strzelecka T.M. and Rill,R.L. (1987) Solid-state 31P NMR studies of DNA liquid crystalline phases. Isotropic to cholesteric transition. J. Am. Chem. Soc., 109, 4513–4518. [Google Scholar]
- 38.Gmelin (1964) Handbuch der Anorganische Chemie. Verlag Chemie, Weinheim, Germany.
- 39.Eggers F. and Funck,T. (1973) Ultrasonic measurements with milliliter liquid samples in the 0.5–100 MHz range. Rev. Sci. Instrum., 44, 969–977. [DOI] [PubMed] [Google Scholar]
- 40.Sarvazyan A.P. (1982) Development of methods of precise ultrasonic measurements in small volumes of liquids. Ultrasonics, 20, 151–154. [Google Scholar]
- 41.Eggers F. and Kaatze,U. (1996) Broad-band ultrasonic measurement techniques for liquids. Meas. Sci. Technol., 7, 1–19. [Google Scholar]
- 42.Kupke D.W. and Beams,J.W. (1972) Magnetic densimetry: partial specific volume and other applications. Methods Enzymol., 26, 74–107. [DOI] [PubMed] [Google Scholar]
- 43.Millero F.J. (1972) The partial molal volumes of electrolytes in aqueous solutions. In Horne,R.A. (ed.), Water and Aqueous Solutions. Wiley-Interscience, New York, NY, pp. 519–564.
- 44.Barnatt S. (1952) The velocity of sound in electrolytic solutions. J. Chem. Phys., 20, 278–279. [Google Scholar]
- 45.Owen B.B. and Simons,H.L. (1957) Standard partial molal compressibilities by ultrasonics. I. Sodium chloride and potassium chloride at 25°C. J. Phys. Chem., 61, 479–482. [Google Scholar]
- 46.Tinoco I.Jr, Mickols,W. Maestre,M.F. and Bustamante,C. (1987) Absorption, scattering and imaging of biomolecular structures with polarized light. Annu. Rev. Biophys. Biophys. Chem., 16, 319–349. [DOI] [PubMed] [Google Scholar]
- 47.Livolant F. and Maestre,M.F. (1988) Circular dichroism microscopy of compact forms of DNA and chromatin in vivo and in vitro: cholesteric liquid-crystalline phases of DNA and single dinoflagellate nuclei. Biochemistry, 27, 3056–3068. [DOI] [PubMed] [Google Scholar]
- 48.Damaschun H., Damaschun,G., Becker,M., Buder,E., Misselwitz,R. and Zirwer,D. (1978) Study of DNA–spermine interactions by use of small-angle and wide-angle X-ray scattering and circular dichroism. Nucleic Acids Res., 10, 3801–3809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shin Y.A., Feroli,S.L. and Eichhorn,G.L. (1986) Ψ compaction of poly[d(AT)]·poly[d(AT)]. Biopolymers, 25, 2133–2148. [DOI] [PubMed] [Google Scholar]
- 50.Vorlichkova M., Sklenar,V. and Kypr,J. (1983) Salt-induced conformational transition of poly[d(A-T)].poly[d(A-T)]. J. Mol. Biol., 166, 85–92. [DOI] [PubMed] [Google Scholar]
- 51.Vorlichkova M., Johnson,W.C.Jr and Kypr,J. (1994) Vacuum-UV CD spectrum of the X-form of double-stranded poly(dA-dT). Biopolymers, 34, 299–301. [Google Scholar]
- 52.Maniatis T., Venable,J.H.J. and Lerman,L.S. (1974) The structure of Ψ DNA. J. Mol. Biol., 84, 37–64. [DOI] [PubMed] [Google Scholar]
- 53.Marquet R., Wyart,A. and Houssier,C. (1987) Influence of DNA length on spermidine-induced condensation. Importance of the bending and stiffening of DNA. Biochim. Biophys. Acta, 909, 165–172. [DOI] [PubMed] [Google Scholar]
- 54.Matulis D., Rouzina,I. and Bloomfield,V.A. (2000) Thermodynamics of DNA binding and condensation: isothermal titration calorimetry and electrostatic mechanism. J. Mol. Biol., 296, 1053–1063. [DOI] [PubMed] [Google Scholar]
- 55.Shio H., Ogawa,T. and Yoshihashi,H. (1955) Measurement of the amount of bound water by ultrasonic interferometer. J. Am. Chem. Soc., 77, 4980–4982. [Google Scholar]
- 56.Lown D.A., Thirsk,H.R. and Wynne-Jones,L. (1968) Effect of pressure on ionization equilibria in water at 25°C. Trans. Faraday Soc., 64, 2073–2080. [Google Scholar]
- 57.Millero F.J., Ward,G.K., Lepple,F.K. and Hoff,E.V. (1974) Isothermal compressibility of aqueous sodium chloride, magnesium chloride, sodium sulfate and magnesium sulfate solutions from 0 to 45°C at 1 atm. J. Phys. Chem., 78, 1636–1643. [Google Scholar]
- 58.Lo Surdo A. and Millero,F.J. (1980) Apparent molal volumes and adiabatic compressibilities of aqueous transition metal chlorides at 25°C. J. Phys. Chem., 84, 710–715. [Google Scholar]
- 59.Buckin V.A. (1988) Hydration of nucleic bases on dilute aqueous solutions. Apparent molar adiabatic and isothermal compressibilities, apparent molar volumes and their temperature slopes at 25°C. Biophys. Chem., 29, 283–292. [DOI] [PubMed] [Google Scholar]
- 60.Kankia B.I., Funck,T., Uedaira,H. and Buckin,V.A. (1997) Volume and compressibility effects in the formation of metal–EDTA complexes. J. Sol. Chem., 26, 877–888. [Google Scholar]
- 61.Marky L.A. and Kupke,D.W. (2000) Enthalpy–entropy compensations in nucleic acids: contribution of electrostriction and structural hydration. Methods Enzymol., 323, 419–441. [DOI] [PubMed] [Google Scholar]
- 62.Buckin V.A., Sarvazyan,A.P. and Kharakoz,D.P. (1989) Water around biological molecules. In Deryagin,B.V., Churayev,N.V. and Ovcharenko,F.D. (eds), Water in Disperse Systems. Khimiya, Moscow, Russia, pp. 45–63.
- 63.Chalikian T.V., Sarvazyan,A.P. and Breslauer,K.J. (1994) Hydration and partial compressibility of biological compounds. Biophys. Chem., 51, 89–109. [DOI] [PubMed] [Google Scholar]
- 64.Sstezowski J.J., Countryman,R. and Hoard,J.L. (1973) Structure of the ethylenediaminetetraacetatoaquomagnesate(II) ion in a crystalline sodium salt. Comparative stereochemistry of the seven-coordinate chelates of magnesium(II), manganese(II) and iron(III). Inorg. Chem., 12, 1749–1754. [Google Scholar]
- 65.Barnett B.L. and Uchtman,V.A. (1979) Structural investigations of calcium-binding molecules. 4. Calcium binding to aminocarboxylates. Crystal structures of Ca(CaEDTA)·7H2O and Na(CaNTA). Inorg. Chem., 18, 2674–2678. [Google Scholar]
- 66.Gessner R.V., Quigley,G.J., Wang,A.H.-J., van der Marel,G.A., van Boom,J.H. and Rich,A. (1985) Structural basis for stabilization of Z-DNA by cobalt hexaammine and magnesium cations. Biochemistry, 24, 237–240. [DOI] [PubMed] [Google Scholar]
- 67.Brennan R.G., Westhof,E. and Sundaralingam,M. (1986) Structure of a Z-DNA with two different backbone chain conformations. Stabilization of the decadeoxyoligonucleotide d(CGTACGTACG) by [Co(NH3)6]3+ binding to the guanine. J. Biomol. Struct. Dyn., 3, 649–665. [DOI] [PubMed] [Google Scholar]
- 68.Nunn C.M. and Neidle,S. (1996) The high resolution crystal structure of the DNA decamer d(AGGCATGCCT). J. Mol. Biol., 253, 340–351. [DOI] [PubMed] [Google Scholar]
- 69.Rudisser S. and Tinoco,I.Jr (2000) Solution structure of cobalt(III)hexammine complexed to the GAAA tetraloop and metal-ion binding to G·A mismatches. J. Mol. Biol., 295, 1211–1223. [DOI] [PubMed] [Google Scholar]