Abstract
A challenge for circulating tumor cell (CTC)-based diagnostics is the development of simple and inexpensive methods that reliably detect the diverse cells that make up CTCs. CTC-derived nucleases are one category of proteins that could be exploited to meet this challenge. Advantages of nucleases as CTC biomarkers include: (1) their elevated expression in many cancer cells, including cells implicated in metastasis that have undergone epithelial-to-mesenchymal transition; and (2) their enzymatic activity, which can be exploited for signal amplification in detection methods. Here, we describe a diagnostic assay based on quenched fluorescent nucleic acid probes that detect breast cancer CTCs via their nuclease activity. This assay exhibited robust performance in distinguishing breast cancer patients from healthy controls, and it is rapid, inexpensive, and easy to implement in most clinical labs. Given its broad applicability, this technology has the potential to have a substantive impact on the diagnosis and treatment of many cancers.
Keywords: cancer, circulating tumor cells, diagnostic nucleic acids, nucleases, diagnostic markers, breast cancer, liquid biopsy
Introduction
Breast cancer (BCa) is the most common cancer diagnosed in women, with approximately 247,000 new cases reported in 2016, and is the second leading cause of death from cancer among women.1 Death from BCa is a result of metastatic disease, which remains largely incurable despite advances in chemotherapeutic drugs with a 5-year survival rate of only 26%.1 Early and robust detection of hematogenous circulating tumor cells (CTCs) holds the promise of greatly reducing the morbidity and mortality from metastatic BCa.2 CTCs are rare cancer cells detected in the circulation of cancer patients.3 CTC-negative patients present improved survival of up to 14 months compared with CTC-positive patients.4, 5 Based on these findings, CTCs are thought to provide a means to diagnose the likelihood for metastatic spread6 and assess response to therapy in advanced and early-stage disease settings.5, 7, 8 However, the accuracy, robustness, and ease of implementation of current CTC tests are insufficient for this approach to have its anticipated impact in clinical practice. Diagnostic tests that robustly detect CTCs, while being simple to operate and inexpensive, would: (1) enable the identification of women at greater risk of developing metastatic disease; (2) allow oncologists to aggressively treat high-risk patients while their tumor burden remains small; (3) identify low-risk patients who would not benefit from further treatments, thereby sparing them the harmful side effects of systemic therapies; and (4) enable the monitoring of the effectiveness of therapy. However, the accuracy, robustness, and ease of implementation of current CTC tests are insufficient for this approach to have its anticipated impact in clinical practice.
Despite the obvious importance of CTCs, incorporation into standard screening and treatment guidelines has been challenging because of several factors.9 First, CTCs are extremely rare in comparison to hematologic cells (about one tumor cell per 1 billion blood cells), making their isolation difficult. Second, a lack of biomarkers that capture and measure the diverse cells that collectively make up CTCs limits the sensitivity of diagnostic assays and results in inaccurate interpretation of disease state. Although there are no US Food and Drug Administration (FDA)-approved CTC diagnostic assays, a CTC enumeration test by CELLSEARCH was recently approved by the FDA. This test relies on an antibody capture agent that binds the cell-surface epithelial cell marker (epithelial cell adhesion molecule [EpCAM]) coupled to an antibody detection agent that binds an intracellular epithelial cell marker (cytokeratin).10 Although breast carcinomas, which are of epithelial origin, express high levels of both EpCAM and cytokeratin, EpCAM and cytokeratin expression are lost when cancer cells enter the bloodstream and begin colonizing distant sites. This process, known as the epithelial-to-mesenchymal transition (EMT), is thus characterized by loss of epithelial cell markers (e.g., EpCAM, E-cadherin, and cytokeratin) and gain of mesenchymal cell markers (e.g., N-cadherin and vimentin).11 Indeed, it is estimated that less than 30% of BCa cells that undergo metastasis express EpCAM.12, 13, 14 As such, detection of two or more CTCs in 7.5 mL of blood was reported in only ∼37% of CTC-positive BCa patient samples analyzed by the CELLSEARCH approach.8, 15, 16, 17, 18 The paucity of molecular markers for screening those CTCs primed for metastatic spread is a weakness of this and other antibody-based capture and analysis methods (e.g., ApoCell).15, 19 Other CTC enumeration and diagnostic methods under development, which are not dependent on molecular markers for capture and identification of CTCs, rely on microfluidic or filter capture technologies followed by an RNA or DNA purification step prior to downstream analysis (e.g., amplification/detection of CTC-specific DNA or RNA sequences with next-generation sequencing [NGS] and microarray technologies).19, 20, 21 Although these methods capture CTCs of both epithelial and mesenchymal type, drawbacks include long processing times, difficulty of implementation in clinical pathology laboratory tests (requiring microfluidic and NGS expertise and infrastructure), and high cost per sample (kits to perform isolation and RNA hybridization and amplification steps). Other fee-for-service technologies like those offered by Epic Sciences, SRI International, and CELLSEARCH have similar drawbacks and require samples to be shipped to the outsourcing company, which often results in loss of sample viability.15, 19
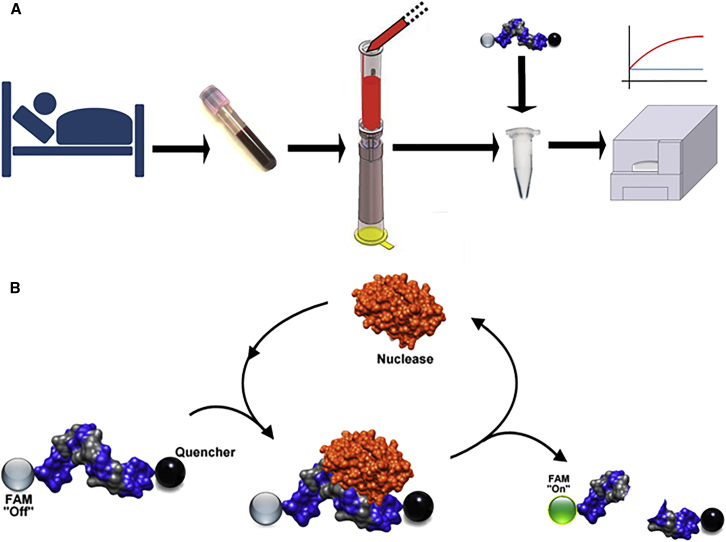
The goal of this study was to perform in stage IV BCa patients (with expected positive CTC counts) proof-of-concept studies of an emerging diagnostic technology, nuclease-activated probe (NucAP), which exploits cancer-derived nucleases as signal-amplifying biomarkers. This novel nuclease-based approach is coupled with state-of-the-art CTC capture technology from SCREENCELL, which enables the capture of both epithelial and mesenchymal cancer cells from blood based on cell size (Figure 1A). Advantages of nucleases for the detection of CTCs include: (1) their elevated expression in most cancer cells, including cells implicated in metastasis that have undergone EMT; and (2) their enzymatic activity, which can be exploited for signal amplification in detection methods (Figure 1B). Additional advantages of this diagnostic assay include its simplicity (which can facilitate on-site implementation), low cost, and broad applicability (because of the widespread expression of nucleases in other cancers).
Figure 1.
Nuclease-Activated Probe Assay for the Detection of Circulating Tumor Cells in Blood
(A) Freshly drawn patient blood was filtered using a microfiltration column device to capture cancer cells. Cancer cells were then lysed in nuclease buffer, and lysates were incubated with nuclease-activated probes. Probe fluorescence indicates the presence of cancer cells in blood. The assay was typically carried out in less than 1 hr from the time of blood draw to the fluorescence readout, and thus provides a rapid turn-around for point-of-care diagnostics. (B) Schematic of the enzymatic nature of the nuclease-activated probe assay. The sequences of the three probes (dsDNA, ssDNA, and ssRNA) used in this study are shown in the Materials and Methods. All three probes are flanked by a FAM fluorophore (5′ terminus) and a pair of fluorescence quenchers (3′ terminus).
Due to their central role in DNA repair and chromosomal stability, nucleases are essential for life and are overexpressed in most cancer cells (even those cells that have undergone EMT), including BCa cells, highlighting their broad potential as biomarkers for cancer detection.22, 23, 24, 25, 26, 27 In this study, we confirmed that nuclease gene expression is elevated in BCa cell lines compared with whole blood and that, unlike EpCAM expression, it is ubiquitously present in BCa cell lines and patient tumor samples. Based on these findings, we developed and characterized three different nuclease-activated probes that act as global substrates (are digested and activated) for nucleases overexpressed in BCa. After optimizing conditions to detect CTCs in blood with these probes, we evaluated their ability to detect CTCs in patient blood samples and to thereby provide a simple, rapid, and robust diagnostic readout.
Results
Assay Sensitivity
We previously described the development of chemically modified, nuclease-activated probes that are specifically digested (activated) by bacterial nucleases.28, 29 Recently, we optimized the nuclease probe methodology to enable detection of attomolar (10−18 molar) levels of a bacterial nuclease in the human plasma.30 Given the high sensitivity of this technology, we reasoned that nuclease-activated probes could be useful in detecting low numbers of cancer cells in blood. Figure 1 outlines the workflow of the assay presented here.
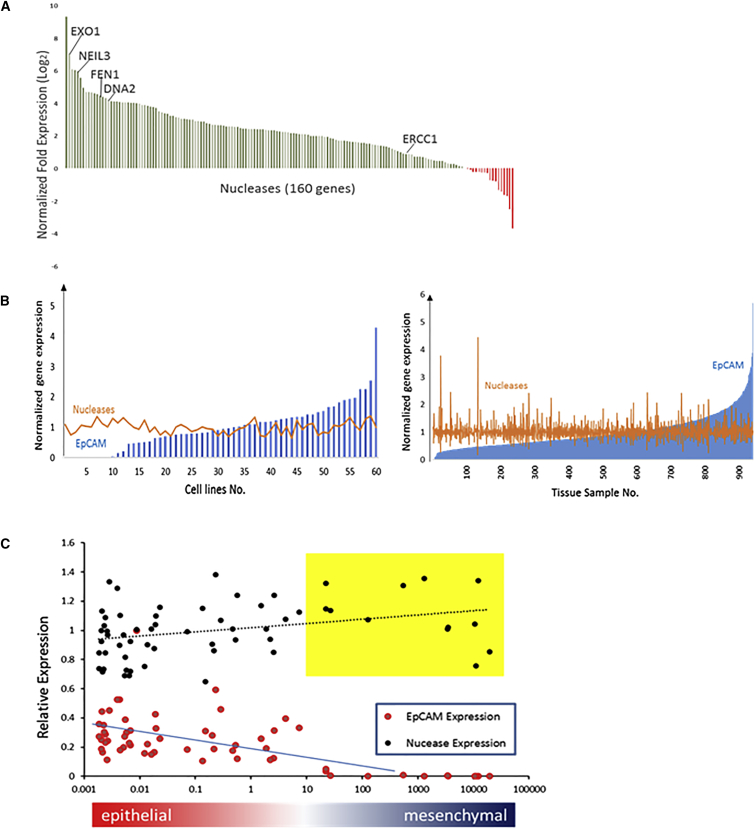
Analysis of gene expression data from the European Molecular Biology Laboratories – European Bioinformatics Institute (EMBL-EBI) database31, 32 revealed that 143 out of 160 genes that encode putative nucleases are upregulated in BCa cell lines compared with whole blood, including cell lines that lack EpCAM expression (Figure 2A; Table S1). The expression of nucleases that have been implicated in cancer progression (EXO1, NEIL3, FEN1, DNA2, and ERCC1)23, 24, 33, 34, 35, 36, 37 was also higher in cancer cells versus blood. In addition, nuclease gene expression is elevated in the clear majority of BCa cell lines (Figure 2B, left panel) and in analyzed patient tumor samples (Figure 2B, right panel). This result is in contrast with EpCAM gene expression, which varies greatly based on the nature of the sample analyzed (Figure 2B). More importantly, unlike EpCAM, nuclease expression is retained in those cells that undergo EMT (Figure 2C). Together, these findings suggested that nuclease activity could be employed as a means of detecting cancer cells in blood.
Figure 2.
Nuclease Gene Expression in BCa Cell Lines and Patient Samples
(A) BCa nuclease gene expression data were obtained from the EMBL-EBI database. Nuclease gene expression in BCa cells was normalized to that in the blood. Normalized fold expression is defined as the average mRNA expression of every nuclease gene across 60 BCa cell lines divided by the expression of the same genes in blood. Nucleases known to be upregulated in CTCs are highlighted (i.e., EXO1, NIEL3, FEN1, DNA2, and ERCC1).61 (B) Left panel: mRNA expression of EpCAM and nuclease genes from 60 different cell lines was normalized before plotting. Normalized EpCAM expression: EpCAM mRNA expression for each cell line was normalized to the average EpCAM mRNA expression level detected across these 60 cell lines (blue bars). Normalized nuclease gene expression: the sum of all 160 nucleases in each cell line was normalized to the average value across all 60 cell lines (orange line). Right panel: an analogous analysis was carried out with data from BCa patient tissues (n = 941) from The Cancer Genome Atlas (TCGA). (C) Nuclease expression in breast cancer cell lines during epithelial-to-mesenchymal transition (EMT). 60 breast cancer cell lines were ranked based on the expression of epithelial (EpCAM, cytokeratin 19, and E-cadherin) and mesenchymal (N-cadherin and vimentin) markers. Expression of EpCAM and nucleases (average expression of 161 nuclease genes) for each cell line was plotted. Yellow box: breast cancer cell lines with little-to-no EpCAM expression that are missed by EpCAM immune capture methods.
To detect nuclease activity, we screened a pool of chemically modified, nuclease-activated oligonucleotide probes (nuclease pool previously described in Hernandez et al.28, 29) and identified three distinct nuclease probes (double-stranded DNA [dsDNA], single-stranded DNA [ssDNA], and 2′fluoro [2′F]-RNA) that are digested by nucleases expressed in human BCa cell lines. The sequences of the probes are shown in the Materials and Methods. The dsDNA probe has a self-complementary sequence that forms a duplex DNA oligo. The ssDNA probe is a DNA oligo. The 2′F-RNA probe is a single-stranded probe with 2′F modification of all pyrimidines in the sequence. All three probes are flanked by a fluorescein amidite (FAM) fluorophore (5′ terminus) and a pair of fluorescence quenchers (3′ terminus).
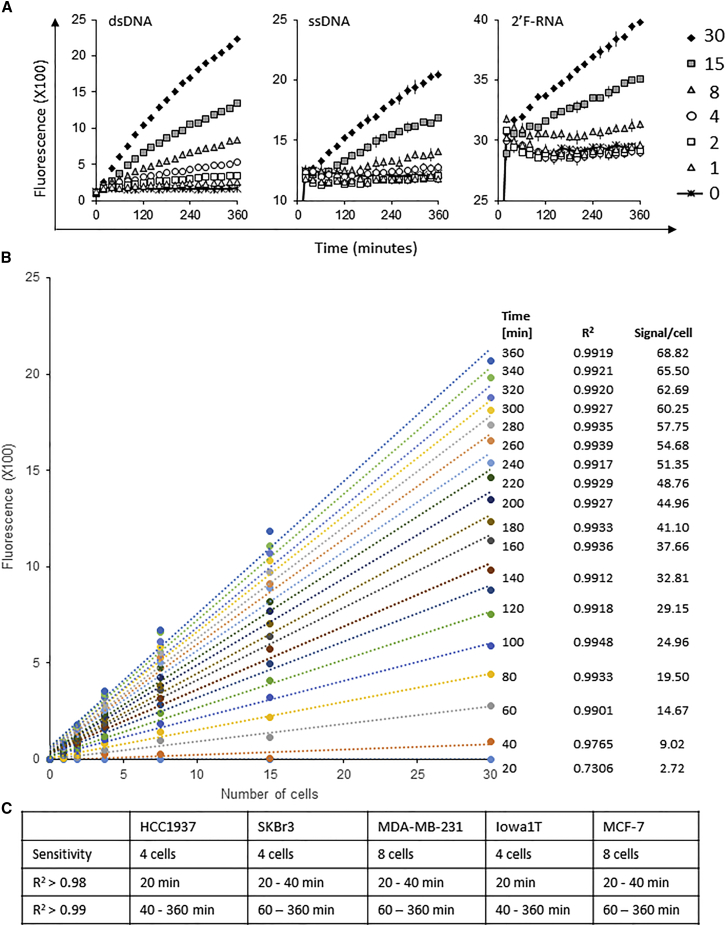
First, we optimized assay conditions, which included components of the probe digestion buffer (e.g., Mg+2 and Ca+2 concentration, pH) (Figure S1A) and the concentration of the probes in the digestion reaction (Figures S1B and S1C). Fluorescence intensity, due to probe digestion, was monitored for a total of 6 hr. Alkaline conditions (pH 8–10) were optimal for all three probes tested (data shown only for ssDNA probe) (Figure S1A). Ten millimolar Mg+2 were found to be optimal for digestion, whereas no requirement for Ca+2 in the digestion buffer was observed (Figure S1A). Furthermore, a small amount of probe (2.5 pmol corresponding to a final concentration of 250 nM) yielded the greatest activity when incubated with low numbers of BCa cells (Figure S1C). Based on the optimal assay conditions (optimized digestion buffer: 10 mM MgCl2, 50 mM NaCl, and 100 mM Tris-HCl [pH 9.0], 1 mM DTT, and 1% Triton X-100; probe concentration: 250 nM), we proceeded to determine the sensitivity of the assay for detecting nuclease activity in BCa cells (Figure 3). Varying amounts of SKBr3 BCa cells (0–30 cells) were lysed in optimized digestion buffer and incubated with the three nuclease-activated probes for a total of 6 hr. Sensitivity was approximately four cancer cells for the dsDNA and eight cancer cells for the ssDNA and the 2′F-RNA probe (Figure 3A). We also noted that optimal fluorescence intensities over background for the three probes varied based on detection time. For example, while the ssDNA probe could reliably predict the presence of eight cancer cells in buffer at 150 min, the dsDNA and 2′F-RNA probes did so for four and eight cells, respectively, at incubation times of less than 60 min. The dsDNA probe also had the strongest correlation between signal intensity and number of cancer cells in buffer. Importantly, the fluorescence signal intensity of the dsDNA probe displayed a strong linear correlation within the range of 1–30 cancer cells in an assay sample of 10 μL for incubation times >40 min (Figure 3B). To determine the broad potential of the dsDNA probe, we also evaluated lysates of low cell numbers (0–30 cells) generated from other BCa cell lines as shown for SKBr3 cells. Similarly, strong signal correlations to cell numbers and sensitivities were observed (Figure 3C). Together, these data indicate high sensitivity (four cancer cells in buffer) of the nuclease-activated probe assay with the dsDNA probe for an assay run time of as little as 40 min.
Figure 3.
Nuclease-Activated Probe Assay Sensitivity
(A) The sensitivity of the nuclease-activated probe assay was measured using SkBr3 BCa cells. Nuclease-activated probes were incubated at 37°C for 6 hr with lysates derived from varying amounts of SKBr3 BCa cells (0–30 cancer cells). Fluorescence was measured every 20 min over the course of 6 hr using a microplate reader. (B) Signals derived from the digestion of the dsDNA nuclease-activated probe showed a linear correlation in the range of 1–30 cells (data based on 3–100 cells/mL fluid evaluated in the assay). Trend lines are given for all single-measurement time points in the standard assay procedure (0–360 min with 20 min increments). Coefficient of determination (R2) was >0.99 for time points >60 min. A linear correlation was found for several BCa cell lines tested. Shown is a representative plot from an experiment with the SKBr3 BCa cell line. By investigating lysates from six different cell numbers (1, 2, 4, 8, 15, and 30), a linear regression plot was used to assess signal-to-cell number correlation. (C) Additional BCa cell lines were examined for the sensitivity and correlation of assay signal to cell number as shown in (A) and (B) for SKBr3 cells. The table sums up the sensitivity levels and R2 values at the different time points of the 6 hr assay.
Nuclease-Activated Probe Assay Specificity
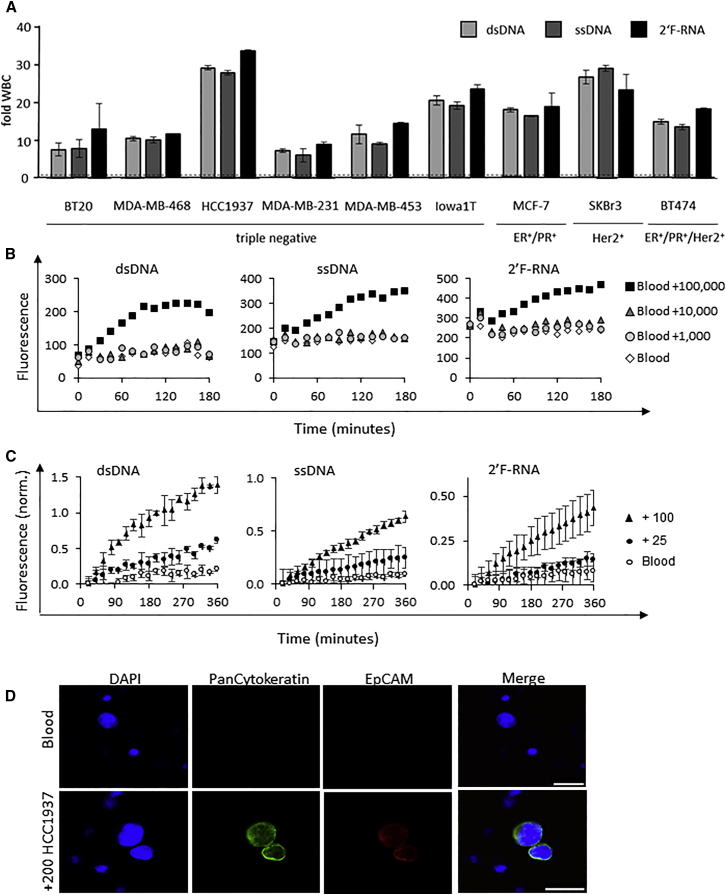
Next, we determined the specificity of the nuclease-activated probe assay for BCa subtypes (e.g., triple-negative versus hormone-positive versus Her2-positive). We compared lysates of various BCa cell lines, broadly categorized as triple-negative or hormone and/or Her2-positive BCa, with the three nuclease probes (Figure 4A). The fluorescence intensities of the BCa cell lysates for each probe were normalized to those of white blood cells (WBCs) from healthy donors (Figure 4A). Importantly, in contrast with WBCs, all BCa cell lines activate the nuclease probes, including BCa cell lines that are reported to have low-to-no EpCAM expression (e.g., MDA-MB-231 and MBA-MB-468).38, 39 Although no clear difference is observed between BCa subtypes with the tested cell lines, these data suggest that the nuclease probes can broadly detect diverse BCa subtypes (Figure 4A).
Figure 4.
Nuclease-Activated Probe Assay Specificity
(A) Nuclease-activated probes (dsDNA, ssDNA, and 2′F-RNA) were incubated with lysates of 100 white blood cells isolated from the blood of a healthy donor or various BCa cell lines (BT20, MDA-MB-468, HCC1937, MDA-MB-231, MDA-MB-453, Iowa1T, MCF7, SKBr-3, and BT474). Fold WBC is defined as the fluorescence signal generated from each individual BCa cell line divided by that generated by the white blood cells. Fluorescence was measured in a microplate reader every 20 min for a total of 6 hr. The 4-hr time point is plotted. (B) Sensitivity of detection of cancer cells in blood without prior cancer cell capture and/or enrichment step. A defined amount of healthy donor blood (about 1 mL) with the addition of increasing amounts of MDA-MB-453 cells (1–100,000 cells per 1 mL of blood) was subjected to the nuclease-activated probe assay using the three nuclease-activated probes (dsDNA, ssDNA, and 2′F-RNA). (C) Nuclease-activated probe assay sensitivity. Blood samples from a healthy donor were spiked with either 100 or 25 HCC1937 cancer cells per milliliter of blood, and samples were evaluated using the nuclease-activated probe assay. Blood, not spiked with cancer cells, served as a control (blood). The signal intensity was normalized to the background fluorescence of the corresponding probe. (D) Blood samples from a healthy donor with and without the addition of 200 HCC1937 cancer cells per milliliter of blood. Cancer cells in blood were immunostained with mouse anti-human PanCytokeratin (green) and rat anti-EpCAM (red). Scale bars: 25 μm.
Although cancer cells had at least a 10-fold higher nuclease activity, the data in Figure 4A also suggested that the nuclease activity signal generated from blood was high, potentially increasing the background signal and affecting the overall sensitivity of the nuclease-activated probe assay. Thus, we next assessed the sensitivity of the nuclease-activated probe assay in blood (Figure 4B). In contrast to Figure 3A, the data in Figure 4B demonstrated that the sensitivity of the assay in the presence of blood was relatively low due to the non-specific activity in blood, requiring greater than 10,000 BCa cells per 1 mL of blood. Based on these data, we reasoned that a cancer cell capture and/or enrichment step performed prior to the nuclease assay detection step was warranted.
CTC Capture Methods
Several methods for the capture of CTCs have been described previously.19, 40 Filter-based methods that capture cancer cells based on size (e.g., ISET and SCREENCELL; Rarecells) are easy to use and do not rely on biomarker expression, which often limits their application.41 Initially, we compared two commercially available microfiltration methods (ISET and SCREENCELL filters) for their ability to filter out red blood cells (RBCs) and WBCs based on cell size (Figure S2). As shown in Figure S2A, the use of the SCREENCELL filter resulted in a lower background signal from blood compared with the use of the ISET filter. The difference in the signal between the two microfilters was due to the number of WBCs retained on the filter. We next re-evaluated the sensitivity of the nuclease-activated probe assay when combined with a capture and/or enrichment step that includes the SCREENCELL microfiltration unit (Figure 4C). As shown in Figure 4C, the addition of the capture and/or enrichment step resulted in a 400-fold increase in sensitivity (25 versus 10,000 BCa cells in 1 mL of blood). To confirm that the fluorescence signal obtained from the SCREENCELL filter unit was indeed from BCa cells captured onto the filter membrane, we performed immunofluorescence for cancer cell markers (EpCAM and cytokeratin) (Figure 4D). These data revealed that the sensitivity of the nuclease-activated probe assay can be greatly enhanced by prior capture and/or enrichment of cancer cells.
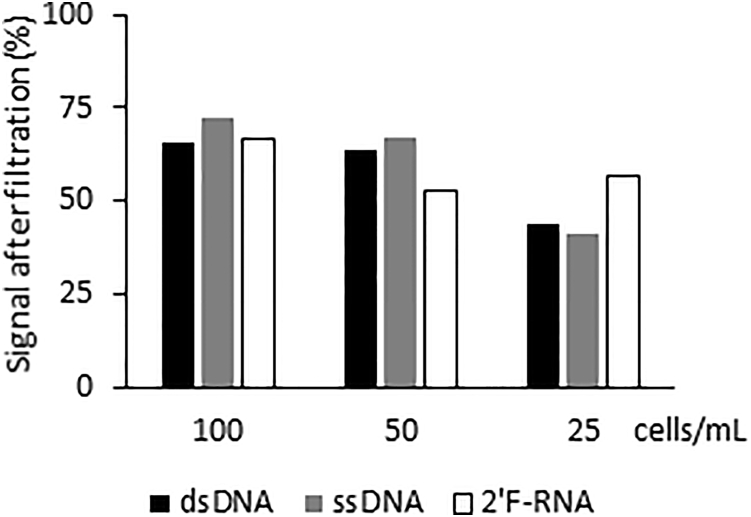
We then determined the percentage of cancer cells retained on the SCREENCELL filtration unit as a function of the number of cancer cells present in the blood (Figure 5). Defined numbers of HCC1937 BCa cells (100, 50, and 25 cells/mL) were either directly lysed in nuclease digestion buffer or were subjected to the SCREENCELL capture and/or enrichment step prior to digestion and quantification of nuclease activity. Retention efficiency of the cancer cells on the microfilters varied with cell number. The greater the concentration of cancer cells in suspension (PBS) was, the higher the percentage of recovery for all three probes tested (ranges for the three probes are provided: 100 cells/mL = 66%–72% retention; 50 cells/mL = 53%–67% retention; 25 cells/mL = 41%–57% retention) (Figure 5; Figure S3). Because the diameter of the cancer cell dictates how well it will be retained by the filter unit, we evaluated the percentage of retention using a different cancer cell line presenting a smaller diameter (e.g., SKBr3) (Figure S4a). As expected, the percentage of recovery was reduced by approximately 25% for cells with a smaller diameter (comparing the retention efficiency for dsDNA probe of HCCC1937 and SKBr3 cells) (Figure 5; Figure S4A). To confirm that the fluorescent signal obtained from these filters was indeed due to the retention of SKBr3 (EpCAM+ BCa cell line) cells onto the filter unit, we performed both qRT-PCR (Figure S4B) and immunohistochemistry (Figure S4C) for established cancer cell markers (EpCAM, cytokeratin 19, plastin 3, and Her2).
Figure 5.
Cancer Cell Retention and/or Capture Efficiency Using the SCREENCELL Filters
Signal intensity (%) of lysates from cells filtered with SCREENCELL filters compared with lysates of the same number of cells without filtration. Lysates were incubated with the nuclease-activated probes, and fluorescence measurements were obtained as described above. Varying number of HCC1937 cells (0–100 cells) were either directly lysed in nuclease lysis buffer or subjected to the filtration capture and/or enrichment step.
Confirmation of Assay Robustness in Patient Samples
Initially, blood samples from different healthy donors (n = 9) were evaluated using the SCREENCELL filtration method prior to performing nuclease probe assay to assess potential variability in the background signal from donor to donor (Figure S2B). Overall, the average signals from healthy donor blood had an SEM <4%. The earlier assay run times exhibited even lower SEMs (<3% for time points <140 min, <2% for time points <40 min) compared with the longer assay run times (Figure S2B). In addition, no difference was observed in the background signal from the same donor sampled on different days (Figure S2C; data for three different donors are shown). These data revealed the effectiveness of the SCREENCELL filtration unit at removing excess blood cells from the blood sample and confirmed that the background signal from blood in the nuclease-activated probe assay is low.
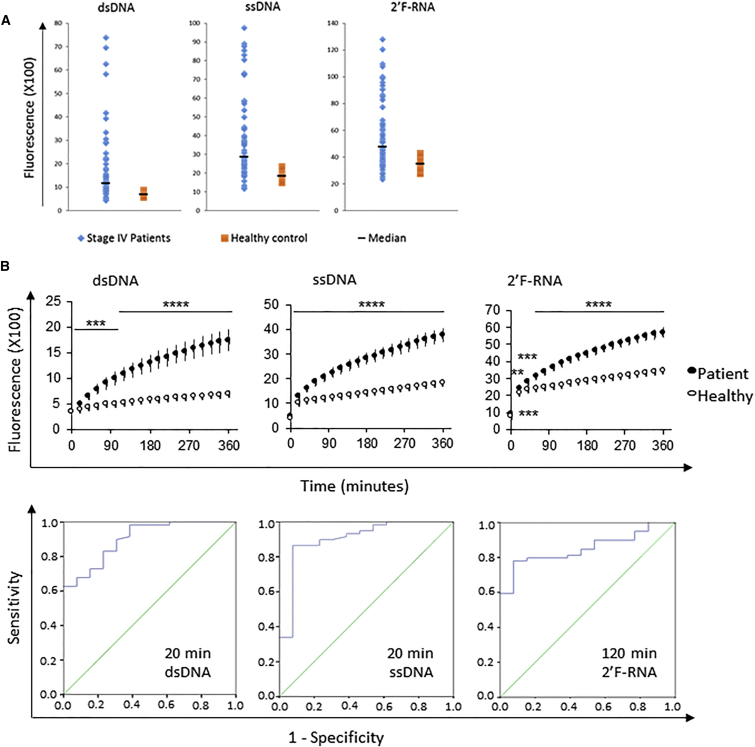
To test the ability of this assay to detect CTCs in blood of patients with BCa, we compared the nuclease activity from blood of BCa patients with established CTCs in blood (stage IV) (n = 29) and healthy donors (n = 15). We detected higher nuclease activity in the blood of patients with BCa with all three probes tested (Figure 6A shows a dot plot for the 6 hr time point). We then investigated the entire time course of the assay run and found that at the final assay run time (0–6 h), signals from patient samples were 4.5-fold (for dsDNA), 3.4-fold (for ssDNA), or 2.7-fold (for 2′F-RNA) higher than those of healthy donor blood samples (Figure 6B, top panels). We also determined the assay run time that was the best predictor of cancer cells present in blood based on fluorescence intensity (Figure 6B, bottom panels). A 20 min assay run time was found to be the best predictor for the two DNA probes. An assay run time of 120 min was found to be the best predictor for the RNA probe. Receiver operating characteristic (ROC) curves were generated for each time point of the nuclease probe assay for each probe. Each ROC curve estimates probe sensitivity and specificity over the range of fluorescence intensity cutoffs that could be used to classify patients as diseased or non-diseased. The accuracy of the nuclease probe assay as an indicator of disease status (samples from patients with BCa versus healthy donors) was evaluated by measuring the area under the ROC curve. Specifically, area under the curve (AUC) estimates can be interpreted as the probability that fluorescence intensity for a patient with BCa is higher than that for a healthy donor. A maximum AUC value of 1.0 indicates perfect diagnostic accuracy, and a value of 0.5 indicates no diagnostic value. The assay run time that best predicted the presence of CTCs in blood (highest AUC values) varied from probe to probe. An assay run time of 20 min was optimal for the dsDNA probe (AUC = 0.902) with a 95% confidence interval (CI) of 0.82–0.96. For the ssDNA probe, an assay run time of 20 min was optimal, with AUC = 0.903 (95% CI 0.80–1.00). For the 2′F-RNA probe, the best predictor time point was at 120 min with AUC = 0.851 (95% CI 0.76–0.94) (Figure 6B, bottom panels). The high AUC values indicated a high degree of diagnostic accuracy, and the exclusion of 0.5 (which represents no diagnostic value) from the CIs significantly indicated that the probes are diagnostic indicators of the disease. ROC curves for the corresponding AUC estimates are included in Figure 6B (bottom panels). The cutoff values for best specificity/sensitivity combination in the ROC curves at the optimal time points are listed in Table 1.
Figure 6.
Detection of Cancer Cells in Blood of Patients with Stage IV BCa
Samples from patients with stage IV BCa were examined using the nuclease-activated probe assay. Fluorescence intensity from the stage IV BCa samples was compared with that of healthy donor samples. (A) Nuclease activity detected after 6 hr from blood of stage IV BCa patients (blue) or healthy donors (orange). The black line indicates the median of the two groups. (B) Plotted are mean values and corresponding SEM for patients (closed circles, n = 29) and healthy donors (open circles, n = 15). Asterisks indicate statistical significance (p values of a two-way ANOVA are described in the depicted table: **p < 0.01; ***p < 0.001; ****p < 0.0001). Statistical analysis of the time points that are the best predictors of the nuclease-activated probe assay. Area under the curve (AUC) plot based on receiver operating characteristic (ROC) analysis of the data. Additionally, Table 1 gives the cutoff values for sensitivity and specificity at these best predictor time points.
Table 1.
Cutoff Values with Best Specificity/Sensitivity Combination in the ROC Curves with Highest AUC Values
| Signal Threshold | Sensitivity/Specificity | Samples above Threshold | True Positive | False Positive | |
|---|---|---|---|---|---|
| dsDNA (20 min) | 400 | 83%/77% | 49/59 (83%) | 28/29 (97%) | 3/13 (23%) |
| ssDNA (20 min) | 1,018 | 86%/92% | 52/60 (87%) | 29/29 (100%) | 1/13 (8%) |
| 2′F-RNA (120 min) | 2,854 | 80%/85% | 47/59 (80%) | 26/29 (90%) | 2/13 (15%) |
ROC curves are shown in Figure 6B.
For each patient and healthy donor blood sample, we also performed whole blood cell counts to confirm that the differences in fluorescence intensity between the two blood sample groups did not result from differences in blood cell numbers between the two groups (e.g., higher numbers of WBCs), but reflected the presence of BCa cells (Figure S5). On average, patient blood samples presented fewer WBCs compared with healthy donor blood samples. These data confirmed that the higher nuclease activity observed in patient samples did not result from higher numbers of WBCs in patient blood, but likely reflected the presence of CTCs in blood.
Discussion
This is the first report of a nuclease-activated probe-based diagnostic assay for the detection of cancer cells in patients. In this study, we show that chemically modified, nuclease-activated probes can robustly detect cancer cells in blood of patients with stage IV BCa (∼96% true positive hits; ∼15% false-positive hits). Based on the National Cancer Institute Surveillance, Epidemiology, and End Results (NCI SEER) database, the BCa prevalence (incidence) for women >18 years of age was ∼2% in 2014 (year of latest report) (https://seer.cancer.gov/statfacts/html/breast.html). At 2% incidence rate, our assay has a positive predictive value (PPV) of 11.9% and a negative predictive value (NPV) of 99.9%. The low PPV indicates that a NucAP assay positive result has a high probability of being a false positive. Thus, our assay is not an optimal screening test for women in the general population but instead a diagnostic test, intended to identify women at high risk for developing metastatic BCa and to potentially spare women with a negative test from the toxic effects of unnecessary therapies.
Because nucleases are ubiquitously expressed27 and their expression is greatly elevated in cancer cells, our approach does not suffer from the drawbacks of current antibody-based capture and analysis methods (e.g., CELLSEARCH and ApoCell) that rely on biomarkers (e.g., EpCAM) whose expression varies in cancer.8, 15, 16 Furthermore, the nuclease-activated probe assay is rapid (the assay can be completed within 1 hr of the initial blood draw), sensitive (can detect less than five BCa cells), cost-effective (under $100 per sample), and easy to operate (requires a common fluorescence plate reader for signal detection). The assay steps include: (1) capture of cancer cells, which is performed within 10 min with commercially available microfiltration units from SCREENCELL, (2) lysis of cancer cells using an optimized nuclease digestion buffer that contains the chemically optimized nuclease probes, and (3) signal detection using standard fluorometry technology.
We previously demonstrated the utility of quenched fluorescent nucleic acid probes for the detection of bacterial nucleases.28, 29 In this initial study, we demonstrated a sensitivity in the low attomolar (10−18 molar) range for the detection of a bacterial nuclease in human plasma.30 In this study, we performed key technical modifications for adapting this technology for the detection of endogenous nucleases in cancer patients. The challenge we faced was to design chemically optimized probes capable of rapid and robust detection of nucleases, which are overexpressed in cancer cells, despite the presence of a diverse mixture of endogenous non-target nucleases present in human blood. To promote selectivity, we used nucleotide chemical modifications and assay conditions (cation composition in assay buffer) that provide resistance to serum nucleases but are susceptible to nucleases overexpressed in BCa cells. Assay conditions and probes with these modifications that are sensitive to nucleases in BCa cell lysates provide the foundation of our NucAP assay.
The release of CTCs from the primary tumor into the bloodstream is thought to give rise to the formation of distant metastases that are central to the often devastating outcomes of this complex illness.4, 42, 43, 44, 45 In recent years, CTCs have been extensively studied because they are readily accessible and afford a non-invasive “blood biopsy” of potential metastatic cells.42, 45 The ability to detect, quantify, and characterize CTCs would afford several advantages in the clinic. They may be advantageous in screening, staging, monitoring of overall and specific chemotherapy responses, and potentially differentiating tumor types. However, a notable challenge for developing broadly useful CTC-based diagnostic tests is the development of an easy-to-operate method that is sufficiently sensitive and capable of capturing the molecular heterogeneity of CTCs present in the circulation.6, 16, 19 The current absence of methods that solve these problems is a critical barrier to the clinical applicability of this promising approach. Here, we overcome many of the limitations of current CTC detection assays through the development of chemically modified, nuclease-activated probes that are specifically digested (i.e., activated) by nucleases expressed in BCa cells. Importantly, we demonstrate the clinical relevance of our approach in blood samples from patients with stage IV BCa (Figure 6).
There are currently no approved diagnostic tests for CTCs. CELLSEARCH, which is currently the only FDA-approved CTC enumeration method (used for BCa), is an immunomagnetic capture and/or enrichment method that relies on targeting a marker specific to epithelial cells, the EpCAM.46, 47 The CELLSEARCH approach captures CTCs from blood with magnetic particles coated with anti-EpCAM antibodies and automates their imaging using flow cytometry. CELLSEARCH defines CTCs as cells that display a DAPI-stained nucleus and express EpCAM and cytokeratins, while being negative for the pan-leukocyte marker CD45. Although the CELLSEARCH approach has been validated in clinical studies and allowed more thorough studies of the clinical relevance of CTCs, it has several limitations, including inherent lack of sensitivity and limited applicability.16, 19, 48 These limitations stem primarily from the loss of EpCAM expression (during the EMT) in those BCa cells that are implicated in metastasis.12, 13, 14, 19, 49, 50 Similarly, our analysis of gene expression data available from the EMBL-EBI and The Cancer Genome Atlas (TCGA)51 databases indicated that EpCAM expression can also vary substantially in BCa cell lines and patient samples (derived from primary epithelial tumors), whereas nucleases are overexpressed across all the BCa cell lines and patient samples analyzed (Figure 2B). Furthermore, unlike EpCAM, nuclease expression is retained in those cells that undergo EMT (Figure 2C).
Nucleases and their activity have been proposed as valuable prognostic markers for cancer.52, 53, 54, 55, 56, 57, 58 For example, the expression of ERCC1, a DNA repair enzyme with nuclease activity, has been shown to be elevated in BCa CTCs that are resistant to chemotherapy.59 Similarly, the expression of several other nuclease genes that belong to the class of DNA repair enzymes has also been shown to be elevated in cancer stem cells, which are thought to lead to metastasis.60, 61 Several of these genes (e.g., EXO1, NEIL3, FEN1, DNA2, and ERCC1) were the top hits in our bioinformatics analysis of BCa gene expression databases (Figure 2A; Table S1). A notable advantage of nucleases versus most alternative biomarkers is that their enzymatic activity can be used for signal amplification; this often enables very high detection sensitivity.
In previous studies, we took advantage of the unique properties that distinguish bacterial nucleases from mammalian nucleases, to develop chemically optimized nuclease probes that are preferentially digested by bacterial nucleases versus mammalian nucleases. We demonstrated the sensitivity of our nuclease-activated probe technology by detecting attomolar (10−18 molar) levels of a bacterial nuclease in the human plasma.30 In the present study, we sought to exploit the potential of the nuclease-activated probe technology to diagnose cancer. The challenge here was to reliably distinguish between mammalian nucleases expressed in cancer cells versus those expressed in leukocytes. Although we determined that nuclease gene expression is elevated in BCa compared with whole blood (Figure 2A), the sheer number of WBCs is still likely to overcome any signals from the cancer cells (Figure 4A versus 4B). Fortunately, the tantalizing possibility that CTCs can serve as a liquid biopsy to diagnose the likelihood of metastatic spread or to assess treatment outcome has prompted intensive efforts focused on the development of novel high-performance CTC capture methods, which provided a solution to this problem.62, 63, 64, 65
Although improvements in affinity-capture methods (e.g., microfluidics) offer significant improvements in the sensitivity and specificity of CTC capture and analysis compared with the gold standard (CELLSEARCH), these methods have an obvious drawback that limits their use: the difficulty of implementation requiring complex technical expertise and infrastructure.19, 66, 67 Next-generation size-based separation approaches are emerging as a promising alternative to this problem. Size-based separation approaches leverage the difference in size of tumor cells versus the normal cell types detected in blood. Many tumor cells have diameters that are significantly larger than normal WBCs or RBCs, thus enabling their capture with high-precision filtration. A clear advantage of the size filtration methods is the ease of operation and implementation. In our proof-of-concept study, we evaluated two different microfiltration methods for the capture of CTCs (Figure S2). We found that the SCREENCELL microfiltration unit was superior to the Rarecells (ISET) filtration device and displayed a significantly lower normal blood cell retention rate, resulting in an overall lower assay background signal (Figure S2). While the fraction of cancer cells recovered from blood with the SCREENCELL microfilter unit positively correlates with the number of cancer cells present in blood and was lower in samples with fewer cancer cells (Figure 5), the effective capture of BCa cells of small diameter was also possible (e.g., SKBr3) (Figure S3). Importantly, the sensitivity of the nuclease-activated probe assay was increased by 400-fold with the addition of the SCREENCELL capture and/or enrichment step (Figures 4B and 4C), enabling the robust detection of cancer cells in patient blood (Figure 6). Based on recent estimates of CTC concentrations in blood of patients with BCa, as expected, this level of sensitivity enabled the robust detection of cancer cells in patient blood (Figure 6). These data are also consistent with the notion that previous estimates of CTC concentrations (e.g., as low as 1–5 cells per 1 mL of blood), which are derived from data obtained with EpCAM-based CTC capture methodology (CELLSEARCH), are gross underestimates.13, 14, 50, 68
In summary, we describe a novel protocol for the detection of CTCs that overcomes many of the limitations of current CTC detection assays and provides a promising avenue for clinical implementation of a robust diagnostic tool for BCa in combination with mammography. Our two-step assay (step 1 = capture; step 2 = detection), which combines novel advances in CTC capture technology with the nuclease-activated probe technology, is easily performed within 1 hr and requires minimal technical expertise and infrastructure. Finally, because the expression of nucleases is upregulated in many cancers, the technology can be readily extended to develop diagnostic assays for other cancers in need of better diagnostic options, including pancreatic, lung, and ovarian cancers. Although the initial intended uses of the NucAP assay are as a surveillance test or a therapy treatment response tool in patients with known BCa, future studies will explore refinements that might allow development of a related screening test.
Materials and Methods
Patient Population
This study was reviewed and approved by The University of Iowa Internal Review Board (IRB 201505719). Patients with stage IV BCa, ages 18–90, hemoglobin levels ≥10.5 g/dL, with no history of blood transfusions in the prior month provided informed consent at the Holden Comprehensive Cancer Center to contribute extra blood samples and clinical data to the study. Any patient blood samples were de-identified by using a code ID. Clinical data on age, gender, and information about disease and treatments were abstracted from the subjects’ electronic medical record linked to the blood samples with a code ID. Written informed consent was also obtained from all participants (healthy donors and patients).
Cell Culture
Cell lines used in this study include BT20 (catalog number [Cat. No.] HTB-19; ATCC, Manassas, VA, USA), BT474 (Cat. No. HTB-20; ATCC), HCC1937 (Cat. No. CRL-2336; ATCC), K562 (Cat. No. CCL-243; ATCC), MCF-7 (Cat. No. HTB-22; ATCC), MDA-MB-231 (Cat. No. HTB-26; ATCC), MDA-MB-453 (Cat. No. HTB-131; ATCC), MDA-MB-468 (Cat. No. HTB-132; ATCC), Iowa1T (Cat. No. CRL-3292; ATCC), SKBr3 (Cat. No. HTB-30; ATCC). All cell lines were cultured under the conditions recommended by the vendor.
Patient Samples
A maximum of about 20 mL of blood at three time points was provided by the subjects at the Clinical Cancer Center. Blood was collected in portions of 6 mL in EDTA-K2-containing vacutainers and was pooled before use.
Oligonucleotide Probes
The oligonucleotide probes used in this study were obtained from Integrated DNA Technologies (Coralville, IA, USA). The sequences of the probes were as follows: dsDNA: 5′-FAM-CTACGTAG-ZEN-RQ-3′; ssDNA: 5′-FAM-TCTCGTACGTAC-ZEN-RQ-3′; and 2′F RNA: 5′-FAM-UFCFUFCFGUFACFGUFACF-ZEN-RQ-3′. The 2′F RNA probe includes 2′-fluoro-modified pyrimidines (CF and UF); the purines are unmodified DNA nucleotides. ZEN and RQ are the ZEN and Iowa Black RQ fluorescence quenchers, respectively, of IDT.
Lysis Buffer Optimization
The nuclease activity in cell lysates was measured in buffers containing different concentrations of MgCl2 (0, 2, 5, and 10 mM) and CaCl2 (0, 2, 5, and 10 mM), and different pH levels (pH 7, 8, 9, and 10). SKBr3 or MDA-MB-231 BCa cells were lysed in a solution of PBS plus 1% Triton X-100. Buffers were exchanged using 3.5K MWCO 96-well microdialysis plates (product #88262; Pierce, Thermo Scientific, Rockford, IL, USA) by dialyzing 50 μL of cell lysate against the various buffers. The dialysis buffer was exchanged three times, with dialysis occurring for 2 hr with two buffer exchanges, and a third dialysis buffer exchange occurring overnight. The composition of the optimized lysis buffer was 10 mM MgCl2, 150 mM NaCl, and 10 mM Tris-HCl (pH 9.0), 1 mM DTT, 1% Triton X-100, and 1/10 tablet of protease inhibitor (product #05 892 791 001; Roche) per milliliter.
BCa Cell Line Nuclease Activity Screening
Lysates of 100 BT20, MDA-MB-468, HCC1937, MDA-MB-231, MDA-MB-453, Iowa1T, MCF7, SKBr3, or BT474 cells in optimized lysis buffer were incubated with 2.5 pmol of ssDNA, dsDNA, or 2′F-RNA probe at 37°C for 6 hr. Fluorescence (due to probe digestion) was measured every 20 min using a Synergy Mx Multi-Mode Microplate Reader. The fold activity of each BCa cell line lysate was compared with that of lysates of 100 K562 cells.
ISET Filtration Capture Method
Three milliliters of whole blood spiked with BCa cells was centrifuged at 750 × g for 10 min at room temperature, and plasma was separated from the RBC pellet containing leukocytes and cancer cells. The RBCs in the pellet were lysed by resuspension in 10 mL of 0.42% NH4Cl and rocking for 10 min at room temperature. The leukocytes and cancer cells were pelleted by centrifugation at 750 × g for 10 min, and the supernatant containing the lysed RBCs was removed. The pelleted leukocytes and cancer cells were washed twice by resuspension in PBS and pelleted by centrifugation at 750 × g for 10 min. Following the lysis and washing steps, the cell samples were resuspended in 1.5 mL of PBS. The cell samples were then filtered using pre-wet polycarbonate membranes containing 8-μm diameter pores (Maine Manufacturing; product #4663G49 purchased from Thomas Scientific, Swedesboro, NJ, USA). The polycarbonate membranes were cut into 3 cm × 3 cm squares and placed on a 25-mm diameter Easy Pressure Syringe Filter Holder (Pall; Cat. No. FMFNL1050). The membranes were initially wetted by placing 1 mL of PBS onto the membrane and applying a vacuum to pull the liquid through the membrane, which allowed the membrane to be stabilized onto the filter holder before filtering the resuspended cells. After filtration of 1.5 mL of cells, the membrane was washed twice with 1 mL of PBS and by applying gentle vacuum to pull the liquid through the membrane. The membrane was then placed into a 1.5 mL Eppendorf tube, fully submerged into 200 μL of optimized lysis buffer, vortexed for 30 s, and frozen at −80°C. For the nuclease activity assays, 10 μL of cell lysate plus 1 μL of 2.5 μM ssDNA, dsDNA, or 2′F RNA probe was placed into a well in a black 384-well flat-bottom plate (product #264705; Nunc, Thermo Fisher Scientific, Rochester, NY, USA). Three replicate wells were used per cell lysate and probe combination. Digestion of the probe was monitored by measuring fluorescence every 20 min in a Synergy Mx Multi-Mode Microplate Reader for 6 hr at 37°C.
SCREENCELL Filtration Capture Method
Cancer cells were isolated from blood using filters purchased from SCREENCELL (Sarcelles, France). SCREENCELL MB-V2 kits were used for obtaining CTCs for the nuclease activity assays. Filtration of the whole blood samples was performed per the manufacturer’s instructions immediately after drawing the blood in K2 EDTA vacutainers (BD, Franklin Lakes, NJ, USA). Briefly, for the MB-V2 filters, 6 mL of blood was placed in a 15 mL RNase-/DNase-free conical tube and 1 mL of the SCREENCELL LC Buffer was added to the blood, which was then inverted five times and incubated at room temperature for 5 min. The blood plus LC Buffer was then placed in the top of the MB-V2 filter, which was then pressed onto the vacutainer included with the kit to pull the blood through the filter. After the blood reached the appropriate level (indicated by a yellow line on the filtration module), 1.6 mL of PBS was added to rinse the filter. Once the PBS had flowed through the filter, the capsule filter was ejected into the Eppendorf tube included with the kit, and 66.7 μL of optimized lysis buffer was added to the filter, which was then capped, vortexed, and incubated on ice for 5 min. The Eppendorf tube containing the capsule filter was then centrifuged at 3,000 × g for 2 min to collect the lysate in the bottom of the Eppendorf tube. This was repeated two more times, resulting in a final lysate volume of 200 μL. Before centrifuging the third time, the Eppendorf tubes containing the capsule filter were frozen at −80°C to maximize lysis of the cells on the filter and were stored until the nuclease-activated probe assay was performed. Upon thawing the lysates, the Eppendorf tube containing the capsule filter was vortexed and centrifuged at 3,000 × g a final time for 2 min. For the nuclease-activated probe activity assays, 10 μL of cell lysate plus 1 μL of 2.5 μM ssDNA, dsDNA, or 2′F-RNA probe was placed into a well in a black 384-well flat-bottom plate (product 264705; Nunc, Thermo Fisher Scientific). Triplicate wells were used per cell lysate and probe combination. Degradation of the probe was monitored by measuring the fluorescence every 20 min in a Synergy Mx Multi-Mode Microplate Reader for 6 hr at 37°C.
RNA Isolation from BCa Cell Lines and Whole Blood
For RNA extraction of BCa cells in cell culture, TRIzol Reagent (15596-026; Ambion; Life Technologies, Carlsbad, CA, USA) was directly added to pelleted cells. For whole blood, 1 mL of freshly drawn blood, collected in a collection tube containing EDTA (367856; BD Medical), was centrifuged at 2,500 × g for 10 min, and the interphase layer (“buffy coat” containing WBCs) between the upper plasma phase and the lower RBC phase was collected. After three washes with 500 μL of chilled PBS (centrifugation at 3,500 × g for 2 min), the resulting cell pellet was resuspended in 1 mL of chilled 0.42% NH4Cl solution and incubated with constant rocking on ice for 20 min to lyse RBCs. WBCs were then pelleted at 3,500 × g for 2 min and washed with PBS (3,500 × g for 2 min).
Total RNA was extracted by using TRIzol Reagent following the manufacturer’s instructions. Briefly, 1 mL of TRIzol was added to the cell pellet in a 1.5 mL reaction tube (containing WBCs from 1 mL of blood), mixed by pipetting up and down, and incubated at room temperature for 5 min. Then, 200 μL of chloroform was added, vortexed for 15 s, incubated for 3 min on ice, and centrifuged at 12,000 × g for 10 min for phase separation. The aqueous phase was pipetted into a new vessel, and 500 μL of chilled isopropanol was added. Mixing and incubation of the sample for at least 10 min on ice was then followed by centrifugation at 12,000 × g for 10 min to yield the final RNA pellet. After aspiration of the supernatant, the pellet was washed by addition of 300 μL of chilled 70% EtOH and centrifuged at 12,000 × g for 5 min. The dried pellet was resuspended in RNase-free water, and the RNA concentration was determined using a NanoDrop UV-Vis spectrometer. RNA was stored at −20°C until the qRT-PCR was performed.
qRT-PCR
To obtain the cDNA from the RNA from whole blood and whole blood spiked with cancer cell samples, we used SuperScript III Reverse Transcriptase (18080093; Thermo Fisher Scientific) following the manufacturer’s instructions. Briefly, in 20 μL reaction volume, 2 μg of total RNA was incubated in the presence of 1 mM dinucleotide triphosphate (dNTP) and 5 ng/μL random hexamer primers at 65°C for 5 min, then placed on ice for at least 1 min prior to the addition of the cDNA Synthesis Mix that included RT-enzyme and reaction buffer. Temperature conditions during the first strand synthesis were 25°C for 10 min, 50°C for 50 min, followed by 85°C for 5 min. The synthesized cDNA was then quantified and amplified via qPCR using the iQ SYBR Green Supermix (#170-8880; Bio-Rad, Hercules, CA, USA) following the manufacturer’s instructions. Sequences (the annealing temperature was 58°C for all primers) for each primer pair were as follows: β-actin: 5′-CAAGAGATGGCCACGGCTGCT-3′ (forward [fwd]), 5′-TCCTTCTGCATCCTGTCGGCA-3′ (reverse [rev]); CD45: 5′-TGTTTCTTAGGGACACGGCT-3′ (fwd), 5′-CCACCAACTGAAGGCTGAAC-3′ (rev); CK19: 5′-GAGCAGGTCCGAGGTTACTG-3′ (fwd), 5′-GCTCACTATCAGCTCGCACA-3′ (rev); EpCAM: 5′-CATGTGCTGGTGTGTGAACA-3′ (fwd), 5′-CGCGTTGTGATCTCCTTCTG-3′ (rev); ERα: 5′-CCACCAACCAGTGCACCATT-3′ (fwd), 5′-GGTCTTTTCGTATCCCACCTTTC-3′ (rev); Her2: 5′-TGTGTGGGAGCTGATGACTT-3′ (fwd), 5′-TCTTGGCCGACATTCAGAGT-3′ (rev); Plastin3: 5′-CCTTCCGTAACTGGATGAACTC-3′ (fwd), 5′-GGATGCTTCCCTAATTCAACAG-3′ (rev).
Immunostaining of Cancer Cells Captured by SCREENCELL Filtration
ScSCREENCELL CYTO kits were used for immunostaining of cancer cells isolated from the blood. For immunostaining, the protocol provided by SCREENCELL was followed. Briefly, cytofilters used for filtration were washed with 1× Tris-buffered saline (TBS) for 10 min. For antigen retrieval, samples were treated with Target Retrieval Solution (S1700; Dako) at 95°C for 20 min. The cytofilters were allowed to cool to room temperature before rinsing with 1× TBS, followed by a 5 min incubation in permeabilizing buffer (1× TBS, 0.2% Triton X-100). Next, the samples were incubated in 3% BSA at room temperature for 30 min. Cytofilters were subsequently incubated with the following primary antibodies in parallel in 3% BSA at 4°C overnight: anti-Pan Cytokeratin (MA5-17687; Dako) at 1:50 and anti-EpCAM (Ab71916; Abcam) at 1:100. After rinsing with Wash Buffer (1× TBS, 0.05% Tween 20), the samples were stained with the following secondary antibodies in parallel at 1:1,000 in 3% BSA for 1 hr at room temperature: goat anti-mouse Alexa 488 and goat anti-rat Alexa 594 (A11001, A11007; Life Technologies). Finally, cytofilters were rinsed with Wash Buffer and protected with ProLong Diamond Mountant containing DAPI nuclear stain (P36962; Life Technologies). Images were acquired using a Leica SP8 STED at the University of Iowa Central Microscopy Research Facility.
Bioinformatics Analysis of Genes Corresponding to Nuclease Proteins
Curated RNA sequencing data were queried from the RNA sequencing (RNA-seq) of 675 commonly used human cancer cell lines and from the RNA-seq from 53 human tissue samples from the Genotype-Tissue Expression (GTEx) Project datasets located on the EMBL-EBI expression atlas. From these datasets, 160 genes with putative nuclease activity (GO: 0004518) from the Amigo-Gene Ontology Consortium were queried in 60 BCa cell lines and whole blood. The average gene expression for the 160 genes was calculated across the 60 BCa cell lines. To calculate the fold change, we divided the average of each nuclease gene from the BCa cell lines by the corresponding gene’s expression in whole blood. mRNA expression of EpCAM and the 160 nuclease genes from 60 BCa cell lines (675 commonly used human cancer cell lines) or 941 tissue samples (TCGA) were normalized as follows: for EpCAM expression, the EpCAM mRNA expression in each cell line or tissue was normalized to the average EpCAM mRNA expression across all cell lines or tissues. For the nuclease genes, the sum of all 160 genes was calculated across all cell lines or tissues and normalized to (divided by) the average nuclease gene expression for each of the cell lines or tissues.
Statistical Analysis
All values are expressed as means with SDs if not stated otherwise. Comparisons between two groups were performed using GraphPad Prism 6 (Graph Pad, San Diego, CA, USA) by a two-tailed unpaired t test. Differences between two means with *p < 0.05 considered significant, **p < 0.01 very significant, and ***p < 0.001 extremely significant, with ****p < 0.0001 being the highest level of significance. ROC analysis was performed to assess the diagnostic accuracy of the nuclease-activated probes studied. Specifically, the accuracy of probe fluorescence intensity in discriminating between BCa patients and healthy donors was estimated with area under the ROC curve (AUC). AUCs were estimated over the range of assay run times (0–360 min) for each of the three probes (dsDNA, ssDNA, and 2′F-RNA). Estimates are reported along with 95% CIs. ROC analysis results were assessed for statistical significance at the 5% level and obtained with the IBM SPSS statistical software version 23.0.
Data Availability
Gene expression data are available for 675 commonly used human cancer cell lines and RNA-seq from 53 human tissue samples on the EMBL-EBI database (http://www.ebi.ac.uk). The results published here are in part based upon data generated by the TCGA Research Network and are located at https://cancergenome.nih.gov. Raw images are available upon request.
Author Contributions
S.K., D.D.D., G.N.B., K.T.U., K.C.C., E.B., and P.H.G. designed and conducted all aspects of the experiments. K.T.U., S.K., and A.T. provided the human blood samples for the study. A.T. and C.H.C. provided clinical data and clinical insight. J.O.M., H.O., and C.H.C. contributed to the experimental design and edited the manuscript. S.K. drafted the manuscript. P.H.G. oversaw the overall project, designed experiments, was responsible for funding, and drafted and edited the manuscript.
Acknowledgments
The authors would like to thank all donors involved in the study. They also acknowledge the use of the University of Iowa Central Microscopy Research Facility, a core resource supported by the Vice President for Research & Economic Development, the Holden Comprehensive Cancer Center, and the Carver College of Medicine. Additionally, the authors thank Earl Gingrich (University of Iowa) and Sysmex America for technical advice and support on the whole blood cell count method. S.K. was supported by the DFG (Deutsche Forschungsgemeinschaft). D.D.D. was supported by a postdoctoral training grant from the NIH (T32 HL07344). This work was supported by grants from the NIH (R01CA138503 to P.H.G.; AI101391 and AI106738 to J.O.M.), Mary Kay Foundation (9033-12 and 001-09 to P.H.G.), and Roy J Carver Charitable Trust (RJCCT 01-224 to P.H.G.).
Footnotes
Supplemental Information includes five figures and one table and can be found with this article online at http://dx.doi.org/10.1016/j.omtn.2017.08.004.
Contributor Information
Carlos H. Chan, Email: carloshfchan@gmail.com.
Paloma H. Giangrande, Email: paloma-giangrande@uiowa.edu.
Supplemental Information
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2016. CA Cancer J. Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Fehm T., Müller V., Alix-Panabières C., Pantel K. Micrometastatic spread in breast cancer: detection, molecular characterization and clinical relevance. Breast Cancer Res. 2008;10(Suppl 1):S1. doi: 10.1186/bcr1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pantel K., Brakenhoff R.H., Brandt B. Detection, clinical relevance and specific biological properties of disseminating tumour cells. Nat. Rev. Cancer. 2008;8:329–340. doi: 10.1038/nrc2375. [DOI] [PubMed] [Google Scholar]
- 4.Cristofanilli M., Budd G.T., Ellis M.J., Stopeck A., Matera J., Miller M.C., Reuben J.M., Doyle G.V., Allard W.J., Terstappen L.W., Hayes D.F. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N. Engl. J. Med. 2004;351:781–791. doi: 10.1056/NEJMoa040766. [DOI] [PubMed] [Google Scholar]
- 5.Cristofanilli M., Hayes D.F., Budd G.T., Ellis M.J., Stopeck A., Reuben J.M., Doyle G.V., Matera J., Allard W.J., Miller M.C. Circulating tumor cells: a novel prognostic factor for newly diagnosed metastatic breast cancer. J. Clin. Oncol. 2005;23:1420–1430. doi: 10.1200/JCO.2005.08.140. [DOI] [PubMed] [Google Scholar]
- 6.Nadal R., Lorente J.A., Rosell R., Serrano M.J. Relevance of molecular characterization of circulating tumor cells in breast cancer in the era of targeted therapies. Expert Rev. Mol. Diagn. 2013;13:295–307. doi: 10.1586/erm.13.7. [DOI] [PubMed] [Google Scholar]
- 7.Aurilio G., Sciandivasci A., Munzone E., Sandri M.T., Zorzino L., Cassatella M.C., Verri E., Rocca M.C., Nolè F. Prognostic value of circulating tumor cells in primary and metastatic breast cancer. Expert Rev. Anticancer Ther. 2012;12:203–214. doi: 10.1586/era.11.208. [DOI] [PubMed] [Google Scholar]
- 8.Hayes D.F., Cristofanilli M., Budd G.T., Ellis M.J., Stopeck A., Miller M.C., Matera J., Allard W.J., Doyle G.V., Terstappen L.W. Circulating tumor cells at each follow-up time point during therapy of metastatic breast cancer patients predict progression-free and overall survival. Clin. Cancer Res. 2006;12:4218–4224. doi: 10.1158/1078-0432.CCR-05-2821. [DOI] [PubMed] [Google Scholar]
- 9.Bidard F.C., Peeters D.J., Fehm T., Nolé F., Gisbert-Criado R., Mavroudis D., Grisanti S., Generali D., Garcia-Saenz J.A., Stebbing J. Clinical validity of circulating tumour cells in patients with metastatic breast cancer: a pooled analysis of individual patient data. Lancet Oncol. 2014;15:406–414. doi: 10.1016/S1470-2045(14)70069-5. [DOI] [PubMed] [Google Scholar]
- 10.Pantel K., Alix-Panabières C. Detection methods of circulating tumor cells. J. Thorac. Dis. 2012;4:446–447. doi: 10.3978/j.issn.2072-1439.2012.08.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hyun K.A., Koo G.B., Han H., Sohn J., Choi W., Kim S.I., Jung H.I., Kim Y.S. Epithelial-to-mesenchymal transition leads to loss of EpCAM and different physical properties in circulating tumor cells from metastatic breast cancer. Oncotarget. 2016;7:24677–24687. doi: 10.18632/oncotarget.8250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fina E., Callari M., Reduzzi C., D’Aiuto F., Mariani G., Generali D., Pierotti M.A., Daidone M.G., Cappelletti V. Gene expression profiling of circulating tumor cells in breast cancer. Clin. Chem. 2015;61:278–289. doi: 10.1373/clinchem.2014.229476. [DOI] [PubMed] [Google Scholar]
- 13.Gorges T.M., Tinhofer I., Drosch M., Röse L., Zollner T.M., Krahn T., von Ahsen O. Circulating tumour cells escape from EpCAM-based detection due to epithelial-to-mesenchymal transition. BMC Cancer. 2012;12:178. doi: 10.1186/1471-2407-12-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Königsberg R., Obermayr E., Bises G., Pfeiler G., Gneist M., Wrba F., de Santis M., Zeillinger R., Hudec M., Dittrich C. Detection of EpCAM positive and negative circulating tumor cells in metastatic breast cancer patients. Acta Oncol. 2011;50:700–710. doi: 10.3109/0284186X.2010.549151. [DOI] [PubMed] [Google Scholar]
- 15.Hong B., Zu Y. Detecting circulating tumor cells: current challenges and new trends. Theranostics. 2013;3:377–394. doi: 10.7150/thno.5195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sheng Y., Wang T., Li H., Zhang Z., Chen J., He C., Li Y., Lv Y., Zhang J., Xu C. Comparison of analytic performances of Cellsearch and iFISH approach in detecting circulating tumor cells. Oncotarget. 2017;8:8801–8806. doi: 10.18632/oncotarget.6688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iwata H., Masuda N., Yamamoto D., Sagara Y., Sato N., Yamamoto Y., Saito M., Fujita T., Oura S., Watanabe J. Circulating tumor cells as a prognostic marker for efficacy in the randomized phase III JO21095 trial in Japanese patients with HER2-negative metastatic breast cancer. Breast Cancer Res. Treat. 2017;162:501–510. doi: 10.1007/s10549-017-4138-3. [DOI] [PubMed] [Google Scholar]
- 18.Che J., Yu V., Dhar M., Renier C., Matsumoto M., Heirich K., Garon E.B., Goldman J., Rao J., Sledge G.W. Classification of large circulating tumor cells isolated with ultra-high throughput microfluidic Vortex technology. Oncotarget. 2016;7:12748–12760. doi: 10.18632/oncotarget.7220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Green B.J., Saberi Safaei T., Mepham A., Labib M., Mohamadi R.M., Kelley S.O. Beyond the capture of circulating tumor cells: next-generation devices and materials. Angew. Chem. Int. Ed. Engl. 2016;55:1252–1265. doi: 10.1002/anie.201505100. [DOI] [PubMed] [Google Scholar]
- 20.Qian W., Zhang Y., Chen W. Capturing cancer: emerging microfluidic technologies for the capture and characterization of circulating tumor cells. Small. 2015;11:3850–3872. doi: 10.1002/smll.201403658. [DOI] [PubMed] [Google Scholar]
- 21.Gogoi P., Sepehri S., Zhou Y., Gorin M.A., Paolillo C., Capoluongo E., Gleason K., Payne A., Boniface B., Cristofanilli M. Development of an automated and sensitive microfluidic device for capturing and characterizing circulating tumor cells (CTCs) from clinical blood samples. PLoS ONE. 2016;11:e0147400. doi: 10.1371/journal.pone.0147400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xue Y., Marvin M.E., Ivanova I.G., Lydall D., Louis E.J., Maringele L. Rif1 and Exo1 regulate the genomic instability following telomere losses. Aging Cell. 2016;15:553–562. doi: 10.1111/acel.12466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Singh P., Yang M., Dai H., Yu D., Huang Q., Tan W., Kernstine K.H., Lin D., Shen B. Overexpression and hypomethylation of flap endonuclease 1 gene in breast and other cancers. Mol. Cancer Res. 2008;6:1710–1717. doi: 10.1158/1541-7786.MCR-08-0269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen B., Zhang Y., Wang Y., Rao J., Jiang X., Xu Z. Curcumin inhibits proliferation of breast cancer cells through Nrf2-mediated down-regulation of Fen1 expression. J. Steroid Biochem. Mol. Biol. 2014;143:11–18. doi: 10.1016/j.jsbmb.2014.01.009. [DOI] [PubMed] [Google Scholar]
- 25.Jariwala N., Rajasekaran D., Srivastava J., Gredler R., Akiel M.A., Robertson C.L., Emdad L., Fisher P.B., Sarkar D. Role of the staphylococcal nuclease and tudor domain containing 1 in oncogenesis (review) Int. J. Oncol. 2015;46:465–473. doi: 10.3892/ijo.2014.2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blanco M.A., Alečković M., Hua Y., Li T., Wei Y., Xu Z., Cristea I.M., Kang Y. Identification of staphylococcal nuclease domain-containing 1 (SND1) as a Metadherin-interacting protein with metastasis-promoting functions. J. Biol. Chem. 2011;286:19982–19992. doi: 10.1074/jbc.M111.240077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang W. Nucleases: diversity of structure, function and mechanism. Q. Rev. Biophys. 2011;44:1–93. doi: 10.1017/S0033583510000181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hernandez F.J., Huang L., Olson M.E., Powers K.M., Hernandez L.I., Meyerholz D.K., Thedens D.R., Behlke M.A., Horswill A.R., McNamara J.O., 2nd Noninvasive imaging of Staphylococcus aureus infections with a nuclease-activated probe. Nat. Med. 2014;20:301–306. doi: 10.1038/nm.3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hernandez F.J., Stockdale K.R., Huang L., Horswill A.R., Behlke M.A., McNamara J.O., 2nd Degradation of nuclease-stabilized RNA oligonucleotides in Mycoplasma-contaminated cell culture media. Nucleic Acid Ther. 2012;22:58–68. doi: 10.1089/nat.2011.0316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burghardt E.L., Flenker K.S., Clark K.C., Miguel J., Ince D., Winokur P., Ford B., McNamara J.O., 2nd Rapid, culture-free detection of Staphylococcus aureus bacteremia. PLoS ONE. 2016;11:e0157234. doi: 10.1371/journal.pone.0157234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Birney E., Clamp M., Hubbard T. Databases and tools for browsing genomes. Annu. Rev. Genomics Hum. Genet. 2002;3:293–310. doi: 10.1146/annurev.genom.3.030502.101529. [DOI] [PubMed] [Google Scholar]
- 32.Perez-Riverol Y., Alpi E., Wang R., Hermjakob H., Vizcaíno J.A. Making proteomics data accessible and reusable: current state of proteomics databases and repositories. Proteomics. 2015;15:930–949. doi: 10.1002/pmic.201400302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoa N.N., Kobayashi J., Omura M., Hirakawa M., Yang S.H., Komatsu K., Paull T.T., Takeda S., Sasanuma H. BRCA1 and CtIP are both required to recruit Dna2 at double-strand breaks in homologous recombination. PLoS ONE. 2015;10:e0124495. doi: 10.1371/journal.pone.0124495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karanja K.K., Cox S.W., Duxin J.P., Stewart S.A., Campbell J.L. DNA2 and EXO1 in replication-coupled, homology-directed repair and in the interplay between HDR and the FA/BRCA network. Cell Cycle. 2012;11:3983–3996. doi: 10.4161/cc.22215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rolseth V., Krokeide S.Z., Kunke D., Neurauter C.G., Suganthan R., Sejersted Y., Hildrestrand G.A., Bjørås M., Luna L. Loss of Neil3, the major DNA glycosylase activity for removal of hydantoins in single stranded DNA, reduces cellular proliferation and sensitizes cells to genotoxic stress. Biochim. Biophys. Acta. 2013;1833:1157–1164. doi: 10.1016/j.bbamcr.2012.12.024. [DOI] [PubMed] [Google Scholar]
- 36.Sokolenko A.P., Preobrazhenskaya E.V., Aleksakhina S.N., Iyevleva A.G., Mitiushkina N.V., Zaitseva O.A., Yatsuk O.S., Tiurin V.I., Strelkova T.N., Togo A.V., Imyanitov E.N. Candidate gene analysis of BRCA1/2 mutation-negative high-risk Russian breast cancer patients. Cancer Lett. 2015;359:259–261. doi: 10.1016/j.canlet.2015.01.022. [DOI] [PubMed] [Google Scholar]
- 37.Valladares A., Hernández N.G., Gómez F.S., Curiel-Quezada E., Madrigal-Bujaidar E., Vergara M.D., Martínez M.S., Arenas Aranda D.J. Genetic expression profiles and chromosomal alterations in sporadic breast cancer in Mexican women. Cancer Genet. Cytogenet. 2006;170:147–151. doi: 10.1016/j.cancergencyto.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 38.Gorges T.M., Riethdorf S., von Ahsen O., Nastał Y P., Röck K., Boede M., Peine S., Kuske A., Schmid E., Kneip C. Heterogeneous PSMA expression on circulating tumor cells: a potential basis for stratification and monitoring of PSMA-directed therapies in prostate cancer. Oncotarget. 2016;7:34930–34941. doi: 10.18632/oncotarget.9004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gostner J.M., Fong D., Wrulich O.A., Lehne F., Zitt M., Hermann M., Krobitsch S., Martowicz A., Gastl G., Spizzo G. Effects of EpCAM overexpression on human breast cancer cell lines. BMC Cancer. 2011;11:45. doi: 10.1186/1471-2407-11-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dickey D.D., Giangrande P.H. Oligonucleotide aptamers: a next-generation technology for the capture and detection of circulating tumor cells. Methods. 2016;97:94–103. doi: 10.1016/j.ymeth.2015.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Adams D.L., Stefansson S., Haudenschild C., Martin S.S., Charpentier M., Chumsri S., Cristofanilli M., Tang C.M., Alpaugh R.K. Cytometric characterization of circulating tumor cells captured by microfiltration and their correlation to the CellSearch(®) CTC test. Cytometry A. 2015;87:137–144. doi: 10.1002/cyto.a.22613. [DOI] [PubMed] [Google Scholar]
- 42.Alix-Panabières C., Pantel K. Circulating tumor cells: liquid biopsy of cancer. Clin. Chem. 2013;59:110–118. doi: 10.1373/clinchem.2012.194258. [DOI] [PubMed] [Google Scholar]
- 43.Friedlander T.W., Premasekharan G., Paris P.L. Looking back, to the future of circulating tumor cells. Pharmacol. Ther. 2014;142:271–280. doi: 10.1016/j.pharmthera.2013.12.011. [DOI] [PubMed] [Google Scholar]
- 44.Mostert B., Sleijfer S., Foekens J.A., Gratama J.W. Circulating tumor cells (CTCs): detection methods and their clinical relevance in breast cancer. Cancer Treat. Rev. 2009;35:463–474. doi: 10.1016/j.ctrv.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 45.Lianidou E.S., Markou A., Strati A. The role of CTCs as tumor biomarkers. Adv. Exp. Med. Biol. 2015;867:341–367. doi: 10.1007/978-94-017-7215-0_21. [DOI] [PubMed] [Google Scholar]
- 46.Sieuwerts A.M., Kraan J., Bolt J., van der Spoel P., Elstrodt F., Schutte M., Martens J.W., Gratama J.W., Sleijfer S., Foekens J.A. Anti-epithelial cell adhesion molecule antibodies and the detection of circulating normal-like breast tumor cells. J. Natl. Cancer Inst. 2009;101:61–66. doi: 10.1093/jnci/djn419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Onstenk W., Kraan J., Mostert B., Timmermans M.M., Charehbili A., Smit V.T., Kroep J.R., Nortier J.W., van de Ven S., Heijns J.B. Improved circulating tumor cell detection by a combined EpCAM and MCAM CellSearch enrichment approach in patients with breast cancer undergoing neoadjuvant chemotherapy. Mol. Cancer Ther. 2015;14:821–827. doi: 10.1158/1535-7163.MCT-14-0653. [DOI] [PubMed] [Google Scholar]
- 48.Schneck H., Gierke B., Uppenkamp F., Behrens B., Niederacher D., Stoecklein N.H., Templin M.F., Pawlak M., Fehm T., Neubauer H., Disseminated Cancer Cell Network (DCC Net) Duesseldorf EpCAM-independent enrichment of circulating tumor cells in metastatic breast cancer. PLoS ONE. 2015;10:e0144535. doi: 10.1371/journal.pone.0144535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Beije N., Jager A., Sleijfer S. Circulating tumor cell enumeration by the CellSearch system: the clinician’s guide to breast cancer treatment? Cancer Treat. Rev. 2015;41:144–150. doi: 10.1016/j.ctrv.2014.12.008. [DOI] [PubMed] [Google Scholar]
- 50.Fina E., Reduzzi C., Motta R., Di Cosimo S., Bianchi G., Martinetti A., Wechsler J., Cappelletti V., Daidone M.G. Did circulating tumor cells tell us all they could? The missed circulating tumor cell message in breast cancer. Int. J. Biol. Markers. 2015;30:e429–e433. doi: 10.5301/jbm.5000166. [DOI] [PubMed] [Google Scholar]
- 51.Hanauer D.A., Rhodes D.R., Sinha-Kumar C., Chinnaiyan A.M. Bioinformatics approaches in the study of cancer. Curr. Mol. Med. 2007;7:133–141. doi: 10.2174/156652407779940431. [DOI] [PubMed] [Google Scholar]
- 52.Drake W.P., Kopyta L.P., Levy C.C., Mardiney M.R., Jr. Alterations in ribonuclease activities in the plasma, spleen, and thymus of tumor-bearing mice. Cancer Res. 1975;35:322–324. [PubMed] [Google Scholar]
- 53.Drake W.P., Pokorney D.R., Chipman S., Levy C.C., Mardiney M.R., Jr. Elevated ribonuclease activity in the thymus and white blood cells of genetically cancer prone mice. J. Exp. Med. 1975;141:918–923. [PMC free article] [PubMed] [Google Scholar]
- 54.Drake W.P., Schmukler M., Pendergrast W.J., Jr., Davis A.S., Lichtenfeld J.L., Mardiney M.R., Jr. Abnormal profile of human nucleolytic activity as a test for cancer. J. Natl. Cancer Inst. 1975;55:1055–1059. doi: 10.1093/jnci/55.5.1055. [DOI] [PubMed] [Google Scholar]
- 55.Wang H.C., Chiu C.F., Tsai R.Y., Kuo Y.S., Chen H.S., Wang R.F., Tsai C.W., Chang C.H., Lin C.C., Bau D.T. Association of genetic polymorphisms of EXO1 gene with risk of breast cancer in Taiwan. Anticancer Res. 2009;29:3897–3901. [PubMed] [Google Scholar]
- 56.Samadder P., Aithal R., Belan O., Krejci L. Cancer TARGETases: DSB repair as a pharmacological target. Pharmacol. Ther. 2016;161:111–131. doi: 10.1016/j.pharmthera.2016.02.007. [DOI] [PubMed] [Google Scholar]
- 57.Doherty R., Madhusudan S. DNA repair endonucleases: physiological roles and potential as drug targets. J. Biomol. Screen. 2015;20:829–841. doi: 10.1177/1087057115581581. [DOI] [PubMed] [Google Scholar]
- 58.Bartosova Z., Krejci L. Nucleases in homologous recombination as targets for cancer therapy. FEBS Lett. 2014;588:2446–2456. doi: 10.1016/j.febslet.2014.06.010. [DOI] [PubMed] [Google Scholar]
- 59.Gong C., Liu B., Yao Y., Qu S., Luo W., Tan W., Liu Q., Yao H., Zou L., Su F., Song E. Potentiated DNA damage response in circulating breast tumor cells confers resistance to chemotherapy. J. Biol. Chem. 2015;290:14811–14825. doi: 10.1074/jbc.M115.652628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang Q.E. DNA damage responses in cancer stem cells: implications for cancer therapeutic strategies. World J. Biol. Chem. 2015;6:57–64. doi: 10.4331/wjbc.v6.i3.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Maugeri-Saccà M., Bartucci M., De Maria R. DNA damage repair pathways in cancer stem cells. Mol. Cancer Ther. 2012;11:1627–1636. doi: 10.1158/1535-7163.MCT-11-1040. [DOI] [PubMed] [Google Scholar]
- 62.Arya S.K., Lim B., Rahman A.R. Enrichment, detection and clinical significance of circulating tumor cells. Lab Chip. 2013;13:1995–2027. doi: 10.1039/c3lc00009e. [DOI] [PubMed] [Google Scholar]
- 63.Alix-Panabières C., Pantel K. Challenges in circulating tumour cell research. Nat. Rev. Cancer. 2014;14:623–631. doi: 10.1038/nrc3820. [DOI] [PubMed] [Google Scholar]
- 64.Chen P., Huang Y.Y., Hoshino K., Zhang X. Multiscale immunomagnetic enrichment of circulating tumor cells: from tubes to microchips. Lab Chip. 2014;14:446–458. doi: 10.1039/c3lc51107c. [DOI] [PubMed] [Google Scholar]
- 65.Yoon H.J., Kozminsky M., Nagrath S. Emerging role of nanomaterials in circulating tumor cell isolation and analysis. ACS Nano. 2014;8:1995–2017. doi: 10.1021/nn5004277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nagrath S., Sequist L.V., Maheswaran S., Bell D.W., Irimia D., Ulkus L., Smith M.R., Kwak E.L., Digumarthy S., Muzikansky A. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature. 2007;450:1235–1239. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chikaishi Y., Yoneda K., Ohnaga T., Tanaka F. EpCAM-independent capture of circulating tumor cells with a ‘universal CTC-chip’. Oncol. Rep. 2017;37:77–82. doi: 10.3892/or.2016.5235. [DOI] [PubMed] [Google Scholar]
- 68.Weissenstein U., Schumann A., Reif M., Link S., Toffol-Schmidt U.D., Heusser P. Detection of circulating tumor cells in blood of metastatic breast cancer patients using a combination of cytokeratin and EpCAM antibodies. BMC Cancer. 2012;12:206. doi: 10.1186/1471-2407-12-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Gene expression data are available for 675 commonly used human cancer cell lines and RNA-seq from 53 human tissue samples on the EMBL-EBI database (http://www.ebi.ac.uk). The results published here are in part based upon data generated by the TCGA Research Network and are located at https://cancergenome.nih.gov. Raw images are available upon request.