Abstract
Ataxia-telangiectasia mutated (ATM) is a serine/threonine protein kinase with a master regulatory function in the DNA damage response. In this role, ATM commands a complex biochemical network that signals the presence of oxidative DNA damage, including the dangerous DNA double-strand break, and facilitates subsequent repair. Here, we review the current state of knowledge regarding ATM-dependent chromatin remodelling and epigenomic alterations that are required to maintain genomic integrity in the presence of DNA double-strand breaks and/or oxidative stress. We will focus particularly on the roles of ATM in adjusting nucleosome spacing at sites of unresolved DNA double-strand breaks within complex chromatin environments, and the impact of ATM on preserving the health of cells within the mammalian central nervous system.
This article is part of the themed issue ‘Chromatin modifiers and remodellers in DNA repair and signalling’.
Keywords: ataxia-telangiectasia mutated, DNA damage, chromatin, brain, neurodegeneration, oxidative stress
1. Ataxia-telangiectasia mutated and the DNA double-strand break response
(a). Properties and activation of the ataxia-telangiectasia mutated protein kinase
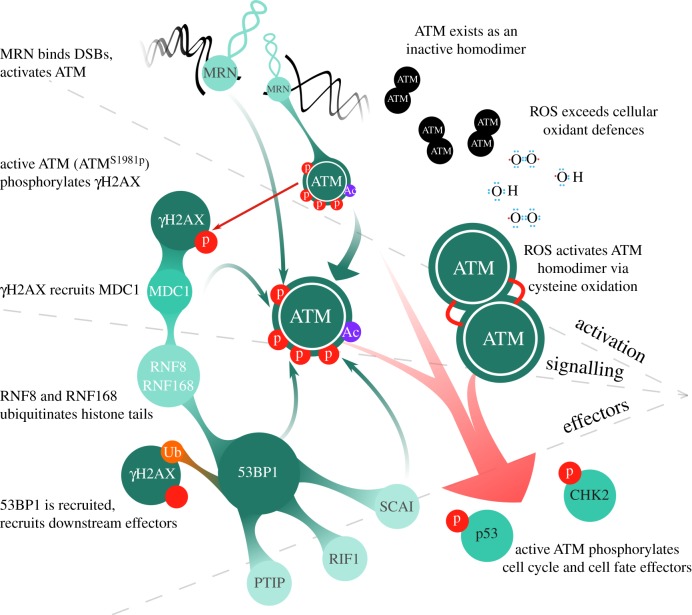
The ataxia-telangiectasia mutated (ATM) protein kinase is a master regulatory factor in the DNA double-strand break (DSB) response. ATM is a large, 3056 amino acid protein encoded by a 160 kb gene on human chromosome 11 [1–3]. ATM is a PI3K-like protein kinase (PIKK), a family that also includes the DNA-dependent protein kinase catalytic subunit DNA-PKcs and the ATM and Rad3-related kinase ATR [1–4]. All PIKKs share four common protein domains, including the FRAP-ATM-TRRAP domain (FAT), the kinase domain, the PIKK regulatory domain and the Fat-C-terminal domain (FATC) [2,4,5]. Much of the N-terminus of these PIKKs, including ATM, is composed of α-helices whose tertiary structures resemble Huntingtin, elongation factor 3, protein phosphatase 2A, TOR1 (HEAT) motif repeats which regulate protein–protein interactions and catalytic activities [2,4,5]. ATM is largely localized to the nucleus where, under physiological conditions, it is catalytically inactive as a homodimer [2,6]. Upon DSB induction, the heterotrimeric Mre11-Rad50-Nbs1 (MRN) complex detects DSB termini and induces ATM trans-autophosphorylation at serine 1981 (ATMS1981p) leading to monomerization (figure 1) [2–4,7]. Early ATM autophosphorylation events additionally form ATMS367p, ATMS1893p and ATMS2996p [4,8,9], and monomerization also depends on acetylation (ac) by the KAT5/Tip60 histone acetyltransferase to form ATMK3016ac [10–12]. ATM monomerization is coupled with interactions between ATM and the NBS1 component of the MRN complex, recruitment to DSB sites and increased protein kinase activity and substrate affinity [2–4,6]. Once active, ATM phosphorylates (serine/threonine)-glutamine (S/T-Q) motifs on more than 700 protein substrates [4]. The multitude of ATM substrates has been reviewed in [3].
Figure 1.
ATM activation and signalling axes. ATM can be activated in the context of a DSB by the MRN complex or by direct oxidation. In response to a DSB, MRN binds DSB ends, and promotes the autophosphorylation and activation of ATMS1981p. When activated, ATM generates Chk2T68p and p53S15p. This promotes recruitment of downstream signalling and effector proteins, including 53BP1, which interacts with ATM and effectors such as RIF1, PTIP and SCAI. In response to oxidative stress, inactive ATM homodimers are directly oxidized and activated, bypassing MRN-dependent autophosphorylation. Downstream targets of ATM activated by ROS are largely unknown, apart from Chk2T68p and p53S15p. (Online version in colour.)
(b). Ataxia-telangiectasia mutated to 53BP1 signal transduction pathways
Once activated by the presence of DSBs, ATM plays an integral intracellular signalling role, effecting alterations to cellular transcription and translation, and regulating cell cycle checkpoint activation (figure 1). In addition to these more well-understood roles, ATM also plays a vital part in modulating the local chromatin environment around DSBs to facilitate DSB signalling and repair. Briefly, ATM targets chromatin surrounding DSBs by phosphorylating S139 of histone variant H2AX to form H2AXS139p, better known as ‘γΗ2AX’ [3]. The BRCT domain-containing protein MDC1 (mediator of DNA-damage checkpoint protein 1) binds directly to γΗ2AX and, through direct MRN–MDC1 interactions, further activates ATM to amplify its response around DSBs. MDC1 also promotes the recruitment and activation of the ubiquitin ligases RNF8 and RNF168, enabling mono- and poly-ubiquitination of histone H2A and H2AX on lysines in nucleosomes surrounding the DSB [13,14]. At the same time, ATM phosphorylates and activates the RNF20-RNF40 ubiquitin ligases, which ubiquitinate K120 of histone H2B within the same region. Ultimately, a critical downstream mediator and effector protein of ATM signalling is 53BP1, which is recruited to sites of DSBs via interactions with dimethylated lysine 20 on histone H4 (H4K20me2) and also ubiquitinated lysine 13 and 15 of histone H2A (H2AK13/K15ub) [15,16]. ATM also phosphorylates 53BP1 on multiple sites to facilitate recruitment of many 53BP1-interacting proteins that play a role in DSB repair, such as RIF1 and PTIP [17]. Through its tandem BRCT domains, 53BP1 also interacts with γH2AX and the Rad50 component of the MRN complex, events that are necessary to recruit and concentrate activated ATM around DSBs [18,19]. Disrupting this 53BP1-BRCT2-γH2AX interaction impairs the ability of activated ATM to accumulate at DSBs, and mutation of the BRCT2 domains in 53BP1 also precludes recruitment of MRN to DSBs [18,19]. 53BP1 is important to concentrate activated ATM at DSBs to facilitate robust phosphorylation of substrates subject to constitutive phosphatase-mediated dephosphorylation [20,21] (discussed in greater detail in §2c). Recently, SCAI (suppressor of cancer cell invasion) was identified as another important 53BP1-dependent effector protein. Using chromatin mass spectrometry and quantitative mass spectrometry analyses, Hansen et al. showed that SCAI is enriched at sites of DSBs in part through an ATM-dependent 53BP1 interaction [22]. Loss of SCAI confers DSB repair defects in both mouse and human cells, and ATM inhibition precludes SCAI interaction with 53BP1 [22]. SCAI was found to interact with multiple heterochromatin-associated proteins such as HP1, and is involved in the repair of heterochromatic DSBs through promoting ATM-dependent signalling events (such as KAP-1S824p) [22]. It remains to be seen whether SCAI influences further downstream DSB-response signalling events.
(c). Ataxia-telangiectasia mutated activation and signalling in response to oxidative stress
Oxidative stress occurs when endogenously or exogenously produced reactive oxygen species (ROS) exceed cellular antioxidant defences [23,24]. These ROS include oxygen molecules and their reactive derivatives (superoxide anions and singlet oxygen), as well as peroxides and hydroxyl radicals [23,24]. ROS can be produced endogenously by processes such as mitochondrial respiration, peroxisome metabolism and inflammatory processes, as well as by exogenous agents including ionizing radiation (IR), ultraviolet light, chemotherapeutics and environmental pollutants [23]. Normally, ROS are scavenged and detoxified through small molecules including uric acid, glutathione, ubiquinone, ascorbic acid, proteins such as haemoglobin, and enzymes that metabolize ROS such as glutathione-S-transferase and superoxide dismutase [25]. Oxidative stress, among many things, produces oxidatively damaged DNA. This type of damage includes base damage (apurinic or apyrimidinic sites), damage to the sugar-phosphate backbone, DNA single-strand breaks (SSBs), DSBs (often from two SSBs in close proximity), inter- and intra-strand crosslinks and protein–DNA adducts [26,27]. These lesions are often clustered and complex, making DNA repair particularly difficult and often requiring the local chromatin environment to be altered (discussed in detail later).
Beginning with the seminal work by Shackelford and co-workers in 2001, ATM has also been shown to be activated by ROS (figure 1) [2,28]. This group showed that ATM-mutated fibroblasts had dysfunctional ROS-activated cell cycle checkpoint responses, and failed to phosphorylate downstream targets in response to ROS treatment [28]. It has since been demonstrated that in response to ROS, ATM phosphorylates downstream targets such as the Chk2 protein kinase and p53 transcription factor [29]. Similarly, CREB is also a direct target for ATM phosphorylation, which serves to regulate CREB-dependent transcriptional activity in response to DNA damage and cellular oxidative stress [30]. In 2010, Guo et al. found that ATM is indeed activated in response to oxidative stress in an MRN-independent manner that is separate from its DSB activation [31]. This activation depends on the oxidation of two cysteine residues in the inactive dimer which form a disulfide bond and activates the normally inactive dimer [2,31]. Once activated by oxidative stress, ATM phosphorylates a specific, but unresolved, set of targets that only partially overlaps with the targets it phosphorylates in the canonical DSB response [2,31]. ATM's role as a ROS sensor in the cell is further supported by the finding that hydrogen peroxide (H2O2) treatment, a robust generator of ROS in the cell, activates ATM and stimulates phosphorylation of its downstream targets [8]. The concentrations of H2O2 used in these studies are low enough to avoid robust DSB generation, and accordingly, this ATM activation occurs in the absence of the MRN complex. Interestingly, targets of ATM in the canonical DSB response, such as KAP-1, remained unphosphorylated in response to H2O2. Taken together, these findings indicate an expansion of the network by which ATM monitors cellular oxidative stress and phosphorylates a specific set of downstream targets independent of those in its DNA damage response (DDR) role to respond to oxidative stress [8]. Accordingly, this activation is attenuated by treatment with the ROS-scavenger N-acetylcysteine or low-oxygen (hypoxic) conditions [32]. This would further indicate that there are distinct cellular responses that are independently controlled through ATM. The existence of an expanded, DNA-independent ATM activation pathway suggests that ATM may function outside the nucleus and, indeed, a recent large-scale phosphoproteomic screen identified cytoplasmic ATM activated by oxidation, with over 2500 unique protein targets [33].
ATM directly and indirectly influences the repair of oxidative DNA damage. Oxidative DNA base damage produced by H2O2 requires the base excision repair (BER) pathway, and the ATM-Chk2 axis phosphorylates the BER factor XRCC1 to facilitate its association with DNA glycosylases that recognize base damage [34]. ATM also plays a role in regulating NHEJ in response to oxidative stress; evidence indicates that treatment of human fibroblasts with H2O2 activates the Ku70/80 heterodimer in an ATM-dependent manner at sites of oxidatively damaged DNA [35,36]. Oxidative base damage can also trap topoisomerase 1 (TOP1) on DNA, creating a TOP1 cleavage complex (TOP1cc). TOP1 catalyses a transient nick in the 3′ phosphodiester backbone of a single DNA strand, and creates a covalent tyrosyl linkage in the process [37,38]. The resulting DNA–protein complex is resolved by tyrosyl-DNA-phosphodiesterase 1 (TDP1), which is, interestingly, regulated by ATM [38]. ATM phosphorylates TDP1 to stabilize it at DSBs produced by the collision of replication and transcription machineries with TOP1cc; this also serves to recruit XRCC1 to facilitate repair of TOP1cc by BER. Additionally, ATM enhances TOP1 proteolysis from DNA which subsequently activates DNA-PK [39]. This DNA-PK activation facilitates ubiquitination and proteasomal turnover of TOP1cc, and enhances mono-ubiquitination of H2A and H2AX to facilitate DSB repair [39]. In addition to BER repair, ATM has also been implicated in mismatch repair (MMR) of oxidatively damaged DNA. In vitro, selenium treatment is a potent generator of ROS, which creates oxidative lesions such as 8-oxoguanine (8-oxoG) [25,40]. The activation of MMR factor MLH1 in response to selenium-generated ROS and the subsequent repair of 8-oxoG lesions depend upon ATM activity [40]. Further, after exposure to doxorubicin, a ROS-generating topoisomerase poison, ATM was demonstrated to phosphorylate MLH1 at serine 406 to facilitate its recruitment to sites of DNA damage and repair [41]. In this way, ATM also regulates MMR in response to oxidative DNA damage.
Collectively, the evidence discussed supports at least two separate mechanisms of ATM activation: (i) in response to DSBs, whereby the MRN complex helps to elicit and sustain ATM activation and (ii) in response to cellular oxidative stress, whereby the MRN complex is not required and direct ATM oxidation elicits activation. In the first, MRN ‘senses’ a DSB and undergoes a conformation change with ATP and DNA binding, which enhances Nbs1 association with the complex, which then in turn recruits ATM via the C-terminal portion of Nbs1. In such a case, ATM activation would necessitate contact with the C-terminal fragment of Nbs1, Rad50-Mre11 dimers and DSB ends. With regards to the second mechanism, two ATM molecules become covalently linked via disulfide oxidation, resulting in a conformational change that may bypass the auto-inhibition of dimeric ATM [2]. Interestingly, ATM-interacting factor ATMIN has been identified as an important interacting factor in the ROS-activated ATM pathway, and binds to the C-terminal region of ATM near the ATM-interacting motif within Nbs1 [42]. Thus, it appears that a competition between NBS1 and ATMIN may regulate the ATM activation pathway choice. Given that many DSBs occur ‘hand-in-hand’ with oxidative stress, studying the interplay between these pathways will be an interesting direction of future inquiry.
2. Ataxia-telangiectasia mutated-dependent chromatin dynamics
(a). Chromatin remodelling within different genomic environments
Chromatin, meaning DNA that is packaged with histone proteins to form nucleosomes, can be roughly subdivided based on transcriptional potential. As a rule, euchromatin is loosely condensed chromatin that is often gene-rich and therefore transcriptionally active, while heterochromatin is tightly condensed chromatin that is typically silenced, gene-poor or transcriptionally inert. Regional nucleosome arrangements are regulated by post-translational modifications that form recruitment platforms for chromatin-modifying enzymes. As DNA damage and repair occur in the context of these highly complex chromatin environments, repair must take place in concert with changes in chromatin structure and organization to maximize the efficiency of repair. These changes to the protein–DNA and protein–protein interactions in chromatin help to alter the chromatin surrounding the area of DNA damage to alleviate chromatin structure barriers, suppress interfering processes (transcription and replication) and allow repair factors to resolve damage. Again, these changes in chromatin organization are carried out through epigenetic post-translational modifications of nucleosome and non-nucleosome proteins, and the work of ATP-dependent chromatin remodellers. Such alterations occur in the context of an ‘access, repair and restore’ model [43], meaning DNA damage is detected, and histone-modifying proteins and chromatin remodellers re-organize local chromatin architecture to allow DNA repair factors to access and repair the DNA. The potential catalogue of histone modifications is expansive, performed by histone acetyltransferases, deacetylases, methyltransferases and demethylases as well as ubiquitin ligases and de-ubiquitinases, histone chaperones, kinases and phosphatases [43]. ATP-dependent chromatin remodellers also use energy from ATP hydrolysis to adjust the spacing between DNA and histones [44]. These enzymes can increase or decrease the length of DNA between nucleosomes, eject or load nucleosomes onto DNA, or exchange histone variants within nucleosomes. Generally, chromatin remodellers can be grouped into four ‘families’ based on similarities in biochemical structure: (i) CHD (chromodomain helicase DNA-binding), (ii) INO80 (inositol requiring), (iii) SWI/SNF (switch/sucrose non-fermentable) and (iv) ISWI (imitation SWI) classes. For a comprehensive review of chromatin remodellers, see [44,45], or for a review of the function of some of these remodellers in the context of DSB repair, see the van Attikum group's review in this issue [46]. The activity of histone-modifying and chromatin remodelling enzymes at sites of DNA damage is tightly regulated. Although many of the processes controlling DNA-damage responsive chromatin remodelling enzymes are still being resolved, it is clear that ATM is key in the regulation of these factors. In this section, we will discuss some of ATM's roles within heterochromatin and euchromatin with regards to orchestrating chromatin remodelling for DSB responses.
(b). Ataxia-telangiectasia mutated and the formation and persistence of γH2AX
The formation of γH2AX foci at DSB sites is one of the first events in DSB repair signalling. H2AX belongs to the histone H2A family, and is phosphorylated redundantly at S139 by ATM, DNA-PK and/or ATR (ATR activity contributing to γH2AX only under conditions of replication stress in S-phase or DSB termini resection in G2) [3,47,48]. γH2AX expands to encompass up to 20–30 megabases of DNA surrounding the DSB lesion. Megabase clusters of γH2AX are easily visualized by immunofluorescence microscopy, and the rate of resolution of these γH2AX foci is a sensitive and accurate surrogate indicator of DSB repair under most conditions [48,49]. In human fibroblasts, H2AX represents approximately 10% of all H2A, but this number varies significantly [48]. For example, in human lymphocytes, only about 2% of total H2A is the H2AX variant [48]. This variation is, in part, due to differences in the regulation of H2AX synthesis and incorporation into nucleosomes, just as the resolution of foci is due, in part, to the removal of H2AX from nucleosomes—the effect of H2AX levels (relative to total H2A) on tissue radiosensitivity and DSB repair kinetics is unknown.
In addition to ATM/DNA-PK/ATR, there are several other events that contribute to γH2AX formation. The BRG1 catalytic subunit of the SWI/SNF chromatin remodelling complex contributes to effective γH2AX formation and, upon DNA damage, ATM phosphorylates BRG1S721 which greatly enhances BRG1 affinity for H2AX-containing nucleosomes and acetylated histone 3 [46,50]. This localizes BRG1 at DSB sites, where its chromatin remodelling activity contributes to enhanced γH2AX formation and spreading [46,50]. This constitutes another example of how ATM activity can be self-reinforcing through its activity on the chromatin facilitating robust γH2AX signalling. Additionally, knockdown of the Williams syndrome transcription factor (WSTF), a component of the WICH chromatin remodelling complex, indicates that it promotes ATM and MDC1 recruitment to DNA damage sites [51]. WSTF phosphorylates H2AXY142 and it appears, through a currently unclear mechanism, that H2AXY142p influences the kinetics of γH2AX formation and resolution. Interestingly, the Eya1/3 phosphatases which target H2AXY142p are themselves ATM/ATR substrates and their phosphorylation may cue their activity at γH2AX, believed to promote the repair response over the apoptotic response under genotoxic stress [52]. Further indication of γH2AX regulation was revealed in quiescent cells, where H2AX is expressed constitutively, but is poly-ubiquitinated by the E3 ubiquitin ligase HUWE1 and is subject to proteasomal degradation [53]. However, upon DSB formation, SIRT6 and the ISWI chromatin remodeller SNF2H are rapidly recruited, and HUWE1 dissociates from chromatin to stabilize and incorporate H2AX at DSBs leading to γH2AX formation [53]. ATM is required for H2AX stabilization, and may directly regulate HUWE1 to facilitate its dissociation from H2AX [53]. Taken together, these findings indicate that γH2AX regulation is emerging as a much more dynamic process than once thought.
(c). Ataxia-telangiectasia mutated-dependent chromatin remodelling events in heterochromatin
While the majority of DSBs are repaired (ultimately) without a need for ATM activity, the kinase plays an essential role in the repair of DSBs induced within heterochromatin and, when inactive or absent, the majority of heterochromatic DSBs (encompassing 10–25% of total lesions) fail to be repaired (figure 2a) [16,54]. Central to this pathway is the phosphorylation (by ATM) of the transcriptional co-repressor and heterochromatin building factor KAP-1 on serine 824 [12,20,21,55,56]. ATM-dependent KAP-1S824p initially forms in a pan-nuclear manner but, over time, occurs in a localized and concentrated manner around DSBs within heterochromatin, as 53BP1 interactions with γH2AX and the Rad50 component of the MRN complex localize ATM protein kinase activity at persisting lesions [19–21,46,55]. As stated earlier, interactions between the tandem BRCT domains of 53BP1 and both Rad50 and γH2AX enable the hyper-accumulation of active ATM at DSBs [19,20]. This abundance of ATM activity is necessary to ‘overwhelm’ the constitutive phosphatase activity of PP4 towards KAP-1S824p, which normally serves to remove KAP-1S824p as the DDR winds down following DSB rejoining [21]. KAP-1 is also phosphorylated on serine 473 by Chk2, a downstream target of ATM, and this post-translational modification, apart from modulating the expression of KAP-1-related stress genes, appears to exert an influence over G2/M checkpoint signalling [21].
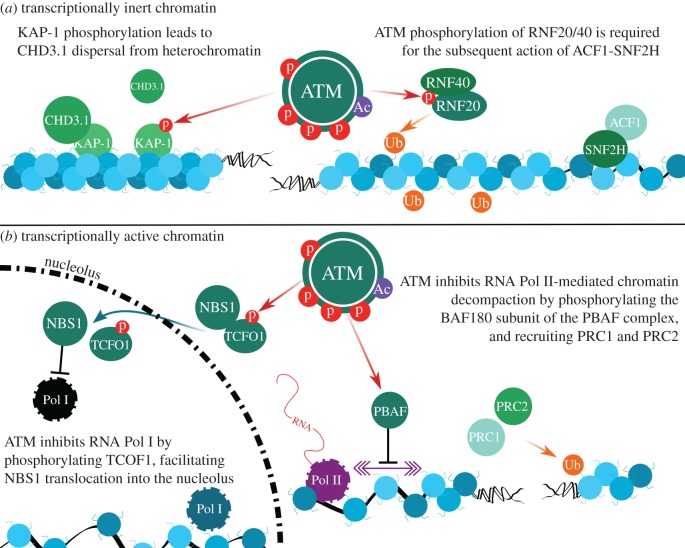
Figure 2.
ATM-dependent signalling events around DSBs in different chromatin environments. (a) ATM's role in DSB repair within transcriptionally silent heterochromatin involves KAP-1 phosphorylation (KAP-1S824p), promoting CHD3.1 dispersal. ATM-dependent RNF20-RNF40 activity, at the same time, promotes H2BK120ub and ACF1-SNF2H recruitment to adjust nucleosome spacing to make chromatin amenable to DSB repair. (b) In response to nucleolar DSBs (i.e. transcriptionally active regions) ATM generates TCOFS1199p, which facilitates NBS1 translocation into the nucleolus. Once there, NBS1 inhibits RNA Pol I globally, enabling DSB repair by NHEJ. Outside the nucleolus, RNA Pol II activity is attenuated by ATM, via BAF180S948p phosphorylation, precluding chromatin decompaction during transcription. ATM-dependent PRC1/2 recruitment facilitates H2AK119ub, which augments this process. (Online version in colour.)
ATM-dependent KAP-1S824p enables DSB repair in heterochromatin by exerting a direct influence on CHD-class chromatin remodellers, specifically CHD3 isoform 1 (CHD3.1) [46,57]. CHD3.1 is associated with transcriptional repression and chromatin compaction, as a core component of mammalian nucleosome remodelling and deacetylase (NuRD) complexes (reviewed in [44]). CHD3.1 is retained in heterochromatin through an interaction between its extreme C-terminal SUMO-interacting motif (SIM) and SUMO-modified KAP-1 found in heterochromatin regions. The formation of KAP-1S824p perturbs the interaction between the CHD3.1 SIM and SUMOylated KAP-1, triggering CHD3.1 dispersal from heterochromatin [16,46,56,58,59]. This event is associated with the DSB-, ATM- and KAP-1S824p-dependent chromatin relaxation event first described by the Shiloh group [55]. More recently, the dispersal of the nucleosome-compacting activity of CHD3.1 was found to be only partly responsible for adjusting chromatin relaxation around a heterochromatic DSB. Indeed, the activity of the ISWI-class chromatin remodeller ACF1-SNF2H is required once CHD3.1 is dispersed in order to actually adjust nucleosome spacing into a configuration amenable for repair [46,57]. ACF1-SNF2H is recruited to chromatin (vacated by CHD3.1) in a pathway requiring ATM-dependent RNF20-RNF40 phosphorylation and RNF20-RNF40-dependent H2BK120ub [46,57,60].
Interestingly, SNF2H, which is a catalytic component of a variety of remodelling enzyme complexes, is also recruited to DSB-associated chromatin via the ATM-dependent phosphorylation of the Rsf-1 subunit of the RSF complex [61]. Rsf-1 augments checkpoint signalling and cell survival after DNA damage, and promotes accumulation of homologous recombination (HR) repair factors Rad51 and RPA at DSBs. Given that the majority of HR in G2 phase is known to occur in heterochromatic lesions [62], then this pathway probably also contributes to heterochromatic DSB repair. In addition to this, Rsf-1 can recruit the centromere-associated proteins CENPS and CENPX to DSBs in an ATM-dependent manner, which may further help to prevent unwound DNA from re-coiling while repair takes place [46,63].
Another substrate of ATM that is relevant to HR-mediated repair is the CHD-class chromatin remodeller CHD4. CHD4, similar to (but mutually exclusive from) CHD3.1, is a catalytic subunit within NuRD complexes, a multi-subunit, transcriptionally repressive complex that combines chromatin remodelling with histone deacetylase and histone demethylase action [44,45,64]. CHD4 is recruited rapidly to DSBs in a poly-ADP ribose polymerase (PARP)-dependent manner and plays an important role in DSB repair [46,64]. In addition, the NuRD complex in part regulates the G1–S checkpoint by deacetylating p53 after DNA damage [43,64]. CHD4 has multiple ATM phosphorylation sites [64,65], of which CHD4S1349p facilitates its retention and stabilization at IR-induced DSBs after its recruitment by PARP [65].
(d). Ataxia-telangiectasia mutated-dependent chromatin remodelling in transcriptionally active chromatin
In contrast with the role that ATM plays in DSB repair in heterochromatin, emerging evidence suggests it is equally important to the repair of DSBs in transcriptionally active euchromatin (figure 2b). An excellent model for looking at transcriptionally active euchromatin is nucleolar ribosomal DNA, which is some of the most highly transcribed DNA in the genome [66], and is one such unique chromatin setting where ATM acts to preserve genomic integrity. ATM mediates transcriptional silencing in response to DNA damage to prevent collision of the transcription machinery with DNA repair factors. ATM coordinates silencing of RNA Pol I, the primary rRNA polymerase, in response to DNA damage in the nucleolus by signalling through Nbs1 and MDC1 to facilitate displacement and nucleolar exit of RNA Pol I [67]. In response to DNA damage outside the nucleolus, ATM phosphorylates TCOF1S1199 (commonly known as treacle) to facilitate the translocation of Nbs1 from the nucleoplasm to the nucleolus and facilitate RNA Pol I inhibition [68,69]. Recent work from the Greenberg group shows that rapid repair of DNA damage via NHEJ preserves rDNA transcription in the nucleolus, but persistent DNA damage activates a large-scale ATM-dependent transcriptional silencing and nucleolar reorganization event [66]. This reorganization shuttles DSBs into nucleolar ‘caps’, which facilitate the recognition and repair of DSBs. Thus, ATM regulates transcriptional silencing to preserve genomic stability, and this silencing induces reorganization of nucleolar proteins that facilitate chromatin reorganization in the nucleolus for effective DSB repair.
In contrast with RNA Pol I, global RNA Pol II activity is not silenced upon induction of DSBs. The Greenberg laboratory designed an assay to monitor transcription of a reporter gene in the vicinity of a Fok1 nuclease-generated DSB [70]. DSB-induced ATM activity blocked the normal chromatin decompaction accompanying RNA Pol II activity, silencing reporter genes proximal to DSBs in an ATM and H2A ubiquitination-dependent manner [70]. Interestingly, this transcriptional silencing process required the action of the SWI/SNF-B (PBAF) complex—specifically, the BAF180 subunit of this complex. BAF180 is phosphorylated at serine 948 by ATM, enabling the PBAF complex to elicit transcriptional silencing in cis to DSBs, and rapid repair of DSBs within transcriptionally active regions via NHEJ [71]. Additionally, ATM also promotes the recruitment of polycomb repressive complexes PRC1 and PRC2, which, together with RNF8, ubiquitinate H2AK119 [71]. Depletion of PRC2 compromises DSB repair and leads to IR sensitivity [72]. Altogether, this activity silences transcription around DSBs and facilitates rapid repair to preclude the complex damage that can occur when transcription encounters DNA repair machinery. This work highlights a role for ATM in a third type of DSB repair, distinct from the classical ‘fast’ ATM-independent euchromatic and the ‘slow’ ATM-dependent heterochromatic DSB repair: namely DSB repair around active transcription sites.
3. Ataxia-telangiectasia mutated and oxidative DNA damage in the central nervous system
(a). Ataxia-telangiectasia and the neurological phenotype
Ataxia telangiectasia (A-T) is an autosomal recessive disorder characterized by mutations in the ATM gene that often lead to protein instability, or catalytically insufficient ATM protein (figure 3) [73–75]. The clinical presentation of A-T is typified by the early onset of progressive cerebellar ataxia, choreoathetosis, cognitive symptoms such as progressive dysarthria, oculocutaneous telangiectasia, immunodeficiency and lymphoid tumours, sterility and a heightened susceptibility to bronchopulmonary disease [74]. On a cellular level, A-T patients show marked cellular radiosensitivity, chromosomal instability and cell cycle checkpoint defects [73,74]. The onset of A-T symptoms occurs in early childhood, and patients are often wheelchair-bound by progressive cerebellar ataxia by the time they reach adolescence [74,75]. This cerebellar ataxia is accompanied by a progressive thinning of the molecular layer of the cerebellum due to the loss of Purkinje neurons [75–77]. Classically, the focus on the neurodegeneration in A-T has been directed at the cerebellum, due to the striking cerebellar ataxia in A-T. This neurodegeneration, however, is not confined to the cerebellum and, upon close inspection, and specifically in older patients, neurodegeneration is found in the dentate and olivary nuclei, the basal ganglia, throughout the cerebrum, and in the brainstem [78]. The complete neurological phenotypes of A-T have been explored elsewhere [79] and, here, we will focus on the emerging ATM-dependent defects in neuronal chromatin remodelling and epigenetic signalling that may underlie (some of) the central nervous system (CNS) symptoms of A-T.
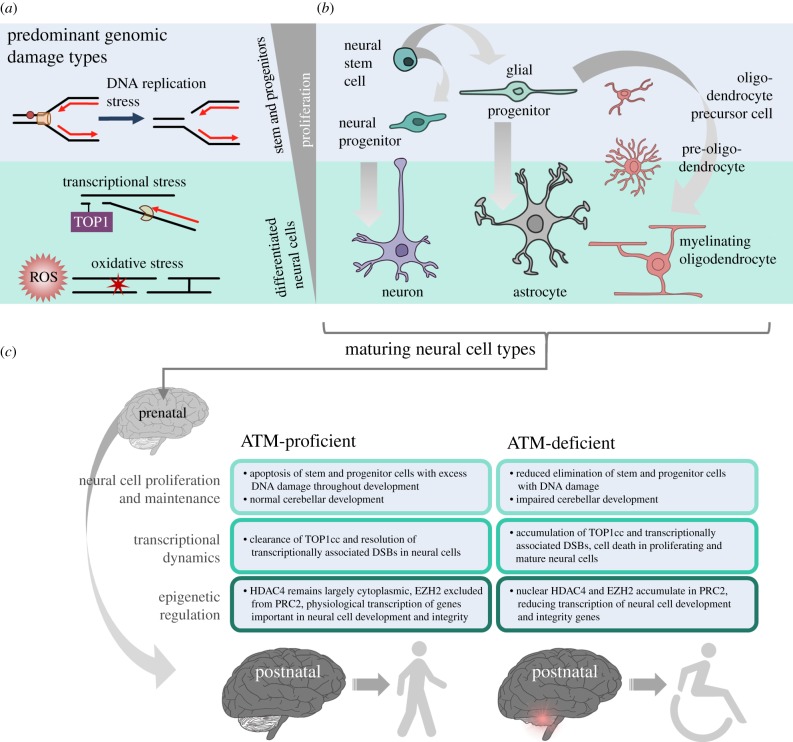
Figure 3.
DNA damage repair in different neural cell types, and ATM-associated neurological phenotypes. (a) At different neurodevelopmental stages, cells encounter different sources of DNA damage: replication stress predominates in rapidly proliferating stem and progenitor cells, while genomic damage from high transcriptional load and ROS are predominant in post-mitotic neural cells. (b) Neurodevelopment involves proliferation, migration and differentiation of neural stem cells down multiple neural cell lineages. Neural stem cells generate neural progenitors that differentiate into neurons that populate the brain, primarily prenatally. Glial cells, including astrocytes and oligodendrocytes, are primarily formed in the early postnatal years from glial progenitor cells. (c) ATM mutation predisposes the cerebellum to rapid postnatal degeneration through defects in proliferation maintenance, transcriptional dynamics and epigenetic regulation which likely underlie the progressive ataxia observed in ataxia-telangiectasia patients. (Online version in colour.)
The marked neurological phenotypes that arise from mutations in DDR factors demonstrate a critical role for DNA repair in the nervous system. Since many of these neurological phenotypes overlap with those of A-T, the field has long thought that these must be due to the deficits in the DDR that comes with dysfunctional ATM. The loss of ATM is different from loss of other DDR factors, however, as there is no overt dysfunction in corticogenesis and early brain development, but rather, deficits in neuronal maintenance in the cerebellum, brain stem and spinal cord. These unique symptoms set A-T apart from other diseases of the DDR in its lack of microcephaly and subtle, if any, cognitive deficits [80]. Notably, the cerebellar ataxia observed in humans is largely absent in mouse models of A-T [81–83]. It is likely that the pleiotropic A-T phenotype is the result of the systemic loss of function across the many roles of ATM critical for proper neuronal differentiation, maturation and integrity. Traditionally, investigation into the CNS phenotypes of A-T has been hindered by challenges in growing and maintaining neuronal cultures relative to other proliferative or transformed cell lines, and interrogating the molecular mechanisms that underpin A-T in a neural cell type-specific manner. Investigation has previously focused on the canonical role of ATM in the signalling and repair of DSBs in mature neurons. These DSBs are often thought to be a product of ROS produced by neuronal metabolism and, indeed, neurons are very oxidatively stressed cells [78]. As discussed earlier, ATM plays a key role in orchestrating a cellular response to this oxidative stress and, so, an expectation that A-T phenotypes are uniquely the result of DDR defects omits the exceptional role ATM plays in countering oxidative stress.
(b). Ataxia-telangiectasia mutated and oxidative stress in the central nervous system
The brain consumes up to 20% of inspired oxygen. As such, it is remarkably metabolically active with relatively low levels of antioxidants and ROS-countering molecules [84]. This makes the brain susceptible to oxidative stress and, given the post-mitotic status and required longevity of many neural cells, protection against the damaging effects of ROS is important. There are many lines of evidence to suggest that exaggerated oxidative stress contributes strongly to the neurodegeneration in A-T. First, A-T cells and tissues display abnormally high levels of oxidative stress. Studies have identified differential levels of redox markers in the brains, and particularly the cerebella, of Atm−/− mice and higher levels of ROS [85], oxidative DNA damage and lipid oxidation products in A-T patient brain tissue [86]. Second, A-T cells and tissues are especially sensitive to ROS-inducing agents (such as hydrogen peroxide and ionizing radiation) and have impaired DNA damage checkpoint responses, persistent low-level DNA damage and a lowered apoptosis threshold [28,36,87]. Finally, treatment of A-T cells and tissues with antioxidants can improve cell and tissue survival. For example, treatment of cultured Purkinje neurons and Atm−/− mice with isoindoline nitroxide increases cell survival in vitro and reduces the oxidative burden in the brain (particularly in the cerebellum) [88]. Treatment of Atm−/− mice with N-acetylcysteine is also chemoprotective, and increases longevity of A-T mice [89,90].
Interestingly, in mouse models of A-T the cerebellar Purkinje and granule cells appear to be uniquely high in oxidative stress and sensitive to ROS-induced death. Many previous studies have shown significantly increased ROS levels in cerebellar Purkinje cells of Atm−/− mice relative to wild-type mice, with evidence of ROS-damaged proteins, lipids and DNA at baseline [85,91,92]. Heightened ROS levels and ROS-induced DNA damage are also found in A-T patient lymphocytes [86]. To some extent, markers of oxidative stress are also found in other brain areas subject to degeneration in A-T such as the striatum and the cortex, but these findings are inconsistent. One thing is clear, however: the cerebellum, the brain area with the greatest neuropathological changes in A-T, is rich in and sensitive to ROS in A-T. What causes these aberrancies in this brain region in the absence of ATM remains largely unknown. Interestingly, the glial cells that make up much of neural cells are also affected by ATM loss. In vitro, cultured Atm−/− astrocytes display increased levels of oxidative stress, and in vivo, astrocyte-like Bergmann glia of the cerebellum mirror this finding [93]. Neurons depend on astrocytes for antioxidant support and, indeed, ATM loss impairs antioxidant glutathione metabolism and secretion which compromises cerebellar neuron survival and outgrowth [94]. Moreover, white matter abnormalities have been noted in A-T patients, which suggest the myelinating oligodendrocytes of the CNS are affected by ATM loss. Several neuroimaging studies have identified white matter-deficient areas across the A-T brain, and some more recent studies have shown degeneration of major white matter tracts in young A-T patients [95–97]. As oligodendrocytes are also highly oxidatively stressed cells due to several physiological factors [98] (high metabolism, high peroxisome content, high iron content and low antioxidant levels), it is likely that the ATM-oxidative stress axis is also at play here. Thus, the innate susceptibility of the cerebellum to oxidative damage and stress probably predisposes it to degeneration without ATM, which carefully regulates the ROS defence in cells. The underlying mechanisms that sensitize the cerebellum to oxidative stress are poorly understood; however, emerging evidence suggests that ROS-induced DNA damage may interact uniquely with transcriptional and epigenetic processes in neurons, particularly those of the cerebellum.
(c). Neuronal transcription-associated DNA damage requires ataxia-telangiectasia mutated for repair
DNA damage occurs as a by-product of active transcriptional processes in neurons, and ATM appears to be necessary to both prevent and repair this damage. On average, the brain has three to five times more actively transcribed genes than any other tissue [99], and neural cells, particularly neurons, are rich in transcriptional processes. While TOP1 poisons such as topotecan and camptothecin can trap TOP1 on DNA and create an irreversible TOP1cc, it is important to note that oxidative stress-induced DNA lesions such as 8-oxoguanine and SSBs can also trap TOP1 on DNA and produce irreversible TOP1cc [100,101]. In a seminal publication in 2009, Yves Pommier's group showed that ATM is activated by, and important for the repair of TOP1cc-asociated DNA damage [101]. In this study, treatment of rat cortical neurons with camptothecin created DSBs when the transcriptional machinery collided with TOP1cc, activating ATM that was found to be necessary to signal and repair these transcriptionally associated DSBs in post-mitotic neurons [101].
ATM functions beyond ‘just’ signalling TOP1cc-associated DSBs, and there is strong evidence that it even regulates TOP1 itself to promote resolution of transcription-associated DNA damage and prevent genome instability. Atm-null neurons have significantly higher levels of TOP1cc-associated DNA damage, which is decreased by inhibiting transcription or in response to treatment with ROS scavengers N-acetylcysteine and mannitol [102]. Katyal et al. [100] have examined TOP1cc-associated DNA damage in an age-specific context in mice, and found that Atm-null mice show high levels of TOP1cc at the peak of neurogenesis, E12.5, which decrease as neurogenesis finishes at E18.5. However, mature Atm-null mouse cerebellar granule neurons show further increased levels of TOP1cc-associated DNA damage relative to wild-type cerebellar neurons, indicating age-specific and brain region-specific roles for ATM signalling. Katyal et al. [100] showed that ATM regulates TOP1 proteasomal turnover in a protein kinase-independent manner, while orchestrating a DDR response to repair these TOP1cc through its kinase activity. Moreover, the phenotypic similarity of A-T to that of spinocerebellar ataxia with axonal neuropathy 1 (SCAN1), caused by a defect in TDP1, further strengthens the idea that ATM plays a critical role in maintaining genome stability by regulating TOP1 during transcription. In the case of A-T, without ATM to regulate TOP1 turnover and facilitate the signalling and repair of TOP1cc, neuronal integrity would be compromised with every transcriptional collision in mature neurons, and every replicative and transcriptional collision in the developing brain. It is also important to note that Purkinje neurons express very high levels of TOP1 in both healthy and A-T individuals [103]. This could also account for the anatomical specificity of the neurodegenerative phenotype in A-T.
As discussed in §§1c and 3b, ATM is highly involved in modulating ROS levels in cells to protect from oxidative DNA damage [31,104]. In A-T, without ATM to suppress chemical modification of DNA by ROS, this oxidative damage would exacerbate TOP1cc levels as neurodevelopment and neuronal maintenance requires high levels of DNA replication and transcription. If cerebellar neurons are particularly rich in TOP1cc and in oxidative stress then ATM-deficient cerebellar neurons, unable to modulate ROS levels and repair TOP1cc, would incur extensive DNA damage and undergo cell death. As discussed in §2d, ATM inhibits RNA Pol I [67] and, without ATM inhibition of transcription in response to DNA damage, these transcriptional collisions between RNA polymerases and TOP1cc would be far more common, compromising genomic stability further. Thus, a model whereby ATM deficiency sensitizes neurons to oxidative DNA damage, which accumulates over time and exacerbates TOP1cc-associated DNA damage, may underlie the profound progressive neurodegeneration seen in A-T.
In addition to TOP1cc-associated DNA damage, transcription-associated DSBs occur spontaneously in specific genes in neural stem and progenitor cells. The recent development of novel high-throughput genome-wide translocation sequencing has allowed the identification of DSBs clustered around transcriptional start sites of highly expressed genes, which are prone to translocate and cause large-scale genomic rearrangements [105,106]. Such DSBs and translocation events occur preferentially in long, actively transcribed genes related to neural function and integrity, and are exacerbated in repair-compromised backgrounds such as ATM-null mice [105,106]. These DSBs are likely the result of a combination of factors: without ATM to regulate TOP1cc levels, transcription and replication (in proliferating neural stem and progenitor cells) machinery probably collides with these covalently linked complexes to form DSBs and SSBs in close proximity. Without ATM to modulate ROS levels in the cell, enhanced oxidative stress would also likely contribute to these DSBs. Ultimately, without ATM to facilitate the signalling and repair of the DSBs, translocations and genome instability will occur, which could result in neuronal death. Additionally, ATM-deficient neural stem and progenitor cells with genomic rearrangements can evade apoptosis, likely resulting in neurodegeneration in later years as genomic damage increases due to ROS levels and transcriptional stress.
In a similar role to its involvement in the clearance of TOP1cc, ATM is also required for the repair of topoisomerase 2β (TOP2β)-blocked DSBs. These lesions also come about when adjacent DNA lesions produced by IR or ROS trap TOP2β on DNA, creating a TOP2β cleavage complex, which is a DSB covalently linked to TOP2β. In a series of experiments using double Tdp2 and Atm knockout mouse embryonic fibroblasts (MEFs), the Cortés-Ledesma group have shown these cells to be exquisitely sensitive to the TOP2 poison etoposide, and to have high levels of genome instability relative to wild-type MEFs after etoposide treatment [107]. Interestingly, this role of ATM in repairing these TOP2βcc is independent of chromatin context. Emerging evidence suggests a physiological role for TOP2β-mediated DSBs, particularly in neurodevelopment. Neuronal activation has been shown to create DSBs both in vitro and in vivo [108]. Exploration of a novel environment or optogenetic stimulation results in increased DSBs in the mouse hippocampus, indicating a role for neuronal DSBs in learning and memory [108]. Indeed, these DSBs may be the result of topoisomerase 2β (TOP2β) [109]. The Tsai group showed that, in response to neuronal activation, TOP2β creates DSBs near promoters of early neural response genes such as Fos, Npas4 and Egr1, facilitating RNA Pol II access to early neural response gene promoters normally constrained by topological organization within chromatin [109]. These early transcription events then go on to activate genes involved in synapse development and modulation, and neurite outgrowth. Thus, the DSBs play a critical role in activity-dependent changes in neuronal circuitry but still require ATM to facilitate proper signalling and repair to prevent large-scale neuronal genomic rearrangements and apoptosis. Thus, without ATM, these TOP2β-induced ‘physiological’ DSBs may become pathogenic. It is important to note that the early postnatal period leading up to the onset of the neurological symptoms of A-T is marked by an immense amount of neuronal arbourization, synaptogenesis, activity-dependent synaptic strengthening and synaptic pruning [110]. Therefore, the role for these physiological DSBs is much larger than in an adult brain. It is possible that without ATM to facilitate signalling and repair, this DNA damage persists and accumulates, and contributes to the neurological decline that begins in A-T late childhood.
(d). Neuron-specific roles for ataxia-telangiectasia mutated in epigenetic modulation
There is increasing evidence that ATM also regulates neuron-specific epigenetic changes. Recently, the Herrup group has identified novel epigenetic roles for ATM that may, in part, explain the cerebellar specificity for the neurodegeneration in A-T. The class IIA histone deacetylase HDAC4 is highly expressed in the brain and in cerebellar Purkinje neurons in particular [111]. HDAC4-null mice show progressive postnatal loss of Purkinje cells and concomitant cerebellar atrophy, which can be rescued by HDAC4 add-back [112]. Physiologically, HDAC4 remains in the cytoplasm of Purkinje neurons in a phosphorylation-dependent manner. In A-T cerebellar neurons, however, HDAC4 relocalizes to the neuronal nucleus, where it is free to suppress MEF2 and CREB-dependent transcription of genes important in neuronal function (such as Egr3, Fos and Nrxn1/3), and directly deacetylate histones to further repress transcription of genes important in neuronal function and synaptic maintenance (Nrxn1/3, Bdnf, Grin2a) [111]. Nuclear relocalization of HDAC4 in A-T facilitates neuronal death and aberrant cell cycle re-entry, which can be reversed by the addition of a cytoplasmic-specific mutant of HDAC4. In Atm−/− mice, a cytoplasmic HDAC4 mutant can correct the subtle cerebellar and behavioural symptoms present. The aberrant HDAC4 localization in A-T is produced by an overactive PP2A phosphatase activity that dephosphorylates HDAC4 promoting nuclear relocalization; phosphomimetic HDAC4 mutants restore cytoplasmic HDAC4 levels and rescue behavioural and cellular defects in Atm−/− mice [111].
On a similar note, the Herrup group went on to show that the EZH2 histone methyltransferase is an ATM substrate, and that EZH2 hyperactivity contributes to the anatomical specificity of A-T neurodegeneration. EZH2 is a component of the multi-subunit polycomb repressive complex 2 (PRC2), which represses transcription via increasing H3K27me3 [113]. Cerebellar granule and Purkinje neurons from A-T patients and Atm−/− mice display aberrantly high H3K27me3 and, indeed, high levels of EZH2 with enhanced H3K27 binding [113]. ATM phosphorylation of EZH2 blocks its assembly into PRC2, and indirectly promotes its degradation [113]. As with HDAC4, the gene targets of aberrant EZH2 activity (i.e. downregulated transcription in A-T) include genes critical for neuronal function and synaptic development such as Slit1 and Nlgn1, as well as cell cycle genes of the CDKN2A/B locus [113]. Also similar to HDAC4, EZH2 gene targets in A-T are not only significantly over-expressed, but shifted with only a 5% overlap between Atm wild-type and Atm−/− genotypes [113]. Notably, HDAC4 and EZH2 gene targets increase in A-T from 2% to 5% overlap, encompassing neuronal and synaptic maintenance and integrity genes [111,113]. Knocking down EZH2 in cerebellar A-T neurons prevents etoposide-induced neuronal death, improves dendritic arbourization in cerebellar neurons, prevents neurodegeneration and corrects behavioural abnormalities in Atm−/− mice [113]. Importantly, knocking down high EZH2 levels in cerebellar A-T neurons also prevents re-entry of cells into the cell cycle, a common occurrence with post-mitotic neurons after DNA damage without functional ATM that rapidly leads to cell death [114].
ATM is also essential for modulating HDAC1 activity during the DDR. More specifically, ATM signalling triggers the recruitment of the SIRT1 NAD+-dependent lysine deacetylase to DSBs, which, in turn, recruits and activates more ATM in a feedback loop [115]. SIRT1 then deacetylates HDAC1, promoting an HDAC activity required for NHEJ [115]. DNA damage enhances the SIRT1 and HDAC1 association and, without ATM, SIRT1 is recruited to DSBs with slower kinetics and fails to deacetylate HDAC1 [115]. Increased SIRT1 activity (via NAD+ supplementation or SIRT1 activators) ameliorates canonical A-T phenotypes in mice, Caenorhabditis elegans and neurons [116]. The Bohr group [116] has demonstrated that SIRT1 activity and intracellular NAD+ stores are significantly reduced in the absence of ATM. The group also tied aberrant SIRT1 and NAD+ levels to mitochondrial dysfunction. Mitochondrial dysfunction has been reported in A-T before, indicated by reduced mitochondrial membrane polarization [117,118], increased ROS levels [85,118] and increased mitochondrial DNA damage [116,119]. Restoring SIRT1 activity and NAD+ levels corrects mitochondrial dysfunction, decreases Purkinje cell death and cerebellar atrophy, increases CREB signalling and BDNF levels, improves NHEJ efficiency and increases lifespan in mouse models of A-T [116]. Traditionally, Atm−/− mice do not display gross cerebellar atrophy and overt motor defects. However, more recent analyses such as the one done by the Bohr group shows that, indeed, these mice do display canonical A-T phenotypes. These mice display subtle motor defects evident in more rigorous rotarod tests, and subtle cognitive deficits as well. These mice also exhibit Purkinje cell death as they age, and mild cerebellar atrophy. These phenotypes become particular prominent the longer these mice live, and supplementing NAD+ or restoring SIRT1 promotes the extended lifespans in the mice helpful in detecting these phenotypes. Along a similar line, ATM has also been shown to regulate DNA ligase III levels in mitochondria, facilitating mitochondrial DNA repair [119]. In its absence, ligase III levels are reduced and mitochondrial DNA damage is increased, which, together with decreased SIRT1 and NAD+ levels, may underpin mitochondrial dysfunction in A-T.
Altered DNA methylation may also pre-dispose cerebellar neurons to degeneration in A-T. Mouse model A-T Purkinje cells have aberrantly low 5-hydroxymethylcytosine (5-hmC), an epigenetic mark catalysed by ten–eleven translocation (TET) proteins that facilitates differential transcriptional regulation in a cell type-specific manner within the brain [120,121]. TET1 is a DNA damage-sensitive ATM substrate and, without ATM, TET1 activity is reduced, particularly after DNA damage [120]. TET1 knockdown can recapitulate A-T phenotypes of Purkinje cell cycle re-entry and death in vitro and in vivo, and TET1 overexpression in the cerebellum can rescue these, as well as behavioural deficits, in vitro and in vivo [120]. Finally, the 5-hmC enrichment sites in mouse models of A-T are shifted in the cerebellum, and are particularly enriched in regulatory elements in cerebellar development genes [120]. This is particularly salient, given the recent findings by the Hatten group that chromatin remodelling and epigenetic signalling are particularly important in cerebellar development. The Hatten group recently showed that the Tet genes, among many other chromatin remodelling and epigenetic signalling factors, undergo a marked increase in expression during cerebellar granule cell development [121]. This corresponds to an increase in 5-hmC levels in cerebellar neurons, and an increase in the expression of axon guidance and ion channel genes [121]. Knockdown of TET1 by RNA interference abrogated granule cell arbourization and circuit formation in the cerebellum [121]. Hence, there is a clear need for TET-mediated epigenetic signalling in the developing cerebellum which is ATM-related and particularly critical after DNA damage.
(e). A word on ataxia-telangiectasia mutated in neurodevelopment
It is worth noting that ATM plays an important role during neurodevelopment in activating DNA damage checkpoints and apoptosis to eliminate neural precursors and neural cells with damaged DNA [75]. This way, ATM helps to ensure the long-term viability of neural cells early in neurodevelopment, and is particularly critical as neural stem cells and progenitors are undergoing rapid proliferation and replication stress-induced DNA damage is high [75]. While much of neurodevelopment takes place embryonically, a significant portion also takes place postnatally. Glial proliferation, migration and differentiation occur in early postnatal years, as does cerebellar development. Indeed, cerebellar development is a protracted process, and continues well past the first postnatal year in humans [122].
The cerebellum contains 70–80% of neurons that make up the human brain, and is also rich in astrocyte variants such as Bergmann glia, and other glial cells that have tightly regulated functional associations with the granule and Purkinje neurons [123,124]. Granule cells make up the vast majority of the cerebellum and, remarkably, 85% of these neurons are generated in the first postnatal year, along with a large number of Purkinje neurons and glial cells [122]. This rapid proliferation of granule cells peaks around postnatal month 8 in the external granule cell layer of the developing cortex, and these post-mitotic cells migrate inwards to the internal layer where they mature to form synapses [125]. No other part of the brain has as many cells that are generated as rapidly postnatally as the cerebellum and, crucially, large-scale chromatin changes are necessary for this rapid postnatal cerebellar development [125]. By tracking chromatin state and gene expression in mouse cerebella up to postnatal day 60, the West group showed that massive changes to chromatin accessibility facilitated the transcription of neuronal gene expression during granule cell proliferation, which peaks at postnatal day 4.5 in mice, as well as in the post-mitotic phase of cerebellum development [125]. These chromatin dynamics continued to postnatal day 60, late in mouse maturation and roughly equivalent to early adulthood in humans [125].
Given the importance of ATM to the long-term viability of neuronal cells during development, and its prominent role in chromatin remodelling, ATM deficiency could predispose cerebellar cells to degeneration soon after they are generated in the early postnatal period. In addition to the role that ATM plays in repairing TOP1cc in the cerebellum and regulating neuronal gene expression through epigenetic modulation, it is possible that the temporal specificity of A-T may be due to the timeline of cerebellar development and maturation. Ataxia symptoms that appear in childhood roughly correspond to the cerebellar degeneration that would occur due to abnormal development and degeneration hastened by neural cells with unrepaired DNA damage from early in development, as well as ongoing DNA damage by transcriptional processes and ROS.
4. Conclusion
As ATM was identified in the early 1990s as a protein kinase targeting p53, the network of proteins that ATM phosphorylates has expanded to hundreds, spanning a multitude of pathways beyond the DDR. Unsurprisingly, ATM's roles and targets within the DDR are among its most well characterized, among which γH2AX is now widely used as an indirect readout of DSB repair. It is interesting that this classical target of ATM has additional ATM-dependent complexity in its regulation. Apart from γH2AX, the ATM-dependent signal transduction cascade continues to grow in intricacy and complexity, with new effector proteins still being identified, such as RIF1, PTIP and, more recently, SCAI. While the ATM : KAP-1 : CHD3.1 axis has been well characterized as integral to heterochromatic DSB repair, this work was followed by many additional novel avenues to investigate chromatin remodelling events, most recently in transcriptionally active chromatin.
In contrast with the DDR, the role of ATM in regulating oxidative stress is more mysterious. An ever-increasing body of evidence implicates it as an important regulator of the oxidative stress response, directing a signalling network that has yet to be fully elucidated. How this network responds to ROS, overlaps with or diverges from the DDR will be interesting, and challenging, to disentangle. The clearest indication of ATM function in response to oxidative stress lies in the neurodegenerative phenotype associated with A-T, as we have discussed in detail. Indeed, the CNS is subject to higher levels of oxidative stress than many other tissues within the human body and, without ATM, cells fare much worse under oxidative stress. In addition to this, the heightened transcriptional demand in neurons, and the novel roles for ATM in responding to transcriptionally associated DNA damage, may predispose neurons to degeneration. Finally, while there has been recent excitement over how ATM mutation might specifically affect neurons of the cerebellum, there is an underappreciated role for ATM in the development and maintenance of glial cells. Glial cells make up more of the brain than neurons, and are more dynamic than we have ever thought in terms of their interactions with other glial cells and also neurons. In particular, emerging neuroimaging which points to white matter abnormalities in A-T patients is a promising foray into the role of ATM in the glial cell. A greater understanding of ATM's role will probably lead to innovative measures to treat A-T, which may be applicable to other DDR-associated and/or chromatin remodelling diseases.
Data accessibility
This article has no additional data.
Competing interests
We declare we have no competing interests.
Funding
N.D.B. is supported by a Vanier Canada Graduate Scholarship, an Alberta Innovates Health Solutions MD-PhD Studentship and an Izaak Walton Killam Graduate Scholarship. S.M. was supported by an Achievers in Medical Sciences award. A.A.G. is the Canada Research Chair for Genome Damage and Instability Disease and this work was undertaken, in part, thanks to funding from the Canada Research Chairs programme. This work in the A.A.G. laboratory was supported by the Canadian Institutes of Health Research.
References
- 1.Liu M, Hu W. 2011. Functional role of ATM in the cellular response to DNA damage. Front. Chem. Sci. Eng. 5, 179–187. ( 10.1007/s11705-009-0268-4) [DOI] [Google Scholar]
- 2.Paull TT. 2015. Mechanisms of ATM activation. Annu. Rev. Biochem. 84, 711–738. ( 10.1146/annurev-biochem-060614-034335) [DOI] [PubMed] [Google Scholar]
- 3.Shiloh Y, Ziv Y. 2013. The ATM protein kinase: regulating the cellular response to genotoxic stress, and more. Nat. Rev. Mol. Cell Biol. 14, 197–210. ( 10.1038/nrm3546) [DOI] [PubMed] [Google Scholar]
- 4.Awasthi P, et al. 2015. ATM and ATR signaling at a glance. J. Cell Sci. 128, 4255–4262. ( 10.1242/jcs.169730) [DOI] [PubMed] [Google Scholar]
- 5.Lempiäinen H, Halazonetis TD. 2009. Emerging common themes in regulation of PIKKs and PI3Ks. EMBO J. 28, 3067–3073. ( 10.1038/emboj.2009.281) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bakkenist CJ, Kastan MB. 2003. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature 421, 499–506. ( 10.1038/nature01368) [DOI] [PubMed] [Google Scholar]
- 7.Bakkenist CJ, Kastan MB. 2015. Chromatin perturbations during the DNA damage response in higher eukaryotes. DNA Repair (Amst.) 36, 8–12. ( 10.1016/j.dnarep.2015.09.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kozlov SV, et al. 2011. Autophosphorylation and ATM activation: additional sites add to the complexity. J. Biol. Chem. 286, 9107–9119. ( 10.1074/jbc.M110.204065) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun Y, Xu Y, Roy K, Price BD. 2007. DNA damage-induced acetylation of lysine 3016 of ATM activates ATM kinase activity. Mol. Cell. Biol. 27, 8502–8509. ( 10.1128/MCB.01382-07) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaidi A, Jackson SP. 2013. KAT5 tyrosine phosphorylation couples chromatin sensing to ATM signalling. Nature 498, 70–74. ( 10.1038/nature12201) [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 11.Sun Y, Jiang X, Chen S, Fernandes N, Price BD. 2005. A role for the Tip60 histone acetyltransferase in the acetylation and activation of ATM. Proc. Natl Acad. Sci. USA 102, 13 182–13 187. ( 10.1073/pnas.0504211102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chiu L-Y, Gong F, Miller KM. 2017. Bromodomain proteins: repairing DNA damage within chromatin. Phil. Trans. R. Soc. B 372, 20160286 ( 10.1098/rstb.2016.0286) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakada S. 2016. Opposing roles of RNF8/RNF168 and deubiquitinating enzymes in ubiquitination-dependent DNA double-strand break response signaling and DNA-repair pathway choice. J. Radiat. Res. 57, i33–i40. ( 10.1093/jrr/rrw027) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goodarzi AA, Jeggo PA. 2013. The repair and signaling responses to DNA double-strand breaks. Adv. Genet. 82, 1–45. ( 10.1016/B978-0-12-407676-1.00001-9) [DOI] [PubMed] [Google Scholar]
- 15.Botuyan MV, Lee J, Ward IM, Kim JE, Thompson JR, Chen J, Mer G. 2006. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell 127, 1361–1373. ( 10.1016/j.cell.2006.10.043) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dhar S, Gursoy-Yuzugullu O, Parasuram R, Price BD. 2017. The tale of a tail: histone H4 acetylation and the repair of DNA breaks. Phil. Trans. R. Soc. B 372, 20160284 ( 10.1098/rstb.2016.0284) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Panier S, Boulton SJ. 2014. Double-strand break repair: 53BP1 comes into focus. Nat. Rev. Mol. Cell Biol. 15, 7–18. ( 10.1038/nrm3719) [DOI] [PubMed] [Google Scholar]
- 18.Lee DH, et al. 2014. Dephosphorylation enables the recruitment of 53BP1 to double-strand DNA breaks. Mol. Cell 54, 512–525. ( 10.1016/j.molcel.2014.03.020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baldock RA, Day M, Wilkinson OJ, Cloney R, Jeggo PA, Oliver AW, Watts FZ, Pearl LH. 2015. ATM localization and heterochromatin repair depend on direct interaction of the 53BP1-BRCT2 domain with γH2AX. Cell Rep. 13, 2081–2089. ( 10.1016/j.celrep.2015.10.074) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Noon AT, Shibata A, Rief N, Löbrich M, Stewart GS, Jeggo PA, Goodarzi AA. 2010. 53BP1-dependent robust localized KAP-1 phosphorylation is essential for heterochromatic DNA double-strand break repair. Nat. Cell Biol. 12, 177–184. ( 10.1038/ncb2017) [DOI] [PubMed] [Google Scholar]
- 21.Lee D-H, Goodarzi AA, Adelmant GO, Pan Y, Jeggo PA, Marto JA, Chowdhury D. 2012. Phosphoproteomic analysis reveals that PP4 dephosphorylates KAP-1 impacting the DNA damage response. EMBO J. 31, 2403–2415. ( 10.1038/emboj.2012.86) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hansen RK, et al. 2016. SCAI promotes DNA double-strand break repair in distinct chromosomal contexts. Nat. Cell Biol. 18, 1357–1366. ( 10.1038/ncb3436) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Finkel T, Holbrook NJ. 2000. Oxidants, oxidative stress and the biology of ageing. Nature 408, 239–247. ( 10.1038/35041687) [DOI] [PubMed] [Google Scholar]
- 24.Jena NR. 2012. DNA damage by reactive species: mechanisms, mutation and repair. J. Biosci. 37, 503–507. ( 10.1007/s12038-012-9218-2) [DOI] [PubMed] [Google Scholar]
- 25.Yan S, Sorrell M, Berman Z. 2014. Functional interplay between ATM/ATR-mediated DNA damage response and DNA repair pathways in oxidative stress. Cell. Mol. Life Sci. 71, 3951–3967. ( 10.1007/s00018-014-1666-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cooke MS, Evans MD, Dizdaroglu M, Lunec J. 2003. Oxidative DNA damage: mechanisms, mutation, and disease. FASEB J. 17, 1195–1214. ( 10.1096/fj.02-0752rev) [DOI] [PubMed] [Google Scholar]
- 27.Berquist BR, Wilson DM. 2012. Pathways for repairing and tolerating the spectrum of oxidative DNA lesions. Cancer Lett. 327, 61–72. ( 10.1016/j.canlet.2012.02.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shackelford RE, Innes CL, Sieber SO, Heinloth AN, Leadon SA, Paules RS. 2001. The ataxia telangiectasia gene product is required for oxidative stress-induced G1 and G2 checkpoint function in human fibroblasts. J. Biol. Chem. 276, 21 951–21 959. ( 10.1074/jbc.M011303200) [DOI] [PubMed] [Google Scholar]
- 29.Kurz EU, Douglas P, Lees-Miller SP. 2004. Doxorubicin activates ATM-dependent phosphorylation of multiple downstream targets in part through the generation of reactive oxygen species. J. Biol. Chem. 279, 53 272–53 281. ( 10.1074/jbc.M406879200) [DOI] [PubMed] [Google Scholar]
- 30.Kim SH, Trinh AT, Larsen MC, Mastrocola AS, Jefcoate CR, Bushel PR, Tibbetts RS. 2016. Tunable regulation of CREB DNA binding activity couples genotoxic stress response and metabolism. Nucleic Acids Res. 44, 9667–9680. ( 10.1093/nar/gkw643) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guo Z, Kozlov S, Lavin MF, Person MD, Paull TT. 2010. ATM activation by oxidative stress. Science 330, 517–521. ( 10.1126/science.1192912) [DOI] [PubMed] [Google Scholar]
- 32.Bencokova Z, Kaufmann MR, Pires IM, Lecane PS, Giaccia AJ, Hammond EM. 2009. ATM activation and signaling under hypoxic conditions. Mol. Cell. Biol. 29, 526–537. ( 10.1128/MCB.01301-08) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kozlov SV, Waardenberg AJ, Engholm-Keller K, Arthur JW, Graham ME, Lavin M. 2016. Reactive oxygen species (ROS)-activated ATM-dependent phosphorylation of cytoplasmic substrates identified by large-scale phosphoproteomics screen. Mol. Cell. Proteomics 15, 1032–1047. ( 10.1074/mcp.M115.055723) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chou W-C, Wang H-C, Wong F-H, Ding S, Wu P-E, Shieh S-Y, Shen C-Y. 2008. Chk2-dependent phosphorylation of XRCC1 in the DNA damage response promotes base excision repair. EMBO J. 27, 3140–3150. ( 10.1038/emboj.2008.229) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kühne C, Tjörnhammar ML, Pongor S, Banks L, Simoncsits A. 2003. Repair of a minimal DNA double-strand break by NHEJ requires DNA-PKcs and is controlled by the ATM/ATR checkpoint. Nucleic Acids Res. 31, 7227–7237. ( 10.1093/nar/gkg937) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jong HL, Kyung HK, Morio T, Kim H. 2006. Ataxia-telangiectasia-mutated-dependent activation of Ku in human fibroblasts exposed to hydrogen peroxide. Ann. NY Acad. Sci. 1091, 76–82. ( 10.1196/annals.1378.056) [DOI] [PubMed] [Google Scholar]
- 37.McKinnon PJ. 2016. Topoisomerases and the regulation of neural function. Nat. Rev. Neurosci. 11, 673–679. ( 10.1038/nrn.2016.101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Das BB, Antony S, Gupta S, Dexheimer TS, Redon CE, Garfield S, Shiloh Y, Pommier Y. 2009. Optimal function of the DNA repair enzyme TDP1 requires its phosphorylation by ATM and/or DNA-PK. EMBO J. 28, 3667–3680. ( 10.1038/emboj.2009.302) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cristini A, Park JH, Capranico G, Legube G, Favre G, Sordet O. 2015. DNA-PK triggers histone ubiquitination and signaling in response to DNA double-strand breaks produced during the repair of transcription-blocking topoisomerase I lesions. Nucleic Acids Res. 44, 1161–1178. ( 10.1093/nar/gkv1196) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qi Y, Schoene NW, Lartey FM, Cheng WH. 2010. Selenium compounds activate ATM-dependent DNA damage response via the mismatch repair protein hMLH1 in colorectal cancer cells. J. Biol. Chem. 285, 33 010–33 017. ( 10.1074/jbc.M110.137406) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Romeo F, Falbo L, Di Sanzo M, Misaggi R, Faniello MC, Viglietto G, Cuda G, Costanzo F, Quaresima B. 2011. BRCA1 is required for hMLH1 stabilization following doxorubicin-induced DNA damage. Int. J. Biochem. Cell Biol. 43, 1754–1763. ( 10.1016/j.biocel.2011.08.011) [DOI] [PubMed] [Google Scholar]
- 42.Zhang T, Penicud K, Bruhn C, Loizou JI, Kanu N, Wang ZQ, Behrens A. 2012. Competition between NBS1 and ATMIN controls ATM signaling pathway choice. Cell Rep. 2, 1498–1504. ( 10.1016/j.celrep.2012.11.002) [DOI] [PubMed] [Google Scholar]
- 43.Price BD, D'Andrea AD. 2013. Chromatin remodeling at DNA double-strand breaks. Cell 152, 1344–1354. ( 10.1016/j.cell.2013.02.011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stanley FKT, Moore S, Goodarzi AA. 2013. CHD chromatin remodelling enzymes and the DNA damage response. Mutat. Res. Fundam. Mol. Mech. Mutagen. 750, 31–44. ( 10.1016/j.mrfmmm.2013.07.008) [DOI] [PubMed] [Google Scholar]
- 45.Saha A, Wittmeyer J, Cairns BR. 2006. Chromatin remodelling: the industrial revolution of DNA around histones. Nat. Rev. Mol. Cell Biol. 7, 437–447. ( 10.1038/nrm1945) [DOI] [PubMed] [Google Scholar]
- 46.Rother MB, van Attikum H. 2017. DNA repair goes hip-hop: SMARCA and CHD chromatin remodellers join the break dance. Phil. Trans. R. Soc. B 372, 20160285 ( 10.1098/rstb.2016.0285) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bönisch C, Hake SB. 2012. Histone H2A variants in nucleosomes and chromatin: more or less stable? Nucleic Acids Res. 40, 10 719–10 741. ( 10.1093/nar/gks865) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bonner WM, Redon CE, Dickey JS, Nakamura AJ, Sedelnikova OA, Solier S, Pommier Y. 2008. γH2AX and cancer. Nat. Rev. Cancer 8, 957–967. ( 10.1038/nrc2523) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Löbrich M, Shibata A, Beucher A, Fisher A, Ensminger M, Goodarzi AA, Barton O, Jeggo PA. 2010. γH2AX foci analysis for monitoring DNA double-strand break repair: strengths, limitations and optimization. Cell Cycle 9, 662–669. ( 10.4161/cc.9.4.10764) [DOI] [PubMed] [Google Scholar]
- 50.Kwon S-J, Park J-H, Park E-J, Lee S-A, Lee H-S, Kang SW, Kwon J. 2015. ATM-mediated phosphorylation of the chromatin remodeling enzyme BRG1 modulates DNA double-strand break repair. Oncogene 34, 303–313. ( 10.1038/onc.2013.556) [DOI] [PubMed] [Google Scholar]
- 51.Xiao A, et al. 2009. WSTF regulates the H2A.X DNA damage response via a novel tyrosine kinase activity. Nature 457, 57–62. ( 10.1038/nature07668) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cook PJ, Ju BG, Telese F, Wang X, Glass CK, Rosenfeld MG. 2009. Tyrosine dephosphorylation of H2AX modulates apoptosis and survival decisions. Nature 458, 591–596. ( 10.1038/nature07849) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Atsumi Y, et al. 2015. ATM and SIRT6/SNF2H mediate transient H2AX stabilization when DSBs form by blocking HUWE1 to allow efficient γH2AX foci formation. Cell Rep. 13, 2728–2740. ( 10.1016/j.celrep.2015.11.054) [DOI] [PubMed] [Google Scholar]
- 54.Goodarzi AA, Noon AT, Deckbar D, Ziv Y, Shiloh Y, Löbrich M, Jeggo PA. 2008. ATM signaling facilitates repair of DNA double-strand breaks associated with heterochromatin. Mol. Cell 31, 167–177. ( 10.1016/j.molcel.2008.05.017) [DOI] [PubMed] [Google Scholar]
- 55.Ziv Y, et al. 2006. Chromatin relaxation in response to DNA double-strand breaks is modulated by a novel ATM- and KAP-1 dependent pathway. Nat. Cell Biol. 8, 870–876. ( 10.1038/ncb1446) [DOI] [PubMed] [Google Scholar]
- 56.Christopher CP, Laetitia D, Grzegorz Z, Irene C. 2017. And yet, it moves: nuclear and chromatin dynamics of a heterochromatic double-strand break. Phil. Trans. R. Soc. B 372, 20160291 ( 10.1098/rstb.2016.0291) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Klement K, Luijsterburg MS, Pinder JB, Cena CS, Del Nero V, Wintersinger CM, Dellaire G, van Attikum H, Goodarzi AA. 2014. Opposing ISWI- and CHD-class chromatin remodeling activities orchestrate heterochromatic DNA repair. J. Cell Biol. 207, 717–733. ( 10.1083/jcb.201405077) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Goodarzi AA, Kurka T, Jeggo PA. 2011. KAP-1 phosphorylation regulates CHD3 nucleosome remodeling during the DNA double-strand break response. Nat. Publ. Gr. 18, 831–839. ( 10.1038/nsmb.2077) [DOI] [PubMed] [Google Scholar]
- 59.Garvin AJ, Morris JR. 2017. SUMO, a small, but powerful, regulator of double-strand break repair. Phil. Trans. R. Soc. B 372, 20160281 ( 10.1098/rstb.2016.0281) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Moyal L, et al. 2011. Requirement of ATM-dependent monoubiquitylation of histone H2B for timely repair of DNA double-strand breaks. Mol. Cell 41, 529–542. ( 10.1016/j.molcel.2011.02.015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Min S, Jo S, Lee HS, Chae S, Lee JS, Ji JH, Cho H. 2014. ATM-dependent chromatin remodeler Rsf-1 facilitates DNA damage checkpoints and homologous recombination repair. Cell Cycle 13, 666–677. ( 10.4161/cc.27548) [DOI] [PubMed] [Google Scholar]
- 62.Beucher A, et al. 2009. ATM and Artemis promote homologous recombination of radiation-induced DNA double-strand breaks in G2. EMBO J. 28, 3413–3427. ( 10.1038/emboj.2009.276) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pessina F, Lowndes NF. 2014. The RSF1 histone-remodelling factor facilitates DNA double-strand break repair by recruiting centromeric and Fanconi anaemia proteins. PLoS Biol. 12, e1001856 ( 10.1371/journal.pbio.1001856) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Polo SE, Kaidi A, Baskcomb L, Galanty Y, Jackson SP. 2010. Regulation of DNA-damage responses and cell-cycle progression by the chromatin remodelling factor CHD4. EMBO J. 29, 3130–3139. ( 10.1038/emboj.2010.188) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Urquhart AJ, Gatei M, Richard DJ, Khanna KK. 2011. ATM mediated phosphorylation of CHD4 contributes to genome maintenance. Genome Integr. 2, 1 ( 10.1186/2041-9414-2-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Harding SM, Boiarsky JA, Greenberg RA. 2015. ATM dependent silencing links nucleolar chromatin reorganization to DNA damage recognition. Cell Rep. 13, 251–259. ( 10.1016/j.celrep.2015.08.085) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kruhlak M, et al. 2007. The ATM repair pathway inhibits RNA polymerase I transcription in response to chromosome breaks. Nature 447, 730–734. ( 10.1038/nature08110) [DOI] [PubMed] [Google Scholar]
- 68.Ciccia A, Huang J-W, Izhar L, Sowa ME, Harper JW, Elledge SJ. 2014. Treacher Collins syndrome TCOF1 protein cooperates with NBS1 in the DNA damage response. Proc. Natl Acad. Sci. USA 111, 18 631–18 636. ( 10.1073/pnas.1422488112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Larsen DH, et al. 2014. The NBS1–treacle complex controls ribosomal RNA transcription in response to DNA damage. Nat. Cell Biol. 16, 792–803. ( 10.1038/ncb3007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shanbhag NM, Rafalska-Metcalf IU, Balane-Bolivar C, Janicki SM, Greenberg RA. 2010. ATM-dependent chromatin changes silence transcription in cis to DNA double-strand breaks. Cell 141, 970–981. ( 10.1016/j.cell.2010.04.038) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kakarougkas A, Ismail A, Chambers AL, Riballo E, Herbert AD, Kunzel J, Lobrich M, Jeggo PA, Downs JA. 2014. Requirement for PBAF in transcriptional repression and repair at DNA breaks in actively transcribed regions of chromatin. Mol. Cell 55, 723–732. ( 10.1016/j.molcel.2014.06.028) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Campbell S, Ismail IH, Young LC, Poirier GG, Hendzel MJ. 2013. Polycomb repressive complex 2 contributes to DNA double-strand break repair. Cell Cycle 12, 2675–2683. ( 10.4161/cc.25795) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lavin MF, Shiloh Y. 1997. The genetic defect in ataxia-telangiectasia. Annu. Rev. Immunol 15, 177–202. ( 10.1146/annurev.immunol.15.1.177) [DOI] [PubMed] [Google Scholar]
- 74.Lavin MF. 2008. Ataxia-telangiectasia: from a rare disorder to a paradigm for cell signalling and cancer. Nat. Rev. Mol. Cell Biol. 9, 759–769. ( 10.1038/nrm2514) [DOI] [PubMed] [Google Scholar]
- 75.McKinnon PJ. 2004. ATM and ataxia telangiectasia. EMBO Rep. 5, 772–776. ( 10.1038/sj.embor.7400210) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Farina L, Uggetti C, Ottolini A, Martelli A, Bergamaschi R, Sibilla L, Zappoli F, Egitto MG, Lanzi G. 1994. Ataxia-telangiectasia: MR and CT findings. J. Comput. Assist. Tomogr. 18, 724–727. ( 10.1097/00004728-199409000-00008) [DOI] [PubMed] [Google Scholar]
- 77.Tavani F, Zimmerman RA, Berry GT, Sullivan K, Gatti R, Bingham P. 2003. Ataxia-telangiectasia: the pattern of cerebellar atrophy on MRI. Neuroradiology 45, 315–319. ( 10.1007/s00234-003-0945-9) [DOI] [PubMed] [Google Scholar]
- 78.Barzilai A, Rotman G, Shiloh Y. 2002. ATM deficiency and oxidative stress: a new dimension of defective response to DNA damage. DNA Repair (Amst.) 1, 3–25. ( 10.1016/S1568-7864(01)00007-6) [DOI] [PubMed] [Google Scholar]
- 79.Biton S, Barzilai A, Shiloh Y. 2008. The neurological phenotype of ataxia-telangiectasia: solving a persistent puzzle. DNA Repair (Amst.) 7, 1028–1038. ( 10.1016/j.dnarep.2008.03.006) [DOI] [PubMed] [Google Scholar]
- 80.Hoche F, et al. 2014. Cognitive phenotype in ataxia-telangiectasia. Pediatr. Neurol. 51, 297–310. ( 10.1016/j.pediatrneurol.2014.04.027) [DOI] [PubMed] [Google Scholar]
- 81.Chiesa N, Barlow C, Wynshaw-Boris A, Strata P, Tempia F. 2000. Atm-deficient mice Purkinje cells show age-dependent defects in calcium spike bursts and calcium currents. Neuroscience 96, 575–583. ( 10.1016/S0306-4522(99)00581-3) [DOI] [PubMed] [Google Scholar]
- 82.Xu Y, Ashley T, Brainerd EE. 1996. Targeted disruption of ATM leads to growth retardation, chromosomal fragmentation during meiosis, immune defects, and thymic lymphoma. Genes Dev. 10, 2411–2422. ( 10.1101/gad.10.19.2411) [DOI] [PubMed] [Google Scholar]
- 83.Elson A, Wang Y, Daugherty CJ, Morton CC, Zhou F, Campos-Torres J, Leder P. 1996. Pleiotropic defects in ataxia-telangiectasia protein-deficient mice. Proc. Natl Acad. Sci. USA 93, 13 084–13 089. ( 10.1073/pnas.93.23.13084) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Uttara B, Singh AV, Zamboni P, Mahajan RT. 2009. Oxidative stress and neurodegenerative diseases: a review of upstream and downstream antioxidant therapeutic options. Curr. Neuropharmacol. 7, 65–74. ( 10.2174/157015909787602823) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kamsler A, Daily D, Hochman A. 2001. Increased oxidative stress in ataxia telangiectasia evidenced by alterations in redox state of brains from ATM-deficient mice. Cancer Res. 1, 1849–1854. [PubMed] [Google Scholar]
- 86.Reichenbach J, Schubert R, Schindler D, Müller K, Böhles H, Zielen S. 2002. Elevated oxidative stress in patients with ataxia telangiectasia. Antioxid. Redox Signal. 4, 465–469. ( 10.1089/15230860260196254) [DOI] [PubMed] [Google Scholar]
- 87.Takao N, Li Y, Yamamoto KI. 2000. Protective roles for ATM in cellular response to oxidative stress. FEBS Lett. 472, 133–136. ( 10.1016/S0014-5793(00)01422-8) [DOI] [PubMed] [Google Scholar]
- 88.Chen P, Peng C, Luff J, Spring K, Watters D, Bottle S, Furuya S, Lavin MF. 2003. Oxidative stress is responsible for deficient survival and dendritogenesis in Purkinje neurons from ataxia-telangiectasia mutated mutant mice. J. Neurosci. 23, 11 453–11 460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Reliene R, Schiestl RH. 2008. Experimental antioxidant therapy in ataxia telangiectasia. Clin. Med. Oncol. 2, 431–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Reliene R, Schiestl RH. 2007. Antioxidants suppress lymphoma and increase longevity in ATM-deficient mice. J. Nutr. 137, 229S–232S. [DOI] [PubMed] [Google Scholar]
- 91.Quick KL, Dugan LL. 2001. Superoxide stress identifies neurons at risk in a model of ataxia-telangiectasia. Ann. Neurol. 49, 627–635. ( 10.1002/ana.1005) [DOI] [PubMed] [Google Scholar]
- 92.Barlow C, Dennery P, Shigenaga M, Smith M, Morrow J, Roberts L, Wynshaw-Boris A, Levine R. 1999. Loss of the ataxia-telangiectasia gene product causes oxidative damage in target organs. Proc. Natl Acad. Sci. USA 96, 9915–9919. ( 10.1073/pnas.96.17.9915) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Liu N, Stoica G, Yan M, Scofield VL, Qiang W, Lynn WS, Wong PKY. 2005. ATM deficiency induces oxidative stress and endoplasmic reticulum stress in astrocytes. Lab. Invest. 85, 1471–1480. ( 10.1038/labinvest.3700354) [DOI] [PubMed] [Google Scholar]
- 94.Campbell A, Bushman J, Munger J, Noble M, Proschel C, Mayer-Proschel M. 2016. Mutation of ataxia-telangiectasia mutated is associated with dysfunctional glutathione homeostasis in cerebellar astroglia. Glia 64, 227–239. ( 10.1002/glia.22925) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Firat AK, Karakaş HM, Firat Y, Yakinci C. 2005. Quantitative evaluation of brain involvement in ataxia telangiectasia by diffusion weighted MR imaging. Eur. J. Radiol. 56, 192–196. ( 10.1016/j.ejrad.2005.04.009) [DOI] [PubMed] [Google Scholar]
- 96.Sahama I, Sinclair K, Fiori S, Pannek K, Lavin M, Rose SE. 2014. Altered corticomotor-cerebellar integrity in young ataxia telangiectasia patients. Mov. Disord. 29, 1289–1298. ( 10.1002/mds.25970) [DOI] [PubMed] [Google Scholar]
- 97.Sahama I, Sinclair K, Fiori S, Doecke J, Pannek K, Reid L, Lavin M, Rose S. 2015. Motor pathway degeneration in young ataxia telangiectasia patients: a diffusion tractography study. NeuroImage Clin. 9, 206–215. ( 10.1016/j.nicl.2015.08.007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tse K, Herrup K. 2016. DNA damage in the oligodendrocyte lineage and its role in brain aging. Mech. Ageing Dev. 161, 37–50. ( 10.1016/j.mad.2016.05.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hetman M, Vashishta A, Rempala G. 2010. Neurotoxic mechanisms of DNA damage: focus on transcriptional inhibition. J. Neurochem. 114, 1537–1549. ( 10.1111/j.1471-4159.2010.06859.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Katyal S, et al. 2014. Aberrant topoisomerase-1 DNA lesions are pathogenic in neurodegenerative genome instability syndromes. Nat. Neurosci. 17, 813–821. ( 10.1038/nn.3715) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sordet O, et al. 2009. Ataxia telangiectasia mutated activation by transcription- and topoisomerase I-induced DNA double-strand breaks. EMBO Rep. 10, 887–893. ( 10.1038/embor.2009.97) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Alagoz M, Chiang SC, Sharma A, El-Khamisy SF. 2013. ATM deficiency results in accumulation of DNA-topoisomerase I covalent intermediates in neural cells. PLoS ONE 8, e58239 ( 10.1371/journal.pone.0058239) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gorodetsky E, Calkins S, Ahn J, Brooks PJ. 2007. ATM, the Mre11/Rad50/Nbs1 complex, and topoisomerase I are concentrated in the nucleus of Purkinje neurons in the juvenile human brain. DNA Repair (Amst.) 6, 1698–1707. ( 10.1016/j.dnarep.2007.06.011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ito K, et al. 2004. Regulation of oxidative stress by ATM is required for self-renewal of haematopoietic stem cells. Nature 431, 997–1002. ( 10.1038/nature02989) [DOI] [PubMed] [Google Scholar]
- 105.Wei PC, Chang AN, Kao J, Du Z, Meyers RM, Alt FW, Schwer B. 2016. Long neural genes harbor recurrent DNA break clusters in neural stem/progenitor cells. Cell 164, 644–655. ( 10.1016/j.cell.2015.12.039) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Schwer B, Wei P-C, Chang AN, Kao J, Du Z, Meyers RM, Alt FW. 2016. Transcription-associated processes cause DNA double-strand breaks and translocations in neural stem/progenitor cells. Proc. Natl Acad. Sci. USA 113, 2258–2263. ( 10.1073/pnas.1525564113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Álvarez-Quilón A, Serrano-Benítez A, Lieberman JA, Quintero C, Sánchez-Gutiérrez D, Escudero LM, Cortés-Ledesma F. 2014. ATM specifically mediates repair of double-strand breaks with blocked DNA ends. Nat. Commun. 5, 3347 ( 10.1038/ncomms4347) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Suberbielle E, Sanchez PE, Kravitz AV, Wang X, Ho K, Eilertson K, Devidze N, Kreitzer AC, Mucke L. 2013. Physiologic brain activity causes DNA double-strand breaks in neurons, with exacerbation by amyloid-β. Nat. Neurosci. 16, 613–621. ( 10.1038/nn.3356) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Madabhushi R, et al. 2015. Activity-induced DNA breaks govern the expression of neuronal early-response genes. Cell 161, 1592–1605. ( 10.1016/j.cell.2015.05.032) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Jiang X, Nardelli J. 2015. Cellular and molecular introduction to brain development. Neurobiol. Dis. 92, 3–17. ( 10.1016/j.nbd.2015.07.007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Li J, Chen J, Ricupero CL, Hart RP, Schwartz MS, Kusnecov A, Herrup K. 2012. Nuclear accumulation of HDAC4 in ATM deficiency promotes neurodegeneration in ataxia telangiectasia. Nat. Med. 18, 783–790. ( 10.1038/nm.2709) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Majdzadeh N, Wang L, Morrison BE, Bassel-Duby R, Olson EN, D'Mello SR. 2008. HDAC4 inhibits cell-cycle progression and protects neurons from cell death. Dev. Neurobiol. 68, 1076–1092. ( 10.1002/dneu.20637) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Li J, Hart RP, Mallimo EM, Swerdel MR, Kusnecov AW, Herrup K. 2013. EZH2-mediated H3K27 trimethylation mediates neurodegeneration in ataxia-telangiectasia. Nat. Neurosci. 16, 1745–1753. ( 10.1038/nn.3564) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Herrup K, Yang Y. 2007. Cell cycle regulation in the postmitotic neuron: oxymoron or new biology? Nat. Rev. Neurosci. 8, 368–378. ( 10.1038/nrn2124) [DOI] [PubMed] [Google Scholar]
- 115.Dobbin MM, et al. 2013. SIRT1 collaborates with ATM and HDAC1 to maintain genomic stability in neurons. Nat. Neurosci. 16, 1008–1015. ( 10.1038/nn.3460) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Fang EF, et al. 2016. NAD+ replenishment improves lifespan and healthspan in ataxia telangiectasia models via mitophagy and DNA repair. Cell Metab. 24, 566–581. ( 10.1016/j.cmet.2016.09.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ambrose M, Goldstine JV, Gatti RA. 2007. Intrinsic mitochondrial dysfunction in ATM-deficient lymphoblastoid cells. Hum. Mol. Genet. 16, 2154–2164. ( 10.1093/hmg/ddm166) [DOI] [PubMed] [Google Scholar]
- 118.D'Souza AD, Parish IA, Krause DS, Kaech SM, Shadel GS. 2012. Reducing mitochondrial ROS improves disease-related pathology in a mouse model of ataxia-telangiectasia. Mol. Ther. 21, 42–48. ( 10.1038/mt.2012.203) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sharma NK, Lebedeva M, Thomas T, Kovalenko OA, Stumpf JD, Shadel GS, Santos JH. 2014. Intrinsic mitochondrial DNA repair defects in ataxia telangiectasia. DNA Repair (Amst.) 13, 22–31. ( 10.1016/j.dnarep.2013.11.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Jiang D, Zhang Y, Hart RP, Chen J, Herrup K, Li J. 2015. Alteration in 5-hydroxymethylcytosine-mediated epigenetic regulation leads to Purkinje cell vulnerability in ATM deficiency. Brain 138, 3520–3536. ( 10.1093/brain/awv284) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zhu X, Girardo D, Govek EE, John K, Mellén M, Tamayo P, Mesirov JP, Hatten ME. 2016. Role of Tet1/3 genes and chromatin remodeling genes in cerebellar circuit formation. Neuron 89, 100–112. ( 10.1016/j.neuron.2015.11.030) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kiessling MC, Büttner A, Butti C, Müller-Starck J, Milz S, Hof PR, Frank HG, Schmitz C. 2014. Cerebellar granule cells are generated postnatally in humans. Brain Struct. Funct. 219, 1271–1286. ( 10.1007/s00429-013-0565-z) [DOI] [PubMed] [Google Scholar]
- 123.Herculano-Houzel S. 2012. The remarkable, yet not extraordinary, human brain as a scaled-up primate brain and its associated cost. Proc. Natl Acad. Sci. USA 109, 10 661–10 668. ( 10.1073/pnas.1201895109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Walløe S, Pakkenberg B, Fabricius K. 2014. Stereological estimation of total cell numbers in the human cerebral and cerebellar cortex. Front. Hum. Neurosci. 8, 508 ( 10.3389/fnhum.2014.00508) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Frank CL, et al. 2015. Regulation of chromatin accessibility and Zic binding at enhancers in the developing cerebellum. Nat. Neurosci. 18, 647–656. ( 10.1038/nn.3995) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This article has no additional data.