Abstract
The accessibility of eukaryotic genomes to the action of enzymes involved in transcription, replication and repair is maintained despite the organization of DNA into nucleosomes. This access is often regulated by the action of ATP-dependent nucleosome remodellers. The INO80 class of nucleosome remodellers has unique structural features and it is implicated in a diverse array of functions, including transcriptional regulation, DNA replication and DNA repair. Underlying these diverse functions is the catalytic activity of the main ATPase subunit, which in the context of a multisubunit complex can shift nucleosomes and carry out histone dimer exchange. In vitro studies showed that INO80 promotes replication fork progression on a chromatin template, while in vivo it was shown to facilitate replication fork restart after stalling and to help evict RNA polymerase II at transcribed genes following the collision of a replication fork with transcription. More recent work in yeast implicates INO80 in the general eviction and degradation of nucleosomes following high doses of oxidative DNA damage. Beyond these replication and repair functions, INO80 was shown to repress inappropriate transcription at promoters in the opposite direction to the coding sequence. Here we discuss the ways in which INO80's diverse functions help maintain genome integrity.
This article is part of the themed issue ‘Chromatin modifiers and remodellers in DNA repair and signalling’.
Keywords: INO80 remodeller, transcription, DNA repair, replication stress, checkpoint, genome instability
1. Introduction
Eukaryotic DNA is tightly packaged into nucleosomes, which are the simplest units of chromatin. The canonical nucleosome contains 147 bp of DNA wrapped twice around an octamer of histones, each containing two histone H2A-H2B heterodimers and a histone H3-H4 heterotetramer. In the presence of linker histones and accessory scaffold proteins, chromatin achieves high levels of compaction, reaching up to 104-fold linear compaction in the condensed mitotic chromosomes of mammalian cells. In interphase, euchromatic domains become transiently decompacted, while the transcription-resistant heterochromatic domains retain their compact state. Despite this folding, both heterochromatin and euchromatin seem to remain largely accessible to factors that mediate essential DNA-based processes like replication, repair and transcription. How do eukaryotic cells reconcile DNA compaction and accessibility?
Chromatin is rarely, if ever, in a static state. Nucleosome turnover, histone replacement, histone variant deposition and the unfolding or shifting of nucleosomes occur constantly. This remodelling of chromatin is not cell-cycle-specific, and can be triggered either by internal signals like cellular differentiation and DNA damage, or by external stimuli that induce new transcriptional states. Chromatin access responds both to the action of ATP-hydrolysing remodellers and to covalent post-translational modifications (PTMs) on histones. The activities of the ATP-dependent remodellers include histone exchange (canonical and variant forms), the eviction of histones or nucleosomes, and the repositioning or sliding of nucleosomes along DNA. In addition, there is accumulating evidence that nucleosome remodellers can facilitate the eviction of non-histone factors from chromatin [1–3].
Nucleosome remodellers are multi-subunit complexes containing an ATPase subunit of the Snf2 (sucrose non-fermenting 2)-type of helicase. Based on the structural characteristics of this subunit, remodellers have been classified in four subfamilies: the SWI/SNF group (which bind acetylated lysines), the ISWI group (containing SANT and SLIDE domains), the CHD group (containing chromodomains that bind methylated lysines) and the INO80 group (which has a large insertion to its ATPase domain) [4–6]. In this article, we focus primarily on the INO80 family of chromatin remodellers, which is present in most species, including budding yeast and humans. The INO80 group includes the INO80 and SWR1 complexes (SRCAP in mammals), which carry a long insertion within the Snf2-ATPase domain that is responsible for the recruitment of the Rvb1/2 helicase, a hexameric subcomplex consisting of two functionally related AAA+ ATPase subunits (Rvb1/Rvb2 in yeast, or Tip49a/b, or pontin/reptin in mammals). Both SWR1 and INO80 remodellers also contain the Arp4-actin dimer and other actin-related proteins (Arp) [4]. INO80 complexes contain Arp5 and Arp8, as well as specialized subunits, like the Ino eighty subunits 2 and 6 (Ies2 and Ies6). The acquisition of divergent subunits in the holo-complex in different species appears to coincide with the expansion of species-specific functions, such as the HMG variant Nhp10 in the budding yeast INO80 complex or the gene-specific factor YY1 in mammalian INO80 [7]. Although both INO80 and SWR1/SRCAP complexes contain the Arp4-actin and Rvb1/Rvb2 subunits, and both contribute to aspects of genome stability, their functions have diverged so strikingly that in some cases they seem to have antagonistic roles [4].
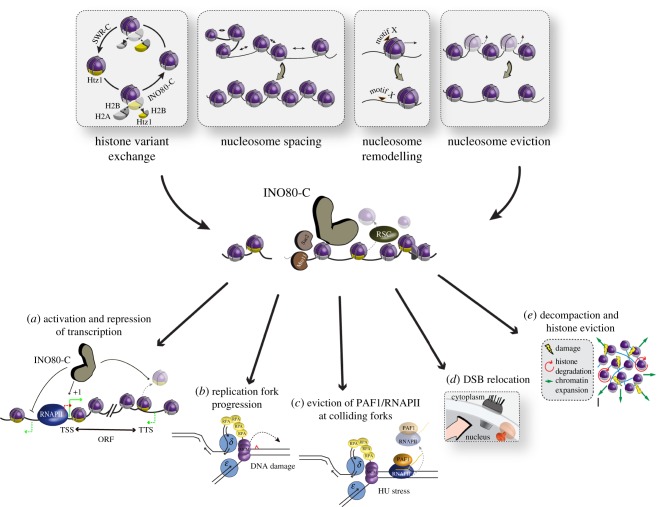
Since its initial discovery in gene activation in response to inositol starvation, INO80 has been implicated in a wide variety of DNA-based transactions, including the regulation of basal and inducible transcription, DNA replication, recombination and the repair of DNA damage (figure 1). Here we provide a comprehensive view of INO80's roles in transcription and replication, and examine its recently reported function in altering chromatin following DNA damage. We discuss the experimental evidence that supports direct involvement of the INO80 complex and, when data are available, the conservation of such functions across species.
Figure 1.
Remodelling activities and cellular functions of the INO80 complex. The INO80 complex mediates several remodelling activities including histone variant exchange (Htz1/H2A.Z removal), nucleosome spacing at genes, nucleosome remodelling/sliding and nucleosome eviction from DNA. (a) At genes, INO80 both activates and represses transcription. On the one hand, it promotes transcription (solid green arrow) at the transcription start site (TSS) by positioning the −1 and +1 nucleosomes and defining the nucleosome free region. On the other hand, it limits pervasive transcription at promoters where it reinforces transcription directionality (repressing antisense RNA; green dashed arrows) at the TSS and at the transcription termination site (TTS) by removing either H2A.Z or by evicting the transcription machinery. INO80 also limits the sense cryptic unstable transcripts (red dashed arrow) at the TSS. (b,c) During S-phase, INO80 (b) promotes replication fork restart at damage sites and (c) solves the problem of replication–transcription collisions by evicting the transcription machinery (RNAPII and the Paf1 complex) during replicative stress. (d) DNA double-strand breaks (DSBs) are repaired either at the nuclear pore complex or on Mps3; the INO80 complex being required for DSB relocalization to Mps3 in S- and G2-phases. (e) In the presence of several DSBs, INO80-C evicts nucleosomes from DNA, leading to chromatin decompaction and global enhancement of chromatin mobility.
2. INO80 and transcription: a puzzling connection
The INO80 ATPase was originally identified in a genetic screen in Saccharomyces cerevisiae for mutants defective in gene activation in response to inositol depletion [8]. Several subsequent studies across different species reported that INO80's chromatin remodelling function promotes the transcription of genes induced by a variety of diverse signalling pathways, such as PHO5 in yeast by phosphate depletion, and GRP78 in human cells by endoplasmic reticulum stress [7,9,10]. Although these early studies argued for a co-activator role in inducible gene expression, transcriptomic data under unstressed conditions showed that a large number of genes have altered transcript levels in the presence or absence of INO80 remodelling function, with almost as many transcripts shifted up as down [11,12]. For instance, in an Affimetrix-based analysis of gene expression comparing ino80 and arp8 deficient strains with an isogenic wild-type background, 1156 genes had at least a 1.5-fold change in steady state mRNA levels (approx. 20% of yeast open reading frames (ORFs)), with 668 being upregulated and 488 showing reduced expression [12]. After exposure to the alkylating agent methyl methanesulfonate (MMS) about 2500 genes showed altered expression in a wild-type background, and only 80 genes failed to respond appropriately, either up or down, in the ino80 mutant [12]. Thus INO80 did not primarily mediate response to stress, but rather affected transcription quite generally. It was therefore proposed that INO80 chromatin remodelling has broad effects on promoters, facilitating both transcriptional activation and repression by modulating the position and composition of nucleosomes at promoters.
This hypothesis has been corroborated by recent studies. RNA-seq in strains lacking either the catalytic Ino80 subunit, or the Arp5-Ies6 core subcomplex, showed misregulation of over 15% of the yeast genome, with roughly half of the affected genes upregulated and the other half downregulated [13], very much in line with earlier work [11,12]. It was then examined whether the changes in transcript levels were a direct or indirect effect of INO80 ablation. Indeed, in Yao et al. [13], the broad effects on transcription could be correlated with the occupancy of Ino80, Arp5 and Ies6 at the +1 nucleosome of the transcriptional start site of the affected genes, arguing strongly for a direct role in transcriptional regulation. Consistently, an elegant genome-wide study showed that INO80 can bind over 90% of budding yeast's gene promoters [14]. Remarkably, transcription expression profiling in HeLa cells following RNA interference (RNAi) against the hINO80 subunit revealed changes in expression of a similar number of human genes (1936 ORFs in total), again split nearly equally between upregulation and downregulation [15].
A thorough analysis of Arp5/Ies6-dependent genes in budding yeast suggested that INO80 has a global effect on metabolic pathways, with most genes involved in glycolysis showing reduced expression and those of the mitochondrial electron transport chain being increased upon loss of INO80 [13]. In addition, the yeast inositol pathway [8] and osmotic-stress-regulated yeast genes [16] are INO80-regulated. Finally, INO80 feeds back to ensure the appropriate expression of its own subunits [12]. Looking beyond yeast, the situation was somewhat different. The Drosophila INO80 was shown to facilitate transcriptional repression of ecdysone-regulated genes during pre-pupal development [17], while loss of the human INO80 affected expression of cell cycle genes, particularly those under control of p53, including the cell-cycle regulator p21Waf1/Cip1 [15]. In the mammalian case, it is unclear whether the cell-cycle effects are direct or indirect, for example, due to an accumulation of DNA damage. While budding yeast INO80 affected transcription broadly in addition to mediating a metabolic/osmotic stress response, in other species the genes under INO80 control fall into specific subclasses. Below we summarize recent advances in a mechanistic understanding of INO80 and transcription.
3. Recruitment of INO80 at genes
Using the ChIP-exo technique to map each of the subunits of INO80 across the genome at near-nucleotide resolution, Pugh and colleagues found that the INO80 complex occupies the nucleosome free regions (NFRs) of the transcription start sites (TSSs) of over 90% of budding yeast promoters [14]. Interestingly, INO80 is also recruited at transcription termination sites (TTSs) [18,19]. Although the role of INO80 at the TTS is unknown, the presence of NFRs at the 3′ end of genes [20] provides further support on the model that INO80 recognizes and preferentially binds DNA that is devoid of nucleosomes. Its recruitment may reflect the ssDNA binding activity attributed to human Arp8 [21]. In line with this model, the association of INO80 to nucleosomes is enhanced by extranucleosomal DNA of at least 20 nucleotides compared to nucleosomes without additional linker DNA [22]. These observations are reminiscent of SWR1 recruitment at NFRs, which also appears to rely on linker DNA [23].
Enrichment of INO80 at promoters in some cases correlates positively with transcriptional activity [16]. During the activation of stress-response genes, its recruitment coincides with that of RNAPII [24]. Consistently, INO80 has been found to interact both with Rpb1 (subunit of RNAPII) and the transcription elongation complex PAF1 [2,3], arguing that the transcription machinery stabilizes, or cooperates with the NFR, to facilitate INO80 recruitment. In contrast to many other ATP-dependent chromatin remodelling complexes, such as mammalian SWI/SNF, INO80 lacks histone-binding motifs, such as CHD1- and bromo-domains. These ‘reader’ motifs generally target complexes or proteins to modified nucleosomes. The only characterized PTM reader motif in the INO80 complex is the YEATS domain of its subunit Taf14. The Taf14 YEATS domain binds to acetylated or crotonylated histone H3K9, both of which are found at sites of active transcription. Whereas Taf14 is implicated in both transcription and the DNA damage response [23,25,26], it is also a subunit of the chromatin remodelling complexes SWI/SNF, RSC, the NuA3 histone acetyltransferase complex, as well as the TFIID and TFIIF general transcription factor complexes. In budding yeast, the INO80-specific subunit Nhp10 appears to have affinity for phospho-H2A-S129, but its function in recruitment in vivo is unclear, and it is not found in INO80 complexes in other species. Instead of recognizing histone modifications, it is speculated that INO80 recruitment either creates the NFR or is dictated by it, after which histone marks and/or specific factors (such as RNAPII) can stabilize the NFR further. Unlike the case with the SWR1 remodeller, no reported INO80 subunit deletion mutant was able to abrogate INO80 binding to chromatin completely. Thus, further work is needed to clarify the mode of INO80 recruitment to chromatin.
4. Chromatin remodelling function of INO80
The purified INO80 complex uses the energy of ATP hydrolysis to mobilize canonical nucleosomes in cis [27,28] and to space them approximately 30 bp apart [22]. To shed light on the contribution of INO80 and other remodellers in nucleosome positioning across the genome, the Korber laboratory developed an in vitro nucleosome reconstitution system [29]. This assay combines yeast genomic DNA, recombinant canonical histones and the four chromatin remodelling complexes RSC, ISW2, INO80 and ISW1a purified from yeast cells [29]. Strikingly, INO80 was the only remodeller tested able to recognize and establish NFRs by itself, and to position correctly the −1 and +1 nucleosomes relative to the TSS (figure 1a) [29]. More specifically, INO80 cooperated with ISWIa to correctly space the nucleosomes downstream to the TSS [29]. These results suggest that INO80 has the intrinsic capability to properly organize the promoter nucleosome architecture, as well as to reposition nucleosomes after their mobilization/destabilization by the elongating transcription machinery. These observations emphasize the crucial and very global role of INO80 discussed above [14].
Apart from its activity on positioning canonical nucleosomes, INO80 also has the capability to exchange nucleosomal histone variant H2A.Z with free H2A in vitro [30], a function that appears to be evolutionarily conserved [14,30–35]. Unlike the SWI/SNF remodeller [36], INO80 does not generally evict octomers at promoters upon gene activation [30], yet together with SWI/SNF, INO80 is necessary for efficient nucleosome remodelling during PHO5 gene activation, which renders the promoter accessible to transcription factors and ensures full PHO5 activation [9,10]. Supporting the notion that INO80-mediated exchange of H2A.Z facilitates complete nucleosome turnover [14], is the finding that the remodelling function of INO80 enhances chromatin mobility of the PHO5 promoter, as monitored by fluorescence microscopy of single locus trajectories [37]. A similar increase in chromatin movement also occurs under other conditions that reduce nucleosome occupancy [38]. Interestingly, the pre-initiation complex of RNAPII also promotes removal of H2A.Z from TSS [39]. Since the interaction of INO80 with the elongating RNAPII machinery [2,3] promotes its recruitment to inducible promoters [16,24], the eviction of H2A.Z by INO80 may take place post-initiation, possibly to modulate the passage of RNAPII through the +1 nucleosome [40,41].
Two recent studies in yeast, one using a strain lacking Ino80 [42] and one that employed an anchor-away strain for conditional depletion of Ino80 from the nucleus [39], reported that the loss of INO80 complex activity did not have an effect on the distribution or the occupancy levels of H2A.Z on chromatin. The underlying reasons for the discrepancies between the different studies on the function of INO80 in H2A.Z eviction are not clear. However, the use of different yeast backgrounds, or a possible residual INO80 activity in the nucleus during the time course of the anchor-away experiment, might account for these discrepancies. It could be that there are multiple mechanisms for H2A.Z eviction which have varying degrees of redundancy in different yeast backgrounds. It should be noted that in some budding yeast backgrounds the deletion of the INO80 gene is lethal, while in others, such as S288C, the ino80 null allele is viable.
5. Regulation of non-coding transcription by INO80
The depletion of INO80 affects the kinetics of both the induction and repression of genes [10,24], yet it seems to have an even more profound effect on the de-repression of non-coding transcripts across the genome. Using an elegant fluorescent reporter system to screen for mutants that upregulate non-coding divergent transcription from a bidirectional promoter, Buratowski and colleagues discovered that INO80 prevents bidirectional transcription at functional promoters [43]. INO80 also enhances transcriptional silencing within heterochromatin [18] and disruption of INO80 leads to extensive, pervasive transcription of long non-coding RNAs (lncRNAs), most of which are degraded by either the 3′–5′ exonuclease activity of the exosome or the 5′–3′ exonuclease Xrn1 (figure 1a) [35].
How does INO80 repress intragenic and pervasive transcription? Unstable non-coding RNAs are regulated both transcriptionally and post-transcriptionally through degradation by the nuclear exosome, or the 5′–3′ exonuclease Xrn1 [44]. Loss of H2A.Z from chromatin suppresses antisense transcription from bidirectional promoters in budding yeast and mouse embryonic stem cells (ESCs) [45]. Therefore, it is possible that the eviction of H2A.Z by INO80 plays a role in transcriptional repression of non-coding RNAs (ncRNAs) [35]. Importantly, INO80 also blocks methylation of H3K79 by Dot1 in vitro and prevents aberrant deposition of H3K79me outside the gene bodies or in heterochromatin [18]. H3K79me is a histone PTM positively associated with transcription elongation; thus its absence may promote transcriptional silencing of cryptic promoters, as in telomeric heterochromatin [46,47].
The impact of INO80 is not limited to antisense transcripts, for inactivation of the INO80 complex also leads to the stabilization of unstable sense ncRNA (cryptic unstable transcripts or CUTs) on a genome-wide level [35]. In wild-type yeast cells, CUTs are very rapidly degraded by Nrd1-Nab3-Sen1 and the exosome, which indicates that INO80 may contribute to the post-transcriptional silencing of non-coding transcription. The function of INO80 in repressing expression of ncRNAs could also reflect the recently characterized role of INO80 in facilitating extraction of RNAPII from chromatin for its subsequent degradation [2]. This occurs in the context of replication fork–transcription complex collisions, where INO80 and the checkpoint kinase Mec1-Ddc2 contribute to the eviction of RNAPII and the PAF1 complex, leading to the transient degradation of the former in yeast treated with hydroxyurea (HU) [3]. Thus, the INO80 complex may reduce pervasive transcription by multiple different mechanisms, e.g., histone removal (H2A.Z), the prevention of histone H3K79 methylation, post-transcriptional degradation or direct eviction of the transcription machinery from the chromatin template (figure 1a,c). Remarkably, two of the initial genes exhibiting INO80-dependent activation, PHO5 and INO1, are regulated by an antisense ncRNA, suggesting that the silencing (ncRNA) and the activating (mRNA) functions of INO80 can occur at the same loci.
6. INO80 in DNA replication
The first evidence suggesting a role of INO80 in genome stability arose from the observation that yeast cells lacking key INO80 subunits were hypersensitive to physical DNA damage such as that provoked by ultraviolet (UV) and ionizing radiation (IR), or MMS and HU, which generate replication stress through different pathways [48]. IR creates single- and double-strand breaks (DSBs) on the DNA template, whereas UV treatment generates thymidine dimers, which secondarily form breaks and ssDNA during the repair process [49]. INO80 is recruited rapidly to a specific HO-induced DSB in yeast [12,50], providing evidence that this remodeller acts directly at sites of DSBs. In this case, INO80 facilitates distinct steps in the subsequent repair process [51].
The role of INO80 at stalled or damaged replication forks was suggested by the fact that strains lacking the remodeller function were very sensitive to HU, a potent inhibitor of the ribonucleotide reductase (RNR), which catalyses the rate-limiting step in the de novo deoxyribonucleotides (dNTP) biosynthesis pathway. HU treatment leads to a drop in intracellular dNTP levels, without completely exhausting the pools [52]. This, in turn, reduces the kinetics of S-phase by slowing fork speed and origin firing [53]. Whereas wild-type cells are able to cope with relatively high levels of replication stress, mutants of the replication machinery, the DNA replication checkpoint, or in replication fork restart pathways are hypersensitive to replication stress [10,54]. Interestingly, replication-stress-induced lethality of ino80 mutants does not stem from an impaired checkpoint response [51] nor a lack of transcriptional response to replication stress [12]. Instead, INO80 may play a direct role in the restart of stalled replication forks.
Support for this hypothesis came from three independent studies in budding yeast that mapped the INO80 complex to about half of the known replication origins in HU-challenged S-phase cells [55–57], as well as during normal S-phase progression [58]. The association of INO80 with origins appears to be linked to replication or early S-phase, because only 4% of the origins were still bound in G2. Unlike most of the S-phase checkpoint effectors, INO80 binds almost equally to early- and late-firing origins (55% versus 45%, respectively) [55,56]. In mammalian cells, INO80 recruitment to replication forks is mediated by ubiquitinylated H2A and the BRCA1-associated protein 1 (BAP1) [58]. Additionally, the yeast INO80 complex physically interacts with the replication protein A (RPA) [59], suggesting that several factors/pathways could favour INO80 binding to the replication forks.
What role does INO80 play at the replisome? Yeast cells deprived of a functional INO80 complex progress more slowly through a normal S-phase [60], and in mammalian cells fork progression is also significantly delayed [61]. The loss of INO80 in mouse embryos perturbs embryonic development, but it is unclear whether this stems from defects in transcription or replication [58]. Instead the best documented phenotype of ino80 mutants in vivo with respect to replication, is their failure to resume replication after an acute treatment with genotoxic drugs (HU and MMS) [55,56,60,61]. Together these observations implicate INO80 in normal DNA replication as well as in the recovery from replication stress.
During replication stress, one of the main functions of the DNA replication checkpoint is to protect stalled replication forks from the formation of toxic recombination intermediates that trigger an irreversible fork collapse [54]. In wild-type cells exposed to HU the replisome remains engaged despite the accumulation of ssDNA. In the presence of MMS, fork reversal and/or translesion synthesis, which entails a switch to error-prone polymerases, ensures fork progression. Depending on the lesion, Rad51-dependent strand invasion is often necessary for fork restart [62,63]. Clearly, the responses of a replication fork to MMS versus HU are very different, yet INO80 appears to be involved in both. On acute fork arrest by HU, the general function of INO80 appears to be downstream of fork maintenance, given that arp8Δ and arp5Δ strains do not exhibit fork collapse in response to HU [55,56]. Unlike the situation with HU, the MMS-induced switch to translesion synthesis is mediated through ubiquitinylation of the proliferating clamp nuclear antigen (PCNA) by the Rad6-Rad18 complex [64]. In this context, an ATPase-dead ino80 mutant failed to recruit Rad18 to an MMS-stalled replisome, thus preventing proper ubiquitinylation of PCNA [56]. The downstream recruitment of the Rad51-dependent recombination machinery was also compromised at MMS-stalled forks [56].
In the presence of HU, the loss of INO80 led to a delayed resumption of fork elongation after removal of the drug, and an increase in stable Mec1-Ddc2 and Rad52 foci in G2-phase, indicating a delayed resumption for fork progression. This could mean that either INO80 is needed to remove complexes that block fork progression, or it could help recruit essential factors for repair and recovery (figure 1b).
Further elucidation of INO80 function at replication forks came from in vitro studies in which the Diffley and Remus laboratories successfully reconstituted efficient DNA replication of naked DNA with purified proteins [65,66]. Upon challenging this ‘minimal’ replication system with a chromatin template, replication initially failed, reflecting a requirement for extra factors. Chromatin template replication was restored by the addition of the histone chaperone FACT, yet at a rate far slower than in vivo [67]. Indeed, efficient replication of the chromatin template was only achieved upon the addition of two ATP-dependent remodellers, INO80 and ISWIa [67]. Neither remodeller could substitute for or replace the histone chaperone FACT, yet normal replisome progression rates were only achieved in the presence of nucleosome remodellers. This is reminiscent of the observations of Shimada et al. [55], and the finding that INO80 promotes the replication of late heterochromatic domains in a normal S-phase in vivo [57].
As INO80 is able to remove nucleosomes around DSBs [68,69], the complex could exhibit the same function ahead of the replication fork. This view is strongly supported by the fact that INO80 ATPase activity is required to promote restart of stalled replication forks [60], and by the observation that loss of both INO80 and the chromatin accessibility complex (CHRAC) increases nucleosome density around replication forks upon MMS treatment [70]. One cannot exclude that INO80 helps process recombination intermediates, or helps remove factors that impair recombination, given that ectopic recombination is strongly reduced upon loss of INO80 function [71]. Finally, the association of INO80 to the replication machinery also seemed to favour other processes such as the establishment of sister chromatid cohesion [72].
7. INO80 promotes removal of RNAPII when replication forks encounter transcription
INO80's ability to remove proteins from chromatin is not restricted to nucleosomes. Indeed, recent studies demonstrated that the INO80 complex also promotes the removal of RNA polymerase II (RNAPII) from chromatin in DNA damage conditions [2] and during HU-induced replication stress [2,3] (figure 1c). The removal of RNAPII required the interaction of INO80 complex with Cdc48, which mediates protein degradation by the 26S proteasome [2]. This newly identified role was shown to occur during DNA replication where the replisome and the transcription machinery collide, thereby jeopardizing completion of genome duplication [73]. In this context, INO80 appeared to bind both RNAPII [2] and the RNAPII-associated complex PAF1 [3]. Chromatin immunoprecipitation studies performed in mutants argued that both INO80 and Paf1 are needed to achieve an efficient removal of the transcription complex, and its transient degradation, at sites where the replisome collides with highly transcribed genes [3]. In this context, it is relevant to note that INO80 triggers the proteasome-dependent degradation of histones following Zeocin®-induced DNA damage as well [38], and that both events require activation of the Mec1-Ddc2 checkpoint kinase (ATR-ATRIP). This function of INO80 might be specifically activated under stress conditions, given that several subunits of the complex are phosphorylated by the checkpoint kinase Mec1/ATR [74–76]. The requirement of Mec1 for the degradation of RNAPII and histones argues that this event may be an integral part of the DNA damage or DNA replication checkpoint response. Whether the checkpoint kinases modulate INO80 subunit composition or its ligands at these sites is unknown. Clearly, further work is necessary to clarify how the Mec1-INO80-Paf1 triad helps remove RNAPII from chromatin.
8. INO80 and chromatin mobility after DNA damage
Coupled with the dynamics of nucleosomes are the much longer range dynamics of the chromatin fibre within the nucleus. Fluorescence microscopy studies showed that several processes elicit long range chromatin movement: the shift of a transcribed gene to the nuclear pore [77], the clustering of active replication forks during S-phase [78], and the transfer of difficult to repair DSBs or collapsed replication forks to the nuclear envelope [71,79]. Moreover, if the sister chromatid is not available for repair by homologous recombination (HR) a search for an ectopic donor sequence ensues, which also requires chromatin mobility. Whether chromatin movement during this homology search is rate limiting or not for ectopic recombination remains unclear.
Intriguingly, using a budding yeast system in which a DSB is induced near a fluorescently tagged locus, it was shown that this DSB moves more than the same locus uncleaved [80,81]. A similar damage-induced increase in mobility was observed at exposed mammalian telomeres [82]. In yeast, enhanced mobility required the damage checkpoint kinase Mec1/ATR, and once a threshold of damage was reached, the enhanced mobility was seen to propagate across the genome affecting the dynamics of undamaged loci in a Rad53-dependent manner [81,83]. In addition, a functional INO80 was required for DSB-induced mobility of chromatin, both in cis and in trans [83–85]. Consistent with this observation, the recruitment of INO80 to an undamaged locus was sufficient to increase local chromatin mobility [37] in a manner requiring the ATPase activity of Ino80. The same increase in mobility was not observed upon the targeting of other ATP-dependent remodellers, like SWI/SNF or SWR1 [37].
A recent mass spectroscopy and nucleosome mapping study brought some insight into the mechanism behind enhanced chromatin mobility [38]. It was shown that about 30% of the four core histones are degraded upon Zeocin®- or IR-induced DNA damage in a checkpoint- and INO80-dependent manner [38]. The reduced histone density induced chromatin decompaction and increased both the flexibility of the chromatin fibre and its dynamics, a reflection of decreased local constraint (figure 1e). A similar mechanism has been documented in mammalian cells upon UV-induced damage, even if in this case histones were displaced, in an INO80-independent manner, from the site of the damage rather than degraded [86]. The differences observed between yeast and mammalian cells could either reflect the nature of the damage (Zeocin® induces mostly single- and double-strand breaks, whereas UV induces thymidine dimers) or divergence in the repair process across evolution. Although the exact role of INO80 in chromatin mobility is far from understood, the fact that nucleosome eviction and histone degradation are involved links it to functions previously ascribed to this ATP-dependent remodeller.
The function of INO80-dependent chromatin mobility may be many-fold, yet it could also simply be an inadvertent side effect of the end-resection and chromatin remodelling events that occur during DSB repair. On the other hand, chromatin dynamics may facilitate relocation to subnuclear sites that favour specific repair outcomes, or which harbour factors that bias repair towards one pathway over another. In the case of endonuclease-induced persistent DSBs, there are at least two distinct sites at the nuclear periphery to which INO80 can bind, and each has a different impact on repair. In yeast, DSBs are recruited either to the nuclear pore complex (NPC) or to Mps3, a SUN-domain protein anchored in the nuclear envelope [87]. Both NPC and Mps3 binding require the deposition of the histone variant Htz1/H2A.Z at the site of the break by the ATP-dependent remodeller Swr1 in yeast [71,88]. DSB association with Mps3 is favoured in S- and G2-phases of the cell cycle, requires INO80, the recombination factor Rad51 which binds to single-stranded DNA generated by resection, and the SMC5/6-Mms21 SUMO-ligase (figure 1d) [71]. Recruitment to the NPC occurs in G1- as well as S-phase, and requires a poly-SUMO chain deposited by the SUMO-ligase Siz2. This, in turn, recruits the SUMO-dependent ubiquitin ligase complex Slx5-Slx8 [79,89], which is necessary for interaction with the Nup84 subcomplex of the NPC. Artificial targeting of SUMO-fusions to an undamaged chromatin template showed that mono-SUMOylation promotes Mps3 anchorage whereas poly-SUMOylation and Slx5 recruitment favour relocation to the NPC [90]. Changes in local tethering forces between and involving nucleosomes are likely to be important for damage relocation.
While the mechanisms remain somewhat enigmatic, it is nonetheless clear that the two anchorage pathways favour different repair outcomes. Persistent DSB association to the NPC promotes alternative recombination pathways such as microhomology-mediated recombination or break-induced replication (BIR) to the detriment of Rad51-dependent canonical recombination events [90,91]. This extends to the repair of eroded telomeres by ectopic recombination [92], and the recovery from fork collapse at triplet nucleotide repeats [93]. Intriguingly, the only ATP-dependent remodeller involved in nuclear pore binding is SWR1, as INO80 inactivation does not alter any aspects of DSB anchorage to NPC. This function of SWR1 may involve H2A-Z deposition, and is thus not limited to DSBs, but may affect gene promoters [94], telomeres and centromeres [95]. On the other hand, Mps3-binding prevents illegitimate recombination processes, thus limiting unequal sister chromatid recombination and loss of genetic information [71]. Mps3 binding may sequester DSBs that were unable to find a proper homologous template during homology search. Interestingly, Mps3 anchoring depends on DNA end-resection, restricting this pathway to S- and G2-phases, when exonucleases are active.
Whereas INO80 is one of several ATP-dependent chromatin remodellers involved in promoting end-resection at DSBs in yeast [68], SWR1 and Htz1/H2A.Z seem to inhibit resection and to promote the loading of the NHEJ-Ku70/80 complex both in yeast [68] and in mammals [96]. Intriguingly, subunits of the INO80 complex, TIP49 (human) and the Rvb1/2 (yeast), exhibit 3′ to 5′ helicase activity that unwinds 3′ ssDNA overhangs in vitro, which is consistent with a function of INO80 in the processing of resected DNA ends [97]. This is consistent with the observation that ino80 mutants are not defective in canonical NHEJ repair assay, yet exhibit partial defects in specific HR-mediated repair assay [68,88]. The loss of INO80 does not alter spontaneous HR events at the MAT locus, but delays the rate of HR upon MMS treatment [98]. The main difference between the repair of the MAT locus by gene conversion and MMS-induced HR is the involvement of the checkpoint. Given that Mec1 directly phosphorylates several subunits of the INO80 complex under damage conditions, this modification may restrict or alter the contribution of INO80 [75,76].
9. INO80 in development and disease
The INO80 complex contributes to transcription regulation, DNA replication and DNA damage repair, three fundamental processes that are required for proper embryonic development and for cell integrity in an adult organism. Several studies in the last few years have highlighted the importance of INO80 in mammalian development and disease. For instance, during the generation of germ cells in meiosis, hundreds of DSBs are induced and subsequently repaired to allow exchange of genetic material between homologous chromosomes. In mice, INO80 is expressed in developing spermatocytes at the early stages of meiotic prophase I [99], and its conditional inactivation induces meiotic arrest and a failure to repair DSBs generated during meiotic recombination [99]. Interestingly, Ino80 knockout mice exhibit early embryonic lethality [100], and other studies suggest that INO80 has a role in the establishment and maintenance of pluripotency in ESCs [101]. This may be related to its role in transcription, as INO80 facilitates the recruitment of Mediator and RNAPII to the promoters of the pluripotency-network genes Oct4, Nanog, Sox2, Klf4 and Esrrb, promoting their expression in ESCs [101].
INO80 also promotes activation of enhancers inducible by the oestrogen receptor (ERα). The recruitment of INO80 at ERα-dependent enhancers is impaired by ubiquitinylation of H2B at K120 by RNF20/RNF4 (H2Bub1), which prevents eviction of H2A.Z and represses transcriptional activation [34]. As H2Bub1 facilitates methylation of H3K79 [102,103], it is of interest to investigate potential crosstalk between INO80 and H2Bub1 in the Dot1L-dependent methylation of H3K79.
Recently, increased expression of HsINO80 has been functionally associated with tumour progression. INO80 is over-expressed in BRAF- and NRAS-mutated melanoma cancer cells [104], as well as in anaplastic thyroid carcinoma stem cells (ATC-CSCs) [105] and in cervical cancer samples [16]. Whether its overexpression reflects the role of INO80 in the survival of replication stress or in gene expression is unclear, yet the downregulation of INO80 impaired melanoma cancer cell growth and tumorigenesis [104]. Interestingly, INO80 occupies and activates the super-enhancers (SEs) of certain oncogenes, apparently by increasing chromatin accessibility and promoting recruitment of Mediator to these sites [104]. Given the similarities between promoters and enhancers [106], it is expected that INO80 represses the production of bidirectional non-coding RNAs at enhancers (eRNAs) and SEs [107], particularly given the increase in aberrant lncRNAs found in cancer cell lines [108]. In the context of cancer development, the fact that INO80 controls the level of spurious transcription (figure 1a) may impact non-coding RNAs. Consistently, the knockdown of INO80 attenuated stem-cell-specific properties of the aforementioned cancer cells, including their ability to form tumours [16,105]. This suggests that the role of INO80 in cancer cells is linked to its stem-cell-promoting function.
Finally, besides cancer, INO80 the proteasome and the RNAPII machinery have all been associated with progression of Alzheimer's disease [109], raising the possibility that degradation of RNAPII by INO80 [2] is involved in neuronal cell functions. The multiplicity of roles in chromatin dynamics ascribed to INO80 makes it almost inevitable that INO80-mediated remodelling and protein eviction impact a range of human pathologies. Underlying this, however, is the fundamental question of how INO80 moves, evicts and exchanges histones and non-histone factors, to facilitate DNA-based enzymatic events (figure 1). In many cases there is redundancy of INO80 with other remodellers, which makes the identification of INO80-specific functions even more difficult. In order to understand its disease links, it will be crucial to determine the crosstalk of INO80 with checkpoint kinases and other stress signalling pathways.
Acknowledgement
We thank C.-B. Gerhold, A. Seeber, M. Hauer and K. Shimada for critical comments on the manuscript.
Authors' contributions
All three authors contributed to the writing of the review.
Competing interests
We declare we have no competing interests.
Funding
The Gasser laboratory acknowledges support from the Swiss National Science Foundation, the Novartis Research Foundation and Human Frontiers Science Program. J.P. thanks the ARC and EMBO for a long-term postdoctoral fellowship.
References
- 1.Sinha M, Watanabe S, Johnson A, Moazed D, Peterson CL. 2009. Recombinational repair within heterochromatin requires ATP-dependent chromatin remodeling. Cell 138, 1109–1121. ( 10.1016/j.cell.2009.07.013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lafon A, Taranum S, Pietrocola F, Dingli F, Loew D, Brahma S, Bartholomew B, Papamichos-Chronakis M. 2015. INO80 chromatin remodeler facilitates release of RNA polymerase II from chromatin for ubiquitin-mediated proteasomal degradation. Mol. Cell 60, 784–796. ( 10.1016/j.molcel.2015.10.028) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Poli J, et al. 2016. Mec1, INO80, and the PAF1 complex cooperate to limit transcription replication conflicts through RNAPII removal during replication stress. Genes Dev. 30, 337–354. ( 10.1101/gad.273813.115) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gerhold CB, Gasser SM. 2014. INO80 and SWR complexes: relating structure to function in chromatin remodeling. Trends Cell Biol. 24, 619–631. ( 10.1016/j.tcb.2014.06.004) [DOI] [PubMed] [Google Scholar]
- 5.Clapier CR, Cairns BR. 2009. The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 78, 273–304. ( 10.1146/annurev.biochem.77.062706.153223) [DOI] [PubMed] [Google Scholar]
- 6.Flaus A, Martin DM, Barton GJ, Owen-Hughes T. 2006. Identification of multiple distinct Snf2 subfamilies with conserved structural motifs. Nucleic Acids Res. 34, 2887–2905. ( 10.1093/nar/gkl295) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cai Y, et al. 2007. YY1 functions with INO80 to activate transcription. Nat. Struct. Mol. Biol. 14, 872–874. ( 10.1038/nsmb1276) [DOI] [PubMed] [Google Scholar]
- 8.Ebbert R, Birkmann A, Schuller HJ. 1999. The product of the SNF2/SWI2 paralogue INO80 of Saccharomyces cerevisiae required for efficient expression of various yeast structural genes is part of a high-molecular-weight protein complex. Mol. Microbiol. 32, 741–751. ( 10.1046/j.1365-2958.1999.01390.x) [DOI] [PubMed] [Google Scholar]
- 9.Steger DJ, Haswell ES, Miller AL, Wente SR, O'Shea EK. 2003. Regulation of chromatin remodeling by inositol polyphosphates. Science 299, 114–116. ( 10.1126/science.1078062) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barbaric S, Luckenbach T, Schmid A, Blaschke D, Horz W, Korber P. 2007. Redundancy of chromatin remodeling pathways for the induction of the yeast PHO5 promoter in vivo. J. Biol. Chem. 282, 27 610–27 621. ( 10.1074/jbc.M700623200) [DOI] [PubMed] [Google Scholar]
- 11.Mizuguchi G, Shen X, Landry J, Wu WH, Sen S, Wu C. 2004. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 303, 343–348. ( 10.1126/science.1090701) [DOI] [PubMed] [Google Scholar]
- 12.van Attikum H, Fritsch O, Hohn B, Gasser SM. 2004. Recruitment of the INO80 complex by H2A phosphorylation links ATP-dependent chromatin remodeling with DNA double-strand break repair. Cell 119, 777–788. ( 10.1016/j.cell.2004.11.033) [DOI] [PubMed] [Google Scholar]
- 13.Yao W, King DA, Beckwith SL, Gowans GJ, Yen K, Zhou C, Morrison AJ. 2016. The INO80 complex requires the Arp5-Ies6 subcomplex for chromatin remodeling and metabolic regulation. Mol. Cell. Biol. 36, 979–991. ( 10.1128/MCB.00801-15) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yen K, Vinayachandran V, Pugh BF. 2013. SWR-C and INO80 chromatin remodelers recognize nucleosome-free regions near +1 nucleosomes. Cell 154, 1246–1256. ( 10.1016/j.cell.2013.08.043) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cao L, et al. 2015. Negative regulation of p21Waf1/Cip1 by human INO80 chromatin remodeling complex is implicated in cell cycle phase G2/M arrest and abnormal chromosome stability. PLoS ONE 10, e0137411 ( 10.1371/journal.pone.0137411) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klopf E, Schmidt HA, Clauder-Munster S, Steinmetz LM, Schuller C. 2016. INO80 represses osmostress induced gene expression by resetting promoter proximal nucleosomes. Nucleic Acids Res. 45, 3752–3766. ( 10.1093/nar/gkw1292) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neuman SD, Ihry RJ, Gruetzmacher KM, Bashirullah A. 2014. INO80-dependent regression of ecdysone-induced transcriptional responses regulates developmental timing in Drosophila. Dev. Biol. 387, 229–239. ( 10.1016/j.ydbio.2014.01.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xue Y, et al. 2015. The Ino80 complex prevents invasion of euchromatin into silent chromatin. Genes Dev. 29, 350–355. ( 10.1101/gad.256255.114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yen K, Vinayachandran V, Batta K, Koerber RT, Pugh BF. 2012. Genome-wide nucleosome specificity and directionality of chromatin remodelers. Cell 149, 1461–1473. ( 10.1016/j.cell.2012.04.036) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mavrich TN, Ioshikhes IP, Venters BJ, Jiang C, Tomsho LP, Qi J, Schuster SC, Albert I, Pugh BF. 2008. A barrier nucleosome model for statistical positioning of nucleosomes throughout the yeast genome. Genome Res. 18, 1073–1083. ( 10.1101/gr.078261.108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Osakabe A, et al. 2014. DNA binding properties of the actin-related protein Arp8 and its role in DNA repair. PLoS ONE 9, e108354 ( 10.1371/journal.pone.0108354) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Udugama M, Sabri A, Bartholomew B. 2011. The INO80 ATP-dependent chromatin remodeling complex is a nucleosome spacing factor. Mol. Cell. Biol. 31, 662–673. ( 10.1128/MCB.01035-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ranjan A, Mizuguchi G, FitzGerald PC, Wei D, Wang F, Huang Y, Luk E, Woodcock CL, Wu C. 2013. Nucleosome-free region dominates histone acetylation in targeting SWR1 to promoters for H2A.Z replacement. Cell 154, 1232–1245. ( 10.1016/j.cell.2013.08.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klopf E, Paskova L, Sole C, Mas G, Petryshyn A, Posas F, Wintersberger U, Ammerer G, Schuller C. 2009. Cooperation between the INO80 complex and histone chaperones determines adaptation of stress gene transcription in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 29, 4994–5007. ( 10.1128/MCB.01858-08) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Andrews FH, et al. 2016. The Taf14 YEATS domain is a reader of histone crotonylation. Nat. Chem. Biol. 12, 396–398. ( 10.1038/nchembio.2065) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shanle EK. et al 2015. Association of Taf14 with acetylated histone H3 directs gene transcription and the DNA damage response. Genes Dev. 29, 1795–1800. ( 10.1101/gad.269977.115) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shen X, Ranallo R, Choi E, Wu C. 2003. Involvement of actin-related proteins in ATP-dependent chromatin remodeling. Mol. Cell 12, 147–155. ( 10.1016/S1097-2765(03)00264-8) [DOI] [PubMed] [Google Scholar]
- 28.Jin J, et al. 2005. A mammalian chromatin remodeling complex with similarities to the yeast INO80 complex. J. Biol. Chem. 280, 41 207–41 212. ( 10.1074/jbc.M509128200) [DOI] [PubMed] [Google Scholar]
- 29.Krietenstein N, Wal M, Watanabe S, Park B, Peterson CL, Pugh BF, Korber P. 2016. Genomic nucleosome organization reconstituted with pure proteins. Cell 167, 709–721. ( 10.1016/j.cell.2016.09.045) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Papamichos-Chronakis M, Watanabe S, Rando OJ, Peterson CL. 2011. Global regulation of H2A.Z localization by the INO80 chromatin-remodeling enzyme is essential for genome integrity. Cell 144, 200–213. ( 10.1016/j.cell.2010.12.021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alatwi HE, Downs JA. 2015. Removal of H2A.Z by INO80 promotes homologous recombination. EMBO Rep. 16, 986–994. ( 10.15252/embr.201540330) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rosonina E, Yurko N, Li W, Hoque M, Tian B, Manley JL. 2014. Threonine-4 of the budding yeast RNAP II CTD couples transcription with Htz1-mediated chromatin remodeling. Proc. Natl Acad. Sci. USA 111, 11 924–11 931. ( 10.1073/pnas.1412802111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chambers AL, Ormerod G, Durley SC, Sing TL, Brown GW, Kent NA, Downs JA. 2012. The INO80 chromatin remodeling complex prevents polyploidy and maintains normal chromatin structure at centromeres. Genes Dev. 26, 2590–2603. ( 10.1101/gad.199976.112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Segala G, Bennesch MA, Pandey DP, Hulo N, Picard D. 2016. Monoubiquitination of histone H2B blocks eviction of histone variant H2A.Z from inducible enhancers. Mol. Cell 64, 334–346. ( 10.1016/j.molcel.2016.08.034) [DOI] [PubMed] [Google Scholar]
- 35.Alcid EA, Tsukiyama T. 2014. ATP-dependent chromatin remodeling shapes the long noncoding RNA landscape. Genes Dev. 28, 2348–2360. ( 10.1101/gad.250902.114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dechassa ML, Sabri A, Pondugula S, Kassabov SR, Chatterjee N, Kladde MP, Bartholomew B. 2010. SWI/SNF has intrinsic nucleosome disassembly activity that is dependent on adjacent nucleosomes. Mol. Cell 38, 590–602. ( 10.1016/j.molcel.2010.02.040) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Neumann FR, Dion V, Gehlen LR, Tsai-Pflugfelder M, Schmid R, Taddei A, Gasser SM. 2012. Targeted INO80 enhances subnuclear chromatin movement and ectopic homologous recombination. Genes Dev. 26, 369–383. ( 10.1101/gad.176156.111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hauer MH, et al. 2017. Histone degradation in response to DNA damage enhances chromatin dynamics and recombination rates. Nat. Struct. Mol. Biol. 24, 99–107. ( 10.1038/nsmb.3347) [DOI] [PubMed] [Google Scholar]
- 39.Tramantano M, Sun L, Au C, Labuz D, Liu Z, Chou M, Shen C, Luk E. 2016. Constitutive turnover of histone H2A.Z at yeast promoters requires the preinitiation complex. eLife 5, e14243 ( 10.7554/eLife.14243) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hintermair C, et al. 2012. Threonine-4 of mammalian RNA polymerase II CTD is targeted by polo-like kinase 3 and required for transcriptional elongation. EMBO J. 31, 2784–2797. ( 10.1038/emboj.2012.123) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weber CM, Ramachandran S, Henikoff S. 2014. Nucleosomes are context-specific, H2A.Z-modulated barriers to RNA polymerase. Mol. Cell 53, 819–830. ( 10.1016/j.molcel.2014.02.014) [DOI] [PubMed] [Google Scholar]
- 42.Jeronimo C, Watanabe S, Kaplan CD, Peterson CL, Robert F. 2015. The histone chaperones FACT and Spt6 restrict H2A.Z from intragenic locations. Mol. Cell 58, 1113–1123. ( 10.1016/j.molcel.2015.03.030) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marquardt S, Escalante-Chong R, Pho N, Wang J, Churchman LS, Springer M, Buratowski S. 2014. A chromatin-based mechanism for limiting divergent noncoding transcription. Cell 157, 1712–1723. ( 10.1016/j.cell.2014.04.036) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tisseur M, Kwapisz M, Morillon A. 2011. Pervasive transcription – lessons from yeast. Biochimie 93, 1889–1896. ( 10.1016/j.biochi.2011.07.001) [DOI] [PubMed] [Google Scholar]
- 45.Rege M, et al. 2015. Chromatin dynamics and the RNA exosome function in concert to regulate transcriptional homeostasis. Cell Rep. 13, 1610–1622. ( 10.1016/j.celrep.2015.10.030) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kitada T, Kuryan BG, Tran NN, Song C, Xue Y, Carey M, Grunstein M. 2012. Mechanism for epigenetic variegation of gene expression at yeast telomeric heterochromatin. Genes Dev. 26, 2443–2455. ( 10.1101/gad.201095.112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nguyen AT, Zhang Y. 2011. The diverse functions of Dot1 and H3K79 methylation. Genes Dev. 25, 1345–1358. ( 10.1101/gad.2057811) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shen X, Mizuguchi G, Hamiche A, Wu C. 2000. A chromatin remodelling complex involved in transcription and DNA processing. Nature 406, 541–544. ( 10.1038/35020123) [DOI] [PubMed] [Google Scholar]
- 49.Downs JA, Allard S, Jobin-Robitaille O, Javaheri A, Auger A, Bouchard N, Kron SJ, Jackson SP, Côté J. 2004. Binding of chromatin-modifying activities to phosphorylated histone H2A at DNA damage sites. Mol. Cell 16, 979–990. ( 10.1016/j.molcel.2004.12.003) [DOI] [PubMed] [Google Scholar]
- 50.Morrison AJ, Highland J, Krogan NJ, Arbel-Eden A, Greenblatt JF, Haber JE, Shen X. 2004. INO80 and γ-H2AX interaction links ATP-dependent chromatin remodeling to DNA damage repair. Cell 119, 767–775. ( 10.1016/j.cell.2004.11.037) [DOI] [PubMed] [Google Scholar]
- 51.Morrison AJ. 2017. Genome maintenance functions of the INO80 chromatin remodeller. Phil. Trans. R. Soc. B 372, 20160289 ( 10.1098/rstb.2016.0289) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Koc A, Wheeler LJ, Mathews CK, Merrill GF. 2004. Hydroxyurea arrests DNA replication by a mechanism that preserves basal dNTP pools. J. Biol. Chem. 279, 223–230. ( 10.1074/jbc.M303952200) [DOI] [PubMed] [Google Scholar]
- 53.Poli J, Tsaponina O, Crabbe L, Keszthelyi A, Pantesco V, Chabes A, Lengronne A, Pasero P. 2012. dNTP pools determine fork progression and origin usage under replication stress. EMBO J. 31, 883–894. ( 10.1038/emboj.2011.470) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yeeles JTP, Poli J, Marians KJ, Pasero P. 2013. Rescuing stalled or damaged replication forks. Cold Spring Harb. Perspect. Biol. 5, a012815 ( 10.1101/cshperspect.a012815) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shimada K, Oma Y, Schleker T, Kugou K, Ohta K, Harata M, Gasser SM. 2008. Ino80 chromatin remodeling complex promotes recovery of stalled replication forks. Curr. Biol. 18, 566–575. ( 10.1016/j.cub.2008.03.049) [DOI] [PubMed] [Google Scholar]
- 56.Falbo KB, et al. 2009. Involvement of a chromatin remodeling complex in damage tolerance during DNA replication. Nat. Struct. Mol. Biol. 16, 1167–1172. ( 10.1038/nsmb.1686) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vincent JA, Kwong TJ, Tsukiyama T. 2008. ATP-dependent chromatin remodeling shapes the DNA replication landscape. Nat. Struct. Mol. Biol. 15, 477–484. ( 10.1038/nsmb.1419) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee HS, Lee SA, Hur SK, Seo JW, Kwon J. 2014. Stabilization and targeting of INO80 to replication forks by BAP1 during normal DNA synthesis. Nat. Commun. 5, 5128 ( 10.1038/ncomms6128) [DOI] [PubMed] [Google Scholar]
- 59.Au TJ, Rodriguez J, Vincent JA, Tsukiyama T. 2011. ATP-dependent chromatin remodeling factors tune S phase checkpoint activity. Mol. Cell. Biol. 31, 4454–4463. ( 10.1128/MCB.05931-11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Papamichos-Chronakis M, Peterson CL. 2008. The Ino80 chromatin-remodeling enzyme regulates replisome function and stability. Nat. Struct. Mol. Biol. 15, 338–345. ( 10.1038/nsmb.1413) [DOI] [PubMed] [Google Scholar]
- 61.Vassileva I, Yanakieva I, Peycheva M, Gospodinov A, Anachkova B. 2014. The mammalian INO80 chromatin remodeling complex is required for replication stress recovery. Nucleic Acids Res. 42, 9074–9086. ( 10.1093/nar/gku605) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bjergbaek L, Cobb JA, Tsai-Pflugfelder M, Gasser SM. 2005. Mechanistically distinct roles for Sgs1p in checkpoint activation and replication fork maintenance. EMBO J. 24, 405–417. ( 10.1038/sj.emboj.7600511) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lambert S, et al. 2010. Homologous recombination restarts blocked replication forks at the expense of genome rearrangements by template exchange. Mol. Cell 39, 346–359. ( 10.1016/j.molcel.2010.07.015) [DOI] [PubMed] [Google Scholar]
- 64.Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. 2002. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419, 135–141. ( 10.1038/nature00991) [DOI] [PubMed] [Google Scholar]
- 65.Yeeles JT, Janska A, Early A, Diffley JF. 2017. How the eukaryotic replisome achieves rapid and efficient DNA replication. Mol. Cell 65, 105–116. ( 10.1016/j.molcel.2016.11.017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Devbhandari S, Jiang J, Kumar C, Whitehouse I, Remus D. 2017. Chromatin constrains the initiation and elongation of DNA replication. Mol. Cell 65, 131–141. ( 10.1016/j.molcel.2016.10.035) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kurat CF, Yeeles JT, Patel H, Early A, Diffley JF. 2017. Chromatin controls DNA replication origin selection, lagging-strand synthesis, and replication fork rates. Mol. Cell 65, 117–130. ( 10.1016/j.molcel.2016.11.016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.van Attikum H, Fritsch O, Gasser SM. 2007. Distinct roles for SWR1 and INO80 chromatin remodeling complexes at chromosomal double-strand breaks. EMBO J. 26, 4113 ( 10.1038/sj.emboj.7601835) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tsukuda T, Fleming AB, Nickoloff JA, Osley MA. 2005. Chromatin remodelling at a DNA double-strand break site in Saccharomyces cerevisiae. Nature 438, 379–383. ( 10.1038/nature04148) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lee L, Rodriguez J, Tsukiyama T. 2015. Chromatin remodeling factors Isw2 and Ino80 regulate checkpoint activity and chromatin structure in S phase. Genetics 199, 1077–1091. ( 10.1534/genetics.115.174730) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Horigome C, Oma Y, Konishi T, Schmid R, Marcomini I, Hauer MH, Gasser SM. 2014. SWR1 and INO80 chromatin remodelers contribute to DNA double-strand break perinuclear anchorage site choice. Mol. Cell 55, 626–639. ( 10.1016/j.molcel.2014.06.027) [DOI] [PubMed] [Google Scholar]
- 72.Ogiwara H, Enomoto T, Seki M. 2007. The INO80 chromatin remodeling complex functions in sister chromatid cohesion. Cell Cycle 6, 1090–1095. ( 10.4161/cc.6.9.4130) [DOI] [PubMed] [Google Scholar]
- 73.Hamperl S, Cimprich KA. 2016. Conflict resolution in the genome: how transcription and replication make it work. Cell 167, 1455–1467. ( 10.1016/j.cell.2016.09.053) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Morrison AJ, et al. 2007. Mec1/Tel1 phosphorylation of the INO80 chromatin remodeling complex influences DNA damage checkpoint responses. Cell 130, 499–511. ( 10.1016/j.cell.2007.06.010) [DOI] [PubMed] [Google Scholar]
- 75.Hustedt N, et al. 2015. Yeast PP4 interacts with ATR homolog Ddc2-Mec1 and regulates checkpoint signaling. Mol. Cell 57, 273–289. ( 10.1016/j.molcel.2014.11.016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bastos de Oliveira FM, et al. 2015. Phosphoproteomics reveals distinct modes of Mec1/ATR signaling during DNA replication. Mol. Cell 57, 1124–1132. ( 10.1016/j.molcel.2015.01.043) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Taddei A, Van Houwe G, Hediger F, Kalck V, Cubizolles F, Schober H, Gasser SM. 2006. Nuclear pore association confers optimal expression levels for an inducible yeast gene. Nature 441, 774–778. ( 10.1038/nature04845) [DOI] [PubMed] [Google Scholar]
- 78.Chagin VO, et al. 2016. 4D Visualization of replication foci in mammalian cells corresponding to individual replicons. Nat. Commun. 7, 11231 ( 10.1038/ncomms11231) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nagai S, et al. 2008. Functional targeting of DNA damage to a nuclear pore-associated SUMO-dependent ubiquitin ligase. Science 322, 597–602. ( 10.1126/science.1162790) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dion V, Kalck V, Horigome C, Towbin BD, Gasser SM. 2012. Increased mobility of double-strand breaks requires Mec1, Rad9 and the homologous recombination machinery. Nat. Cell Biol. 14, 502–509. ( 10.1038/ncb2465) [DOI] [PubMed] [Google Scholar]
- 81.Mine-Hattab J, Rothstein R. 2012. Increased chromosome mobility facilitates homology search during recombination. Nat. Cell Biol. 14, 510–517. ( 10.1038/ncb2472) [DOI] [PubMed] [Google Scholar]
- 82.Lottersberger F, Karssemeijer RA, Dimitrova N, de Lange T. 2015. 53BP1 and the LINC complex promote microtubule-dependent DSB mobility and DNA repair. Cell 163, 880–893. ( 10.1016/j.cell.2015.09.057) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Seeber A, Dion V, Gasser SM. 2013. Checkpoint kinases and the INO80 nucleosome remodeling complex enhance global chromatin mobility in response to DNA damage. Genes Dev. 27, 1999–2008. ( 10.1101/gad.222992.113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Strecker J, Gupta GD, Zhang W, Bashkurov M, Landry MC, Pelletier L, Durocher D. 2016. DNA damage signalling targets the kinetochore to promote chromatin mobility. Nat. Cell Biol. 18, 281–290. ( 10.1038/ncb3308) [DOI] [PubMed] [Google Scholar]
- 85.Amitai A, Seeber A, Gasser SM, Holcman D. 2017. Visualization of chromatin decompaction and break site extrusion as predicted by statistical polymer modeling of single-locus trajectories. Cell Rep. 18, 1200–1214. ( 10.1016/j.celrep.2017.01.018) [DOI] [PubMed] [Google Scholar]
- 86.Adam S, et al. 2016. Real-time tracking of parental histones reveals their contribution to chromatin integrity following DNA damage. Mol. Cell 64, 65–78. ( 10.1016/j.molcel.2016.08.019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Seeber A, Gasser SM. 2017. Chromatin organization and dynamics in double-strand break repair. Curr. Opin Genet. Dev. 43, 9–16. ( 10.1016/j.gde.2016.10.005) [DOI] [PubMed] [Google Scholar]
- 88.Papamichos-Chronakis M, Krebs JE, Peterson CL. 2006. Interplay between Ino80 and Swr1 chromatin remodeling enzymes regulates cell cycle checkpoint adaptation in response to DNA damage. Genes Dev. 20, 2437–2449. ( 10.1101/gad.1440206) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ryu T, Spatola B, Delabaere L, Bowlin K, Hopp H, Kunitake R, Karpen GH, Chiolo I. 2015. Heterochromatic breaks move to the nuclear periphery to continue recombinational repair. Nat. Cell Biol. 17, 1401–1411. ( 10.1038/ncb3258) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Horigome C, Bustard DE, Marcomini I, Delgoshaie N, Tsai-Pflugfelder M, Cobb JA, Gasser SM. 2016. PolySUMOylation by Siz2 and Mms21 triggers relocation of DNA breaks to nuclear pores through the Slx5/Slx8 STUbL. Genes Dev. 30, 931–945. ( 10.1101/gad.277665.116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Burgess RC, Rahman S, Lisby M, Rothstein R, Zhao X. 2007. The Slx5-Slx8 complex affects sumoylation of DNA repair proteins and negatively regulates recombination. Mol. Cell. Biol. 27, 6153–6162. ( 10.1128/MCB.00787-07) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Churikov D, Charifi F, Eckert-Boulet N, Silva S, Simon MN, Lisby M, Géli V. 2016. SUMO-dependent relocalization of eroded telomeres to nuclear pore complexes controls telomere recombination. Cell Rep. 15, 1242–1253. ( 10.1016/j.celrep.2016.04.008) [DOI] [PubMed] [Google Scholar]
- 93.Su XA, Dion V, Gasser SM, Freudenreich CH. 2015. Regulation of recombination at yeast nuclear pores controls repair and triplet repeat stability. Genes Dev. 29, 1006–1017. ( 10.1101/gad.256404.114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Krogan NJ, et al. 2003. A Snf2 family ATPase complex required for recruitment of the histone H2A variant Htz1. Mol. Cell 12, 1565–1576. ( 10.1016/S1097-2765(03)00497-0) [DOI] [PubMed] [Google Scholar]
- 95.Krogan NJ, et al. 2004. Regulation of chromosome stability by the histone H2A variant Htz1, the Swr1 chromatin remodeling complex, and the histone acetyltransferase NuA4. Proc. Natl Acad. Sci. USA 101, 13 513–13 518. ( 10.1073/pnas.0405753101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Xu Y, Ayrapetov MK, Xu C, Gursoy-Yuzugullu O, Hu Y, Price BD. 2012. Histone H2A.Z controls a critical chromatin remodeling step required for DNA double-strand break repair. Mol. Cell 48, 723–733. ( 10.1016/j.molcel.2012.09.026) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Papin C, Humbert O, Kalashnikova A, Eckert K, Morera S, Kas E, Grigoriev M. 2010. 3'- to 5' DNA unwinding by TIP49b proteins. FEBS J. 277, 2705–2714. ( 10.1111/j.1742-4658.2010.07687.x) [DOI] [PubMed] [Google Scholar]
- 98.Kawashima S, Ogiwara H, Tada S, Harata M, Wintersberger U, Enomoto T, Seki M. 2007. The INO80 complex is required for damage-induced recombination. Biochem. Biophys. Res. Commun. 355, 835–841. ( 10.1016/j.bbrc.2007.02.036) [DOI] [PubMed] [Google Scholar]
- 99.Serber DW, Runge JS, Menon DU, Magnuson T. 2016. The mouse INO80 chromatin-remodeling complex is an essential meiotic factor for spermatogenesis. Biol. Reprod. 94, 8 ( 10.1095/biolreprod.115.135533) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Min JN, Tian Y, Xiao Y, Wu L, Li L, Chang S. 2013. The mINO80 chromatin remodeling complex is required for efficient telomere replication and maintenance of genome stability. Cell Res. 23, 1396–1413. ( 10.1038/cr.2013.113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wang L, et al. 2014. INO80 facilitates pluripotency gene activation in embryonic stem cell self-renewal, reprogramming, and blastocyst development. Cell Stem Cell 14, 575–591. ( 10.1016/j.stem.2014.02.013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Weake VM, Workman JL. 2008. Histone ubiquitination: triggering gene activity. Mol. Cell 29, 653–663. ( 10.1016/j.molcel.2008.02.014) [DOI] [PubMed] [Google Scholar]
- 103.Shilatifard A. 2006. Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu. Rev. Biochem. 75, 243–269. ( 10.1146/annurev.biochem.75.103004.142422) [DOI] [PubMed] [Google Scholar]
- 104.Zhou B, et al. 2016. INO80 governs superenhancer-mediated oncogenic transcription and tumor growth in melanoma. Genes Dev. 30, 1440–1453. ( 10.1101/gad.277178.115) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sheng W, Chen Y, Gong Y, Dong T, Zhang B, Gao W. 2016. miR-148a inhibits self-renewal of thyroid cancer stem cells via repressing INO80 expression. Oncol. Rep. 36, 3387–3396. ( 10.3892/or.2016.5203) [DOI] [PubMed] [Google Scholar]
- 106.Kim TK, Shiekhattar R. 2015. Architectural and functional commonalities between enhancers and promoters. Cell 162, 948–959. ( 10.1016/j.cell.2015.08.008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hah N, Danko CG, Core L, Waterfall JJ, Siepel A, Lis JT, Kraus WL. 2011. A rapid, extensive, and transient transcriptional response to estrogen signaling in breast cancer cells. Cell 145, 622–634. ( 10.1016/j.cell.2011.03.042) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lin Z, et al. 2015. Long noncoding RNA: its partners and their roles in cancer. Neoplasma 62, 846–854. ( 10.4149/neo_2015_103) [DOI] [PubMed] [Google Scholar]
- 109.Kikuchi M, Ogishima S, Miyamoto T, Miyashita A, Kuwano R, Nakaya J, Tanaka H, Csermely P. 2013. Identification of unstable network modules reveals disease modules associated with the progression of Alzheimer's disease. PLoS ONE 8, e76162 ( 10.1371/journal.pone.0076162) [DOI] [PMC free article] [PubMed] [Google Scholar]