Abstract
Nucleosome remodelling (NR) regulates transcription in an ATP-dependent manner, and influences gene expression required for development and cellular functions, including those involved in anti-cancer and anti-ageing processes. ATP-utilizing chromatin assembly and remodelling factor (ACF) and Brahma-associated factor (BAF) complexes, belonging to the ISWI and SWI/SNF families, respectively, are involved in various types of DNA repair. Suppression of several BAF factors makes U2OS cells significantly sensitive to X-rays, UV and especially to cisplatin, and these BAF factors contribute to the accumulation of repair proteins at various types of DNA damage and to DNA repair. Recent cancer genome sequencing and expression analysis has shown that BAF factors are frequently mutated or, more frequently, silenced in various types of cancer cells. Thus, those cancer cells are potentially X-ray- and especially cisplatin-sensitive, suggesting a way of optimizing current cancer therapy. Recent single–stem cell analysis suggests that mutations and epigenetic changes influence stem cell functionality leading to cellular ageing. Genetic and epigenetic changes in the BAF factors diminish DNA repair as well as transcriptional regulation activities, and DNA repair defects in turn negatively influence NR and transcriptional regulation. Thus, they build negative feedback loops, which accelerate both cellular senescence and transformation as common and rare cellular events, respectively, causing cellular ageing.
This article is part of the themed issue ‘Chromatin modifiers and remodellers in DNA repair and signalling’.
Keywords: chromatin remodeling, nucleosome remodeling, DNA repair, BAF complex, cancer therapy, cellular aging
1. Introduction
DNA interacts closely with histones and other proteins to build chromatin, often in a very compact configuration. To utilize the genetic information held in DNA for transcription and replication, chromatin structure needs to be transiently relaxed and remodelled and then to recover, in a process that is called chromatin remodelling (CR). DNA damage of different types is produced in DNA, even within a compact chromatin structure, where DNA repair proteins are inaccessible, and DNA repair does not occur until chromatin structure relaxes. If DNA damage is not repaired due to inaccessible chromatin structure, replication encounters unrepaired DNA damage, which enhances the possibility of genome instability, mutation, cellular senescence and cell death.
CR has a fundamental role in transcriptional regulation, and its mechanisms have been extensively analysed in transcription research. There are two types of CR: one initiated by various types of histone modification and the other by ATP-dependent mobilization of nucleosomes; the latter is called nucleosome remodelling (NR) [1–3], which will be discussed in terms of DNA repair in this review. The functions of NR are nucleosome sliding, histone exchange and ejection of nucleosomes containing a target sequence, by utilizing ATP. NR also recovers the correct nucleosome spacing after the completion of transcription and replication. In many cellular processes, both enzymatic modification of histones and NR occur in a combined manner to regulate chromatin structure [4]. Accumulating evidence suggests that NR supports the repair of DNA damage. Therefore, NR is important for transcriptional regulation and DNA repair. The high frequency of mutation and silencing of SWI/SNF NR factors recently found in cancer cells suggests frequent genetic and epigenetic changes of NR genes and proteins. As NR, DNA repair and transcriptional regulation all influence each other, we discuss the possibility that defects in NR influence the integrity of genome and cellular functions, leading to acceleration of both cellular senescence and transformation as common and rare cellular events, respectively, and that they cause cellular ageing.
2. Nucleosome remodelling influences DNA damage response and DNA repair
NR complexes are divided into four families, SWI/SNF, ISWI, CHD and INO80, each of which harbours a family-specific ATPase; BRM or BRG1 (SWI/SNF), SNF2H (ISWI), CHD3 (CHD) and INO80 (INO80). In each family there are many protein complexes consisting of an ATPase, core and variant factors, targeting specific DNA sequences or chromatin for the regulation of transcription in certain cellular tissues or at specific developmental stages. NR is also necessary in DNA replication for the process of opening up and reconstituting chromatin, before and after DNA synthesis, respectively. The influence of NR complexes or factors belonging to different subfamilies on various DNA repair systems has been reported in mammalian cells [5–11]. The accumulation of DNA repair proteins at sites of DNA damage can be influenced by chromatin itself due to the inability of repair proteins to access DNA damage. If knockdown of an NR subunit decreases the access of a DNA repair protein to its substrate DNA damage, thus influencing repair of the damage and making cells sensitive to the DNA-damaging agent, the NR subunit and possibly its NR complex may be judged to be involved in the repair process. However, questions remain as to which NR complex contributes to DNA repair, which NR factors are involved in the repair, which DNA repair is supported by NR, where and when NR is required for DNA repair and how important is the contribution of NR to cellular resistance against DNA damage. The data reviewed here indicate that NR plays significant roles in DNA damage repair and cellular resistance to various types of DNA damage.
3. DNA damage response of ACF/CHRAC complex of the ISWI family
The ISWI family of ATP-NR harbours SNF2H as the ATPase, building an ATP-utilizing chromatin assembly and remodelling factor (ACF) complex with ACF1. There are two supporting factors influencing transcriptional regulation, CHRAC15 and CHRAC17, which are known as subunits in the Polymerase ε complex and these build the CHRAC complex with SNF2H and ACF1. All the subunits of CHRAC accumulate at DNA damage, including DSB for example, and interact via ACF1 with KU proteins when cells are treated with bleomycin; in addition, they are required for cellular resistance to X-rays, where DNA damage–induced interaction between ACF1 and KU may initiate at least a part of the repair process and NR [11]. ISWI contributes to non-homologous end joining (NHEJ), homologous recombination and nucleotide excision repair (NER), suggesting a general role in DNA repair [7,11]. Artemis-dependent NHEJ of DSB in heterochromatin requires ACF1-SNF2H with the aid of RNF20-mediated chromatin relaxation [12], suggesting a role for ISWI NR in DNA repair in heterochromatin. Another paper has reported that the deacetylase sirtuin 6 (SIRT6) recruits SNF2H to DSB for CR to prepare for DSB repair [13]. Cooperation between deacetylation of histone H3K56 and NR by SNF2H may contribute to efficient DSB repair in heterochromatin.
4. DNA repair by nucleosome remodelling complex belonging to SWI/SNF family
NR complex or subunits belonging to the SWI/SNF family have been reported to be involved in various types of DNA repair processes in various species [9,14–16]. The Brahma-associated factors (BAF) complex consists of more than 13 subunits in mammalian cells and its precise components differ in a tissue- and developmental stage–dependent manner [17,18]. A BAF-like SWI/SNF complex, PBAF, has also been reported to be required for the DNA damage response and repair [14]. Recently, whole-cell sequencing of cancer genomes has revealed that a number of genes encoding the subunits of BAF and PBAF are frequently mutated in various types of cancer cells. The subunits of NR complexes in the SWI/SNF family turned out to be mutated in around 20% of human cancers of various tumour types (see below) [19]. Therefore, knowing which BAF factors are involved in DNA repair and which DNA repair is under the influence of the repair-related BAF complex becomes extremely important for understanding DNA repair within cells. Furthermore, it suggests a possible relationship between the high frequency of cancer with DNA repair defects, and it may lead to an efficient cancer therapy because of the repair defects in cancer cells. Another interesting question is the difference in NR and its mechanisms between transcription and DNA repair. DNA damage is produced anywhere in the genome, whereas DNA is transcribed only at certain places, in a sequence- and cellular condition–dependent manner. How DNA damage is recognized and repaired in chromatin is a long-standing key question in DNA repair research. Here we present our recent analysis of the BAF factors that influence the DNA damage response and repair, including some new data on the cellular response to UV for discussion regarding the role of NR in DNA repair and beyond.
5. BAF factors are required for the recruitment of KU proteins at DSB and NHEJ activity
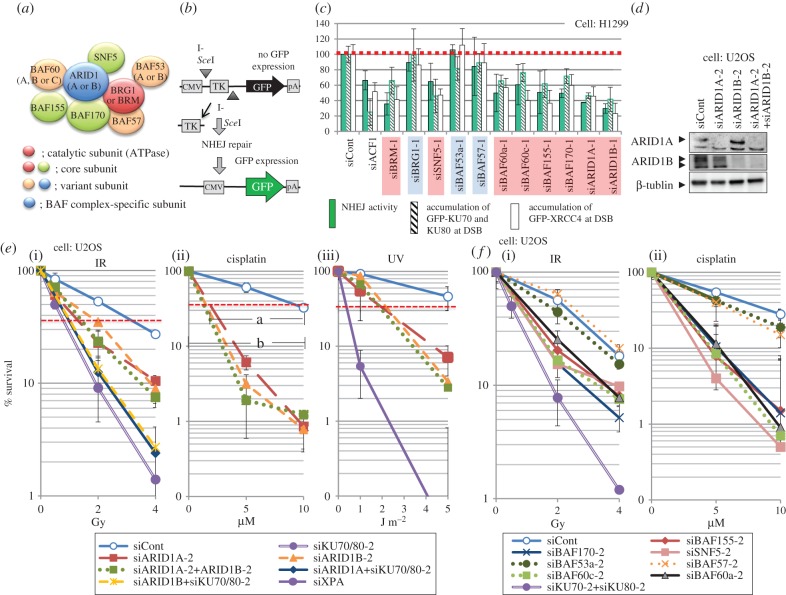
Figure 1 shows our recent analysis of BAF factors at their commitment to DNA repair and cellular resistance to various treatments with an addition of UV-survival curves for ARID1A or/and ARID1B-suppressed U2OS cells [20]. A model for BAF complex showing interacting subunits is presented in figure 1a, in which only those factors characterized in our assays are indicated. There are theoretically a large number of different BAF complexes containing different combinations of the components [17]. The major subunits in the BAF complexes are: the catalytic ATPase core subunit, either BRG1 or BRM; three core subunits present in almost every BAF complex, BAF170, BAF155 and SNF5 (BAF47); and several subunits that vary in each complex, including BAF-specific variant subunits, ARID1A (BAF250a) or ARID1B (BAF250b), which are thought to be mutually exclusively present in the complexes for transcriptional regulation.
Figure 1.
Influence of suppression of BAF factors on NHEJ of DSB and sensitivity to X-rays, cisplatin and UV, modified from Watanabe et al. [20]: (a) schematic complex model of the BAF complex and factors characterized here. (b) Assay for NHEJ activity in living human cells. This assay unit was integrated in the genome of the H1299 cell line and NHEJ activity was measured as the rate of GFP expression after I-SceI was expressed in the cells [21]. (c) Relative influence of suppression of BAF factors on NHEJ activity, accumulation of GFP-KU proteins and GFP-XRCC4 at laser-irradiated sites in H1299 cells. NHEJ activity and accumulation of GFP-tagged proteins at laser-irradiated sites obtained by control siRNA were set as 100 (no effect), relative values in the cells treated with siRNA for each BAF factor were evaluated. H1299 cell expresses all the BAF factors tested here except BRG1. The siRNA with suppressed NHEJ is marked in red, while that without the influence is in blue. siACF1 was used as positive control. (d) Western blots for ARID1A and/or ARID1B suppression in U2OS cell. (e) Colony-forming ability of U2OS cells treated with siRNA directed to ARID1A or/and ARID1B after X-Ray irradiation ((i)), cisplatin treatment ((ii)) or UV irradiation ((iii), new data). For explanation of a and b in the middle survival curve see text. (f) Colony-forming ability of U2OS cells treated with siRNA directed to other BAF factors after X-ray irradiation ((i)) and cisplatin treatment ((ii)).
Our approach was to use two independent sets of presumed off-target free siRNAs for each subunit and the influence of the suppression of each subunit on the repair of DSB was analysed in regard to NHEJ activity, accumulation of co-expressed GFP-tagged KU70 and KU80, and GFP-tagged XRCC4 at laser micro-irradiation sites, the latter of which corresponds to the accumulation of endogenous KU protein at laser-induced DSB. NHEJ activity was determined as the expression rate of GFP in an H1299 cell line harbouring an NHEJ assay system integrated within its genome sequence to measure the frequency of NHEJ repair of I-SceI-induced DSBs [21] (figure 1b). Figure 1c shows the results of the three assays for H1299 cells with suppressed expression of a BAF factor, using one set of siRNAs for each subunit. As H1299 cells express all the subunits tested except BRG1, knockdown of BRG1 expression represents a further negative control in addition to the control siRNA treatment. Recently, it was reported that BRG1 has a specific role in the DSB response and repair. ATM activated by DSB phosphorylates BRG1, which binds γH2AX-containing nucleosomes and stimulates γH2AX formation and NHEJ of DSB, whereby no ATPase activity is required [22]. It is, therefore, not a NR function. The results obtained here with H1299 cells represent the DSB response and repair, where BRM was used as the ATPase for CR, and indicate that in addition to the BRG1-specific function in response to DSB, there is BAF complex contributing to DSB repair. The results obtained with H1299 coincide very well with the high cellular sensitivity obtained with U2OS cells expressing both BRG1 and BRM (see below). Going back to figure 1c, the results of the three different assays were quite similar for each subunit. Importantly, we obtained exactly the same results by using another set of siRNAs (not shown here but see Watanabe et al. [20]). The siBRG1 negative control provided the same results as those obtained with siControl, siBAF53a or siBAF57, whereas the results of other subunit deletions showed between 30 and 60% of the activity obtained with the controls and were similar to the result obtained with a positive control for ACF1, which has been discussed above.
Besides the ATPase and three core factors, the two BAF-specific variant factors, ARID1A and ARID1B, have emerged as components required for DSB repair (figure 1c, far right two siRNAs). ARID1A (BAF250a) and ARID1B (BAF250b) are proteins of around 240 kDa and contain a large disordered region harbouring an ARID domain and a carboxy-terminal armadillo-like helical domain (figure 2a). The name ARID is derived from A–T Rich Interaction Domain, which is conserved in the human ARID family consisting of 15 proteins [23]. Since both ARID1A and ARID1B proteins interact with transcriptional activators and bind to DNA, their roles are thought to be the recruitment of the BAF complex to the target sequence or chromatin for transcriptional regulation [24,25]. We obtained the results of the ARID1A and ARID1B suppression experiments by using the second siRNA sets, and a third siRNA, targeting the 3′-UTR of each gene, and further confirmed the results of KU-accumulation at DSB and its complementation with wild-type cDNA expression of either ARID1A or ARID1B. Because of the interdependent protein stability within the BAF complex, knockdown of ARID1A reduces the expression of BAF155, which further reduces the expression of SNF5, but ARID1B does not influence other BAF factor so far analysed. Knockdown of either ARID1A or ARID1B does not influence the expression of ARID1B or ARID1A, respectively, for H1299 as well as for U2OS (figure 1d) [20]. Cellular resistance to X-rays, shown in figure 1e(i), indicates that either ARID1A or ARID1B knockdown provides U2OS cells with almost the same per cent survival as the double knockdown, which is milder than that obtained with KU knockdown. KU and ARID1A or ARID1B knockdown did not increase the sensitivity obtained by KU knockdown alone. These data suggest independent roles for ARID1A and ARID1B in the classic NHEJ pathway of DSB repair, which are different from their roles in transcriptional regulation, where they are thought to be mutually exclusive components in the complex for transcriptional regulation. These BAF factors may build a specific complex for the DNA damage response or they may belong to different complexes required for the DNA damage response. We have no answer to this question yet.
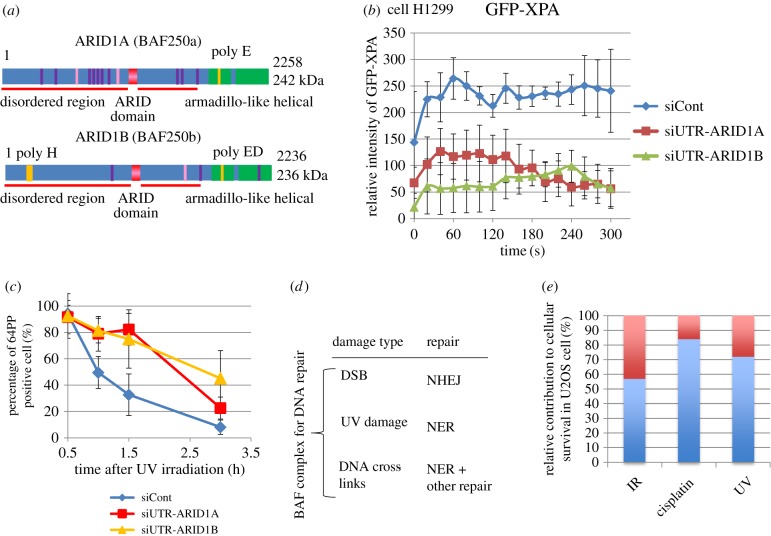
Figure 2.
Contribution of ARID1A and ARID1B to DNA damage response and cellular survival to DNA damage: (a) Domains of ARID1A and ARID1B. (b) Accumulation of GFP-XPA at laser-irradiated site in cells depleted with ARID1A or ARID1B in contrast to control siRNA treatment. (c) Repair of (6–4) photoproducts in U2OS cells depleted either with ARID1A or ARID1B after UV irradiation (new data). U2OS cells were irradiated with 254 nm UV and (6–4) photoproducts were measured with antibody after UV irradiation. (d) Contribution of the BAF complex to DNA repair, damage type and repair. (e) Relative contribution of DNA repair-related BAF to the survival of U2OS cell after X-rays, cisplatin and UV irradiation based on figure 1e: it was determined by the dose increment obtained with the presence of ARID1A and ARID1B (a) divided by the dose for control to achieve 37% survival (b) ×100.
Suppression of the expression of other BAF factors conferred X-ray sensitivity on U2OS cells when the BAF factors required for NHEJ and KU accumulation in figure 1c were depleted (figure 1f(i)), whereas suppression of the BAF factors, BAF57 or BAF53a, which were negative in NHEJ assays in figure 1c, did not also influence cellular resistance to X-rays. Thus, the results of the NHEJ assays using H1299 cells in figure 1c exactly coincide with the results of X-ray sensitivity using U2OS cells, strongly suggesting that the BAF factors function in the recruitment of KU protein at DSB and contribute to NHEJ. The X-rays' sensitivity obtained by the suppression of the BAF factor was milder than that obtained with KU suppression, but the depletion of each of the BAF factors provided cells with the same sensitivity to X-rays (figure 1f(i)), suggesting that they are working as a complex in a process or on the same repair pathway with equivalent contribution.
6. The BAF factors required for NHEJ are necessary for cisplatin resistance
Besides being necessary for resistance to X-rays, the BAF factors required for NHEJ are also necessary for cisplatin resistance. Suppression of ARID1A or ARID1B provided U2OS cells with the same sensitivity of cells to cisplatin as the double knock-down (figure 1e (ii)). It has been reported that BRM- or BRG1-depleted cells are sensitive to cisplatin [26]. In our assay, the BAF-depleted cells are significantly more sensitive to cisplatin than to X-rays, or in other words, resistance to cisplatin is more BAF-dependent than resistance to X-rays. The same is true for the other BAF factors required for X-ray resistance (figure 1f(ii)). Suppression of ARID1A or ARID1B expression provided cells with sensitivity to UV in addition (figure 1e(iii)), suggesting their involvement in NER, which partly explains the sensitivity to cisplatin in cells depleted with the ARID1 protein. It has been reported that a human cell line lacking BRG1 expression is sensitive to UV and, after exogenous expression of BRG1, UV resistance was recovered [9]. Whether BRG1 may play an additional role in the UV damage response is not yet clear.
Suppression of the expression of either ARID1 gene leads to a reduced accumulation of GFP-tagged XPA at UV damage and slows the repair of UV-induced (6–4) photoproducts (figure 2b and c, respectively). These data may explain the UV sensitivity induced by knockdown of ARID1A or ARID1B (figure 1e(iii)). While (6–4) photoproducts are recognized first by XPC-RAD23B [27], accumulation of GFP-XPC is hardly influenced by suppression of ARID protein expression, rather disturbed by ARID1B protein (not shown), suggesting that ARID1 proteins may support the process between XPC and XPA. It was reported previously that downregulation of BRG1 and BRM did not affect the recruitment of XPC to cisplatin DNA lesions, but affected ERCC1 recruitment [26]. This ERCC1 recruitment to cisplatin may have followed XPA. As DDB1 with CR activity supports XPC to recognize UV-induced DNA damage, XPC may not need NR.
7. Significant contribution of the BAF complex for repair to cellular resistance
Thus, the BAF complex for DNA repair contributes to NHEJ, NER and cisplatin resistance (figure 2d). Not only the variety of the repair, to which the BAF complex contributes, but also the extent of the survival increase by the NR is surprising. When the doses resulting in 37% survival (one lethal lesion per cell) after X-rays, cisplatin or UV irradiation of U2OS cells are compared between control and ARID1A and ARID1B double knocked-down cells (red dotted lines in figure 1e), the contribution of the BAF factors to the repair of DNA damage is measured as the dose increase (dose increment) needed in the control compared with the BAF factor knockdown, as a proportion of the dose (a/b×100 in figure 1e(ii) for cisplatin) for a 37% survival rate. The dose increment was 56% for X-rays, 70% for UV and 85% for cisplatin (figure 2d), which corresponds almost exactly to the amount of lesions repaired by the contribution of the BAF, if the amount of lesions increases linearly with the dose, as was the case as judged from the survival curves. Because of the incomplete nature of gene silencing, the actual influence of the BAF complex on cellular survival may be greater than suggested by the data presented above. The significant contribution of the BAF factors to cellular resistance of U2OS cells to various types of DNA damage suggests that the BAF complex is required before several DNA repair processes begin, and that it operates in a larger area of chromatin, possibly more than that of heterochromatin, which remains to be determined. Cisplatin produces both intra- and inter-DNA cross-links, the latter of which requires a multi-step repair process, including NER and other repair activities [28], which may be dependent on the BAF. The high sensitivity of BAF-defective cells to cisplatin may explain why it is still often useful for cancer therapy, which has not been well understood.
8. Mutation of BAF factors and their silencing: their relation to cancer and cancer therapy
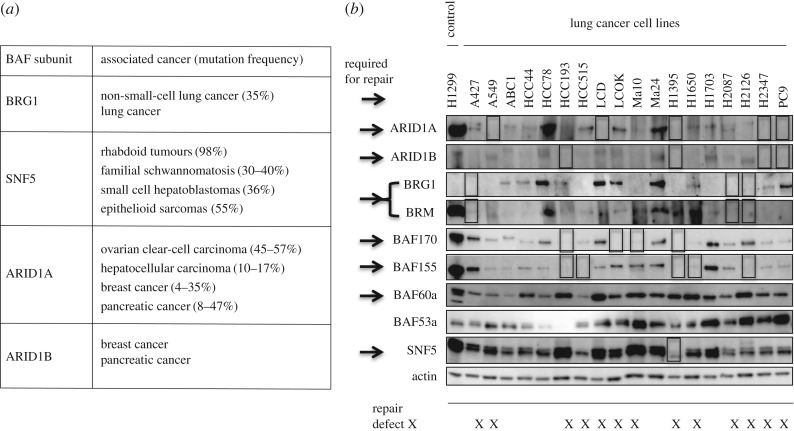
Recent whole-genome sequencing of cancer cells has identified a number of CR factors that are highly mutated in various types of cancer cells. Mutations of SWI/SNF subunits are found to be widespread across human cancers. The subunits of BAF complexes including PBAF complex, in which ARID1A or ARID1B is replaced by BAF200/BAF180/BRD7 complex, consisting of 29 genes encoding 15 subunits, are mutated in around 20% of human cancers [19]. Figure 3a depicts BAF factors and associated frequent cancers, taken from reviews [29,30]. There must be specific anti-oncogenic roles for each BAF subunit to explain why mutations in BAF factors are so strongly biased toward specific types of cancer formation [30]. ARID1A targets SWI/SNF complex to transcriptional enhancer regions for gene activation, whereas ARID1B does not replace the role of the enhancer activity of ARID1A, suggesting that the ARID1A-specific gene regulation prevents cancer [31]. As ARID1A and ARID1B make a similar contribution to DNA repair and cellular resistance to X-rays and cisplatin, their role in DNA repair does not explain the significantly greater number of mutations in ARID1A in specific cancers. However, there are many examples of preference in specific cancer caused by DNA repair deficiency, such as mismatch repair in colorectal cancer and BRCA1/2 in breast cancer and, therefore, careful examination of any influence of repair defect on the tumourigenesis is necessary. Within the BAF components tested here, mutation in either BAF57 or BAF53a, which does not contribute to DNA repair (figure 1), has barely been reported in cancer cells [32,33].
Figure 3.
Mutation and silencing of the BAF factors related to DNA repair in cancer cells: (a) BAF mutations in cancer [29], (b) Western blotting of lung cancer cell lines using antibodies against the BAF factors characterized in this paper modified from Watanabe et al. [20]. Apparent loss of expression is marked with a square. BAF factors required for DNA repair are indicated by arrows.
In addition to genetic mutation, BAF subunits are very frequently silenced in cancer cells [34]. In a majority of lung cancer cell lines we found loss of detectable expression in a number of BAF factors, which are required for DNA repair [20] (figure 3b). Of 18 lung cancer cell lines 13 cell lines indicated with X in figure 3b are lacking at least one of the BAF factors required for DNA repair, predicting repair defects. Furthermore, there are cell lines lacking several BAF factors expression simultaneously. Therefore, a majority of these cancer cells are repair-deficient due to mutated or silenced BAF factors related to DNA repair. While multiple silencing of the expression of SWI/SNF factors in cancer cells may be achieved by loss of an interaction partner, as shown previously [20], it explains only a part of the silencing occurring in cancer cells, suggesting a mechanism of multiple silencing of various BAF factors. One mutation or silencing of a subunit may cause silencing of the other and accompany loss of several functions of BAF complexes, which may cause repair deficiency and abnormality in transcriptional regulation. It is known that many cancer cells are sensitive to X-rays or cisplatin treatment at the early phase of cancer, for example, at the first chemotherapy, but cells become resistant later on. The resistance may be due to a mutation in p53 leading to resistance to apoptosis. While two reports published in 2011 suggested a mutually exclusive relationship between ARID1A and TP53 mutations in ovarian clear-cell carcinoma and gastric cancer [35,36], in pancreatic cancer all the SWI/SNF mutated cancer carried TP53 mutations, which suggests that mutual exclusivity may correlate with tumour subtypes [37] and tumour stage. However, these data are very encouraging, as ARID1A-deficient cancer cells may be sensitive to DNA damage. Since the silencing as well as mutation in the BAF factors required for DNA repair can be detected in cancer tissues, sensitivity of cancer cells to cisplatin or X-rays can be predicted before therapy begins. Intensive treatment of cancer with cisplatin at the first instance for cells lacking the BAF factor may help to eliminate cancer cells or delay cancer development.
9. Accumulation of DNA damage and cell killing
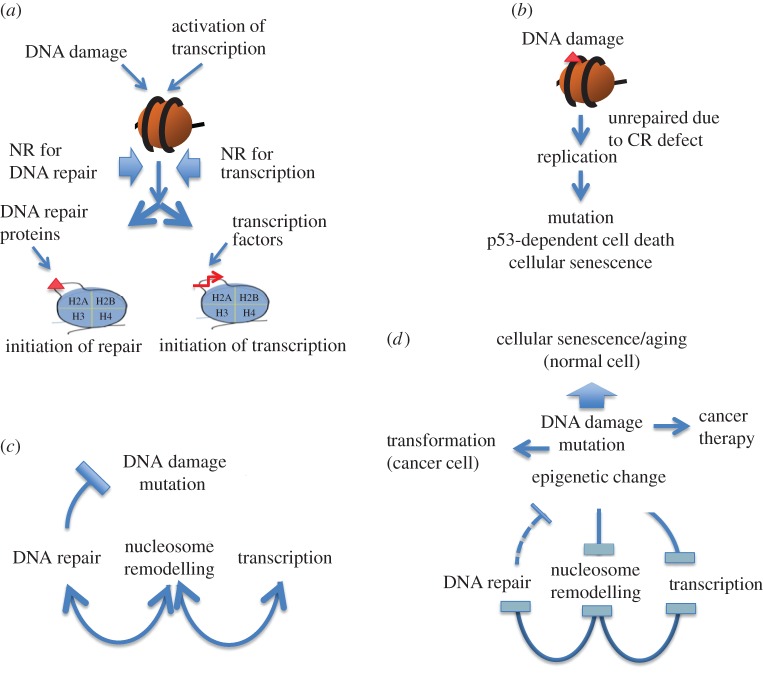
NR and its factors function in transcriptional regulation as well as in DNA repair (figure 4a). Although BAF factors for transcriptional regulation differ from those for DNA repair, there are many common factors in both functions and one mutation in a gene encoding such a common factor inactivates both functions. Judging from the data presented in figures 1 and 2, the majority of DNA damage produced by X-rays, UV or cisplatin requires NR for efficient DNA repair and for survival of cells. The amount of DNA damage left unrepaired increases in cells with a deficient BAF complex for DNA repair and accumulates especially in resting or slowly growing cells like stem cells, oocyte cells or aged cells before cells begin to replicate, suggesting a source of mutation and epigenetics changes. It has been reported that mammalian SWI/SNF NR complex prevents apoptosis after DNA damage [38]. We think that the BAF complex rescues cells from DNA damage by supporting DNA repair and minimizing apoptosis caused by unrepaired DNA damage at a replication site (figure 4b).
Figure 4.
Functionally interacting pathways of NR, DNA repair and transcriptional regulation and their negative feedback loops via ageing: (a) NR complexes responding to DNA damage and to transcriptional activation are different. (b) DNA damage unrepaired due to NR defect causes mutation, p53-dependent cell death and cellular senescence. (c) NR supports DNA repair as well as transcriptional regulation. DNA repair suppresses the increase in DNA damage and mutation. (d) Cellular ageing increases DNA damage, mutation rate and epigenetic changes, which repress NR leading to decreased DNA repair activity and transcriptional regulation activity. These changes further suppress NR activity, leading to negative feedback loops among NR, DNA repair and transcription, a model for fundamental cellular ageing.
10. Relationships among nucleosome remodelling, DNA repair and transcription and cellular ageing
There are mutually supportive relationships among NR, transcriptional regulation and DNA repair processes (figure 4c). NR supports DNA repair and DNA repair suppresses mutation frequency. NR and transcription cooperate for optimum regulation of gene expression. Genetic and epigenetic changes in genes and proteins of NR factors increase with age as a result of constitutive oxidative stresses within the cell. Chromatin modifications are also induced during repair of DNA damage [39]. Concerning the relationship between NR and transcriptional regulation related to cellular ageing, it has been reported that the SWI/SNF complex interacts with and regulates the transcription factor DAF-16/FOXO, which regulates the stress response and contributes to cellular longevity in Caenorhabditis elegans [40]. Defective NR increases unrepaired DNA damage, which furthermore suppresses both NR and transcriptional activity. Recent studies on stem cells suggest that mutations and epigenetic changes influence stem cell functionality leading to cellular ageing [41]. Epigenetic changes in CR factors have been thought to cause cellular ageing [42–44], but the relationship between the epigenetic changes and ageing-dependent genome instability or mutation has remained unknown. While cellular senescence is a model for cellular ageing [45], the way in which genetic or epigenetic damage accumulates with age, the basis for cellular ageing, has also remained elusive.
11. Negative feedback loops in NR, DNA repair and transcriptional regulation accelerate cellular transformation and ageing
A recent review has proposed nine tentative ‘hallmarks’ of ageing, which include genomic instability, telomere attrition, epigenetic alterations, loss of proteostasis, deregulated nutrient sensing, mitochondrial dysfunction, cellular senescence, stem cell exhaustion and altered intercellular communication, and emphasized their interconnected contribution to ageing [46]. Genome-wide changes and their link to the alteration of molecular complexes and cellular networks are thought to be important for interconnectedness of the hallmarks leading to age-related phenotypic consequences [47]. Genetic and epigenetic changes in the BAF factors diminish DNA repair as well as transcriptional regulation activities, and DNA repair defects negatively influence NR and transcriptional regulation, representing a typical undermining cross talk between genome, epigenome and complexes for transcriptional regulation (figure 4d). While DNA damage unrepaired due to defective NR causes genome instability and mutation leading to cancer, high frequency of NR defects and repair defects in cancer cells is useful for cancer therapy. Thus, NR, DNA repair and transcriptional regulation build negative feedback loops, which accelerate the genomic and epigenomic defects leading to both cellular senescence in normal cells and transformation of cancer cells as common and rare cellular events, respectively.
Acknowledgments
We thank Dr Shirley McCready for editing the text and Dr Penny Jeggo for discussion.
Data accessibility
This article has no additional data.
Authors' contributions
R.W. contributed to figures 1–3, S.K. contributed to the acquisition of data in figure 2, A.M. in figure 2 and A.U. in figure 1. A.Y. contributed to the conception and design of this work and wrote the paper.
Competing interests
We have no competing interests.
Funding
This work was supported by JSPS KAKENHI grant number 15H01737 to A.Y.
References
- 1.Becker PB, Workman JL. 2013. Nucleosome remodeling and epigenetics. Cold Spring Harb. Perspect. Biol. 5, a017905 ( 10.1101/cshperspect.a017905) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berger ND, Stanley FKT, Moore S, Goodarzi AA. 2017. ATM-dependent pathways of chromatin remodelling and oxidative DNA damage responses. Phil. Trans. R. Soc. B 372, 20160283 ( 10.1098/rstb.2016.0283) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rother MB, van Attikum H. 2017. DNA repair goes hip-hop: SMARCA and CHD chromatin remodelers join the break dance. Phil. Trans. R. Soc. B 372, 20160285 ( 10.1098/rstb.2016.0285) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Attikum H, Gasser SM. 2009. Crosstalk between histone modifications during the DNA damage response. Trends Cell Biol. 19, 207–217. ( 10.1016/j.tcb.2009.03.001) [DOI] [PubMed] [Google Scholar]
- 5.Jeggo PA, Downs JA. 2014. Roles of chromatin remodellers in DNA double strand break repair. Exp. Cell Res. 329, 69–77. ( 10.1016/j.yexcr.2014.09.023) [DOI] [PubMed] [Google Scholar]
- 6.Stanley FK, Moore S, Goodarzi AA. 2013. CHD chromatin remodelling enzymes and the DNA damage response. Mutat. Res. 750, 31–44. ( 10.1016/j.mrfmmm.2013.07.008) [DOI] [PubMed] [Google Scholar]
- 7.Aydin OZ, Vermeulen W, Lans H. 2014. ISWI chromatin remodeling complexes in the DNA damage response. Cell Cycle 13, 3016–3025. ( 10.4161/15384101.2014.956551) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Czaja W, Mao P, Smerdon MJ. 2012. The emerging roles of ATP-dependent chromatin remodeling enzymes in nucleotide excision repair. Int. J. Mol. Sci. 13, 11 954–11 973. ( 10.3390/ijms130911954) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gong F, Fahy D, Liu H, Wang W, Smerdon MJ. 2008. Role of the mammalian SWI/SNF chromatin remodeling complex in the cellular response to UV damage. Cell Cycle 7, 1067–1074. ( 10.4161/cc.7.8.5647) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hinz JM, Czaja W. 2015. Facilitation of base excision repair by chromatin remodeling. DNA Repair 36, 91–97. ( 10.1016/j.dnarep.2015.09.011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lan L, et al. 2010. The ACF1 complex is required for DNA double-strand break repair in human cells. Mol. Cell 40, 976–987. ( 10.1016/j.molcel.2010.12.003) [DOI] [PubMed] [Google Scholar]
- 12.Klement K, Luijsterburg MS, Pinder JB, Cena CS, Del Nero V, Wintersinger CM, Dellaire G, van Attikum H, Goodarzi AA. 2014. Opposing ISWI- and CHD-class chromatin remodeling activities orchestrate heterochromatic DNA repair. J. Cell Biol. 207, 717–733. ( 10.1083/jcb.201405077) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Toiber D, et al. 2013. SIRT6 recruits SNF2H to DNA break sites, preventing genomic instability through chromatin remodeling. Mol. Cell 51, 454–468. ( 10.1016/j.molcel.2013.06.018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brownlee PM, Meisenberg C, Downs JA. 2015. The SWI/SNF chromatin remodelling complex: its role in maintaining genome stability and preventing tumourigenesis. DNA Repair 32, 127–133. ( 10.1016/j.dnarep.2015.04.023) [DOI] [PubMed] [Google Scholar]
- 15.Euskirchen G, Auerbach RK, Snyder M. 2012. SWI/SNF chromatin-remodeling factors: multiscale analyses and diverse functions. J. Biol. Chem. 287, 30 897–30 905. ( 10.1074/jbc.R111.309302) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hara R, Sancar A. 2002. The SWI/SNF chromatin-remodeling factor stimulates repair by human excision nuclease in the mononucleosome core particle. Mol. Cell. Biol. 22, 6779–6787. ( 10.1128/MCB.22.19.6779-6787.2002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ho L, Crabtree GR. 2010. Chromatin remodelling during development. Nature 463, 474–484. ( 10.1038/nature08911) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hodges C, Kirkland JG, Crabtree GR. 2016. The many roles of BAF (mSWI/SNF) and PBAF complexes in cancer. Cold Spring Harb. Perspect. Med. 6, a026930 ( 10.1101/cshperspect.a026930) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kadoch C, Crabtree GR. 2015. Mammalian SWI/SNF chromatin remodeling complexes and cancer: mechanistic insights gained from human genomics. Sci. Adv. 1, e1500447 ( 10.1126/sciadv.1500447) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Watanabe R, Ui A, Kanno S, Ogiwara H, Nagase T, Kohno T, Yasui A. 2014. SWI/SNF factors required for cellular resistance to DNA damage include ARID1A and ARID1B and show interdependent protein stability. Cancer Res. 74, 2465–2475. ( 10.1158/0008-5472.CAN-13-3608) [DOI] [PubMed] [Google Scholar]
- 21.Ogiwara H, Ui A, Otsuka A, Satoh H, Yokomi I, Nakajima S, Yasui A, Yokota J, Kohno T. 2011. Histone acetylation by CBP and p300 at double-strand break sites facilitates SWI/SNF chromatin remodeling and the recruitment of non-homologous end joining factors. Oncogene 30, 2135–2146. ( 10.1038/onc.2010.592) [DOI] [PubMed] [Google Scholar]
- 22.Kwon SJ, Park JH, Park EJ, Lee SA, Lee HS, Kang SW, Kwon J. 2015. ATM-mediated phosphorylation of the chromatin remodeling enzyme BRG1 modulates DNA double-strand break repair. Oncogene 34, 303–313. ( 10.1038/onc.2013.556) [DOI] [PubMed] [Google Scholar]
- 23.Patsialou A, Wilsker D, Moran E. 2005. DNA-binding properties of ARID family proteins. Nucleic Acids Res. 33, 66–80. ( 10.1093/nar/gki145) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chandler RL, Brennan J, Schisler JC, Serber D, Patterson C, Magnuson T. 2013. ARID1a-DNA interactions are required for promoter occupancy by SWI/SNF. Mol. Cell. Biol. 33, 265–280. ( 10.1128/MCB.01008-12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu RC, Wang TL, Shih Ie M.. 2014. The emerging roles of ARID1A in tumor suppression. Cancer Biol. Ther. 15, 655–664. ( 10.4161/cbt.28411) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kothandapani A, Gopalakrishnan K, Kahali B, Reisman D, Patrick SM. 2012. Downregulation of SWI/SNF chromatin remodeling factor subunits modulates cisplatin cytotoxicity. Exp. Cell Res. 318, 1973–1986. ( 10.1016/j.yexcr.2012.06.011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sugasawa K. 2016. Molecular mechanisms of DNA damage recognition for mammalian nucleotide excision repair. DNA Repair 44, 110–117. ( 10.1016/j.dnarep.2016.05.015) [DOI] [PubMed] [Google Scholar]
- 28.Roy U, Scharer OD. 2016. Involvement of translesion synthesis DNA polymerases in DNA interstrand crosslink repair. DNA Repair 44, 33–41. ( 10.1016/j.dnarep.2016.05.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilson BG, Roberts CW. 2011. SWI/SNF nucleosome remodellers and cancer. Nat. Rev. Cancer 11, 481–492. ( 10.1038/nrc3068) [DOI] [PubMed] [Google Scholar]
- 30.Tang Y, et al. 2017. Linking long non-coding RNAs and SWI/SNF complexes to chromatin remodeling in cancer. Mol. Cancer 16, 42 ( 10.1186/s12943-017-0612-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mathur R, Alver BH, San Roman AK, Wilson BG, Wang X, Agoston AT, Park PJ, Shivdasani RA, Roberts CWM. 2017. ARID1A loss impairs enhancer-mediated gene regulation and drives colon cancer in mice. Nat. Genet. 49, 296–302. ( 10.1038/ng.3744) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lomeli H, Castillo-Robles J. 2016. The developmental and pathogenic roles of BAF57, a special subunit of the BAF chromatin-remodeling complex. FEBS Lett. 590, 1555–1569. ( 10.1002/1873-3468.12201) [DOI] [PubMed] [Google Scholar]
- 33.Biegel JA, Busse TM, Weissman BE. 2014. SWI/SNF chromatin remodeling complexes and cancer. Am. J. Med. Genet. C 166C, 350–366. ( 10.1002/ajmg.c.31410) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reisman D, Glaros S, Thompson EA. 2009. The SWI/SNF complex and cancer. Oncogene 28, 1653–1668. ( 10.1038/onc.2009.4) [DOI] [PubMed] [Google Scholar]
- 35.Wang K, et al. 2011. Exome sequencing identifies frequent mutation of ARID1A in molecular subtypes of gastric cancer. Nat. Genet. 43, 1219–1223. ( 10.1038/ng.982) [DOI] [PubMed] [Google Scholar]
- 36.Guan B, Wang TL, Shih Ie M. 2011. ARID1A, a factor that promotes formation of SWI/SNF-mediated chromatin remodeling, is a tumor suppressor in gynecologic cancers. Cancer Res. 71, 6718–6727. ( 10.1158/0008-5472.CAN-11-1562) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shain AH, Pollack JR. 2013. The spectrum of SWI/SNF mutations, ubiquitous in human cancers. PLoS ONE 8, e55119 ( 10.1371/journal.pone.0055119) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park JH, Park EJ, Hur SK, Kim S, Kwon J. 2009. Mammalian SWI/SNF chromatin remodeling complexes are required to prevent apoptosis after DNA damage. DNA Repair 8, 29–39. ( 10.1016/j.dnarep.2008.08.011) [DOI] [PubMed] [Google Scholar]
- 39.O'Hagan HM. 2014. Chromatin modifications during repair of environmental exposure-induced DNA damage: a potential mechanism for stable epigenetic alterations. Environ. Mol. Mutagen 55, 278–291. ( 10.1002/em.21830) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Riedel CG, et al. 2013. DAF-16 employs the chromatin remodeller SWI/SNF to promote stress resistance and longevity. Nat. Cell Biol. 15, 491–501. ( 10.1038/ncb2720) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goodell MA, Rando TA. 2015. Stem cells and healthy aging. Science 350, 1199–1204. ( 10.1126/science.aab3388) [DOI] [PubMed] [Google Scholar]
- 42.Sen P, Shah PP, Nativio R, Berger SL. 2016. Epigenetic mechanisms of longevity and aging. Cell 166, 822–839. ( 10.1016/j.cell.2016.07.050) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lazarus J, Mather KA, Thalamuthu A, Kwok JB. 2015. Genetic factors and epigenetic mechanisms of longevity: current perspectives. Epigenomics 7, 1339–1349. ( 10.2217/epi.15.80) [DOI] [PubMed] [Google Scholar]
- 44.Benayoun BA, Pollina EA, Brunet A. 2015. Epigenetic regulation of ageing: linking environmental inputs to genomic stability. Nat. Rev. Mol. Cell Biol. 16, 593–610. ( 10.1038/nrm4048) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wiley CD, Campisi J. 2016. From ancient pathways to aging cells-connecting metabolism and cellular senescence. Cell Metab. 23, 1013–1021. ( 10.1016/j.cmet.2016.05.010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.López-Otín C, Blasco MA, Partridge L, Serrano M, Kroemer G. 2013. The hallmarks of aging. Cell 153, 1194–1217. ( 10.1016/j.cell.2013.05.039) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Veitia RA, Govindaraju DR, Bottani S, Birchler JA. 2016. Aging: somatic mutations, epigenetic drift and gene dosage imbalance. Trends Cell Biol. 27, 299–310. ( 10.1016/j.tcb.2016.11.006) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This article has no additional data.