Abstract
There is considerable evidence that males will increase the number of sperm ejaculated in response to sperm competition risk. However, whether they have the capacity to adjust seminal fluid components of the ejaculate has received less attention. Male crickets (Teleogryllus oceanicus) have been shown to adjust the viability of sperm in their ejaculate in response to sperm competition risk. Here we show that socially mediated plasticity in sperm viability is probably due, at least in part, to male adjustments in the protein composition of the seminal fluid. Seven seminal fluid protein genes were found to have an increased expression in males exposed to rival calls. Increased expression of these genes was correlated with increased sperm viability in whole ejaculates, and gene knockdown confirmed that at least one of these proteins promotes sperm viability. Our results lend support for recent theoretical models that predict complex responses in male allocation to seminal fluid composition in response to sperm competition risk.
Keywords: strategic ejaculation, seminal fluid, Teleogryllus oceanicus
1. Introduction
It is now widely appreciated that females, like males, can increase their reproductive success by accepting more than one mating partner [1–3]. Multiple mating by females (polyandry) has wide-ranging evolutionary consequences [4,5], not least the selection imposed on males through sperm competition [6] and cryptic female choice [7].
When females mate with more than one male, the sperm from these males must compete to fertilize a limited supply of ova. Among species, increased risk (the probability that females will mate with one other male) and intensity (the average number of males mated) of sperm competition are both predicted to favour the evolution of increased expenditure on the ejaculate [8]. Within species, however, males are predicted to increase their immediate allocation of ejaculate reserves to individual females as the risk of sperm competition increases, but to decrease their allocation with increasing intensity of sperm competition because the payoff per unit allocation will decline as the number of competing males increases [8]. Considerable research effort has been made to test these theoretical predictions. Thus, there is taxonomically widespread evidence of evolutionary associations between the strength of selection from sperm competition and testes mass, productivity, and sperm form and function (reviewed in [9,10]). Within species, males have been found to allocate greater numbers of sperm to females when they perceive the presence of sperm competition rivals in their environment [11,12]. Empirical tests of sperm competition theory have thus focused primarily on the production and allocation of sperm. However, sperm represent only one component of the ejaculate.
Sperm are delivered to the female bathed in a cocktail of seminal fluid proteins (sfps) derived from the male accessory glands. Seminal fluid proteins have significant effects on sperm function, including the promotion of motility, capacitation and fertilization efficacy [13]. They also protect sperm within the female tract, and can profoundly affect female reproductive behaviour and physiology [9,13–15]. Given their role in promoting male fertilization success we might expect that sfps, like sperm, should be subject to selection from sperm competition [9]. Indeed, seminal fluid proteins include the most rapidly diverging proteins known, and comparative studies suggest that sfp divergence is correlated with the strength of selection from sperm competition [16–20]. Within species, studies of fish with alternative reproductive tactics have reported greater velocity of sperm produced by sneaker males, with differences in sperm performance in at least one species being due to differences in seminal fluid [21]. A handful of studies have reported phenotypic plasticity in sperm velocity in response to social cues to sperm competition [22,23] or female quality [24,25], which might also be due to plasticity in seminal fluid composition [26,27].
Theoretical models are beginning to be developed in order to predict how males should allocate their seminal fluids in response to sperm competition [28–30]. The broad predictions arising from these models make intuitive sense, in that males are predicted to increase their allocation to sfps that enhance fertilization success and/or decrease their allocation to sfps that enhance female fecundity when they face sperm competition. However, tests of these predictions are currently limited to just two species, Drosophila melanogaster and house mice Mus musculus. Male D. melanogaster transfer a sfp (ovulin) that stimulates females to manufacture and lay eggs, and a second sfp (sex peptide) that inhibits female remating. When males mate with previously mated females they reduce their allocation of ovulin, though not sex peptide, because reproduction is already triggered in these females by their previous mates [31]. Social manipulations have also found that males exposed to rivals during copulation transfer more sex peptide and ovulin to females than males copulating in isolation [32], but exposure to rivals prior to mating results in a decreased expression of Acp26a, the gene responsible for ovulin production, and a third sfp gene, Acp62f, implicated in defensive sperm competition success [33]. In house mice, exposure to rival male scent during sexual development was found to increase the abundance of three sfps in the seminal vesicles (SVS6, SVS5 and CEACAM 10), supporting the idea that sfp production can exhibit socially cued phenotypic plasticity [34]. Though the effect of these sfps on competitive fertilization success is yet to be fully understood, CEACAM has been reported to enhance sperm motility [35]. These studies have been possible because Drosophila and house mice are both well characterized genomically. Functionally characterizing individual seminal fluid proteins in non-model organisms remains challenging [27]. Nonetheless, here we document socially cued phenotypic plasticity in sfp gene expression, and identify those sfps that contribute to sperm fertilization competency in a non-model organism, the Australian field cricket Teleogryllus oceanicus.
When exposed to the sexual advertisement calls of rival males during their development, male T. oceanicus produce ejaculates with a greater proportion of viable sperm compared with males reared in acoustic isolation [36]. Moreover, manipulating male perceptions of sperm competition intensity, by the perfuming of females with cuticular hydrocarbons from increasing numbers of rival males, has the effect of reducing the proportion of viable sperm in a male's ejaculate [37]. These studies show that males can make finely tuned adjustments to the quality of their ejaculates in response to the risk and intensity of sperm competition (see also [38,39]). Interestingly, sperm viability has a greater impact on a male's competitive fertilization success [40] than does the number of sperm [41], and the reciprocal transfer of sperm between the seminal fluid fractions of rival ejaculates suggests that variation in seminal fluid composition might underlie the strategic adjustments in sperm viability seen in this species [42]. Proteomic analyses have identified at least 21 different proteins in the seminal fluid of T. oceanicus [43], and eleven of these proteins were found to increase in abundance in ejaculates as males mature sexually [44]. Both sperm viability [40] and competitive fertilization success [45] also increase with male age, suggesting these eleven sfps as candidate proteins for the strategic adjustment of seminal fluid composition, and thus sperm viability, in response to sperm competition. Here we examined sperm viability and quantified sfp gene expression for males exposed to the calls of rival males during their development, and compared these with males reared in acoustic isolation. We show that the expression of seven sfp genes vary in response to sperm competition risk and use RNA interference to identify which of these sfps are responsible for the strategic adjustments in sperm viability found in this cricket.
2. Material and methods
(a). Social manipulation
Animals used in this study were taken from a large outbred stock derived originally from a fruit plantation in Carnarvon Western Australia, and seeded annually with newly caught individuals. First instar nymphs were taken at hatching and assigned randomly to one of two treatments. The first treatment group was exposed throughout their development to the calls of conspecifics via five boxes containing approximately 100 adult sexually active crickets, placed within a 2 m radius of the experimental crickets. Crickets were thus exposed continually to both the calling and courtship calls of conspecifics at a sound intensity of 70–80 dB. The second treatment group was raised in acoustic isolation (silence) [36].
Crickets were initially raised en masse at 29°C on a 12 : 12 h light : dark cycle with constant access to cat chow and water. At the seventh nymphal instar, when sex can first be determined, females were discarded and males housed individually in boxes (7 × 7 × 5 cm) separated from each other by cardboard divisions that prevented visual contact. Crickets were then monitored daily until they emerged as adults. Upon emergence the sound-producing forewings of males in both treatments were removed to prevent them from producing their own calls.
(b). Sperm viability
Fourteen days after the adult molt, a spermatophore was removed from the genital pouch of 32 males from each treatment and assessed for sperm viability. The spermatophore was placed on a slide with 20 µl of Beadle saline (128.3 mM NaCl, 4.7 mM KCl and 23 mM CaCl2). The evacuating tube was severed with fine scissors and the sperm were allowed to completely evacuate over a period of 10 min. The ejaculate was mixed gently with the saline on the slide and 5 µl transferred to a second slide. Five microlitres of SYBR 14 (1 mM), diluted 1 : 50 with Beadle saline, was added and the sample incubated in the dark for 10 min. Two microlitres of propidium iodide (2.4 mM) was then added and incubated for a further 10 min. The sperm were then viewed using a fluorescence microscope, and the number of live (stained green with SYBR 14) and dead (stained red with propidium iodide) sperm in the first 500 sperm counted was used to estimate sperm viability (the proportion of live sperm in the sample). After assessment of ejaculate quality crickets were frozen at −20°C. The proportion of viable sperm in an ejaculate was square-root-arcsine transformed for statistical analysis.
(c). Gene expression
Each cricket was thawed and the accessory gland removed, weighed and placed in RNAlater (Life Technologies), stored overnight at 4°C and then placed at −20°C. Using the PureLink RNA mini kit (Life Technologies), RNA was extracted from the entire accessory gland following the manufacturer's instructions. The tissue was disrupted using a micropestle (Interpath) and then a homogenizer (Life Technologies). RNA was removed using an on-column PureLink DNAse treatment (Life Technologies). The RNA yield was quantified using a Qubit 2.0 fluorometer (Life Technologies). The elution volume for the RNA was 100 µl and yields ranged from 366 to 1200 µg ml−1 per sample. Two micrograms of each RNA sample were converted to cDNA in a 20 µl reaction volume using the high capacity RNA-to-cDNA kit (Life Technologies), following the manufacturer's instructions.
Ten seminal fluid proteins and one sperm protein were selected for gene expression assays based on their observed changes in abundance in the seminal fluid with age [44]. We used actin as the reference gene (Isotig01761 from the T. oceanicus sequence database lodged with Genbank under the project number SRA044883.2 [46]). The six TaqMan gene expression custom assays (Life Technologies) designed previously [44] were used, and in addition, a further six assays were designed using the online Custom TaqMan assay design tool. Forward, reverse and reporter sequences for the 12 assays can be found in electronic supplementary material, table S1. In order to determine a suitable starting concentration of cDNA for the new gene expression assays, a standard curve of cDNA was prepared from one of the acoustic isolation samples, to include 100, 10, 1 and 0.1 ng µl−1. Gene expression assays were then set up in triplicate for each of the standards and for each of the assays as follows: 1 µl cDNA, 5 µl 2× TaqMan Gene expression master mix (Life Technologies) and 0.5 µl 20× TaqMan custom assay mix in a 10 µl reaction volume. The assays were run in compatible 96-well plates on a StepOne Plus Real-Time PCR system (Life Technologies) using the following cycling conditions: 50°C for 2 min, 95°C for 10 min, and then 40 cycles of 95°C for 15 s and 60°C for 1 min. The efficiency of each assay was assessed with all R2 values greater than 0.994 and slopes from −3.772 to −3.282 corresponding to efficiencies varying from 84% to 102%. As the efficiency for the assay corresponding to ToSfp005 (isotig01832) was low at 84%, the standard curve was repeated with the following concentrations of cDNA: 100, 50, 25 µl−1, 12.5, 6.25 and 3.125 ng µl−1. The R2 value for this assay was 0.994, slope − 3.58 and the efficiency 90%, all within acceptable limits. For 10 of the genes, 10 ng cDNA gave average CT values varying from 19.97 to 26.93, so this was the chosen input amount for all further assays. For ToSfp026 (isotig06304), 10 ng cDNA gave a CT value of 29.36, which is unacceptably high, so this assay required 100 ng of input cDNA to bring the CT value within acceptable limits.
Gene expression assays were carried out on all samples using the cDNA amounts determined above, each assay run in triplicate for each candidate gene and reference gene, and with negative controls (without cDNA) on each plate. Results were analysed by StepOne software v. 2.3 (Life Technologies) and the results exported into DataAssist v. 3.0 software (Life Technologies) for sample comparison using the comparative CT (ΔΔCT) method [47]. The expression of actin did not differ between treatment groups (CT: male calls, 22.74 ± 0.09; silence 22.62 ± 0.10; t61 = 0.94, p = 0.350) so was an appropriate gene to use as a reference gene.
(d). Interference RNA
We explored the function of seven seminal fluid genes that exhibited socially mediated changes in expression using RNAi. Double-stranded RNA was prepared using the Megascript RNAi kit (Thermo Fisher Scientific). The manufacturer recommends dsRNA 400 bp or larger so PCR primers were designed with Geneious v. R6 [48] to give a product of approximately 500 bp (see electronic supplementary material, table S2) using the RNA sequence data for the genes previously identified. The T7 promotor sequence was added to the 5′ end of each primer. Template DNA was then prepared in four identical polymerase chain reactions (PCRs), each 50 µl reaction containing 1 × PCR buffer (10 mM Tris–HCl pH 8.3, 50 mM KCl), 1.5 mM MgCl2, 200 µM of each dNTP, 250 nM of forward primer, 250 nM of reverse primer, 2.5 units of Platinum Taq polymerase and 100 ng cDNA. PCR amplification was performed with cycling conditions as follows: 94°C for 3 min, then 35 cycles of 94°C for 30 s, 60°C for 60 s and 72°C for 60 s, and finally 72°C for 15 min. For genes ToSfp011 (Isotig1709) and ToSfp022 (Isotig444), a two-step PCR reaction was performed as follows: the same concentrations of reagents as above were used with the addition of 10% DMSO and an increase in MgCl2 concentration from 1.5 to 3 mM and cycling conditions of 94°C for 3 min, then 35 cycles of 94°C for 30 s and 68°C for 60 s, and finally 72°C for 5 min (all reagents were from Thermo Fisher Scientific). The four PCR reactions were pooled and purified using the Favorgen PCR clean-up kit (Fisher Biotech) with elution into 40 µl. The DNA was then ethanol precipitated using 1/10 volume of 3 M sodium acetate pH 5.2 and 2 volumes of 100% ethanol. The DNA pellet was resuspended in 20 µl elution buffer and the concentration determined using a Nanodrop 1000. The Megascript RNAi kit was then used to generate dsRNA via T7 RNA transcription using the manufacturer's protocol. Each transcription reaction contained 1 µg template DNA and the transcription time was 4 h. The excess DNA and ssRNA were removed, and the dsRNA purified following the manufacturer's instructions with the final elution in 100 µl. A small amount was diluted 1 in 10 and run on a 3% agarose gel to check the size and integrity of the dsRNA. The concentration was also checked using the Nanodrop 1000. The volume was adjusted to a final concentration of 1 µg µl−1 in elution buffer.
Eighth instar male nymphs were collected from laboratory stock. The nymphs were housed individually in boxes (7 × 7 × 5 cm), and provided with cat chow and water ad libitum. On the first day of the final (ninth) nymphal instar, each cricket was injected with 2 µg dsRNA and checked daily for adult emergence. An average of 15 males (10–18) were injected for each gene. Upon adult emergence, males were reinjected with a further 2 µg dsRNA and left for 4 days. At 4 days of adult age the male was assayed for sperm viability following the protocol outlined above. We were able to obtain spermatophores from 9 to 15 males per gene (mean ± s.e. 12 ± 1). A group of 30 males acted as controls, and were injected with the elution buffer from the Megascript kit. To confirm that gene expression was reduced via the RNAi, we conducted gene expression assays for a subsample of males (n = 5 per gene and control) following the protocols described above.
3. Results
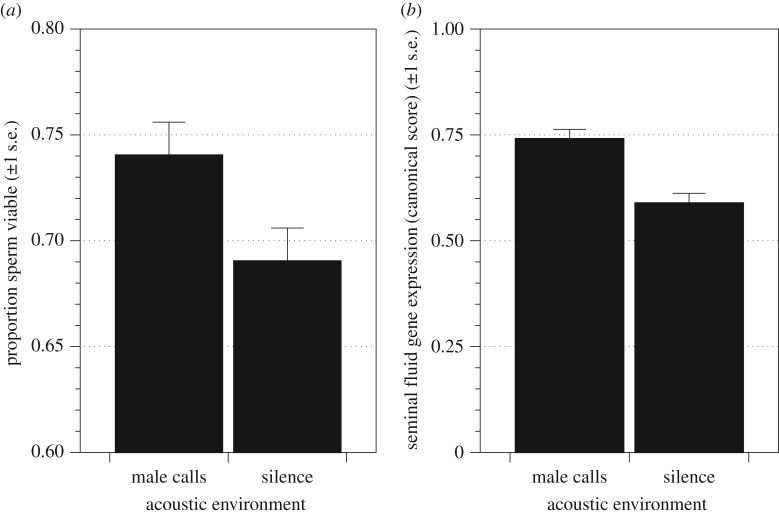
Replicating previous work [36], we found that male T. oceanicus exposed to the calls of rivals during development produced ejaculates containing a higher proportion of viable sperm than males reared in silence (t61 = 2.58, p < 0.012; figure 1a). Increased sperm viability in response to rival calls was accompanied by increased expression of seminal fluid protein genes in the male accessory glands.
Figure 1.
The effects of exposure to rival male calls during development on (a) the mean ± s.e proportion of live sperm in an ejaculate (sperm viability) and (b) the expression of seminal fluid protein genes. In (b) the mean ±s.e. canonical scores extracted from a MANOVA describe the summed expression of 10 seminal fluid and 1 sperm protein gene.
Expression levels for each of the 11 candidate genes were entered into a single MANOVA to test for differences in gene expression between social environments. Accessory gland weight was entered as a covariate. There was a significant effect of social environment on seminal fluid protein gene expression (F1,60 = 19.49, p < 0.001), but the effect of accessory gland weight was not significant (F1,60 = 3.83 p = 0.060). Males exposed to the calls of rivals had higher levels of seminal fluid gene expression than males reared in silence (figure 1b). Univariate tests revealed that 7 of the 11 genes assayed had higher expression in males exposed to rival calls (table 1). To explore how these genes might affect sperm viability we entered their expression levels into a principal components analysis that returned two axes of variation which explained 77% and 11% of the variation in gene expression, respectively (electronic supplementary material, table S3). The first axis was loaded most strongly by the expression of ToSfp017 and to a lesser extent by ToSfp011, and predicted variation in sperm viability in whole ejaculates (effect estimate 0.022 ± 0.010; F1,60 = 4.46, p = 0.039). The second axis was loaded most strongly by the expression of ToSfp005 and to a lesser extent ToSfp027, but this axis did not predict sperm viability (effect estimate 0.004 ± 0.028; F1,60 = 0.02, p = 0.885). We used interference RNA to further explore the function of these seven genes.
Table 1.
Differences in seminal fluid gene expression between males exposed to rival calls during their development and those raised in silence.
| gene | transcript reference | ΔΔCT ± s.e. |
univariate effect |
||
|---|---|---|---|---|---|
| male calls | silence | t61 | p | ||
| ToSfp001 | isotig01262 | 0.185 ± 0.009 | 0.156 ± 0.009 | 2.232 | 0.029 |
| ToSfp005 | isotig01832 | 0.489 ± 0.081 | 0.209 ± 0.083 | 2.415 | 0.019 |
| ToSfp007 | isotig00263 | 0.054 ± 0.021 | 0.050 ± 0.013 | 0.755 | 0.453 |
| ToSfp011 | isotig01709 | 0.923 ± 0.054 | 0.758 ± 0.055 | 2.137 | 0.037 |
| ToSfp017 | isotig05129 | 5.359 ± 0.207 | 4.371 ± 0.211 | 3.343 | 0.001 |
| ToSfp022 | isotig00444 | 0.141 ± 0.033 | 0.046 ± 0.034 | 2.020 | 0.047 |
| ToSfp023 | isotig00811 | 0.521 ± 0.019 | 0.464 ± 0.020 | 2.090 | 0.041 |
| ToSfp024 | isotig02293 | 0.302 ± 0.013 | 0.291 ± 0.014 | 0.566 | 0.573 |
| ToSfp026 | isotig06304 | 0.019 ± 0.004 | 0.023 ± 0.004 | 0.725 | 0.471 |
| ToSfp027 | isotig00169 | 0.896 ± 0.066 | 0.678 ± 0.067 | 2.313 | 0.024 |
| ToSp009 | isotig00560 | 0.048 ± 0.002 | 0.047 ± 0.002 | 0.435 | 0.665 |
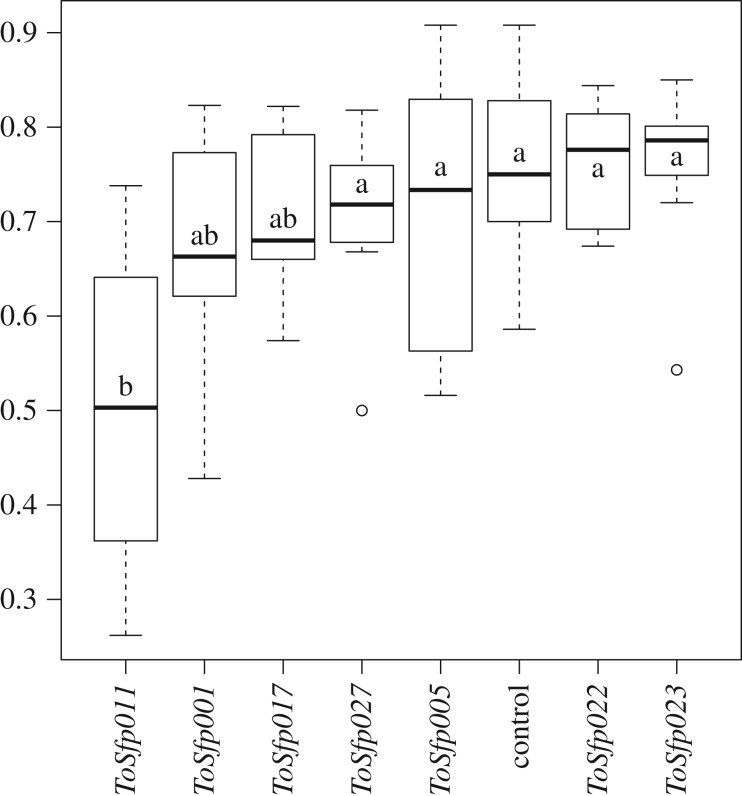
RNAi was successful in knocking down the expression of target genes (table 2). There was significant variation in sperm viability across RNAi treatment groups (F7,70 = 3.35, p = 0.004; figure 2). Knockdown of ToSfp011 significantly reduced sperm viability relative to controls, while knockdown of ToSfp001 and ToSfp0017 resulted in males with sperm viabilities intermediate between ToSfp011 knockdowns and controls. Knockdown of the remaining genes had no discernible effect on sperm viability relative to controls (figure 2).
Table 2.
Seminal fluid gene expression following RNA interference relative to controls.
| gene | ΔΔCT ± s.e. |
|
|---|---|---|
| control | RNAi | |
| ToSfp001 | 0.232 ± 0.045 | 0.005 ± 0.001 |
| ToSfp005 | 0.734 ± 0.425 | 0.017 ± 0.006 |
| ToSfp011 | 0.484 ± 0.080 | 0.007 ± 0.001 |
| ToSfp017 | 4.472 ± 0.306 | 0.033 ± 0.005 |
| ToSfp022 | 0.105 ± 0.056 | 0.003 ± 0.003 |
| ToSfp023 | 0.567 ± 0.054 | 0.075 ± 0.017 |
| ToSfp027 | 0.096 ± 0.006 | 0.037 ± 0.007 |
Figure 2.
Box plots showing the proportion of sperm alive (sperm viability) in ejaculates from males in which seminal fluid gene expression was knocked down using RNAi or control males injected with elution buffer. Statistical analysis conducted on square-root-arcsine transformed proportions. Groups not connected by the same letter (a,b) are significantly different using post hoc Tukey HSD.
4. Discussion
We found socially mediated increases in seminal fluid protein gene expression in the accessory glands of male crickets, T. oceanicus. Socially mediated changes in gene expression have been reported previously, in the context of pre-mating sexual selection. Rapid increases in the expression of genes in various regions of the female brain have been documented in response to social exposure to courting males in fish [49,50], birds [51–53] and insects [54,55], with these changes in gene expression often being of greater magnitude when females experience more attractive males. For males, changes in gene expression in the brain have been found following territory intrusion by rivals in both birds [56] and fish [57]. In male Drosophila melanogaster, the social environment was found to affect the expression of over 500 genes [58–60], half of which change only in response to rival males [60]. Socially mediated plasticity in gene expression was recently reported in the closely related cricket T. commodus using a similar experimental paradigm to that used here [61].
Male T. commodus exposed to rival male calls tend to show accelerated development and a reduced lifespan, but invest less in calling effort than males reared in silence [62–64]. These socially cued changes in life-history strategy were found to be associated with changes in the expression of genes that affect courtship, muscle development and the storage of energy reserves [61]. Interestingly, Kasumovic et al. [61] found changes in the expression of five genes associated with mating and spermatogenesis in male T. commodus exposed to rival calls. Two of these genes, Four wheel drive and spargel, appear to be involved in spermatogenesis and mitochondrial metabolism [65–67], and their increased expression in males exposed to rival male calls might be associated with the general finding that males increase their expenditure on sperm production in response to social cues to sperm competition risk [11,12]. Like T. commodus, male T. oceanicus also show socially mediated plasticity in a host of life-history traits, including immune function [68], cuticular hydrocarbon expression [69], mating behaviour [70] and sperm viability [36]. Our targeted approach examined the expression of 11 sfp genes that were candidates for the socially mediated changes in sperm viability.
We found an increased expression of seven sfp genes in response to rival calls. One of these genes, ToSfp011, showed a 20% increase in expression compared with males reared in silence. This sfp has a close match to a hypothetical accessory gland protein in the field cricket Gryllus pennsylvanicus, but was previously of unknown function [71,72]. When ToSfp011 was knocked down using interference RNA, males produced ejaculates with a 30% reduction in the proportion of sperm that were viable. While increased gene expression is indicative of an increase in protein production, we cannot directly infer that the amount of protein allocated to individual ejaculates is increased. Moreover, if the targeted gene is also expressed in tissues beyond the accessory gland, its knockdown could affect other aspects of male physiology that indirectly influence sperm viability. Nevertheless, increased ToSfp011 and ToSfp017 expression were also associated with increased sperm viability in whole ejaculates when males were exposed to rival calls. We conclude, therefore, that adjustments in the production of this protein may underlie adjustments in sperm viability found in this cricket as males respond to the risk and intensity of sperm competition [36–39].
Knockdown of two additional sfp genes, ToSfp001 and ToSfp017, resulted in sperm viability intermediate between ToSfp011 knockdowns and controls, suggesting that these proteins may also contribute to strategic adjustments in sperm viability. One of these proteins, ToSfp001, is an apyrase with a predicted function in nucleotide metabolism [43]. Apyrases have been shown to play important roles in sperm motility and viability, and to bind tightly to sperm [43]. Indeed ToSfp001 is the same protein as ToSp027 identified from the T. oceanicus sperm proteome [43]. The second protein, ToSfp017, is a hypothetical protein of previously unknown function [43].
For the remaining four sfp genes that showed increased expression on exposure to rival calls, interference RNA had no detectable effect on sperm viability. However, these sfps may serve other functions in postcopulatory male fitness. Two of these proteins, ToSfp022 and ToSfp023, remain of unknown function [44], but the third, ToSfp005, is a dipeptidase [43]. Dipeptidases are involved in the biosynthesis of prostaglandin [73,74]. Studies of T. commodus have found that males transfer prostaglandin synthesizing compounds in the ejaculate, and that prostaglandin induces egg laying and oviposition [75–79]. Fecundity-enhancing seminal fluid proteins are well known in insects [80], with this function being served by ovulin in D. melanogaster [31]. However, in contrast to our finding of increased expression in ToSfp005 in males exposed to rivals, in D. melanogaster exposure to rivals was found to result in a reduced expression of the gene responsible for ovulin production [33].
Four sfp genes that show increased expression with male age were unaffected by the social manipulation. It may be that these genes lack phenotypic plasticity because they encode proteins essential for minimal reproductive function. Alternatively, the seven genes that did show phenotypic plasticity may share some regulatory signal or cell type expression that the remaining four genes do not. Further work is required to fully understand the function of seminal fluid proteins in this and other species [9,27].
In conclusion, our data provide evidence that males will increase their expenditure on sfps that enhance the fertilization capacity of their sperm when faced with an increased risk of sperm competition. Studies of Drosophila have found that males will reduce their allocation of fecundity stimulating sfps in response to rival males, while male house mice exposed to rivals have an increased abundance of several seminal fluid proteins in their seminal vesicles, some of which may enhance sperm motility. Collectively these studies lend growing support for recent theoretical models that predict complex responses in male allocation to seminal fluid composition, responses that depend on the roles that individual seminal fluid proteins play in fecundity enhancement and competitive fertilization success [28–30].
Supplementary Material
Data accessibility
Data are available at the Dryad Digital Repository (http://dx.doi.org/10.5061/dryad.8rq18) [81].
Authors' contributions
L.W.S. designed the study, analysed the data and wrote the manuscript. M.L. conducted the molecular work. Both authors approved the final version of the manuscript.
Competing interests
The authors declare no competing interests.
Funding
This work was funded by the Australian Research Council (DP110104594).
References
- 1.Arnqvist G, Nilsson T. 2000. The evolution of polyandry: multiple mating and female fitness in insects. Anim. Behav. 60, 145–164. (doi:10.1006/anbe.2000.1446) [DOI] [PubMed] [Google Scholar]
- 2.Simmons LW. 2005. The evolution of polyandry: sperm competition, sperm selection, and offspring viability. Ann. Rev. Ecol. Evol. Syst. 36, 125–146. (doi:10.1146/annurev.ecolsys.36.102403.112501) [Google Scholar]
- 3.Slatyer RA, Mautz BS, Backwell PRY, Jennions MD. 2012. Estimating genetic benefits of polyandry from experimental studies: a meta-analysis. Biol. Rev. 87, 1–33. (doi:10.1111/j.1469-185X.2011.00182.x) [DOI] [PubMed] [Google Scholar]
- 4.Pizzari T, Wedell N. 2013. The polyandry revolution. Phil. Trans. R. Soc. B 368, 20120041 (doi:10.1098/rstb.2012.0041) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kvarnemo C, Simmons LW. 2013. Polyandry as a mediator of sexual selection before and after mating. Phil. Trans. R. Soc. B 368, 20120042 (doi:10.1098/rstb.2012.0042) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parker GA. 1970. Sperm competition and its evolutionary consequences in the insects. Biol. Rev. 45, 525–567. (doi:10.1111/j.1469-185X.1970.tb01176.x) [Google Scholar]
- 7.Eberhard WG. 1996. Female control: sexual selection by cryptic female choice. Princeton, NJ: Princeton University Press. [Google Scholar]
- 8.Parker GA, Pizzari T. 2010. Sperm competition and ejaculate economics. Biol. Rev. 85, 897–934. (doi:10.1086/656840) [DOI] [PubMed] [Google Scholar]
- 9.Simmons LW, Fitzpatrick JL. 2012. Sperm wars and the evolution of male fertility. Reproduction 144, 519–534. (doi:10.1530/REP-12-0285) [DOI] [PubMed] [Google Scholar]
- 10.Parker GA. 2016. The evolution of expenditure on testes. J. Zool. 298, 3–19. (doi:10.1111/jzo.12297) [Google Scholar]
- 11.delBarco-Trillo J. 2011. Adjustment of sperm allocation under high risk of sperm competition across taxa: a meta-analysis. J. Evol. Biol. 24, 1706–1714 (doi:10.1111/j.1420-9101.2011.02293.x) [DOI] [PubMed] [Google Scholar]
- 12.Kelly CD, Jennions MD. 2011. Sexual selection and sperm quantity: meta-analyses of strategic ejaculation. Biol. Rev. 86, 863–884. (doi:10.1111/j.1469-185X.2011.00175.x) [DOI] [PubMed] [Google Scholar]
- 13.Poiani A. 2006. Complexity of seminal fluid: a review. Behav. Ecol. Sociobiol. 60, 289–310. (doi:10.1007/s00265-006-0178-0) [Google Scholar]
- 14.Wolfner MF. 2002. The gifts that keep on giving: physiological functions and evolutionary dynamics of male seminal proteins in Drosophila. Heredity 88, 85–93. (doi:10.1038/sj.hdy.6800017) [DOI] [PubMed] [Google Scholar]
- 15.McGraw LA, Suarez SS, Wolfner MF. 2015. On a matter of seminal importance. Bioessays 37, 142–147. (doi:10.1002/bies.201400117) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haerty W, et al. 2007. Evolution in the fast lane: rapidly evolving sex-related genes in Drosophila. Genetics 177, 1321–1335. (doi:10.1534/genetics.107.078865) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karn RC, Clark NL, Nguyen ED, Swanson WJ. 2008. Adaptive evolution in rodent seminal vesicle secretion proteins. Mol. Biol. Evol. 25, 2301–2310. (doi:10.1093/molbev/msn182) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ramm SA, McDonald L, Hurst JL, Beynon RJ, Stockley P. 2009. Comparative proteomics reveals evidence for evolutionary diversification of rodent seminal fluid and its functional significance in sperm competition. Mol. Biol. Evol. 26, 189–198. (doi:10.1093/molbev/msn237) [DOI] [PubMed] [Google Scholar]
- 19.Dorus S, Evans PD, Wyckoff GJ, Choi SS, Lahn BT. 2004. Rate of molecular evolution of the seminal protein gene SEMG2 correlates with levels of female promiscuity. Nat. Genet. 36, 1326–1329. (doi:10.1038/ng1471) [DOI] [PubMed] [Google Scholar]
- 20.Wagstaff BJ, Begun DJ. 2007. Adaptive evolution of recently duplicated accessory gland protein genes in desert drosophila. Genetics 177, 1023–1030. (doi:10.1534/genetics.107.077503) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Locatello L, Poli F, Rasotto MB. 2013. Tactic-specific differences in seminal fluid influence sperm performance. Proc. R. Soc. B 280, 20122891 (doi:10.1098/rspb.2012.2891) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kilgallon SJ, Simmons LW. 2005. Image content influences men's semen quality. Biol. Lett. 1, 235–255. (doi:10.1098/rsbl.2005.0324) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith C.C., Ryan M.J. 2011. Tactic-dependent plasticity in ejaculate traits in the swordtail Xiphophorus nigrensis. Biol. Lett. 7, 733–735. (doi:10.1098/rsbl.2011.0286) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cornwallis CK, Birkhead TR. 2007. Changes in sperm quality and numbers in response to experimental manipulation of male social status and female attractiveness. Am. Nat. 170, 758–770. (doi:10.1086/521955) [DOI] [PubMed] [Google Scholar]
- 25.Leivers S, Rhodes G, Simmons LW. 2014. Context-dependent relationship between a composite measure of men's mate value and ejaculate quality. Behav. Ecol. 25, 1115–1122. (doi:10.1093/beheco/aru093) [Google Scholar]
- 26.Cornwallis CK, O'Conner EA. 2009. Sperm: seminal fluid interactions and the adjustment of sperm quality in relation to female attractiveness. Proc. R. Soc. B 276, 3467–3475. (doi:10.1098/rspb.2009.0807) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perry JC, Sirot L, Wigby S. 2013. The seminal symphony: how to compose an ejaculate. Trends Eco. Evol. 28, 414–422. (doi:10.1016/j.tree.2013.03.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cameron E, Day T, Rowe L. 2007. Sperm competition and the evolution of ejaculate composition. Am. Nat. 169, E158–E172. (doi:10.1086/516718) [DOI] [PubMed] [Google Scholar]
- 29.Alonzo SH, Pizzari T. 2010. Male fecundity stimulation: conflict and cooperation within and between the sexes: model analyses and coevolutionary dynamics. Am. Nat. 175, 174–185. (doi:10.1086/649596) [DOI] [PubMed] [Google Scholar]
- 30.Dhole S, Servedio MR. 2014. Sperm competition and the evolution of seminal fluid composition. Evolution 68, 3008–3019. (doi:10.1111/evo.12477) [DOI] [PubMed] [Google Scholar]
- 31.Sirot LK, Wolfner MF, Wigby S. 2011. Protein-specific manipulation of ejaculate composition in response to female mating status in Drosophila melanogaster. Proc. Natl Acad. Sci. USA 108, 9922–9926. (doi:10.1073/pnas.1100905108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wigby S, Sirot LK, Linklater JR, Buehner N, Calboli FCF, Bretman A, Wolfner MF, Chapman T. 2009. Seminal fluid protein allocation and male reproductive success. Curr. Biol. 19, 751–757. (doi:10.1016/j.cub.2009.03.036) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fedorka KM, Winterhalter WE, Ware B. 2011. Perceived sperm competition intensity influences seminal fluid protein production prior to courtship and mating. Evolution 65, 584–590. (doi:10.1111/j.1558-5646.2010.01141.x) [DOI] [PubMed] [Google Scholar]
- 34.Ramm S, Edward D, Claydon A, Hammond D, Brownridge P, Hurst J, Beynon R, Stockley P. 2015. Sperm competition risk drives plasticity in seminal fluid composition. BMC Biol. 13, 87 (doi:10.1186/s12915-015-0197-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li S-H, Lee RK-K, Hsiao Y-L, Chen Y-H. 2005. Demonstration of a glycoprotein derived from the Ceacam10 gene in mouse seminal vesicle secretions. Biol. Reprod. 73, 546–553. (doi:10.1095/biolreprod.105.039651) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gray B, Simmons LW. 2013. Acoustic cues alter perceived sperm competition risk in the field cricket Teleogryllus oceanicus. Behav. Ecol. 24, 982–986. (doi:10.1093/beheco/art009) [Google Scholar]
- 37.Thomas ML, Simmons LW. 2008. Male-derived cuticular hydrocarbons signal sperm competition intensity and affect ejaculate expenditure in crickets. Proc. R. Soc. B 276, 383–388. (doi:10.1098/rspb.2008.1206) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thomas ML, Simmons LW. 2007. Male crickets adjust the viability of their sperm in response to female mating status. Am. Nat. 170, 190–195. (doi:10.1086/519404) [DOI] [PubMed] [Google Scholar]
- 39.Simmons LW, Denholm A, Jackson C, Levy E, Madon E. 2007. Male crickets adjust ejaculate quality with both risk and intensity of sperm competition. Biol. Lett. 3, 520–522. (doi:10.1098/rsbl.2007.0328) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.García-González F, Simmons LW. 2005. Sperm viability matters in insect sperm competition. Curr. Biol. 15, 271–275. (doi:10.1016/j.cub.2005.01.032) [DOI] [PubMed] [Google Scholar]
- 41.Simmons LW, Wernham J, García-González F, Kamien D. 2003. Variation in paternity in the field cricket Teleogryllus oceanicus: no detectable influence of sperm numbers or sperm length. Behav. Ecol. 14, 539–545. (doi:10.1093/beheco/arg038) [Google Scholar]
- 42.Simmons LW, Beveridge M. 2011. Seminal fluid affects sperm viability in a cricket. PLoS ONE 6, e17975 (doi:17910.11371/journal.pone.0017975) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Simmons LW, Tan Y-F, Millar AH. 2013. Sperm and seminal fluid proteomes of the field cricket Teleogryllus oceanicus: identification of novel proteins transferred to females at mating. Insect. Mol. Biol. 22, 115–130. (doi:10.1111/imb.12007) [DOI] [PubMed] [Google Scholar]
- 44.Simmons LW, Beveridge M, Li L, Tan Y-F, Millar AH. 2014. Ontogenetic changes in seminal fluid gene expression and the protein composition of cricket seminal fluid. Evol. Dev. 16, 101–109. (doi:10.1111/ede.12068) [DOI] [PubMed] [Google Scholar]
- 45.Dowling DK, Nystrand M, Simmons LW. 2010. Maternal effects, but no good or compatible genes for sperm competitiveness in Australian crickets. Evolution 64, 1257–1266. (doi:10.1111/j.1558-5646.2009.00912.x) [DOI] [PubMed] [Google Scholar]
- 46.Bailey NW, Veltsos P, Tan Y-F, Millar AH, Ritchie MG, Simmons LW. 2013. Tissue-specific transcriptomics in the field cricket Teleogryllus oceanicus. G3: Genes|Genomes|Genetics 3, 225–230. (doi:10.1534/g3.112.004341) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schmittgen TD, Livak KJ. 2008. Analysing real-time PCR data by the comparative CT method. Nat. Prot. 3, 1101–1108. (doi:10.1038/nprot.2008.73) [DOI] [PubMed] [Google Scholar]
- 48.Kearse M, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649. (doi:10.1093/bioinformatics/bts199) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cummings ME, Larkins-Ford J, Reilly CRL, Wong RY, Ramsey M, Hofmann HA. 2008. Sexual and social stimuli elicit rapid and contrasting genomic responses. Proc. R. Soc. B 275, 393–402. (doi:10.1098/rspb.2007.1454) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ramsey M, Maginnis T, Wong R, Brock C, Cummings M. 2012. Identifying context-specific gene profiles of social, reproductive, and mate preference behavior in a fish species with female mate choice. Front. Neurosci. 6, 62 (doi:10.3389/fnins.2012.00062) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mello CV, Vicario DS, Clayton DF. 1992. Song presentation induces gene expression in the songbird forebrain. Proc. Natl Acad. Sci. USA 89, 6818–6822. (doi:10.1073/pnas.89.15.6818) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sockman KW, Gentner TQ, Ball GF. 2002. Recent experience modulates forebrain gene-expression in response to mate-choice cues in European starlings. Proc. R. Soc. Lond. B 269, 2479–2485. (doi:10.1098/rspb.2002.2180) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Avey MT, Phillmore LS, MacDougall-Shackleton SA. 2005. Immediate early gene expression following exposure to acoustic and visual components of courtship in zebra finches. Behav. Brain Res. 165, 247–253. (doi:10.1016/j.bbr.2005.07.002) [DOI] [PubMed] [Google Scholar]
- 54.Lawniczak MKN, Begun DJ. 2004. A genome-wide analysis of courting and mating responses in Drosophila melanogaster females. Genome 47, 900–910. (doi:10.1139/g04-050) [DOI] [PubMed] [Google Scholar]
- 55.Immonen E, Ritchie MG. 2012. The genomic response to courtship song stimulation in female Drosophila melanogaster. Proc. R. Soc. B 279, 1359–1365. (doi:10.1098/rspb.2011.1644) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mukai M, Replogle K, Drnevich J, Wang G, Wacker D, Band M, Clayton DF, Wingfield JC. 2009. Seasonal differences of gene expression profiles in song sparrow (Melospiza melodia) hypothalamus in relation to territorial aggression. PLoS ONE 4, e8182 (doi:10.1371/journal.pone.0008182) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sanogo YO, Band M, Blatti C, Sinha S, Bell AM. 2012. Transcriptional regulation of brain gene expression in response to a territorial intrusion. Proc. R. Soc. B 279, 4929–4938. (doi:10.1098/rspb.2012.2087) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Carney G. 2007. A rapid genome-wide response to Drosophila melanogaster social interactions. BMC Genomics 8, 288 (doi:10.1186/1471-2164-8-288) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ellis LL, Carney GE. 2009. Drosophila melanogaster males respond differently at the behavioural and genome-wide levels to Drosophila melanogaster and Drosophila simulans females. J. Evol. Biol. 22, 2183–2191. (doi:10.1111/j.1420-9101.2009.01834.x) [DOI] [PubMed] [Google Scholar]
- 60.Ellis LL, Carney GE. 2011. Socially-responsive gene expression in male Drosophila melanogaster is influenced by the sex of the interaction partner. Genetics 187, 157–169. (doi:10.1534/genetics.110.122754) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kasumovic MM, Chen Z, Wilkins MR. 2016. Australian black field crickets show changes in neural gene expression associated with socially-induced morphological, life-history, and behavioral plasticity. BMC Genomics 17, 827 (doi:10.1186/s12864-016-3119-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kasumovic MM, Hall MD, Try H, Brooks RC. 2011. The importance of listening: juvenile allocation shifts in response to acoustic cues of the social environment. J. Evol. Biol. 24, 1325–1334. (doi:10.1111/j.1420-9101.2011.02267.x) [DOI] [PubMed] [Google Scholar]
- 63.Kasumovic MM, Hall MD, Brooks RC. 2012. The juvenile social environment introduces variation in the choice and expression of sexually selected traits. Ecol, Evol. 2, 1036–1047. (doi:10.1002/ece3.230) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kasumovic MM, Hall MD, Try H, Brooks RC. 2013. Socially cued developmental plasticity affects condition-dependent trait expression. Behav. Ecol. 24, 429–434. (doi:10.1093/beheco/ars180) [Google Scholar]
- 65.Giansanti MG, Belloni G, Gatti M. 2007. Rab11 is required for membrane trafficking and actomyosin ring constriction in meiotic cytokinesis of Drosophila males. Mol. Biol. Cell 18, 5034–5047. (doi:10.1091/mbc.E07-05-0415) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Polevoy G, Wei H-C, Wong R, Szentpetery Z, Kim YJ, Goldbach P, Steinbach SK, Balla T, Brill JA. 2009. Dual roles for the Drosophila PI 4-kinase Four wheel drive in localizing Rab11 during cytokinesis. J. Cell Biol. 187, 847 (doi:10.1083/jcb.200908107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tiefenböck SK, Baltzer C, Egli NA, Frei C. 2009. The Drosophila PGC-1 homologue Spargel coordinates mitochondrial activity to insulin signalling. EMBO J. 29, 171 (doi:10.1038/emboj.2009.330) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bailey NW, Gray B, Zuk M. 2011. Exposure to sexual signals during rearing increases immune defence in adult field crickets. Biol. Lett. 7, 217–220. (doi:10.1098/rsbl.2010.0659) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Thomas ML, Gray B, Simmons LW. 2011. Male crickets alter the relative expression of cuticular hydrocarbons when exposed to different acoustic environments. Anim. Behav. 82, 49–53. (doi:10.1016/j.anbehav.2011.03.023) [Google Scholar]
- 70.Bailey NW, Gray B, Zuk M. 2010. Acoustic experience shapes alternative mating tactics and reproductive investment in male field crickets. Curr. Biol. 20, 845–849. (doi:10.1016/j.cub.2010.02.063) [DOI] [PubMed] [Google Scholar]
- 71.Andrés JA, Maroja LS, Bogdanowicz SM, Swanson WJ, Harrison RG. 2006. Molecular evolution of seminal proteins in field crickets. Mol. Biol. Evol. 23, 1574–1584. (doi:10.1093/molbev/msl020) [DOI] [PubMed] [Google Scholar]
- 72.Andrés JA, Maroja LS, Harrison RG. 2008. Searching for candidate speciation genes using a proteomic approach: seminal proteins in field crickets. Proc. R. Soc. B 275, 1975–1983. (doi:10.1098/rspb.2008.0423) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Trachte GJ, Meixner K, Ferrario CM, Khosla MC. 1990. Prostaglandin production in response to angiotensin-(1–7) in rabbit isolated vasa deferentia. Prostaglandins 39, 385–394. (doi:10.1016/0090-6980(90)90120-k) [DOI] [PubMed] [Google Scholar]
- 74.Jaiswal N, Diz DI, Chappell MC, Khosla MC, Ferrario CM. 1992. Stimulation of endothelial cell prostaglandin production by angiotensin peptides: characterization of receptors. Hypertension 19(2 Suppl.), II49–II55. (doi:10.1161/01.HYP.19.2_Suppl.II49) [DOI] [PubMed] [Google Scholar]
- 75.Loher W, Edson K. 1973. The effect of mating on egg production and release in the cricket Teleogryllus commodus. Entomol. Exp. Appl. 16, 483–490. (doi:10.1111/j.1570-7458.1973.tb00300.x) [Google Scholar]
- 76.Loher W. 1979. The influence of prostaglandin E2 on oviposition in Teleogryllus commodus. Ent. Exp. Appl. 25, 107–119. (doi:10.1111/j.1570-7458.1979.tb02853.x) [Google Scholar]
- 77.Stanley-Samuelson DW, Loher W. 1983. Arachidonic and other long-chain polyunsaturated fatty acids in spermatophores and spermathecae of Teleogryllus commodus: significance in prostaglandin-mediated reproductive behaviour. J. Insect. Physiol. 29, 41–45. (doi:10.1016/0022-1910(83)90104-X) [Google Scholar]
- 78.Stanley-Samuelson DW, Peloquin JJ, Loher W. 1986. Egg-laying in response to prostaglandin injections in the Australian field cricket, Teleogryllus commodus. Physiol. Entomol. 11, 213–219. (doi:10.1111/j.1365-3032.1986.tb00408.x) [Google Scholar]
- 79.Stanley-Samuelson DW, Jurenka RA, Blomquist GJ, Loher W. 1987. Sexual transfer of prostaglandin precursor in the field cricket Teleogryllus commodus. Physiol. Entomol. 12, 347–354. (doi:10.1111/j.1365-3032.1987.tb00760.x) [Google Scholar]
- 80.Simmons LW. 2001. Sperm competition and its evolutionary consequences in the insects. Princeton, NJ: Princeton University Press. [Google Scholar]
- 81.Simmons LW, Lovegrove M. 2017. Data from: Socially cued seminal fluid gene expression mediates responses in ejaculate quality to sperm competition risk Dryad Digital Repository. (http://dx.doi.org/10.5061/dryad.8rq18) [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Simmons LW, Lovegrove M. 2017. Data from: Socially cued seminal fluid gene expression mediates responses in ejaculate quality to sperm competition risk Dryad Digital Repository. (http://dx.doi.org/10.5061/dryad.8rq18) [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
Data are available at the Dryad Digital Repository (http://dx.doi.org/10.5061/dryad.8rq18) [81].