Abstract
Ion-pair, reverse-phase high performance liquid chromatography (HPLC) is a standard analytical platform for separating, purifying and analyzing RNAs. However, a single-nucleotide resolution by using HPLC is currently limited to RNAs shorter than 25 nucleotides (nt). Here we describe a method of separating three RNA aptamers with 57, 58 and 59 nt on an XBridge ion-pair, reverse-phase HPLC column by a single-nucleotide resolution. Under a similar condition, we also show the capability of our method to resolve two structurally different, yet sequence or mass identical 59-nt aptamers. We establish that the optimal condition to achieve a single-nucleotide resolution correlates to 50 °C and zero magnesium concentration in mobile phases. The ion-pairing agent, the buffer and the solvent we use are also compatible for post-HPLC analysis such as mass spectrometry. Therefore, our method provides a new way of detecting, analyzing and separating RNAs by conformation or structure, and extends the ability of separating RNAs that are longer than 25-nt in length by single-nucleotide resolution.
Keywords: RNA purification and separation, PAGE column, HPLC, single-nucleotide resolution
Separation and purification of RNA is generally required not only for using RNA as potential probes and therapeutics [1], but also for studying the structure and function of RNA, such as RNA catalysis [2; 3; 4] and RNA structure determination by NMR [5] or crystallography [6]. This is because an RNA sample is usually heterogeneous. RNA heterogeneity refers to difference in length or size (i.e., the number of nucleotides or nt), sequence, and alternative but coexisting conformations. The RNA heterogeneity originates directly from sample preparation. An RNA sample can be synthesized chemically (e.g., by solid-phase synthesis) or enzymatically (e.g., by in vitro transcription). For instance, during a transcription reaction, perhaps the most commonly practiced means of preparing RNA samples, T7 RNA polymerase often catalyzes nonspecific addition of extra nucleotides to the ends of a desired RNA without the DNA template, thereby producing heterogeneous RNA molecules [7; 8]. Both chemical and enzymatic reactions for synthesizing RNA can also produce early aborted, shorter RNAs [8; 9]. An RNA sample can be isolated from natural sources. For gene expression analysis, for instance, an RNA sample can be extracted from tissues or cells. The RNAs from such a sample may exist as different sequence variants with the same length, possibly introduced by alternative splicing or posttranscriptional editing [10]. More often, the RNAs from such a sample have a range of sizes, such as a sample that contains regulatory RNAs isolated from bacteria (see a review [11]). However, RNAs even with a subtle difference in sequence could have different functions. For instance, in Arabidopsis, both 21-nt and 22-nt microRNAs (miRNA) bind to Argonaute-1 (AGO1) and guide accurate cleavage of target transcripts. Yet, only the 22-nt miRNA-AGO1 complexes are competent to trigger RNA-dependent RNA polymerase 6 mediated siRNA production from a target RNA [12]. Recently, it is shown that the asymmetry in the duplex structure of the 22-nt miRNAs is responsible for triggering the production of secondary siRNAs [13]. Furthermore, RNA with a single, chemically identical sequence could fold into more than one conformation with distinct function [14], and these conformations may not be interconvertible [14; 15; 16]. Therefore, for characterizing the structure and function of RNA, separating and purifying RNA from a mixture to sequence and structure homogeneity is necessary.
Through an ideal separation process, every RNA molecule in an RNA sample should be chemically identical in that the number of the nucleotides in an RNA, the sequence and the chemical identity of the 5ʹ-end and the 3ʹ-end are all the same. To achieve this goal, one of the most technically challenging tasks is to separate and purify RNAs which may differ by a single nucleotide or by an average molecular weight of 321 Dalton (Da) in a single-stranded RNA [17]. A separation based on a single nucleotide difference becomes more difficult when the length of an RNA molecule becomes longer, because the average molecular weight contribution of a single nucleotide relative to its overall molecular weight becomes smaller. Furthermore, a longer RNA tends to have more complex secondary structures [18], and those structures can still remain even at higher temperature [19; 20].
The two most widely used separation and purification techniques for RNAs (and DNAs as well as proteins) are polyacrylamide gel electrophoresis (PAGE) and high-performance liquid chromatography (HPLC). Although PAGE or precisely denaturing PAGE is the principal technique for purifying RNA [21; 22], HPLC, especially ion-pair, reverse-phase HPLC, is preferred. The latter technique is less time consuming, and is more suitable for high throughput analysis [20]. To date, ion-pair, reverse-phase HPLC is a standard platform for separation and quantification of a wide range of labeled/unlabeled, regular/modified RNAs [23], such as siRNAs [24; 25; 26], mRNA [27], and large ribosomal RNA (rRNA) [28]. However, HPLC can be used to separate short RNAs (~20 nt) by single-nucleotide resolution [29; 30; 31]. Separation and purification of single-stranded RNAs up to 1000 nt has been demonstrated but the best resolution is limited to 100 nt fragments [20]. Separation of longer RNAs such as 18S and 28S rRNA (1869 and 5035 nt respectively) is possible but the separation is no longer size dependent [20]. Conformational flexibility of RNAs and sequence compositions have been major sources of difficulty to separate RNA molecules longer than 25 nt with single-nucleotide resolution [32; 33]. An untested hypothesis is whether RNAs longer than 25 nt in length, presumably possessing secondary structures, can be resolved by single nucleotide, using HPLC.
Here we describe a method by which 57–59 nt RNAs can be individually resolved using an ion-pair, reverse-phase HPLC. These samples are all chemically synthesized. To test our ability of resolving RNAs based on conformational flexibility, we also use the enzymatic transcript of the 59 nt RNA. This 59 nt RNA is a potent aptamer or an inhibitor of the -amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptor, a subtype of the ionotropic glutamate receptor family [14]. The 59 nt aptamer, generated by enzymatic transcription, exists in two, non-convertible conformations or structures with identical sequences, which we term AN59-M1 and AN59-M2. The two RNA species must be used together for inhibiting the receptor [14]. The chemically synthesized 59 nt RNA aptamer is a potent inhibitor as well but folds into a structure different from either of the two enzymatic transcripts [14]. Using AN59-M1 and AN59-M2), we show that our method further allows us to resolve the two, structurally different, sequence identical RNAs, using HPLC. In this study, we also use a tubular, continuous-elution PAGE apparatus to purify the AN59-M1 and AN59-M2 under native condition. In fact, the use of HPLC serves also as an analytical tool to verify the quality of the separation of the 59 nt RNA by this tubular PAGE.
Materials and methods
RNA sample preparation
SynAN57-59 RNAs (see Fig. 1) were purchased from Trilink Biotechnology. They were purified by HPLC to remove any contaminants or shorter products before our experiment. AN59-M1 and AN59-M2 were made by in vitro enzymatic transcription using an AN59-glmS DNA template [14], which was constructed to produce a homogenous 3′-end by utilizing the self-catalysis function of the downstream glmS ribozyme [34]. Ambion MEGAshortscript T7 kit [14] was used for enzymatic transcription. The transcription product was incubated with 1 mM glucosamine-6-phosphate, a metabolite that accelerates the cleavage reaction, for 2 hours in a buffer containing 50 mM Tris-HCl (pH 7.4), 10 mM MgCl2 and 200 mM KCl. An LC-ESI (electrospray ionization)-MS experiment was performed on an ESI-Q-TOF II mass spectrometer (Micromass/Waters) to verify that the AN59-M1 and AN59-M2 had identical mass or length [14].
Fig. 1.
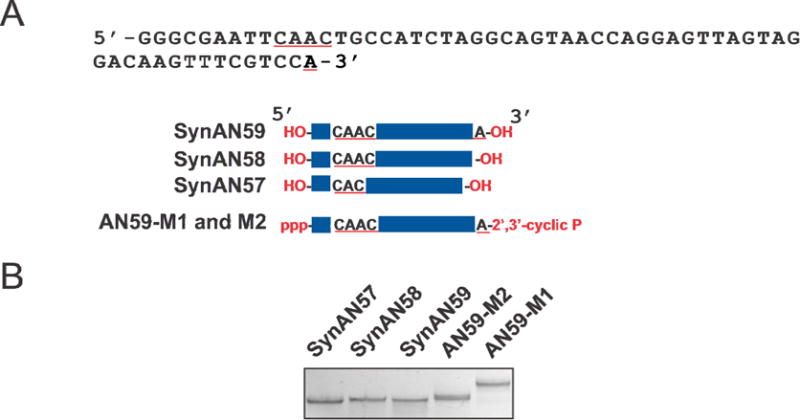
(A) There were five RNAs used in this study, and the longest was a 59-nt RNA termed AN59 whose sequence is shown. Among the five, three were prepared by solid state synthesis, i.e., SynAN57, SynAN58 and SynAN59. Both the 5ʹ and 3ʹ ends of these RNAs have a hydroxyl group. As shown, SynAN58 was constructed without the last nucleotide or the “A” at the 3ʹ end, as compared with SynAN59. SynAN57 was missing another ‘A’ located in the middle of the sequence, as underlined in red. All of the three were inhibitors of AMPA receptors. AN59-M1 and AN59-M2 were two enzymatic transcripts generated from the same transcription reaction. As shown, AN59-M1 and AN59-M2 had identical sequence, length and the 5ʹ as well as the 3ʹ end groups; yet they had different structures. AN59-M1 and AN59-M2 must be used as a pair to inhibit the AMPA receptor channels (see Methods). It should be also noted that AN58-M1 and AN58-M2, lacking the last “A” as shown in the sequence, can be also made, and AN58-M1 and AN58-M2 as a pair are also biologically active. (B) All five RNAs showed different electrophoretic mobility on a 10% native polyacrylamide gel.
Cylindrical polyacrylamide gel preparation
To purify AN59-M1 and AN59-M2, we used a Bio-Rad Prep Cell (Model 491), a continuous-elution PAGE apparatus. The gel casting unit was a 12 cm long, concentric cylinder with outer and inner shell diameter of 37 mm and 28 mm, respectively. Once cast, a tubular PAGE gel had an 8.2 cm2 flat surface. Under the elution chamber was a dialysis membrane with a molecular weight cut off of 5,000 Da. To prepare a 12% tubular PAGE gel, 100 ml of 12% acrylamide/bis-acrylamide (37.5:1) solution in 1× Tris-Borate-EDTA (TBE) buffer were prepared from 30% ProtoGel premix stock (National Diagnostics) at room temperature. The mixture was filtered and degased. Then, 125 μl of 10% ammonium persulfate and 50 μl of tetramethylethylenediamine (TEMED) were added to the gel mixture before it was poured into the Prep Cell tube. Finally, n-butanol was carefully loaded to form a layer in order to barricade the gel from air. The gel was allowed to polymerize over night at room temperature before being assembled into the electrophoresis apparatus. Note that the surface of a tubular gel should be flat in order to achieve high quality separation.
Use of Prep Cell for RNA purification
For separating AN59-M1 from AN59-M2, we loaded ~640 μg of AN59 transcripts dissolved in 1 ml of loading buffer containing 5% glycerol. The electrophoresis was run at 150 V for 20 hours. A peristaltic pump was used to regulate the mobile phase or 1× TBE buffer at 1 ml/minute flow rate. The RNA elution was monitored with a UV detector, and the fractions were collected with a fraction collector (BioFrac, Bio-Rad) at 4 ml/fraction. The fractions were then pooled based on the chromatography trace and concentrated in an Amicon filtration centrifuge tube (3 kDa cut-off, Millipore). The TBE buffer in the eluted samples was exchanged with 10 mM Tris-HCl buffer by spinning in an Amicon filtration tube; the same procedure was repeated two more times. The concentration of the collected sample was determined with a Nanodrop 1000 spectrophotometer (Thermal Fisher Scientific).
HPLC hardware
An XBridge C18 column (Waters, 4.6 × 150 mm dimension, 3.5 μm pore size) mounted on a Waters Breeze HPLC system was used for RNA separation and purification. The column was embedded in the chamber of a Waters Temperature Control Module. The injection loop was 25 μl. A 2487 UV/Vis dual wavelength detector (Waters), set at 254 nm, was used for monitoring elution. Waters Breeze software was used to run all HPLC elution protocols.
RNA separation by ion-pair reverse-phase HPLC
An ion-pair agent was added to the mobile phase to form nonpolar ion pairs between the ion-pairing agent and the negatively charged phosphate backbone of RNA, which led to subsequent adsorption to the neutral, nonpolar surface of the column. Specifically, we used triethylamine (TEA) (HPLC grade, Fluka) (16.3 mM, final concentration) as the ion-pair agent. TEA is shown to have a fast equilibration time with the column [35]. Furthermor 400 mM 1,1,1,3,3,3-hexafluoro-2-propanol (HFIP) (99+%, Sigma) and methanol (see concentration below), the two organic modifiers, were included [33]. The pH of all of the mobile phases was 7.9.
Mobile phase A was the TEA/HFIP buffer and the concentration of TEA and HFIP were described above. Mobile phase B contained 50% methanol in TEA/HFIP buffer. The HPLC column was maintained at 50 °C for all of the purification protocols, unless otherwise noted. Different concentrations of methanol were made in-line by controlling, through the Breeze software, the amount of phase A and phase B flowing through the column. The column was initialized by using 20 ml of 100% acetonitrile (HPLC grade, Pharmco), followed by 20 ml of water (purified from Barnstead, Nanopure Diamond Water Purification System) and then 50 ml of phase A before sample loading. The flow rate was 1 ml/min unless noted otherwise. In general, 1–10 μg of sample was prepared in 25 μl (final volume) of phase A solution for each run and heated to 50 °C before loading. After loading, the sample on the column was washed with 5 ml of 100% phase A and then 15 ml of 70% phase A/30% phase B mixture. A linear gradient elution usually started from 30% phase B and ended at 60%, unless noted otherwise. The flow rate for sample separation was maintained at 0.5 ml/min.
Results
RNA samples
To test our ability of separating RNAs that are longer than 25 nt with single-nucleotide resolution, we used SynAN57, SynAN58 and SynAN59 (Fig. 1A). Due to the chemistry of solid-state RNA synthesis [36], all SynAN57-59 RNAs contained hydroxyl groups at both 5ʹ and 3ʹ ends. We also used AN59-M1 and AN59-M2, which were structurally different but sequence identical [14]. Because AN59-M1 and AN59-M2 were generated from transcription but enzymatically cleaved by the glmS ribozyme whose DNA sequence was linked to the 3ʹ end of the AN59 aptamer sequence [14], AN59-M1 and AN59-M2 had the identical length, as shown by the identical mass/charge ratio observed in mass spectroscopy [14]. However, the ribozyme cleavage reaction generated a 2ʹ,3ʹ-cyclic phosphate group at the 3ʹ end and a triphosphate group at the 5ʹ end of the two RNAs [34] (Fig. 1A). Consequently, AN59-M1 and AN59-M2 had different 5ʹ and 3ʹ ends from SynAN59, although the number of nucleotides and the sequence were all identical.
By electrophoresis on a native 10% polyacrylamide slab gel, the three chemically synthesized 57, 58, and 59 nt long RNAs differing by a single nucleotide could be hardly resolved (Fig. 1B). On the other hand, the electrophoretic mobilities of the 59-nt long AN59-M1 and AN59-M2 RNAs were clearly different, and neither one was running with the same electrophoretic mobility as SynAN59 (Fig. 1B). As we reported earlier, the difference in the electrophoretic mobility between AN59-M1 and AN59-M2 (Fig. 1B) was attributed to the difference in their structures [14]. Because SynAN57-59 differ by either one or two nucleotides and AN59-M1/M2 differ by their structures, the use of these five RNA samples would enable us to test (i) whether RNAs of up to 59 nt in length could be separated with single-nucleotide resolution and (ii) whether two different RNA folds with the same length (i.e., 59 nt) and the same molecular weight could be resolved.
Separation of SynAN57, SynAN58, and SynAN59 by ion-pair reverse-phase HPLC
To explore whether the 57–59 nt RNAs (Fig. 1A) could be resolved individually, we selected a recently developed XBridge C18 reverse-phase column. This column has not been shown to resolve a single nucleotide difference for RNAs of this size range. By trial and error, we established the optimal condition to separate the RNA samples as described above. The elution from an XBridge C18 reverse phase column revealed a distinct retention time for each of the three RNA species (Fig. 2A). When the three RNAs were mixed and run together, they were individually resolved (Fig. 2B). The three peaks that ran individually, as in Fig. 2A, were eluted between 41.2% and 44.4% of phase B, which correspond to 20.6% and 22.2% of methanol, respectively (see the gradient profile in Fig. 2A). The gradient was applied at 20 min of run time with a rate of 0.24% of phase B/minute (Fig. 2B), but the rate decreased to 0.08% of phase B/minute when phase B reached 40%. It should be noted that this optimal separation protocol was achieved at a temperature of 50 ± 0.1 °C (experimentally, all the samples were pre-heated through a Waters Temperature Module before entering the XBridge column, and the column itself was also embedded in the chamber of the same temperature module).
Fig. 2.
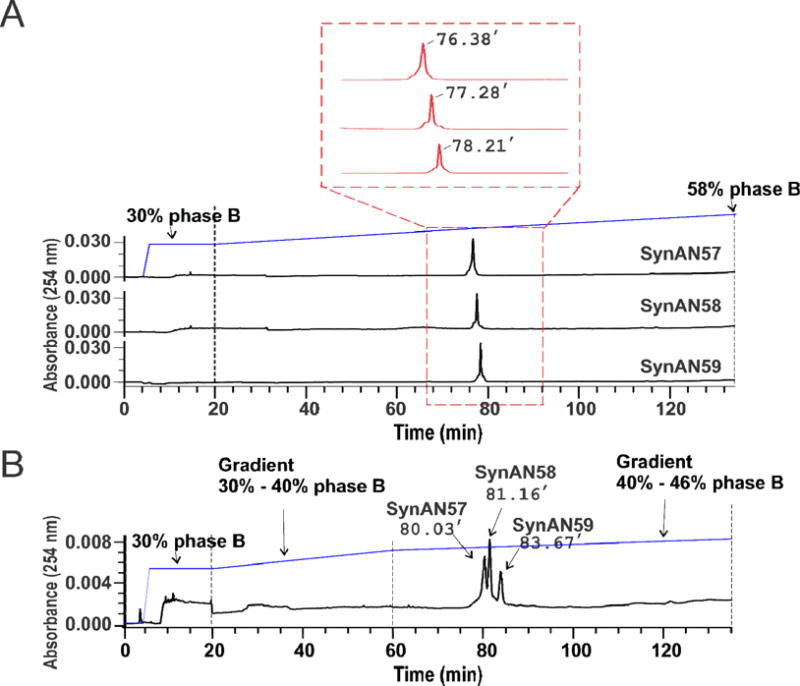
Single-nucleotide separation of RNAs up to 59 nt by HPLC. (A) Individual chromatogram for SynAN57, SynAN58 and SynAN59. Each sample was run through an XBridge C18 reverse-phase column at 50 °C (see detail in Methods). The middle section framed by the red box shows a detailed peak profile and retention times. The running buffer contained 16.3 mM of TEA and 400 mM of HFIP at pH 7.9 with 5% methanol (Phase A) or 30% methanol (Phase B) (see Methods). The gradient started at 30% of Phase B and ended at 58% of Phase B in 115 minutes. Unless otherwise noted, the flow rate of all experiments was kept at 0.5 ml/minute and the temperature of column was maintained at 50 °C. (B) An equal molar SynAN57, SynAN58, and SynAN59 were mixed and run through the same XBridge column. The retention times of three RNAs, from 57-nt to 59-nt, are marked. For elution, two sections of linear gradient were used to improve the separation. The first linear gradient started at 30% of Phase B and ended at 40% in 40 minutes, and the second linear gradient began at 40% of Phase B and ended at 46% of Phase B in 75 minutes. The flow rate was kept at 0.5 ml/minute.
Preparative separation of AN59-M1 from AN59-M2 RNAs by electrophoresis on a cylindrical polyacrylamide gel
We further explored whether two structurally different but sequence identical RNAs could be separated using ion-pair, reverse-phase HPLC. To do this, we needed to prepare pure AN59-M1 and AN59-M2 samples. AN59-M1/M2 could be separated by electrophoresis on a native polyacrylamide slab gel (Fig. 1B). However, to develop a large-scale purification of the same RNA species, we used a continuous-elution PAGE apparatus or Prep Cell (see detail in Methods). In a single run on a 12% native PAGE, 640 μg of AN59 transcripts in 1 mL (final volume) was loaded for separation of AN59-M1 from AN59-M2 (Fig. 3A). As expected, two peaks were revealed in the elution profile of AN59 enzymatic transcripts (Fig. 3A). Peak 1 (fractions 27 to 40), corresponded to AN59-M2, whereas peak 2 (fractions 42–49) corresponded to AN59-M1. We confirmed these peak identities by analytical PAGE of the pooled fractions (Fig. 3B).
Fig. 3.
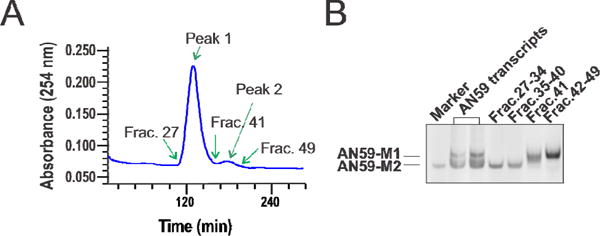
(A) A transcription reaction mixture containing AN59-M1 and AN59-M2 was purified on a preparative PAGE column. The electrophoresis was run at 150V for 20 hours. The elution was driven by a peristaltic pump at a speed of 1 ml/minute, and was pooled at 4 ml/fraction, as monitored by UV absorption (see Methods). (B) T ractions containing AN59-M1 and AN59-M2 were examined on a 10% native PAGE. The “marker” on the gel was an AN59-M2 sample previously purified using a slab PAGE gel.
We also determined the overall recovery yield of the PAGE purification using the Prep Cell. For this experiment, we used three RNA samples, i.e., AN59-M1/AN59-M2 (as shown in the Fig. 3B), SynAN57 and a 96-nt RNA, which is a sequence-unrelated, structurally different RNA molecule (see its sequence in Supplementary Figure S1). Each sample was first purified and then used for yield determination. For a single run, we loaded 600 μg – 1 mg of RNA to the PAGE gel. The amount recovered from the PAGE elution for each RNA sample was first pooled and concentrated with an Amicon filtration centrifuge tube, and then quantified by the UV-Vis measurement. Based on a duplicate run of each of the three samples, we found that the averaged recovery rate of using this preparative PAGE purification method was 78 ± 10%. It should be noted that the yield represents the percentage of the overall recovery, which included gel running, fraction collection and sample concentration.
Effect of magnesium ion on the separation of AN59-M1 and AN59-M2
To date, no method has been reported that uses HPLC to resolve any RNAs of the same length, but longer than 25 nt, solely based on structure [24]. In attempts to separate AN59-M1 from AN59-M2 on HPLC, we first tested the effect of magnesium concentration during loading of the sample. Mg2+ is one of the key factors that affect the folding of an RNA molecule [37]. A potential difference in folding in the presence of Mg2+ ion between the two RNA molecules was expected to give rise to difference in hydrophobic interaction of the two RNA molecules with the stationary phase, thereby generating different retention times. Using AN59-M2 as an example, we observed (Fig. 4) that presence of Mg2+ during loading significantly affected the elution profile and retention time of AN59-M2 (Mg2+ was added only during loading, and neither of the mobile phases A and B contained any Mg2+; furthermore, the AN59-M2 stock was in the Phase A buffer). Specifically, as Mg2+ concentration increased, the peak intensity at 88 min retention time was reduced (Fig. 4) while the UV absorption increased in a more hydrophobic region (>190 min). This result was consistent with the hypothesis that a high Mg2+ concentration promoted RNA folding [37; 38]. Therefore the reduction of the UV absorbance at the earlier elution peak (~88 min) and the concurrent rise of a broad peak at a later time (>190 min) could be attributed to the hypochromic effect [39] in that the RNA species at a higher Mg2+ concentration became more folded and more hydrophobic. Hydrophobic properties of RNAs are generally associated with their secondary structures driven by base stacking [40]. Consequently, these RNAs absorbed to the hydrophobic stationary phase of XBridge C18 column more strongly, therefore exhibiting longer retention times. Based on this experiment, we concluded that not using any Mg2+ ions during sample preparation, loading and running was the best condition for separating AN59-M1 from AN59-M2. Under this condition, virtually all AN59-M2 molecules were de-absorbed from the column at approximately the same time, which allowed us to follow a major and sharper elution peak (Fig. 4).
Fig. 4.
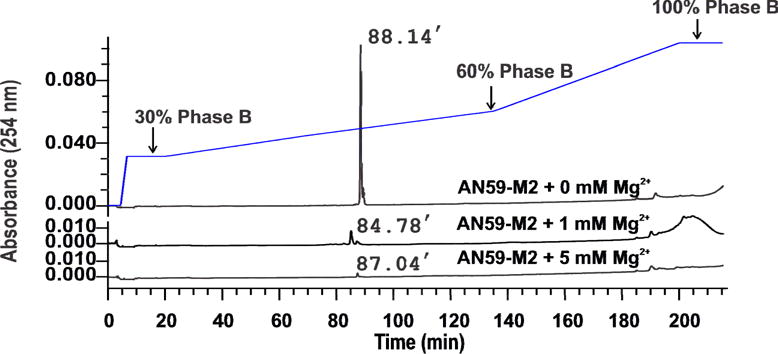
Effect of Mg2+ concentration, i.e., 0 mM, 1 mM and 5 mM as labeled, on the elution of AN59-M2. A sample was prepared in Phase A buffer and was heated at 50 °C for 10 minutes before injection. The linear gradient contained two sections. The first section was from 30% to 60% Phase B in 115 minutes. The second section was from 60% to 100% Phase B in 65 minutes. The flow rate was kept at 0.5 ml/minute.
Effect of temperature on the separation of AN59-M1 and AN59-M2
For separating AN59-M1 from AN59-M2 on an XBridge C18 column, we further investigated the effect of temperature on elution. As shown (Fig. 5), there were two distinctive features in the chromatograms obtained from 35 °C to 65 °C. First, AN59-M1 and AN59-M2 exhibited a major peak at 103.92 min and 103.96 min, respectively, at 35 °C (Fig. 5, the top two panels or the pair of blue and red traces). However, as temperature increased, these two main peaks began to shift leftwards and the two species emerged from the column with shorter retention times (see the panels from top to bottom in Fig. 5). Second, AN59-M2 showed two characteristic peaks (a major, initial peak and a minor, trailing peak). In contrast, AN59-M1 displayed a complex elution profile (e.g., a main peak followed by a series of small peaks at higher retention time); yet the profile became much simpler at higher temperature. These phenomena could be explained on the basis that higher temperature favored a less ordered or less packed RNA structures. Similar to the effect of Mg2+ concentration on the hydrophobicity of an RNA, temperature is also known to affect the hydrophobicity of an RNA in that increasing temperature is expected to render an RNA less ordered or partially denatured. Thus, from 50 °C and higher we were able to elute AN59-M1 or AN59-M2 essentially in a single peak. The fact that AN59-M1 and AN59-M2 each yielded a single band on a polyacrylamide slab gel suggested these multiple peaks, observed in the HPLC chromatograms (as in the red traces of Fig. 5), were most likely due to multiple conformations associated with AN59-M1 rather than some unspecified impurities (Fig. 5). More importantly, those multiple, broad peaks were all associated with low temperature conditions, where more ordered or highly packed structures were favored. These results suggested that AN59-M1 was more hydrophobic overall than AN59-M2 (Fig. 5). Such a conclusion is also consistent with the notion that even at an elevated temperature, RNA secondary structures can still exist [20]. In fact, the presence of secondary structures makes the size-dependent separation of RNA, particularly a large RNA, challenging.
Fig. 5.
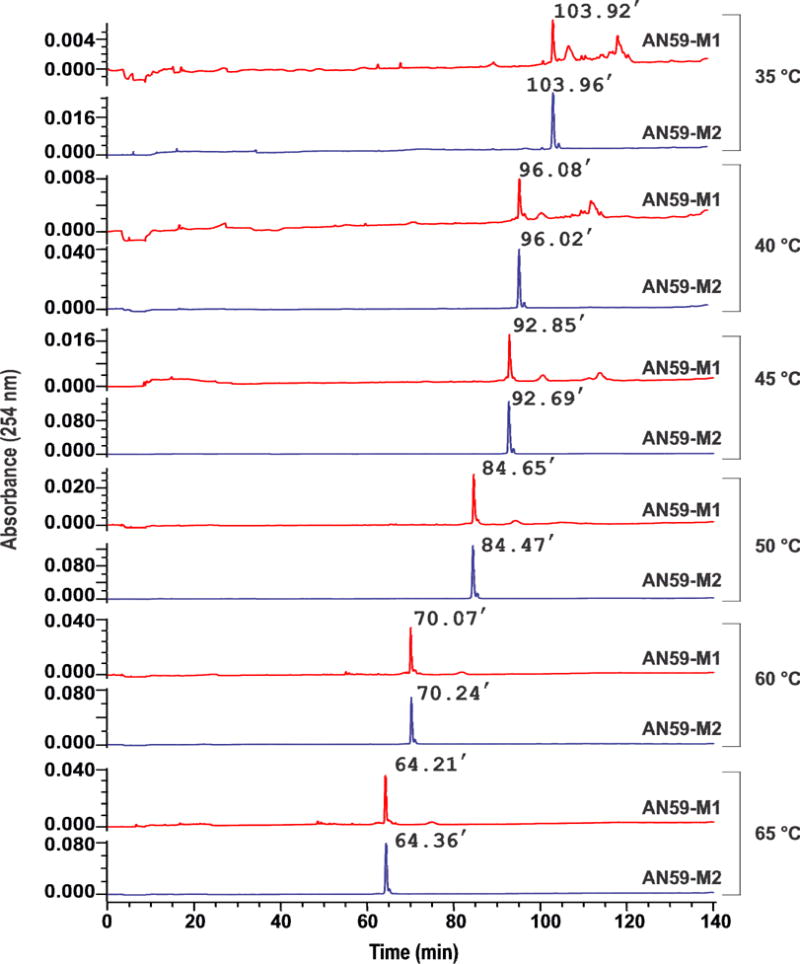
Effect of temperature on the elution of AN59-M1 and AN59-M2 on an XBridge column. The gradient started at 30% of Phase B and ended at 58% of Phase B in 115 minutes. The Mg2+ concentration was zero in all of the experiments.
Using both in-line probing and the selective 2ʹ-hydroxyl acylation analyzed by primer extension (SHAPE) experiments, we have previously shown that both the 58 nt and 59 nt M1 species or AN59-M1/AN58-M1 are indeed more packed than AN59-M2/AN58-M2 [14]. Furthermore, the shift from a more complex elution profile of AN59-M1 at lower tem perature to a simpler profile at higher temperature indicated that there was equilibrium among multiple conformations in the same structural repertoire [14]. In addition, the intensity of absorbance for the main peak associated with AN59-M1 (i.e., the main red peak in Fig. 5) increased with the increase of temperature. The AN59-M2 peak (i.e., the blue peak) also increased, albeit to a lesser extent.
Separation of SynAN59 and AN59-M2 by HPLC
SynAN59 and AN59-M2 have the same sequence and the same number of nucleotides, but differ in the chemical nature of the 5ʹ and 3ʹ ends. Specifically, AN59-M2 has a triphosphate group at the 5ʹ end a cyclic phosphate group at its 3ʹ end. In contrast, SynAN59 has a hydroxyl group at both ends (Fig. 1A). Consequently, AN59-M2 is 302 Da heavier than SynAN59. The difference in molecular weight is almost equal to one C (305 Da) or U (306 Da). Therefore, as a control experiment for single-nucleotide resolution, we further tested if AN59-M2 and SynAN59 could be individually resolved. Using two clean samples as visualized on a PAGE gel (Fig. 6A), we ran the two samples separately (the two upper panels of Fig. 6B) or together (the lower panel of Fig. 6B). It can be seen that SynAN59 and AN59-M2 RNAs were separated clearly from one another. This result (Fig. 6) further demonstrated that the condition we established enabled us to separate RNAs up to 59 nt with single-nucleotide resolution using an XBridge reverse-phase column. The separation of SynAN59 from AN59-M2 was most likely based on the difference in the molecular weight due to the different nature of both the 5ʹ and 3ʹ ends. This is because the difference in retention time between SynAN59 and slightly heavier AN59-M2 is about 1 min (or 78.25 and 79.24 min, respectively, in Fig. 6B). This difference is similar to the relative retention time between SynAN57/SynAN58 and SynAN58/SynAN59 (Fig. 2A). It is also possible that the extra phosphates in AN59-M2 bind to TEA in the buffer, which makes the RNA more hydrophobic and de-absorbs later.
Fig. 6.
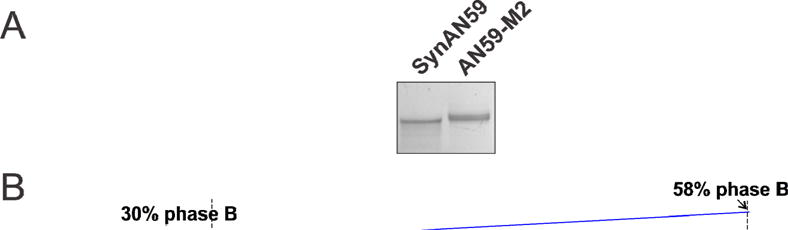
(A) Purified SynAN59 and AN59-M2 were visualized on a 10% native polyacrylamide gel. (B) SynAN59 and AN59-M2 were run through the XBridge column individually (the top two panels in blue color) and in mixture (the bottom trace in red). One linear gradient was used for elution, and it started at 30% of Phase B and ended at 58% of Phase B in 115 minutes.
Discussion
The major aim of our study is to develop a chromatographic method, with the use of ion-pair, reverse-phase HPLC, by which single-nucleotide resolution can be achieved for RNAs with the length up to 59 nt. By the same method, we also aim to resolve conformational difference between the two 59 nt RNA molecules with identical sequence and molecular weight where size-based separation is no longer possible. Using a combination of five RNA samples, we show that these aims have been achieved. The method we have established provides a new way of detecting, analyzing and separating RNAs by conformation or structure, and extends the ability of separating RNAs, by single-nucleotide resolution, that are longer than the 25 nt mark currently achievable by the use of ion-pair, reverse phase HPLC [25; 33; 41].
The samples we have chosen for our method development are RNA aptamers with known function. SynAN58 and SynAN59 and their enzymatic counterparts, AN58-M1/M2 and AN59-M1/M2, together with SynAN57, are all biologically active - they are all potent antagonists of AMPA ion channel receptors [14]. Their inhibitory properties allowed us to check whether a purification procedure destroyed their function. As expected, our RNA samples remained biologically active after purification (data not shown). Furthermore, the size of our samples, i.e., 57–59 nt, is in the range of other RNAs of therapeutic values. In fact, most RNA aptamers, reported thus far, fall into the 5–15 kDa molecular mass range, which corresponds to 15–50 nt in length [42]. Therefore, our method may be useful for identification, purification and characterization of other RNA aptamers.
A separation of RNAs up to 59 nt in length with single-nucleotide resolution has not been previously possible, and the method we have established to accomplish this separation relies on the use of an XBridge C18 reverse-phase column. During this work, we also tested a conventional Waters XTerra C18 column. Although a single-nucleotide separation has also been achieved (data not shown) with the XTerra column, the XBridge column is preferred because it has a longer lifetime. It should be also noted that the buffer we use contains TEA/HFIP (TEA is also an ion-pair agent). TEA/HFIP is a widely used buffer in the analysis of oligonucleotides using HPLC [24; 43; 44]. The use of HFIP is particularly desirable because it is compatible with post-HPLC analysis of RNA species using ESI-MS [24; 43; 44]. Furthermore, HFIP is known to reduce the impact of oligonucleotide hydrophobicity upon retention [33]. Therefore, our buffer and mobile phase components are fully compatible with post-HPLC analysis techniques previously developed with shorter RNAs.
Using ion-pair, reversed-phase HPLC, previous studies show that RNAs can be separated under non-denaturing, partially denaturing, or fully denaturing conditions [23; 28]. We believe that the separation method we present here is a partially denaturing protocol despite the fact that we used no Mg2+ ions and our separation was at elevated temperature, both of which correlated to more of a denatured condition. This is because AN59-M1 and AN59-M2 differ by structure, rather than by length, as previously shown by the LC-ESI-MS experiment [14].
There are limitations in using the method we present here. To achieve the single-nucleotide resolution, we have to use a shallower gradient (Fig. 2), which correlates to a longer run time as compared with existing protocols for smaller RNAs [20]. A steeper or faster gradient program would be possible but only at the expense of a lower resolution. However, a faster elution program can be used satisfactorily if the mixture to be separated is not complex, the peak-to-peak distance between individual RNA pieces is far apart [20] or a single-nucleotide resolution is not required. Similarly, a single-nucleotide resolution also limits the loading capacity on an XBridge C18 reverse-phase column for the purpose of purifying individual RNAs. In fact, when we gradually increase the loading capacity to more than 10 μg per RNA species for SynAN57–59, the separation of one peak from the other becomes less complete (Fig. 3; see additional data in Supplemental Figure 2). Yet even with a higher loading capacity, SynAN57–59 can still be resolved individually (see Supplementary Figure S2). Therefore, our method can be best used for RNA detection with single-nucleotide resolution, rather than for large scale RNA purification. However, a larger loading capacity can be used when an RNA sample is less complex or a single-nucleotide resolution is not required. Alternatively, an RNA sample with the same length can be prepared in large scale so that a stringent separation to achieve single-nucleotide resolution may not be necessary. As we have shown, an RNA sample can be prepared in a pure form by making a plasmid construct that contains the glmS ribozyme sequence [14] for in vitro transcription. This way, a clean RNA can be produced through a post-transcriptional cleavage by the ribozyme.
The second method we used is electrophoresis on a cylindrical polyacrylamide gel to purify two size-identical, yet structurally different RNAs (i.e., AN59-M1 and AN59-M2). Currently, there are two electrophoretic approaches for RNA separation and purification: slab gel and cylindrical (column) gel electrophoresis. The theory of the two electrophoretic operations is identical in that a gel suppresses thermal convection caused by application of an electric field, and also acts as a sieving medium, retarding nucleic acids and thus separating them due to their charges (and shapes/charges in native gel conditions). Practically, however, electrophoresis on a cylindrical polyacrylamide gel may be more advantageous as a preparative method for large scale production of a pure RNA. Using cylindrical PAGE, RNA can be separated by collecting the fraction directly from a detector/eluent. In contrast, the use of polyacrylamide slab gel purification requires an additional recovery step, often known as “crush and soak method” [22]. It should be noted, however, that column electrophoresis does not replace a slab gel. Slab gels are easier to cast and run. Multiple samples can be run simultaneously in separate lanes under the same condition to avoid sample variation. A slab gel further enables one to continue with blot analyses and/or autoradiographic analysis. However, a cylindrical polyacrylamide gel has a larger loading capacity or higher throughput (weight/time). Once a gel is properly cast, it can also be used 3–4 times. In this study, we show that even two closely running, structurally different but sequence identical RNA species can be cleanly resolved using a cylindrical polyacrylamide gel. An overall recovery yield of ~80% in our experiment is similar to the value reported by others by using either cylindrical PAGE [45] or conventional slab PAGE [46].
Supplementary Material
Acknowledgments
We thank Drs Cougzhou Wang, Yan Han and Weimin Pei in our laboratory for testing the inhibitory function of these RNA aptamers during the course of this work. We are grateful to the anonymous reviewers for their helpful comments and suggestions for the manuscript. This work was supported by grants from Department of Defense (to L.N.), National Institutes of Health (to L.N.), and Muscular Dystrophy Association (to L.N.) and a postdoctoral fellowship from Muscular Dystrophy Association (to Z.H.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Subject category: RNA chromatography
References
- 1.Fotin-Mleczek M, Duchardt KM, Lorenz C, Pfeiffer R, Ojkic-Zrna S, Probst J, Kallen KJ. Messenger RNA-based vaccines with dual activity induce balanced TLR-7 dependent adaptive immune responses and provide antitumor activity. Journal of immunotherapy. 2011;34:1–15. doi: 10.1097/CJI.0b013e3181f7dbe8. [DOI] [PubMed] [Google Scholar]
- 2.Fedor MJ, Williamson JR. The catalytic diversity of RNAs. Nature reviews Molecular cell biology. 2005;6:399–412. doi: 10.1038/nrm1647. [DOI] [PubMed] [Google Scholar]
- 3.Cech TR, Zaug AJ, Grabowski PJ. In vitro splicing of the ribosomal RNA precursor of Tetrahymena: involvement of a guanosine nucleotide in the excision of the intervening sequence. Cell. 1981;27:487–96. doi: 10.1016/0092-8674(81)90390-1. [DOI] [PubMed] [Google Scholar]
- 4.Guerrier-Takada C, Gardiner K, Marsh T, Pace N, Altman S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell. 1983;35:849–57. doi: 10.1016/0092-8674(83)90117-4. [DOI] [PubMed] [Google Scholar]
- 5.Thakur CS, Sama JN, Jackson ME, Chen B, Dayie TK. Selective 13C labeling of nucleotides for large RNA NMR spectroscopy using an E. coli strain disabled in the TCA cycle. Journal of biomolecular NMR. 2010;48:179–92. doi: 10.1007/s10858-010-9454-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pallan PS, Egli M. Selenium modification of nucleic acids: preparation of oligonucleotides with incorporated 2ʹ-SeMe-uridine for crystallographic phasing of nucleic acid structures. Nature protocols. 2007;2:647–51. doi: 10.1038/nprot.2007.75. [DOI] [PubMed] [Google Scholar]
- 7.Sastry SS, Ross BM. Nuclease activity of T7 RNA polymerase and the heterogeneity of transcription elongation complexes. J Biol Chem. 1997;272:8644–52. doi: 10.1074/jbc.272.13.8644. [DOI] [PubMed] [Google Scholar]
- 8.Milligan JF, Groebe DR, Witherell GW, Uhlenbeck OC. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic acids research. 1987;15:8783–98. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scaringe SA, Wincott FE, Caruthers MH. Novel RNA Synthesis Method Using 5ʹ-O-Silyl-2ʹ-O-orthoester Protecting Groups. J Am Chem Soc. 1998;120:11820–11821. [Google Scholar]
- 10.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 11.Waters LS, Storz G. Regulatory RNAs in bacteria. Cell. 2009;136:615–28. doi: 10.1016/j.cell.2009.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cuperus JT, Carbonell A, Fahlgren N, Garcia-Ruiz H, Burke RT, Takeda A, Sullivan CM, Gilbert SD, Montgomery TA, Carrington JC. Unique functionality of 22-nt miRNAs in triggering RDR6-dependent siRNA biogenesis from target transcripts in Arabidopsis. Nat Struct Mol Biol. 2010;17:997–1003. doi: 10.1038/nsmb.1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Manavella PA, Koenig D, Weigel D. Plant secondary siRNA production determined by microRNA-duplex structure. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:2461–6. doi: 10.1073/pnas.1200169109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang Z, Pei W, Han Y, Jayaseelan S, Shekhtman A, Shi H, Niu L. One RNA aptamer sequence, two structures: a collaborating pair that inhibits AMPA receptors. Nucleic Acids Res. 2009;37:4022–32. doi: 10.1093/nar/gkp284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Solomatin SV, Greenfeld M, Chu S, Herschlag D. Multiple native states reveal persistent ruggedness of an RNA folding landscape. Nature. 2010;463:681–4. doi: 10.1038/nature08717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhuang X, Kim H, Pereira MJ, Babcock HP, Walter NG, Chu S. Correlating structural dynamics and function in single ribozyme molecules. Science. 2002;296:1473–6. doi: 10.1126/science.1069013. [DOI] [PubMed] [Google Scholar]
- 17.Kibbe WA. OligoCalc: an online oligonucleotide properties calculator. Nucleic acids research. 2007;35:W43–6. doi: 10.1093/nar/gkm234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schuster P, Fontana W, Stadler PF, Hofacker IL. From sequences to shapes and back: a case study in RNA secondary structures. Proceedings Biological sciences/The Royal Society. 1994;255:279–84. doi: 10.1098/rspb.1994.0040. [DOI] [PubMed] [Google Scholar]
- 19.Li PT, Vieregg J, Tinoco I., Jr How RNA unfolds and refolds. Annu Rev Biochem. 2008;77:77–100. doi: 10.1146/annurev.biochem.77.061206.174353. [DOI] [PubMed] [Google Scholar]
- 20.Waghmare SP, Pousinis P, Hornby DP, Dickman MJ. Studying the mechanism of RNA separations using RNA chromatography and its application in the analysis of ribosomal RNA and RNA:RNA interactions. Journal of chromatography A. 2009;1216:1377–82. doi: 10.1016/j.chroma.2008.12.077. [DOI] [PubMed] [Google Scholar]
- 21.Rio DC, Ares M, Jr, Hannon GJ, Nilsen TW. Polyacrylamide gel electrophoresis of RNA. Cold Spring Harbor protocols. 2010;2010:pdb prot5444. doi: 10.1101/pdb.prot5444. [DOI] [PubMed] [Google Scholar]
- 22.Sambrook J, Russell DW. Isolation of DNA Fragments from Polyacrylamide Gels by the Crush and Soak Method. Cold Spring Harbor Protocols. 2006;2006:pdb.prot2936–pdb.prot2936. doi: 10.1101/pdb.prot100479. [DOI] [PubMed] [Google Scholar]
- 23.Azarani A, Hecker KH. RNA analysis by ion-pair reversed-phase high performance liquid chromatography. Nucleic acids research. 2001;29:E7. doi: 10.1093/nar/29.2.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Noll B, Seiffert S, Vornlocher HP, Roehl I. Characterization of small interfering RNA by non-denaturing ion-pair reversed-phase liquid chromatography. Journal of chromatography A. 2011;1218:5609–17. doi: 10.1016/j.chroma.2011.06.057. [DOI] [PubMed] [Google Scholar]
- 25.Levin DS, Shepperd BT, Gruenloh CJ. Combining ion pairing agents for enhanced analysis of oligonucleotide therapeutics by reversed phase-ion pairing ultra performance liquid chromatography (UPLC) Journal of chromatography B, Analytical technologies in the biomedical and life sciences. 2011;879:1587–95. doi: 10.1016/j.jchromb.2011.03.051. [DOI] [PubMed] [Google Scholar]
- 26.Murugaiah V, Zedalis W, Lavine G, Charisse K, Manoharan M. Reversed-phase high-performance liquid chromatography method for simultaneous analysis of two liposome-formulated short interfering RNA duplexes. Analytical biochemistry. 2010;401:61–7. doi: 10.1016/j.ab.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 27.Kariko K, Muramatsu H, Ludwig J, Weissman D. Generating the optimal mRNA for therapy: HPLC purification eliminates immune activation and improves translation of nucleoside-modified, protein-encoding mRNA. Nucleic acids research. 2011;39:e142. doi: 10.1093/nar/gkr695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dickman MJ, Hornby DP. Enrichment and analysis of RNA centered on ion pair reverse phase methodology. Rna. 2006;12:691–6. doi: 10.1261/rna.2278606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wincott F, DiRenzo A, Shaffer C, Grimm S, Tracz D, Workman C, Sweedler D, Gonzalez C, Scaringe S, Usman N. Synthesis, deprotection, analysis and purification of RNA and ribozymes. Nucleic acids research. 1995;23:2677–84. doi: 10.1093/nar/23.14.2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anderson AC, Scaringe SA, Earp BE, Frederick CA. HPLC purification of RNA for crystallography and NMR. Rna. 1996;2:110–7. [PMC free article] [PubMed] [Google Scholar]
- 31.Dickman MJ, Conroy MJ, Grasby JA, Hornby DP. RNA footprinting analysis using ion pair reverse phase liquid chromatography. Rna. 2002;8:247–51. doi: 10.1017/s1355838202012888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garcia S, Liautard JP. Behaviour of macromolecular RNA in reversed-phase HPLC. Journal of chromatographic science. 1983;21:398–404. doi: 10.1093/chromsci/21.9.398. [DOI] [PubMed] [Google Scholar]
- 33.Gilar M, Fountain KJ, Budman Y, Neue UD, Yardley KR, Rainville PD, Russell RJ, 2nd, Gebler JC. Ion-pair reversed-phase high-performance liquid chromatography analysis of oligonucleotides: retention prediction. Journal of chromatography A. 2002;958:167–82. doi: 10.1016/s0021-9673(02)00306-0. [DOI] [PubMed] [Google Scholar]
- 34.Winkler WC, Nahvi A, Roth A, Collins JA, Breaker RR. Control of gene expression by a natural metabolite-responsive ribozyme. Nature. 2004;428:281–6. doi: 10.1038/nature02362. [DOI] [PubMed] [Google Scholar]
- 35.Chuang CZ, Ragan FA, Prasad C. Use of triethylamine as an ion-pairing reagent. Journal of liquid chromatography. 1994;17:2383–2394. [Google Scholar]
- 36.Beaucage SL. Solid-phase synthesis of siRNA oligonucleotides. Current opinion in drug discovery & development. 2008;11:203–16. [PubMed] [Google Scholar]
- 37.Misra VK, Draper DE. The linkage between magnesium binding and RNA folding. Journal of molecular biology. 2002;317:507–21. doi: 10.1006/jmbi.2002.5422. [DOI] [PubMed] [Google Scholar]
- 38.Serra MJ, Baird JD, Dale T, Fey BL, Retatagos K, Westhof E. Effects of magnesium ions on the stabilization of RNA oligomers of defined structures. Rna. 2002;8:307–23. doi: 10.1017/s1355838202024226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cox RA. Conformation of nucleic acids and the analysis of the hypochromic effect. The Biochemical journal. 1970;120:539–47. doi: 10.1042/bj1200539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Friedman RA, Honig B. A free energy analysis of nucleic acid base stacking in aqueous solution. Biophysical journal. 1995;69:1528–35. doi: 10.1016/S0006-3495(95)80023-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Noll B, Seiffert S, Hertel F, Debelak H, Hadwiger P, Vornlocher HP, Roehl I. Purification of small interfering RNA using nondenaturing anion-exchange chromatography. Nucleic acid therapeutics. 2011;21:383–93. doi: 10.1089/nat.2011.0317. [DOI] [PubMed] [Google Scholar]
- 42.Keefe AD, Pai S, Ellington A. Aptamers as therapeutics. Nat Rev Drug Discov. 2010;9:537–50. doi: 10.1038/nrd3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Apffel A, Chakel JA, Fischer S, Lictenwalter K, Hancock WS. Analysis of Oligonucleotides by HPLC-Electrospray Ionization Mass Spectrometry. Analytical chemistry. 1997;69:1320–5. doi: 10.1021/ac960916h. [DOI] [PubMed] [Google Scholar]
- 44.Apffel A, Chakel JA, Fischer S, Lichtenwalter K, Hancock WS. New procedure for the use of high-performance liquid chromatography-electrospray ionization mass spectrometry for the analysis of nucleotides and oligonucleotides. Journal of chromatography A. 1997;777:3–21. [Google Scholar]
- 45.Nguyen TH, Cunningham LA, Hammond KM, Lu Y. High-resolution preparative-scale purification of RNA using the. Prep Cell Analytical biochemistry. 1999;269:216–8. doi: 10.1006/abio.1999.4030. [DOI] [PubMed] [Google Scholar]
- 46.Chen Z, Ruffner DE. Modified crush-and-soak method for recovering oligodeoxynucleotides from polyacrylamide gel. BioTechniques. 1996;21:820–2. doi: 10.2144/96215bm14. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


