Abstract
Intensive study of the organic extract of the marine-derived bacterium Saccharomonospora sp. CNQ-490 has yielded three new α-pyrones, saccharomonopyrones A–C (1–3). The chemical structures of these compounds were assigned from the interpretation of 1D, 2D NMR and mass spectrometry data. Saccharomonopyrone A (1) is the first α-pyrone microbial natural product bearing the ethyl-butyl ether chain in the molecule, while saccharomonopyrones B and C possess unusual 3-methyl and a 6-alkyl side-chain within a 3,4,5,6-tetrasubstituted α-pyrone moiety. Saccharomonopyrone A exhibited weak antioxidant activity using a cation radical scavenging activity assay with an IC50 value of 140 μM.
Keywords: Saccharomonospora sp., marine natural products, α-pyrones
1. Introduction
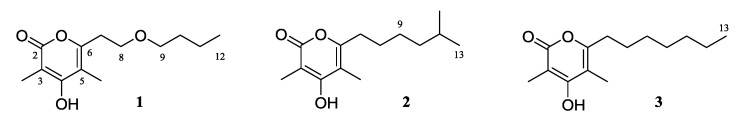
Actinobacteria are known as an abundant source of novel secondary metabolites comprising over 45% of all bioactive microbial metabolites known [1]. The recent discovery of numerous taxonomically unique marine actinomycetes, along with the isolation of structurally unprecedented secondary metabolites from these strains, illustrates marine actinomycetes as a promising source for the discovery of new natural products [2]. Saccharomonospora, a genus in the actinomycete family Pseudonocardiaceae, was first described in 1971 [3,4]. Members of the genus Saccharomonospora are interesting because they originate from diverse habitats and play an important role in the primary degradation of plant material by attacking hemicellulose [5]. Previous chemical investigations of members of this genus have led to the isolation of bioactive secondary metabolites, such as antibiotic AB 65, sakyomicin E, saccharonol A, antimicrobial saccharonol B, and piericidin A3 [6,7]. As part of our ongoing research for new secondary metabolites from marine actinobacteria, a Saccharomonospora bacterial strain CNQ-490 was documented. In a previous study, this strain was found to produce a novel cytotoxic alkaloid, lodopyridone A [8]. Further chemical investigation of this strain has now yielded three new natural products of the α-pyrone class. Herein, we report the isolation and structure elucidation of saccharomonopyrones A–C (1–3) along with their biological activities (Figure 1).
Figure 1.
Structures of saccharomonopyrones A–C (1–3).
2. Results and Discussion
Saccharomonopyrone A (1) was obtained as a white amorphous powder. An high-resolution electrospray ionization mass spectrometry (HR-ESIMS) peak [M + H]+ at m/z 241.1446 (calcd for 241.1434) indicated the molecular formula C13H20O4, which implied four degrees of unsaturation. UV absorption at 288 nm and IR absorptions at 3402, 1672, and 1569 cm−1 indicated the presence of an α-pyrone moiety.
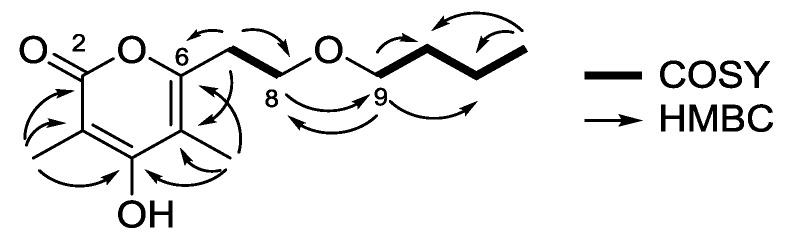
The planar structure of 1 was confidently assigned from the interpretation of 1D and 2D NMR data. Analysis of the 13C NMR (Table 1) and heteronuclear single quantum correlation (HSQC) spectroscopic data identified three oxygenated quaternary sp2 carbons (δC 155.2, 164.3, and 164.5), two quaternary sp2 carbons (δC 97.3 and 107.5), along with two methyl singlets [3-CH3 (δH 1.82; δC 9.1) and 5-CH3 (δH 1.87; δC 9.9)], one methyl triplet (δH 0.83, J = 7.2 Hz; δC 13.6) and five methylene groups [CH2-7 (δH 2.70, J = 6.4 Hz; δC 31.0)/CH2-8 (δH 3.56, J = 6.4 Hz; δC 67.0)/CH2-9 (δH 3.35, J = 6.4 Hz; δC 69.6)/CH2-10 (δH 1.43, m; δC 31.1)/CH2-11 (δH 1.26, m; δC 18.7)]. Interpretation of 1H-1H correlation spectroscopy (COSY) correlations allowed the construction of two spin systems: [H-7 (δH 2.70)/H-8 (δH 3.56)] and [H-9 (δH 3.35)/H-10 (δH 1.43)/H-11 (δH 1.26)/H-12 (δH 0.83)]. Heteronuclear multiple bond correlation (HMBC) correlations from H-8 to C-9 (δC 69.6) and from H-9 to C-8 (δC 67.0), as well as the oxygenated carbon chemical shifts of C-8 and C-9, supported the attachment of two spin systems through an oxygen atom (Figure 2).
Table 1.
1H and 13C NMR spectral data for saccharomonopyrones A–C (1–3) in DMSO-d6 1.
| No. C | (1) | (2) | (3) | |||
|---|---|---|---|---|---|---|
| δC, Type | δH, Mult. 2 (J in Hz) | δC, Type | δH, Mult. 2 (J in Hz) | δC, Type | δH, Mult. 2 (J in Hz) | |
| 2 | 164.5 | 165.1 | 164.6 | |||
| 3 | 97.3 | 97.6 | 97.1 | |||
| 4 | 164.3 | 164.9 | 164.4 | |||
| 5 | 107.5 | 107.0 | 106.5 | |||
| 6 | 155.2 | 158.2 | 157.8 | |||
| 7 | 31.0, CH2 | 2.70, t (6.4) | 30.4, CH2 | 2.47, t (6.5) | 29.9, CH2 | 2.45, t (7.5) |
| 8 | 67.0, CH2 | 3.56. t (6.4) | 27.6, CH2 | 1.51, m 3 | 26.8, CH2 | 1.51, m |
| 9 | 69.6, CH2 | 3.35, t (6.4) | 26.6, CH2 | 1.29, m | 28.3, CH2 | 1.26, m 3 |
| 10 | 31.1, CH2 | 1.43, m | 38.4, CH2 | 1.17, m | 28.3, CH2 | 1.26, m 3 |
| 11 | 18.7, CH2 | 1.26, m | 27.8, CH | 1.51, m3 | 31.1, CH2 | 1.25, m 3 |
| 12 | 13.6, CH3 | 0.83, t (7.2) | 22.9, CH3 | 0.85, d (6.6) | 22.0, CH2 | 1.26, m 3 |
| 13 | 22.9, CH3 | 0.85, d (6.6) | 13.8, CH3 | 0.85, t (7.1) | ||
| 3-CH3 | 9.1, CH3 | 1.82, s | 9.6, CH3 | 1.83, s | 9.1, CH3 | 1.82, s |
| 5-CH3 | 9.9, CH3 | 1.87, s | 10.3, CH3 | 1.88, s | 9.8, CH3 | 1.87, s |
1 300 MHz for 1H NMR, 75 MHz for 13C NMR. 2 Number of attached protons was deduced from 2D NMR analysis. 3 Signals were overlapping.
Figure 2.
COSY and key HMBC correlations for saccharomonopyrone A (1).
The α-pyrone functionality was constructed as 3,5-dimethyl-4-hydroxypyran-2-one with a side chain substituent at C-6 by analysis of 13C chemical shifts and HMBC correlations. HMBC correlations from 3-CH3 to C-2 (δC 164.5), C-3 (δC 97.3), C-4 (δC 164.3), and from 5-CH3 to C-4, C-5 (δC 107.5), C-6 (δC 155.2), indicated the position of the methyl groups on the α-pyrone ring. Lastly, the HMBC cross-peaks from H-7 to C-6 and C-5 supported the connectivity of the α-pyrone ring to the linear chain, thus completing the structure assignment of saccharomonopyrone A (1).
Saccharomonopyrone B (2) was isolated as an amorphous white powder. Its molecular formula C14H22O3 was determined by the observation of a HR-ESIMS ion peak [M + H]+ at m/z 239.1651, (calcd for 239.1642). The infrared (IR) absorption at 1673 cm−1 was assigned to a conjugated ketone functionality. The 1H NMR spectrum of 2 was similar to that of 1 except for the absence of two oxygenated methylene protons and the presence of two methyl doublets. COSY NMR correlations (H-7 (δH 2.47)/H-8 (δH 1.51)/H-9 (δH 1.29)/H-10 (δH 1.17), H-12 (δH 0.85)/H-11 (δH 1.51)/H-13 (δH 0.85)) and HMBC correlations of H-12 and H-13 to C-10 (δC 38.4), and from H-9 (δH 1.29) to C-10, and from H-10 to C-9 (δC 26.6) permitted the construction of the aliphatic chain as a 5-methylhexyl moiety. The linkage of the alkyl moiety to the α-pyrone ring was secured from the HMBC correlation from H-7 to C-6 (δC 158.2), completing the structure assignment of saccharomonopyrone B (2) as 4-hydroxy-3,5-dimethyl-6-(5-methylhexyl)-α-pyrone.
Saccharomonopyrone C (3) was also obtained as a white amorphous powder. The formula of 3 was deduced as C14H22O3 by the observation of the [M + H]+ ion peak at m/z 239.1665 (calcd for 239.1642). Compound 3 had the same molecular formula as 2. The only difference was that the 1H NMR of 3 displayed a methyl triplet. The analysis of 2D NMR spectroscopic data allowed the establishment of 3 as 4-hydroxy-3,5-dimethyl-6-heptyl-α-pyrone.
Pyrones are a well-known class of microbial secondary metabolites and are found to have a wide range of biological activities such as anti-inflammatory [9], anticancer [10], antimicrobial [11,12], anti-obesity [13,14], and antibiotic activities [15]. Previous studies on microbial sources have shown that the main producer of α-pyrones is fungi, but α-pyrones are also produced by plants, animals, and bacteria. Marine bacteria have also yielded α-pyrones with interesting bioactivities, examples of which are antibiotic Sch419560 [15], germicidins, and cytotoxic violapyrones. A recent study also expanded our knowledge for the first time that pyrones can be used as signaling molecules of the bacterial cell-cell communications in the soil bacterium Pseudomonas [16]. Bacteria monitor other bacteria in their living environment by producing and responding to signaling molecules. This led to a strategy to prevent pathogenicity by blocking bacterial communication in their environment [17,18,19].
Saccharomonopyrones A–C (1–3) were tested on various assays such as monoamine oxidase (MAO) inhibitory, acetylcholinesterase (AChE) inhibitory, β-site amyloid precursor protein cleaving enzyme 1 (BACE1), anti-osteoporosis, cytotoxicity, anti-tyrosinase, and antibacterial activities. However, the compounds did not display any significant biological activities in these assays. Interestingly, saccharomonopyrone A (1) showed weak antioxidant activity measuring free radical scavenging activity and cation radical scavenging activity in assays with IC50 values of 911 μM and 140 μM, respectively.
Saccharomonopyrone A (1) has an unusual ethylbutyl ether moiety attached at C-6. There are no reports of an ether moiety attached at C-6 within the α-pyrone class. Some similar molecules with ethyl-methyl sulfide and propyl-methyl sulfide groups were obtained as bioengineered products [20]. A similar compound possessing an ethyl chain with an acetyl end group was obtained during the synthesis of β-polyketones [21]. In addition, another naturally occurring and structurally similar compound is known to possess a propyl chain with a methyl carbonyl end attached at the C-6 position [22]. However, the butoxyethyl side chain in 1 is unprecedented within this class of natural products.
Nocapyrone R and violapyrone B are the most structurally related compounds of saccharomonopyrone B (2) [11,23]. The only difference is that nocapyrone R has a methoxy group at C-3 and violapyrone B has no methyl group on C-5. Violapyrone I, structurally the most similar natural product of saccharomonopyrone C (3), also lacks the methyl group on C-5 [10].
The genus Saccharomonospora (Family Pseudonocardiaceae), with eleven known species to date, has been known since 1971 [24]. Saccharomonospora sp. CNQ-490 is a unique sediment-derived strain which produces the structurally unprecedented lodopyridones A–C [8,25]. Furthermore, by genome mining, this strain has been shown to possess a full biosynthetic pathway to produce a new antibiotic taromycin A through direct cloning and refactoring methods [26]. Saccharomonopyrones A–C, and previously reported natural products, lodopyridones and taromycin A, illustrate the versatile secondary metabolites producing ability of this strain. The application of a recent genetic modification method for the biosynthetic gene cluster of this strain to introduce a methyl group in natural products could lead to the successful production of new secondary metabolites [27].
3. Materials and Methods
3.1. General Experimental Procedures
The UV spectra were recorded in methanol (MeOH) on a UVS-2100 (Scinco, Seoul, Korea). The IR spectra were obtained using a Varian Scimitar Series. The NMR spectra were obtained using a Varian Inova NMR spectrometer (500 and 125 MHz for 1H and 13C NMR, respectively, Varian Inc., Palo Alto, CA, USA), using the signals of the residual solvent as internal references and δH 2.50 and δC 39.5 ppm for dimethyl sulfoxide-d6 (DMSO). High Resolution Mass spectra were determined by a JMS-AX505WA mass spectrometer (Jeol Ltd., Tokyo, Japan). Low-resolution LC-MS data were analyzed using an Agilent Technologies 6120 quadrupole LC/MS system with a reversed-phase column (Phenomenex Luna C18(2) 100 Å, 50 mm × 4.6 mm, 5 μm, Phenomenex, Torrance, CA, USA) at a flow rate of 1.0 mL/min (Agilent Technologies, Santa Clara, CA, USA). Column chromatographic separation was performed using C18 (40–63 μm, ZEOprep 90, Zeochem, Zurich, Switzerland) with a gradient solvent of MeOH and water (H2O). The fractions were purified using reversed-phase HPLC (Waters 600S controller with 996 PDA detector (Waters Corporation, Milford, MA, USA), Phenomenex Luna C18 (250 mm × 10 mm, 5 μm) column (Phenomenex, Torrance, CA, USA)) with a mixture of acetonitrile (ACN) and H2O at flow rate of 2.0 mL/min. All chemicals were purchased from Sigma-Aldrich (Sigma-Aldrich, St. Louis, MO, USA) and used without further purification.
3.2. Strain Isolation and Fermentation
Actinomycete strain Saccharomonospora sp. CNQ-490 was collected from a deep sea sediment sample 2 km west of the Scripps pier, in La Jolla, CA, USA. The 16S rDNA sequence of this strain showed modest identity with the genus Saccharomonospora (accession number: KF301601). Strain CNQ-490 was cultured in 40-L scale using 2.5-L Ultra Yield (Thomson Instrument Company, Oceanside, CA, USA) flasks, each containing 1 L of the medium (10 g/L of soluble starch, 2 g/L of yeast, 4 g/L of peptone, 10 g/L of CaCO3, 20 g/L of KBr, 8 g/L of Fe2(SO4)3·4H2O dissolved in 1000 mL artificial seawater) at 25 °C with shaking at 150 rpm. After 7 days, the broth was extracted with ethyl acetate (added ratio 1:1 of volume). The ethyl acetate layer was separated and dried with anhydrous sodium sulfate. The organic solvent was removed to yield 2.5 g of the organic extract.
3.3. Extraction and Isolation
The crude organic extract was loaded on the C18 resin and fractionated by reversed-phase C18 (40–63 μm, ZEO prep 90) flash chromatography with gradient elution (from 80% of H2O in MeOH to 100% MeOH) to yield eight fractions. Fractions five and six eluted with 40% and 30% of H2O, respectively, were further purified by reversed-phase HPLC (Phenomenex Luna C18 column 250 mm × 10 mm, 5 μm) under isocratic conditions (50% of ACN in H2O) to give 5.7 mg of saccharomonopyrone A (1) (retention time 15.4 min), along with saccharomonopyrones B (2) (6.7 mg, retention time 45.6 min) and C (3) (4.9 mg, retention time 49.6 min).
Saccharomonopyrone A (1): amorphous white solid, UV (MeOH) λmax (log ε) 209 (2.53), 288 (2.49) nm; IR (KBr) νmax 3402, 2941, 1672, 1569 cm−1, 1H and 13C NMR data, Table 1 and Supplementary Materials; HRESIMS m/z 241.1446 [M + H]+ (calcd for C13H21O4, 241.1434).
Saccharomonopyrone B (2): amorphous white solid, UV (MeOH) λmax (log ε) 203 (2.53), 292 (2.32) nm; IR (KBr) νmax 3427, 2940, 1673, 1575 cm−1, 1H and 13C NMR data, Table 1 and Supplementary Materials; HRESIMS m/z 239.1651 [M + H]+ (calcd for C14H23O3, 239.1642).
Saccharomonopyrone C (3): amorphous white solid, UV (MeOH) λmax (log ε) 201 (2.53), 291 (2.20) nm; IR (KBr) νmax 3395, 2930, 1665, 1560 cm−1, 1H and 13C NMR data, Table 1 and Supplementary Materials; HRESIMS m/z 239.1665 [M + H]+ (calcd for C14H23O3, 239.1642).
3.4. Bioactivity Assays
MAO inhibitory assay [28], AChE inhibitory assay [29], BACE1 [25], anti-osteoporosis assay [30], and anti-tyrosinase assay [31] were performed following the previously published methods. Cytotoxicity tests were performed on two human kidney cancer cell lines, A498 and ACHN renal cancers, according to previously published methods [32]. Antibacterial assays were performed against seven bacterial strains including four Gram-positive (Staphylococcus epidermidis ATCC 12228, Kocuria rhizophila ATCC 9341, Bacillus subtilis ATCC 6633, Staphylococcus aureus ATCC 65381) and three Gram-negative (Escherichia coli ATCC 11775, Salmonella typhimurium ATCC 14028, Klebsiella pneumoniae ATCC 4352) strains following a previously published method [25]. The antioxidant activity was performed using the 1,1-diphenyl-2-picrylhydrazyl free radical (DPPH) as described previously with slight modification [33]. The DPPH solution (0.45 mM) was prepared daily in a 20-mL conical tube and kept in the dark at 4 °C. The DPPH solution (120 µL) was added to 60 µL of sample, control, or standard solution in 70% ethanol at different concentrations. The solutions were mixed, covered, and allowed to react in the dark for 15 min; afterward, the absorbance at 517 nm was read. Ascorbic acid was used as a positive control (IC50 21.02 ± 0.82 µM). Data are presented as the mean values ± standard deviation (SD) of three measurements. The free radical scavenging activity of each solution was then calculated as the percent inhibition according to the following equation:
| Scavenging rate (%) = [A (blank) − A (sample)]/A (blank) × 100 |
2,2′-Azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) diammonium salt (ABTS) cation radical scavenging activity of the compounds were tested using the spectroscopic method described by Roberta et al. [34]. The ABTS cation radical (ABTS+) was acquired by reacting 7 mM solution of ABTS with 2.45 mM potassium persulfate reaction. The mixture was left to stand in the dark at room temperature for 12–16 h before use. Prior to the assay, the ABTS radical cation solution was diluted with ethyl alcohol to an absorbance of 0.750 ± 0.05 at 734 nm. The ABTS+ solution was then added to each sample, standard, and control solution. Ascorbic acid was used as a positive control (IC50 13.01 ± 0.21 µM). Data are presented as the mean values ± standard deviation (SD) of the three measurements. The extent of decolorization is calculated as a percent reduction in absorbance. The percentage of cation radical scavenging was computed using the following equation:
| Scavenging rate (%) = [A (blank) − A (sample)]/A (blank) × 100 |
3.5. Statistical Analyses
Statistical analyses for DPPH and ABTS cation radical scavenging activities were performed using GraphPad Prism 5 (GraphPad Software, Inc., La Jolla, CA, USA). The nonlinear regression was used to determine the dose-response inhibition. Results are expressed as means ± standard deviation of three independent experiments.
4. Conclusions
Three new α-pyrones, saccharomonopyrones A–C (1–3), were isolated from the marine sediment-derived bacterium Saccharomonospora sp. strain CNQ-490. Saccharomonopyrones A–C (1–3) are 3,4,5,6-tetra-subtituted α-pyrones. Saccharomonopyrone A is unusual in having an ether moiety attached at C-6, observed for the first time in this class of natural products. Analysis of the tetra-substituted α-pyrone biosynthetic gene cluster from this strain may provide an opportunity to discover diverse α-pyrone analogues.
Acknowledgments
This work was supported by the Marine Biotechnology Program, the Ministry of Oceans and Fisheries, Korea and by the National Research Foundation of Korea grants (NRF-2012M1A5A1054307), the Ministry of Education, Science and Technology, Korea., by Suncheon Research Center for Natural Medicines (to SJN), and by the US National Cancer Institute (NIH) under grant R37 CA044848 (to WF).
Supplementary Materials
The following are available online at www.mdpi.com/1660-3397/15/8/239/s1; 1H, 13C and 2D NMR spectroscopic data of saccharomonopyrones A–C (1–3).
Author Contributions
C.-Y.Y. did large culture, isolated the compounds, and wrote the manuscript. T.C.L. elucidated the chemical structure and wrote the manuscript. T.G.L. performed bioassays and wrote the manuscript. J.L. and I.Y. contributed NMR analysis and surveyed the literature. H.C. did large culture and collected NMR spectroscopic data. J.S.L. and K.-M.L. performed anti-tyrosinase assay. K.-Y.K. and S.-T.Y. performed anti-oxidation assay. H.K. was the project leader for bioassays and analysis. S.-J.N. was the project leader for guiding the experiments of chemical analysis and writing manuscript. W.F. was the project leader and contributed microbial strain and guided the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Bérdy J. Bioactive microbial metabolites. J. Antibiot. 2005;58:1–26. doi: 10.1038/ja.2005.1. [DOI] [PubMed] [Google Scholar]
- 2.Jensen P.R., Mincer T.J., Williams P.G., Fenical W. Marine actinomycete diversity and natural product discovery. Antonie Van Leeuwenhoek. 2005;87:43–48. doi: 10.1007/s10482-004-6540-1. [DOI] [PubMed] [Google Scholar]
- 3.Liu Z., Li Y., Zheng L.-Q., Huang Y.-J., Li W.-J. Saccharomonospora marina sp. nov., isolated from an ocean sediment of the East China Sea. Int. J. Syst. Evol. Microbiol. 2010;60:1854–1857. doi: 10.1099/ijs.0.017038-0. [DOI] [PubMed] [Google Scholar]
- 4.Veyisoglu A., Sazak A., Cetin D., Guven K., Sahin N. Saccharomonospora amisosensis sp. nov., isolated from deep marine sediment. Int. J. Syst. Evol. Microbiol. 2013;63:3782–3786. doi: 10.1099/ijs.0.051516-0. [DOI] [PubMed] [Google Scholar]
- 5.Meier-Kolthoff J.P., Lu M., Huntemann M., Lucas S., Lapidus A., Copeland A., Pitluck S., Goodwin L.A., Han C., Tapia R., et al. Genome sequence of the chemoheterotrophic soil bacterium Saccharomonospora cyanea type strain (NA-134(T)) Stand. Genomic Sci. 2013;9:28–41. doi: 10.4056/sigs.4207886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tamura A., Takeda I. Antibiotic AB-65, a new antibiotic from Saccharomonospora viride. J. Antibiot. 1975;28:395–397. doi: 10.7164/antibiotics.28.395. [DOI] [PubMed] [Google Scholar]
- 7.Singh B., Parshad R., Khajuria R.K., Guru S.K., Pathania A.S., Sharma R., Chib R., Aravinda S., Gupta V.K., Khan I.A., et al. Saccharonol B, a new cytotoxic methylated isocoumarin from Saccharomonospora azurea. Tetrahedron Lett. 2013;54:6695–6699. doi: 10.1016/j.tetlet.2013.09.060. [DOI] [Google Scholar]
- 8.Maloney K.N., Macmillan J.B., Kauffman C.A., Jensen P.R., Dipasquale A.G., Rheingold A.L., Fenical W. Lodopyridone, a structurally unprecedented alkaloid from a marine actinomycete. Org. Lett. 2009;11:5422–5424. doi: 10.1021/ol901997k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee H.-S., An B.-J., Kim H.J., Cho Y.H., Kim D.I., Jang J.Y., Kwak J.H., Lee H.-S., Lee Y.-J., Lee J.S., et al. Anti-Inflammatory Effect of Violapyrones B and C from a Marine-derived Streptomyces sp. Nat. Prod. Sci. 2015;21:251–254. doi: 10.20307/nps.2015.21.4.251. [DOI] [Google Scholar]
- 10.Shin H., Lee H.-S., Lee J., Shin J., Lee M., Lee H.-S., Lee Y.-J., Yun J., Kang J. Violapyrones H and I, New Cytotoxic Compounds Isolated from Streptomyces sp. Associated with the Marine Starfish Acanthaster planci. Mar. Drugs. 2014;12:3283–3291. doi: 10.3390/md12063283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang J., Jiang Y., Cao Y., Liu J., Zheng D., Chen X., Han L., Jiang C., Huang X. Violapyrones A–G, α-pyrone derivatives from Streptomyces violascens isolated from Hylobates hoolock feces. J. Nat. Prod. 2013;76:2126–2130. doi: 10.1021/np4003417. [DOI] [PubMed] [Google Scholar]
- 12.Bertin M.J., Demirkiran O., Navarro G., Moss N.A., Lee J., Goldgof G.M., Vigil E., Winzeler E.A., Valeriote F.A., Gerwick W.H. Kalkipyrone B, a marine cyanobacterial γ-pyrone possessing cytotoxic and anti-fungal activities. Phytochemistry. 2016;122:113–118. doi: 10.1016/j.phytochem.2015.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chu M., Mierzwa R., Xu L., He L., Terracciano J., Patel M., Zhao W., Black T.A., Chan T.-M. Structure of Sch 419560, a novel α-pyrone antibiotic produced by Pseudomonas fluorescens. J. Antibiot. 2002;55:215–218. doi: 10.7164/antibiotics.55.215. [DOI] [PubMed] [Google Scholar]
- 14.Koyama T., Kawazoe Y., Iwasaki A., Ohno O., Suenaga K., Uemura D. Anti-obesity activities of the yoshinone A and the related marine γ-pyrone compounds. J. Antibiot. 2016;69:348–351. doi: 10.1038/ja.2016.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Inuzuka T., Yamamoto K., Iwasaki A., Ohno O., Suenaga K., Kawazoe Y., Uemura D. An inhibitor of the adipogenic differentiation of 3T3-L1 cells, yoshinone A, and its analogs, isolated from the marine cyanobacterium Leptolyngbya sp. Tetrahedron Lett. 2014;55:6711–6714. doi: 10.1016/j.tetlet.2014.10.032. [DOI] [Google Scholar]
- 16.Brachmann A.O., Brameyer S., Kresovic D., Hitkova I., Kopp Y., Manske C., Schubert K., Bode H.B., Heermann R. Pyrones as bacterial signaling molecules. Nat. Chem. Biol. 2013;9:573–578. doi: 10.1038/nchembio.1295. [DOI] [PubMed] [Google Scholar]
- 17.Taga M.E., Bassler B.L. Chemical communication among bacteria. Proc. Natl. Acad. Sci. USA. 2003;100:14549–14554. doi: 10.1073/pnas.1934514100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bassler B.L., Losick R. Bacterially speaking. Cell. 2006;125:237–246. doi: 10.1016/j.cell.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 19.Bassler B.L. Small Talk. Cell. 2002;109:421–424. doi: 10.1016/S0092-8674(02)00749-3. [DOI] [PubMed] [Google Scholar]
- 20.Huitt-Roehl C.R., Hill E.A., Adams M.M., Vagstad A.L., Li J.W., Townsend C.A. Starter unit flexibility for engineered product synthesis by the nonreducing polyketide synthase PksA. ACS Chem. Biol. 2015;10:1443–1449. doi: 10.1021/acschembio.5b00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tian J.-F., Yu R.-J., Li X.-X., Gao H., Guo L.-D., Tang J.-S., Yao X.-S. 1H and 13C NMR spectral assignments of 2-pyrone derivatives from an endophytic fungus of sarcosomataceae. Magn. Reson. Chem. 2015;53:866–871. doi: 10.1002/mrc.4282. [DOI] [PubMed] [Google Scholar]
- 22.Douglas J.L., Money T. Pyrone studies. Linear α-pyrone route to protected β-polyketones. Can. J. Chem. 1968;46:695–700. doi: 10.1139/v68-118. [DOI] [Google Scholar]
- 23.Kim Y., Ogura H., Akasaka K., Oikawa T., Matsuura N., Imada C., Yasuda H., Igarashi Y. Nocapyrones: α- and γ-pyrones from a marine-derived Nocardiopsis sp. Mar. Drugs. 2014;12:4110–4125. doi: 10.3390/md12074110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nonomura H., Ohara Y. Distribution of actinomycetes in soil. X. New genus and species of monosporic actinomycetes. J. Ferment. Techonol. 1971;49:895–903. [Google Scholar]
- 25.Le T.C., Yim C.-Y., Park S., Katila N., Yang I., Song M.C., Yoon Y.J., Choi D.-Y., Choi H., Nam S.-J., et al. Lodopyridones B and C from a marine sediment-derived bacterium Saccharomonospora sp. Bioorg. Med. Chem. Lett. 2017;27:3123–3126. doi: 10.1016/j.bmcl.2017.05.035. in press. [DOI] [PubMed] [Google Scholar]
- 26.Yamanaka K., Reynolds K.A., Kersten R.D., Ryan K.S., Gonzalez D.J., Nizet V., Dorrestein P.C., Moore B.S. Direct cloning and refactoring of a silent lipopeptide biosynthetic gene cluster yields the antibiotic taromycin A. Proc. Natl. Acad. Sci. USA. 2014;111:1957–1962. doi: 10.1073/pnas.1319584111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koryakina I., McArthur J.B., Draelos M.M., Williams G.J. Promiscuity of a modular polyketide synthase towards natural and non-natural extender units. Org. Biomol. Chem. 2013;11:4449–4458. doi: 10.1039/c3ob40633d. [DOI] [PubMed] [Google Scholar]
- 28.Lee H.W., Choi H., Nam S.-J., Fenical W., Kim H. Potent Inhibition of Monoamine Oxidase B by a Piloquinone from Marine-Derived Streptomyces sp. CNQ-027. J. Microbiol. Biotechnol. 2017;27:785–790. doi: 10.4014/jmb.1612.12025. [DOI] [PubMed] [Google Scholar]
- 29.Kim H., Yang I., Patil R.S., Kang S., Lee J., Choi H., Kim M.-S., Nam S.-J., Kang H. Anithiactins A-C, modified 2-phenylthiazoles from a mudflat-derived Streptomyces sp. J. Nat. Prod. 2014;77:2716–2719. doi: 10.1021/np500558b. [DOI] [PubMed] [Google Scholar]
- 30.Kim H., Kim K.-J., Yeon J.-T., Kim S.H., Won D.H., Choi H., Nam S.-J., Son Y.-J., Kang H. Placotylene A, an inhibitor of the receptor activator of nuclear factor-κB ligand-induced osteoclast differentiation, from a Korean sponge Placospongia sp. Mar. Drugs. 2014;12:2054–2065. doi: 10.3390/md12042054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim K., Leutou A.S., Jeong H., Kim D., Seong C.N., Nam S.-J., Lim K.-M. Anti-pigmentary effect of (−)-4-hydroxysattabacin from the marine-derived bacterium Bacillus sp. Mar. Drugs. 2017;15:138. doi: 10.3390/md15050138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leutou A.S., Yang I., Kang H., Seo E.K., Nam S.-J., Fenical W. Nocarimidazoles A and B from a Marine-Derived Actinomycete of the Genus Nocardiopsis. J. Nat. Prod. 2015;78:2846–2849. doi: 10.1021/acs.jnatprod.5b00746. [DOI] [PubMed] [Google Scholar]
- 33.Thomas J.H., Priyadarshini G., Michael T. High-throughput micro plate assays for screening flavonoid content and DPPH-scavenging activity in sorghum bran and flour. J. Sci. Food Agric. 2012;92:2326–2331. doi: 10.1002/jsfa.5633. [DOI] [PubMed] [Google Scholar]
- 34.Roberta R., Nicoletta P., Anna P., Ananth P., Min Y., Catherine R.E. Antioxidant activity applying an improved ABTS radical cation decolorization assay. Free Radic. Biol. Med. 1999;26:1231–1237. doi: 10.1016/s0891-5849(98)00315-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.