Abstract
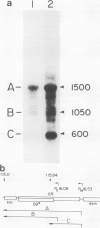
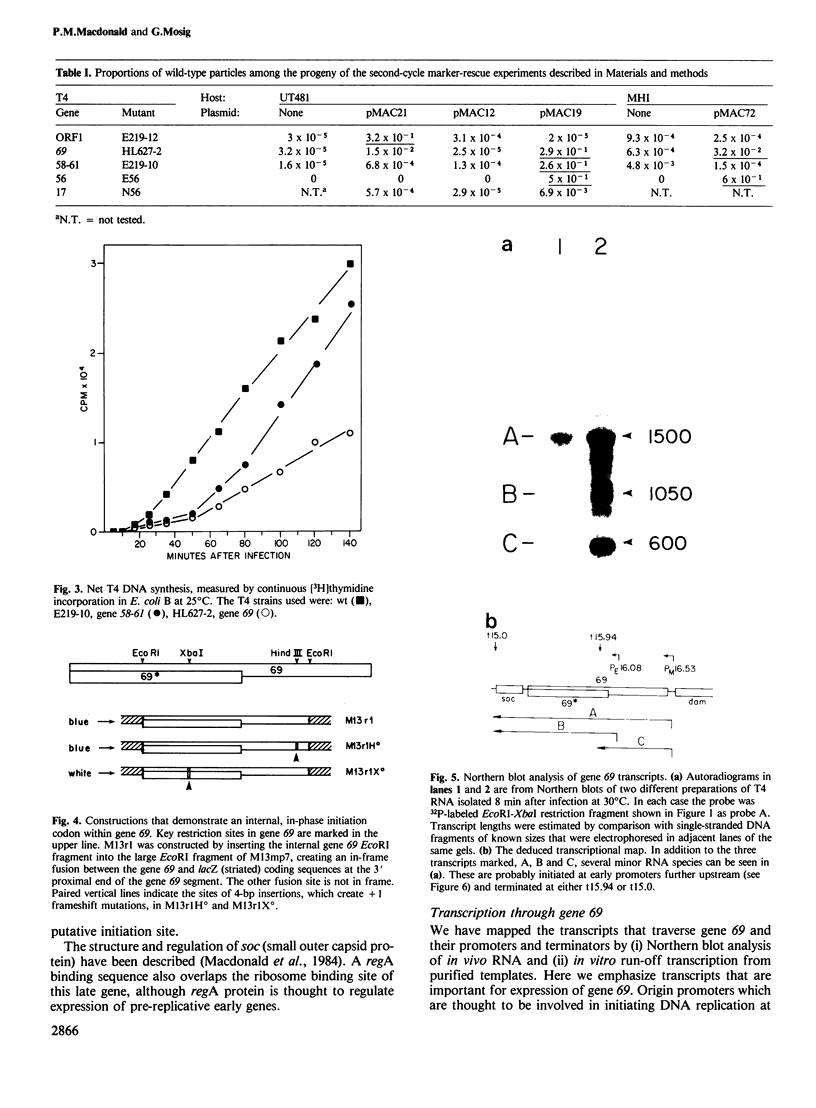
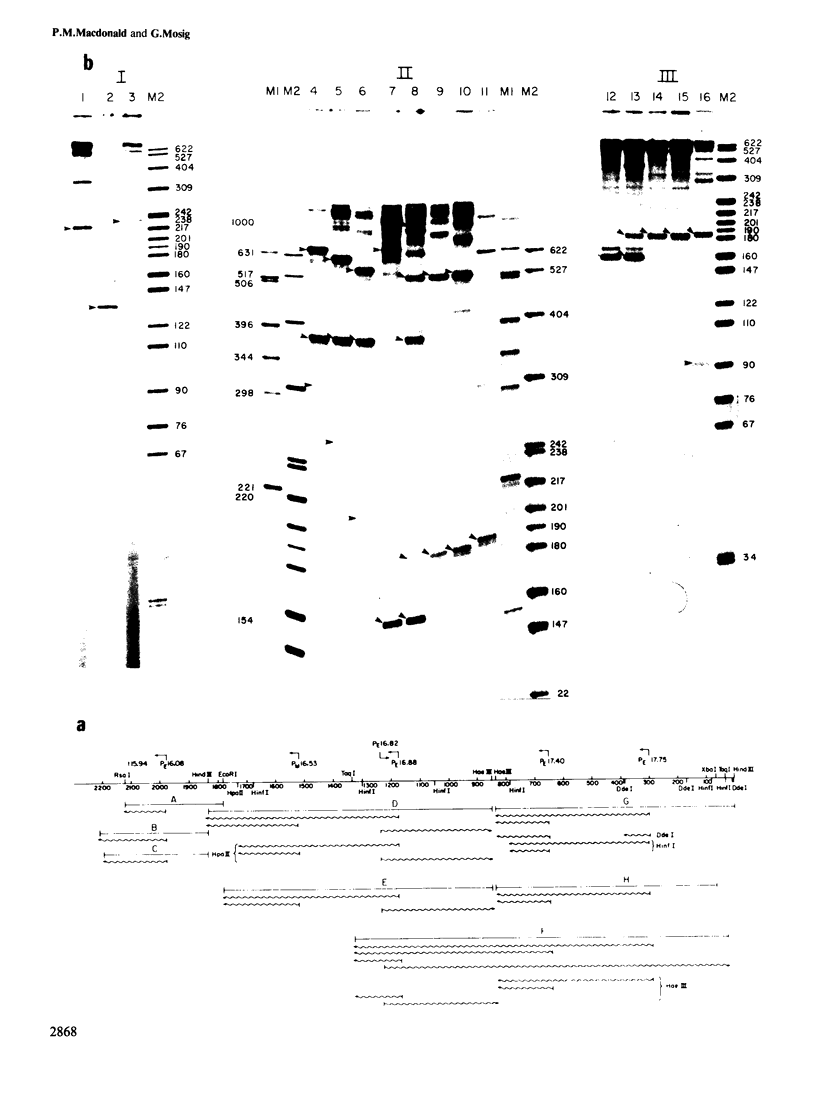
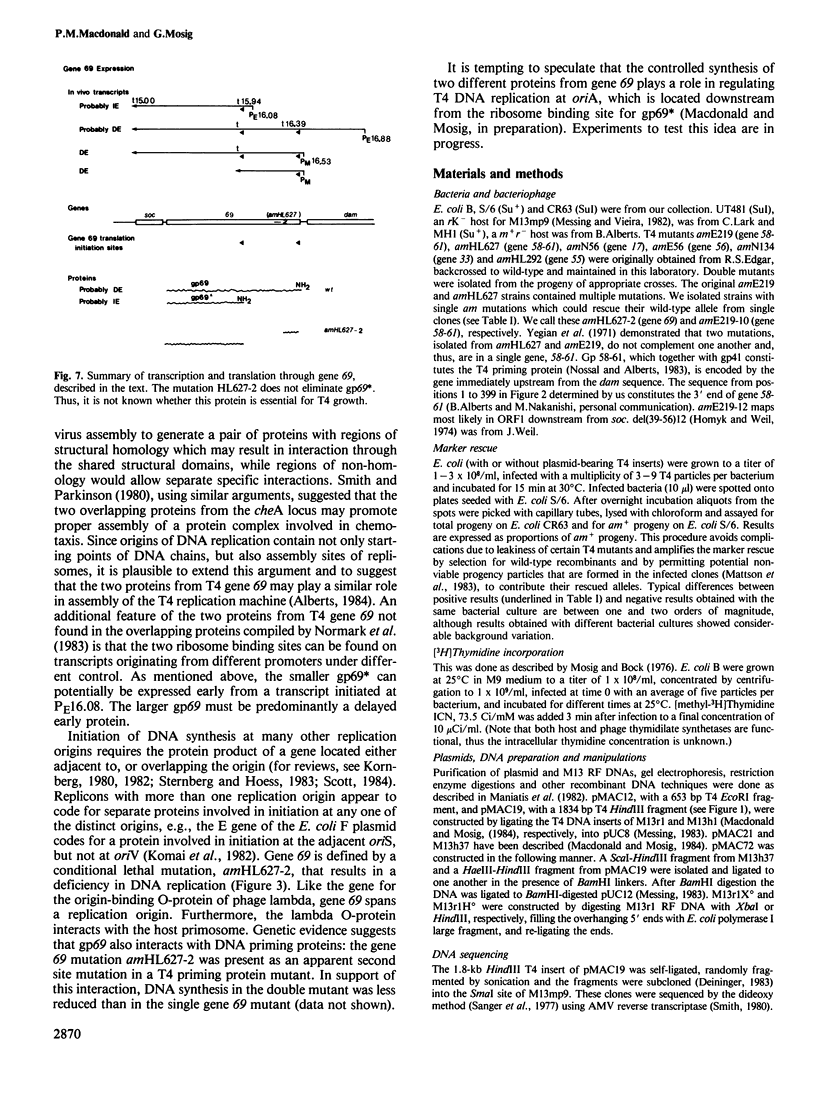
We have determined the DNA sequence and transcription patterns in a 3-kb segment (between 15 and 18 kb on the standard phage T4 map) spanning an origin of DNA replication. A new gene, 69, spans this origin. Gene 69 codes for two overlapping proteins that share a common C-terminal segment. Defective DNA replication in an appropriate amber mutant shows that at least the larger of the two proteins is required for efficient T4 DNA replication. The two proteins coded by gene 69 are expressed from different transcripts that are under different regulation. The smaller protein, gp69*, can be expressed immediately from an Escherichia coli-like promoter, whereas expression of the larger protein, gp69, must be delayed since its middle promoter requires T4 coded proteins, most likely gp mot, for activation. We discuss the possible significance of two overlapping proteins in the assembly of replisomes. Gene 69 is bracketed by the non-essential early gene dam (DNA adenine methylase) and the late gene soc (small outer capsid protein). Transcripts through this region are interdigitated in a complex pattern, which reveals all elements that are thought to be important in regulation of pre-replicative and post-replicative T4 genes.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Chu F. K., Maley G. F., Maley F., Belfort M. Intervening sequence in the thymidylate synthase gene of bacteriophage T4. Proc Natl Acad Sci U S A. 1984 May;81(10):3049–3053. doi: 10.1073/pnas.81.10.3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deininger P. L. Random subcloning of sonicated DNA: application to shotgun DNA sequence analysis. Anal Biochem. 1983 Feb 15;129(1):216–223. doi: 10.1016/0003-2697(83)90072-6. [DOI] [PubMed] [Google Scholar]
- Elliott T., Geiduschek E. P. Defining a bacteriophage T4 late promoter: absence of a "-35" region. Cell. 1984 Jan;36(1):211–219. doi: 10.1016/0092-8674(84)90091-6. [DOI] [PubMed] [Google Scholar]
- Gram H., Liebig H. D., Hack A., Niggemann E., Rüger W. A physical map of bacteriophage T4 including the positions of strong promoters and terminators recognized in vitro. Mol Gen Genet. 1984;194(1-2):232–240. doi: 10.1007/BF00383522. [DOI] [PubMed] [Google Scholar]
- Homyk T., Jr, Weil J. Deletion analysis of two nonessential regions of the T4 genome. Virology. 1974 Oct;61(2):505–523. doi: 10.1016/0042-6822(74)90286-4. [DOI] [PubMed] [Google Scholar]
- Isono K. Computer programs to analyze DNA and amino acid sequence data. Nucleic Acids Res. 1982 Jan 11;10(1):85–89. doi: 10.1093/nar/10.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassavetis G. A., Elliott T., Rabussay D. P., Geiduschek E. P. Initiation of transcription at phage T4 late promoters with purified RNA polymerase. Cell. 1983 Jul;33(3):887–897. doi: 10.1016/0092-8674(83)90031-4. [DOI] [PubMed] [Google Scholar]
- Komai N., Nishizawa T., Hayakawa Y., Murotsu T., Matsubara K. Detection and mapping of six miniF-encoded proteins by cloning analysis of dissected miniF segments. Mol Gen Genet. 1982;186(2):193–203. doi: 10.1007/BF00331850. [DOI] [PubMed] [Google Scholar]
- Linder C. H., Sköld O. Control of early gene expression of bacteriophage T4: involvement of the host rho factor and the mot gene of the bacteriophage. J Virol. 1980 Feb;33(2):724–732. doi: 10.1128/jvi.33.2.724-732.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald P. M., Kutter E., Mosig G. Regulation of a bacteriophage T4 late gene, soc, which maps in an early region. Genetics. 1984 Jan;106(1):17–27. doi: 10.1093/genetics/106.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald P. M., Mosig G. Cloning and physical mapping of an early region of the bacteriophage T4 genome. Genetics. 1984 Jan;106(1):1–16. doi: 10.1093/genetics/106.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattson T., Richardson J., Goodin D. Mutant of bacteriophage T4D affecting expression of many early genes. Nature. 1974 Jul 5;250(461):48–50. doi: 10.1038/250048a0. [DOI] [PubMed] [Google Scholar]
- Mattson T., Van Houwe G., Epstein R. Recombination between bacteriophage T4 and plasmid pBR322 molecules containing cloned T4 DNA. J Mol Biol. 1983 Oct 25;170(2):357–379. doi: 10.1016/s0022-2836(83)80153-3. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Messing J., Vieira J. A new pair of M13 vectors for selecting either DNA strand of double-digest restriction fragments. Gene. 1982 Oct;19(3):269–276. doi: 10.1016/0378-1119(82)90016-6. [DOI] [PubMed] [Google Scholar]
- Mosig G., Bock S. Gene 32 protein of bacteriophage T4 moderates the activities of the T4 gene 46/47-controlled nuclease and of the Escherichia coli RecBC nuclease in vivo. J Virol. 1976 Mar;17(3):756–761. doi: 10.1128/jvi.17.3.756-761.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normark S., Bergström S., Edlund T., Grundström T., Jaurin B., Lindberg F. P., Olsson O. Overlapping genes. Annu Rev Genet. 1983;17:499–525. doi: 10.1146/annurev.ge.17.120183.002435. [DOI] [PubMed] [Google Scholar]
- Price A. R., Warner H. R. Bacteriophage T4-induced deoxycytidine triphosphate-deoxyuridine triphosphate nucleotidohydrolase: its properties and its role during phage infection of Escherichia coli. Virology. 1969 Dec;39(4):882–892. doi: 10.1016/0042-6822(69)90024-5. [DOI] [PubMed] [Google Scholar]
- Pulitzer J. F., Coppo A., Caruso M. Host--virus interactions in the control of T4 prereplicative transcription. II. Interaction between tabC (rho) mutants and T4 mot mutants. J Mol Biol. 1979 Dec 25;135(4):979–997. doi: 10.1016/0022-2836(79)90523-0. [DOI] [PubMed] [Google Scholar]
- Sanger F., Air G. M., Barrell B. G., Brown N. L., Coulson A. R., Fiddes C. A., Hutchison C. A., Slocombe P. M., Smith M. Nucleotide sequence of bacteriophage phi X174 DNA. Nature. 1977 Feb 24;265(5596):687–695. doi: 10.1038/265687a0. [DOI] [PubMed] [Google Scholar]
- Schlagman S. L., Hattman S. Molecular cloning of a functional dam+ gene coding for phage T4 DNA adenine methylase. Gene. 1983 May-Jun;22(2-3):139–156. doi: 10.1016/0378-1119(83)90098-7. [DOI] [PubMed] [Google Scholar]
- Scott J. R. Regulation of plasmid replication. Microbiol Rev. 1984 Mar;48(1):1–23. doi: 10.1016/b978-0-12-048850-6.50006-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw J. E., Murialdo H. Morphogenetic genes C and Nu3 overlap in bacteriophage lambda. Nature. 1980 Jan 3;283(5742):30–35. doi: 10.1038/283030a0. [DOI] [PubMed] [Google Scholar]
- Smith A. J. DNA sequence analysis by primed synthesis. Methods Enzymol. 1980;65(1):560–580. doi: 10.1016/s0076-6879(80)65060-5. [DOI] [PubMed] [Google Scholar]
- Smith R. A., Parkinson J. S. Overlapping genes at the cheA locus of Escherichia coli. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5370–5374. doi: 10.1073/pnas.77.9.5370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg N., Hoess R. The molecular genetics of bacteriophage P1. Annu Rev Genet. 1983;17:123–154. doi: 10.1146/annurev.ge.17.120183.001011. [DOI] [PubMed] [Google Scholar]
- Stitt B. L., Revel H. R., Lielausis I., Wood W. B. Role of the host cell in bacteriophage T4 development. II. Characterization of host mutants that have pleiotropic effects on T4 growth. J Virol. 1980 Sep;35(3):775–789. doi: 10.1128/jvi.35.3.775-789.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stormo G. D., Schneider T. D., Gold L., Ehrenfeucht A. Use of the 'Perceptron' algorithm to distinguish translational initiation sites in E. coli. Nucleic Acids Res. 1982 May 11;10(9):2997–3011. doi: 10.1093/nar/10.9.2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzan M., Leautey J., d'Aubenton-Carafa Y., Brody E. Identification and biosynthesis of the bacteriophage T4 mot regulatory protein. EMBO J. 1983;2(7):1207–1212. doi: 10.1002/j.1460-2075.1983.tb01568.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yegian C. D., Mueller M., Selzer G., Russo V., Stahl F. W. Properties of the DNA-delay mutants of bacteriophage T4. Virology. 1971 Dec;46(3):900–919. doi: 10.1016/0042-6822(71)90090-0. [DOI] [PubMed] [Google Scholar]