Abstract
Background
3-hydroxypalmitoleoyl-carnitine (C16:1-OH) was recently reported to be elevated in acylcarnitine profile of propionic acidemia (PA) or methylmalonic acidemia (MMA) patients during expanded newborn screening (NBS). High levels of C16:1-OH, combined with other hydroxylated long chain acylcarnitines are related to long-chain 3-hydroxyacyl-CoA dehydrogenase deficiency (LCHADD).
Methods
The acylcarnitine profile of two LCHADD patients was evaluated using liquid chromatography-tandem mass spectrometric method. A specific retention time was reported for each hydroxylated long chain acylcarnitine. The same method was applied to some neonatal dried blood spots (DBS) from PA and MMA patients presenting abnormal C16:1-OH concentrations.
Results
The final retention time of the peak corresponding to C16:1-OH in LCHADD patients differed from those in MMA and PA patients. Heptadecanoylcarnitine (C17) has been identified as the novel biomarker specific for PA and MMA patients through high resolution mass spectrometry (Orbitrap) experiments. We found that 21 out of 23 neonates (22 MMA, and 1PA) diagnosed through the Tuscany region NBS program had significantly higher levels of C17 compared to levels detected in controls.
Twenty-three maternal deficiencies (21 vitamin B12 deficiency, 1 homocystinuria and 1 gastrin deficiency) and 82 false positive for propionylcarnitine (C3) results were also analyzed.
Conclusions
This paper reports on the characterization of a novel biomarker able to detect propionate disorders during expanded newborn screening (NBS). The use of this new biomarker may improve the analytical performances of NBS programs especially in laboratories where second tier tests are not performed.
Keywords: propionic acidemia, methylmalonic acidemia, expanded newborn screening, acylcarnitine, heptadecanoylcarnitine
Introduction
Propionic and methylmalonic acidemias are inborn errors of propionate metabolism that originate from the incorrect catabolism of isoleucine, valine, threonine, methionine, odd-chain fatty acids and cholesterol (1).
Propionic acidemia (PA, OMIM 606054) is a result of defective activity of propionyl-CoA carboxylase, an enzyme that catalyzes the conversion of propionyl-CoA to methylmalonyl-CoA. It is caused by mutation in the genes PCCA (OMIM 232000) and PCCB (OMIM 232050) which encode for alpha and beta subunits of propionyl-CoA carboxylase, respectively (2).
Methylmalonic acidemia (MMA) is a genetically heterogeneous disorder caused by either mutations in the MUT gene encoding for the methylmalonyl-CoA mutase (OMIM 609058) (and causing partial, mut (−), or complete mut 0, enzyme deficiency) or a defect in the transport or biosynthesis of MUT coenzyme adenosylcobalamin (AdoCbl). AdoCbl defect are classified according to complementation group in CblA, caused by mutation in the MMAAgene (OMIM 607481), and CblB, caused by mutation in the MMAB gene (OMIM 607568).
Combined methylmalonic acidemia and homocystinuria may be seen in complementation groups CblC (OMIM 277400), CblD (OMIM 277410), and CblF (OMIM 277380). Methylmalonic acidemia may be due to a mutation in the gene CD320 (OMIM 606475) encoding the transcobalamin receptor TCBLR or a mutation in the methylmalonyl-CoA epimerase gene (OMIM 608419) (3, 4).
The clinical manifestations of these disorders include vomiting, lethargy, coma, intermittent ketoacidosis, hyperglycinemia, neutropenia, thrombocytopenia, hyperammonemia and hypoglycemia. The severity of clinical and biochemical phenotype is associated with different genes and/or mutations involved. Diagnosis and treatment of patients with MMA/PA can prevent acute acidosis and metabolic decompensation (5). Expanded newborn screening (NBS) for metabolic diseases by tandem mass spectrometry allows pre-symptomatic detection and early treatment of affected newborns with MMA or PA.
The Acylcarnitine profile of dried blood spot (DBS) samples from newborns with propionate metabolism defect usually shows increased levels of propionylcarnitine (C3). This analyte is used worldwide as a marker for both defects, but it is not specific and causes large numbers of false-positive results (6). In order to improve the specificity of the test, some authors have suggested including the calculation of metabolite ratios C3/C2, C3/C16, C3/Met in the newborn screening panel and to use specific algorithms (7).
In a previous published paper we proposed a second-tier test (2nd TT) able to reveal the presence of 3-OH-propionic or methylmalonic acids on the same dried blood spot. The application of this 2nd TT in all newborns with C3 above the cut off levels has potentially allowed a positive predictive value close to 100% to be achieved (8).
Some newborn screening laboratories are still not performing 2nd TT when C3 is outside the normal ranges; they are therefore forced to keep a relatively high C3 cut off to avoid an unsustainable number of false positive results.
While laboratories performing 2nd TT for C3 have minimized false-positive rate, the risk of false negative results remains because 2nd TT is only applied to initial out-of-range screening results.
Methylmalonic acidemia combined with homocystinuria (CblC defect) can be missed during newborn screening because of the initial low levels of C3 (9, 10) as well as maternal vitamin B12 deficiencies (11).
Several papers have recently reported that in many patients affected by PA or MMA, newborn screening results showed an increased concentration of 3-hydroxyhexadecenoyl (or 3-hydroxypalmitoleoyl) -carnitine (C16:1-OH) in addition to elevated C3 and related ratios, even if no correlation with the diseases’ pathogenesis was recognized at that time (7, 11).
C16:1-OH and other hydroxylated long chain acylcarnitines are well-known markers of long-chain 3-hydroxyacyl-CoA dehydrogenase deficiency (LCHADD) and/or trifunctional protein (TFP) deficiency.
In this paper we provide evidence that the new biomarker involved in PA and MMA is heptadecanoylcarnitine C17 and not C16:1-OH.
Finally, we investigate the diagnostic value of this new biomarker for improving the sensitivity and the specificity of NBS for propionate metabolism disorders.
MATERIALS AND METHODS
Retrospective study population
DBS samples are routinely collected from all neonates born in the Italian regions of Tuscany and Umbria (about 40000/year) for NBS (12). In order to have the highest possible sensitivity, a 2nd TT for free methylmalonic and 3-OH-propionic acid has been performed since 2007 if first screen C3 value is ≥ 3.3 µmol/L. The cutoff value (98th percentile of our population, based on 50000 NBS tests) corresponds to approximately 2 % of 2nd TT per year.
Twenty-three affected neonates were evaluated in this study: 22 with heterogeneous genetic disorders of MMA, one with PA. In addition, a retrospective revaluation was performed on 23 newborns with MMA due to maternal deficiency (21 vitamin B12 deficiencies, one homocystinuria and one cobalamin malabsorption due to gastric atrophy).
Eighty-two false-positives (C3 ≥ 3.3 µmol/L, and negative at 2nd TT) detected during the same period were also evaluated for comparative analysis.
Five controls for each of the index cases (n=635) were taken randomly and blinded in the same analytic batch. We proceeded in the same way for the cases with MMA due to maternal deficiency (n=138). Low birth weight infants (<1800g), newborns receiving parenteral nutrition or a blood transfusion before newborn screening test were excluded.
All experiments were conducted in compliance with Institutional Review Board guidelines of the Meyer Children’s Hospital Ethic Committee.
Standards
C17 and C17-D3 were purchased from GiottoBiotech (Florence, Italy). Stock solutions of chemical and labelled standards at 100 mmol/L were prepared in methanol and stored at- 20°C. Working solutions were prepared daily from stock solutions at final concentration of 0.01 µmol/L. Other reagents, such as HPLC-grade water, formic acid, and methanol, were purchased from Panreac (Barcelona, Spain). Labelled acylcarnitine internal standards were purchased from Cambridge Isotope Laboratories (Andover, MA, USA); a stock solution was made in methanol. The standard acylcarnitine concentrations were in the range 7.6–152 mmol/L.
LC-MS/MS method for C16:1-OH isobaric compound separation
Two DBS (about 6.4–6.8 µL whole blood) for each sample were extracted with 200 µL of methanol for 25 min at room temperature. The extract was then transferred to a clear 1.5 mL vial for LC-MS/MS analysis.
The data were obtained using an API 4000 triple quadrupole mass spectrometer (AB SCIEX, Toronto, Canada) equipped with Turbo Ion Spray source and operating in MRM positive ion mode. The ion spray voltage was set to 5200 V, the gas 1 and gas 2 were set both at 30 and 45 respectively and the temperature was set to 450 °C.
Two MS/MS experiments were performed: an MRM experiment to monitor the transition 414.5>85.1 m/z specific for C16:1-OH acylcarnitine, and a precursor of 85 m/z scan experiment to analyze all acylcarnitines in the range of 300–500 Da.
The chromatographic separation was achieved using an Agilent 1260 Infinity HPLC capillary system (Agilent Technologies, Waldbronn, Germany), operating in gradient mode, coupled with a thermostated autosampler and fully controlled by Analyst Software (Version 1.5.2).
The chromatographic run was performed by using a Synergi Polar column, 4 µm, 150×2 mm (Phenomenex, Torrance, CA) at a flow rate of 200 µL/min; the eluate was directed into the ESI source without splitting. The mobile phase was composed by water +0.1 % formic acid (phase A, pH 2.8) and methanol + 0.1% formic acid (phase B).
Separation was achieved using a linear gradient from 90% to 30% (phase A) over 50 minutes; the conditions were maintained for 15 minutes, after which the mobile phase was returned to the starting conditions within 0.5 minutes and re-equilibrated for 9.5 minutes. The total run time was 75 minutes. The injection volume was 20 µL, and the autosampler temperature was maintained at 4°C throughout the analyses.
Orbitrap experiments
A Nexera liquid chromatograph (Shimadzu, Kyoto, Japan) coupled to a Thermo Q Exactive mass spectrometer (Thermo Fisher Scientific, Waltham, USA) was used for liquid chromatography–mass spectrometry (LC-MS) measurements. The mass spectrometer, operated in positive electrospray (ESI+) mode, was equipped with a heated ESI source (HESI-II, at 200°C) and the ionization voltage was set to 3.5 kV at a capillary temperature of 320°C. The scan event cycle used a full scan mass spectrum (100–1000 Da) at a resolving power of 140.000 (at m/z 200) and three corresponding data dependent MS/MS events acquired at a resolving power of 70.000 using HCD. The three most intense ions detected during full scan MS triggered data dependent scanning. Data dependent scanning was performed with and without the use of a precursor ion list. The MS/MS activation parameters were an isolation width of 4 Da, stepped collision energies of 20, 30, and 35 V and an activation time of 30 milliseconds. The mass accuracy was determined from an external calibration performed the same day. The software Xcalibur 3.0.63 (Thermo Fisher Scientific, Waltham, USA) was used for data acquisition.
C17
The fragmentation patterns of C17 and C17-D3 were recorded by infusing standard solutions (0.1 mg/L) at 10 µL/min, directly in the ESI source of the API 4000. Three specific MRM transitions (414.4>85 m/z, 414.4>355.1 m/z, 414.4>253.1 m/z; 417.4>85 m/z, 417.4>358.1 m/z, 417.4>256.1 m/z, for C17 and C17-D3 respectively) were recorded with experimentally optimized declustering, collision energy and collision exit potential values (42V, 25V and 29V, respectively).
Statistical analysis
We used the STATA 13 (T Stat s.r.l., College Station, Texas) for statistical analysis and descriptive statistics (means, standard deviation) to describe participants’ main variables.
ANOVA and Bartlett’s test were used to analyze the difference between the three groups (MMA/PA cases, negative controls and false-positives C3 ≥ 3.3 µmol/L).
To assess and compare the diagnostic accuracy of the measured biomarkers (C3, C17, C3/C0, C3/C2, C3/C4, C3/C16, C3/Met, methionine) we calculated sensitivity, specificity, area under the receiver operating characteristic (ROC) curve (AUC). We chose the cut-off that maximized the percentage of correctly classified and, in case of equal AUC, sensitivity.
Finally, we have defined a testing strategy combining information from the biomarkers.
When appropriate, confidence intervals (CI) were calculated using likelihood (13). Level of significance was set at 5% two sided.
Results
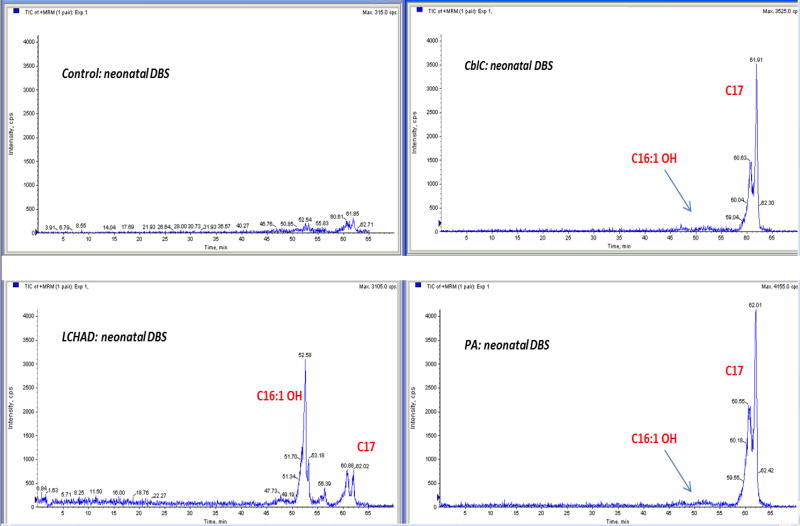
In order to understand the significance of the increased concentration of C16:1-OH observed on DBS of patients with propionate metabolism defect, we performed a chromatographic separation of the extracts obtained from neonatal DBS of controls, MMA, PA and LCHADD patients.
Two MS/MS experiments were monitored during the chromatographic separation: a MRM for the transition 414.5>85.1 m/z and a precursor ion scan of 85 m/z, enabling the measurement of all acylcarnitines in the range 300–500 m/z.
The total ion current of the MRM experiments revealed a practically flat baseline in control samples (two small peaks were detectable at about 60.6 and 61.9 minutes). The profile obtained from LCHADD samples showed one intense peak at retention time (RT) 52.6 minutes (peak 1) and two smaller peaks (about 60.8 and 62.0 minutes, peak 2 and peak 3, respectively) while PA and MMA samples reveals only the two peaks at 60.6 and 62.0 minutes (Figure 1). High resolution LC-Orbitrap MS and MS/MS experiments have demonstrated that peak 1 corresponds to C16:1-OH (as expected in LCHADD patient, data not shown) while peaks 2 and 3 correspond to acylcarnitine C17 (Figure S1). Considering that the pKa of the carboxyl group of acylcarnitines is 3.8 (14), dissociated and undissociated isoforms were present (the elution mixture measured pH 3.25 at approximately RT 60 minutes).
Figure 1.
The LC-MS/MS profile obtained from LCHADD neonatal samples.
Low- and high-resolution MS/MS spectra of C17 chemical standards and related RT corresponded to those found in PA and MMA patients.
Considering that NBS laboratories usually measure the metabolite C16:1-OH/C17 in precursor ion scan mode by using labelled C16 as internal standard, and considering that their concentration and MS signal is significantly different and potentially not accurate, we decided to add new specific MRMs using D3-C17 at 0.01 µmol/L. A precursor ion scan of 85 m/z can be also used by adding D3-C17 as internal standard.
Biomarker concentrations from neonatal DBS were reported for patients (Table 1) and maternal deficiencies (Table 2).
Table 1.
Newborn screening results in infants with proprionate metabolism disorders.
| Diagnosis | C3 | C17 | Met | MMA | C3/C0 | C3/C4 | C3/C2 | C3/C16 | C3/Met |
|---|---|---|---|---|---|---|---|---|---|
| n.v. <3.3a µmol/L |
n.v. <0.09b µmol/L |
n.v. 6–36c µmol/L |
n.v. <2 µmol/L |
n.v. < 0.3d | n.v. < 25.8e | n.v. <0.21f | n.v. <2.2g | n.v. <0.50h | |
| Mut 0 | 4.69 | 0.12 | 20.15 | 258 | 0.87 | 50.38 | 0.41 | 2.13 | 0.23 |
| Mut 0 | 9.84 | 0.26 | 27.41 | 162 | 0.68 | 62.74 | 0.54 | 4.99 | 0.36 |
| Mut 0 | 12.20 | 0.36 | 36.50 | 175 | 0.49 | 48.61 | 0.46 | 4.60 | 0.33 |
| Mut 0 | 8.84 | 0.19 | 23.73 | 159 | 0.67 | 62.67 | 0.68 | 5.20 | 0.37 |
| Mut (−) | 5.75 | 0.06 | 11.41 | 3.0 | 0.28 | 26.36 | 0.24 | 2.13 | 0.50 |
| Mut (−) | 4.86 | 0.20 | 18.32 | 14 | 0.29 | 18.98 | 0.19 | 1.73 | 0.27 |
| Mut (−) | 3.97 | 0.16 | 11.13 | 41 | 0.23 | 29.40 | 0.29 | 2.74 | 0.36 |
| Mut (−) | 5.63 | 0.05 | 16.17 | 4.0 | 0.46 | 19.38 | 0.29 | 1.71 | 0.35 |
| Cbl B | 17.45 | 0.26 | 15.35 | 210 | 0.55 | 96.85 | 0.65 | 6.58 | 1.14 |
| CblD, CblF | 10.25 | 0.16 | 5.77 | 155 | 0.54 | 38.44 | 0.50 | 4.07 | 1.78 |
| CblC | 5.21 | 0.27 | 8.15 | 9 | 0.35 | 17.89 | 0.34 | 1.66 | 0.64 |
| CblC | 6.66 | 0.12 | 6.05 | 21 | 0.36 | 20.06 | 0.36 | 3.70 | 1.10 |
| CblC | 5.27 | 0.17 | 5.11 | 20 | 0.28 | 7.48 | 0.25 | 1.97 | 1.03 |
| CblC | 7.12 | 0.09 | 4.95 | 30 | 0.22 | 41.65 | 0.33 | 4.11 | 1.44 |
| CblC | 6.76 | 0.23 | 6.74 | 41 | 0.34 | 12.26 | 0.40 | 2.55 | 1.00 |
| CblC | 7.04 | 0.14 | 5.57 | 43 | 0.27 | 23.72 | 0.32 | 2.69 | 1.26 |
| CblC | 4.20 | 0.12 | 14.69 | 5.2 | 0.39 | 44.44 | 0.51 | 3.85 | 0.29 |
| CblC | 3.81 | 0.16 | 12.17 | 11 | 0.29 | 12.16 | 0.18 | 2.01 | 0.31 |
| CblC | 11.21 | 0.19 | 11.68 | 17 | 0.29 | 29.38 | 0.32 | 4.33 | 0.96 |
| CblC | 7.99 | 0.21 | 4.91 | 29.5 | 0.78 | 33.35 | 0.36 | 2.74 | 1.63 |
| CblC | 9.00 | 0.21 | 6.57 | 25 | 0.50 | 16.97 | 0.37 | 2.34 | 1.37 |
| CblC | 3.05 | 0.14 | 27.40 | 15 | 0.11 | 14.02 | 0.15 | 1.90 | 0.11 |
| PA | 23.09 | 0.38 | 22.02 | - | 0.28 | 112.30 | 3.34 | 51.98 | 1.05 |
| trans II* | 3.46 | 0.04 | 9.64 | n.a. | 0.35 | 39.68 | 0.37 | 5.33 | 0.36 |
98th percentile;
99.9th percentile;
0.01th – 99.9th percentile;
99.2th percentile;
99.7th percentile;
99.5th percentile;
99.3th percentile;
99.9th percentile
disorder not included in the Tuscany region NBS panel; this patient was not considered in the statistical analysis
Table 2.
Newborn screening results in infants with MMA secondary to maternal vitamin B12 deficiency
| Diagnosis | C3 | C17 | Met | MMA | C3/C0 | C3/C4 | C3/C2 | C3/C16 | C3/Met |
|---|---|---|---|---|---|---|---|---|---|
| n.v. <3.3a µmol/L |
n.v. <0.09b µmol/L |
n.v. 6–36c µmol/L |
n.v. <2 µmol/L |
n.v. < 0.3d | n.v. < 25.8e | n.v. <0.21f | n.v. <2.2g | n.v. <0.50h | |
| Maternal vit B12 def | 6.19 | 0.09 | 9.21 | n.a. | 0.25 | 23.11 | 0.22 | 1.84 | 0.67 |
| Maternal vit B12 def | 3.50 | 0.05 | 12.93 | 2.1 | 0.23 | 8.94 | 0.19 | 1.70 | 0.27 |
| Maternal vit B12 def | 3.83 | 0.07 | 11.30 | 16 | 0.40 | 28.66 | 0.57 | 2.43 | 0.34 |
| Maternal vit B12 def | 5.58 | 0.05 | 13.81 | 2.5 | 0.26 | 15.49 | 0.13 | 1.07 | 0.40 |
| Maternal vit B12 def | 4.42 | 0.06 | 9.44 | 2.5 | 0.21 | 7.32 | 0.16 | 1.07 | 0.47 |
| Maternal vit B12 def | 7.99 | 0.04 | 14.82 | 2.7 | 0.31 | 10.02 | 0.35 | 3.68 | 0.54 |
| Maternal vit B12 def | 2.61 | 0.10 | 8.27 | 3.6 | 0.11 | 16.31 | 0.15 | 1.05 | 0.32 |
| Maternal vit B12 def | 3.32 | 0.07 | 11.69 | 2.3 | 0.28 | 11.34 | 0.14 | 1.06 | 0.28 |
| Maternal vit B12 def | 5.53 | 0.05 | 17.52 | 2.3 | 0.29 | 17.45 | 0.27 | 1.50 | 0.32 |
| Maternal vit B12 def | 3.35 | 0.04 | 10.85 | 2.3 | 0.14 | 15.25 | 0.17 | 2.40 | 0.31 |
| Maternal vit B12 def | 4.41 | 0.09 | 15.52 | 2.0 | 0.23 | 19.76 | 0.17 | 1.53 | 0.28 |
| Maternal vit B12 def | 4.75 | 0.10 | 9.60 | 7.1 | 0.36 | 21.76 | 0.21 | 2.37 | 0.49 |
| Maternal vit B12 def | 5.33 | 0.05 | 18.53 | 2.0 | 0.21 | 14.93 | 0.23 | 2.90 | 0.29 |
| Maternal vit B12 def | 5.30 | 0.05 | 11.27 | 2.0 | 0.32 | 11.58 | 0.22 | 0.28 | 0.47 |
| Maternal vit B12 def | 4.42 | 0.06 | 9.44 | 5.3 | 0.21 | 7.32 | 0.16 | 1.07 | 0.47 |
| Maternal vit B12 def | 4.62 | 0.10 | 17.32 | 5.4 | 0.22 | 15.36 | 0.18 | 1.01 | 0.27 |
| Maternal vit B12 def | 3.22 | 0.13 | 6.00 | 5.4 | 0.34 | 13.43 | 0.21 | 1.89 | 0.54 |
| Maternal vit B12 def | 3.47 | 0.10 | 11.24 | 3.7 | 0.52 | 20.65 | 0.20 | 1.61 | 0.31 |
| Maternal vit B12 def | 4.81 | 0.03 | 11.05 | 7.8 | 0.46 | 37.23 | 0.28 | 2.50 | 0.43 |
| Maternal vit B12 def | 4.83 | 0.15 | 18.15 | 4.0 | 0.31 | 11.64 | 0.37 | 2.09 | 0.27 |
| Maternal vit B12 def | 7.89 | 0.18 | 12.93 | n.a. | 0.22 | 22.36 | 0.23 | 2.23 | 0.61 |
| Gastrin def | 7.25 | 0.09 | 12.43 | n.a. | 0.27 | 24.54 | 0.38 | 4.20 | 0.58 |
| HCY mat | 3.34 | 0.05 | 12.26 | 5.5 | 0.29 | 27.19 | 0.21 | 1.71 | 0.27 |
98th percentile;
99.9th percentile;
0.01th – 99.9th percentile;
99.2th percentile;
99.7th percentile;
99.5th percentile;
99.3th percentile;
9.9th percentil
Statistical analysis
Box-plot of cases, controls and false-positives show the distribution of biomarker concentrations within each group (Figure S2). The group means (ANOVA) and variances (Bartlett-test) were significantly different from each other for all biomarkers except methionine (Met) (Table 3). For this reason we decided to exclude the biomarker Met from the diagnostic analysis.
Table 3.
ANOVA and Bartlett’s test results for all biomarkers.
| ANOVA | Bartlett's test | |||
|---|---|---|---|---|
|
| ||||
| F(2) | P-value | Χ2(2) | P-value | |
| C3 | 573.450 | <0.001 | 692.750 | <0.001 |
| C17 | 454.720 | <0.001 | 459.696 | <0.001 |
| Met | 2.410 | 0.090 | 0.437 | 0.804 |
| C3/C0 | 151.560 | <0.001 | 70.620 | <0.001 |
| C3/C4 | 239.530 | <0.001 | 362.769 | <0.001 |
| C3/C2 | 23.680 | <0.001 | 266.749 | <0.001 |
| C3/C16 | 68.430 | <0.001 | 190<0.001 | <0.001 |
| C3/Met | 74.250 | <0.001 | 240<0.001 | <0.001 |
Table 4 shows the results of diagnostic analysis. The best diagnostic performance for metabolism defects (MMA and PA) was observed by C17 with cut-off set up to 0.092 µmol/L (corresponding to the 99.8 percentile) 91.30% sensitivity (95% CI 72.0%; 98.9%); 99.84% specificity (95% CI 99.1%; 100%) and AUC = 0.956 (95% CI 0.897; 1.000); C3 with cut-off set up to 5.633 µmol/L (corresponding to the 99.93 percentile) 65.22% sensitivity (95% CI 42.7%; 83.6%); 99.51% specificity (95% CI 98.6%; 99.9%) and AUC = 0.824 (95% CI 0.724; 0.923) and C3/C16 ratio with cut-off set up to 2.338 (corresponding to the 99.4 percentile) 65.22% sensitivity (95% CI 42.7%; 83.6%); 98.69% specificity (95% CI 97.4%; 99.4%) and AUC = 0.820 (95% CI 0.720; 0.919).
Table 4.
Cut-off, sensitivity, specificity and AUC for biomarkers of MMA/PA metabolism defects. 95% confidence intervals in parentheses
| Cut-off | Sensitivity | Specificity | AUC | |
|---|---|---|---|---|
| C3 | ≥ 5.633 | 65.22% | 99.51% | 0.824 |
| (42.7%; 83.6%) | (98.6%; 99.9%) | (0.724; 0.923) | ||
| C17 | ≥ 0.092 | 91.30% | 99.84% | 0.956 |
| (72.0%; 98.9%) | (99.1%; 100%) | (0.897; 1.000) | ||
| C3/C0 | ≥ 0.339 | 52.17% | 99.02% | 0.756 |
| (30.6%; 73.2%) | (97.9%; 99.6%) | (0.652; 0.860) | ||
| C3/C4 | ≥ 29.376 | 52.17% | 99.35% | 0.758 |
| (30.6%; 73.2%) | (98.3%; 99.8%) | (0.653; 0.862) | ||
| C3/C2 | ≥ 1.120 | 4.35% | 99.84% | 0.521 |
| (0.11%; 21.9%) | (99.1%; 100%) | (0.478; 0.564) | ||
| C3/C16 | ≥ 2.338 | 65.22% | 98.69% | 0.820 |
| (42.7%; 83.6%) | (97.4%; 99.4%) | (0.720; 0.919) | ||
| C3/Met | ≥ 0.504 | 56.52% | 99.84% | 0.782 |
| (34.5%; 76.8%) | (99.1%; 100%) | (0.678; 0.885) |
Based on these results we explored a combination of biomarkers (Table 5): the best strategy was a parallel testing of C17 and C3 (tests are declared positive if C17 ≥ 0.092 µmol/L or C3 ≥ 5.633 µmol/L) with AUC = 0.997; 95% CI 0.994; 1.000; 100% sensitivity (95% CI 85.2%; 100%), 99.35% specificity (95% CI 98.3%; 99.8%).
Table 5.
Sensitivity, specificity and AUC for a combination of tests for MMA/PA. 95% confidence intervals in parentheses
| Sensitivity | Specificity | AUC | |
|---|---|---|---|
| C17≥0.092 or C3≥5.633 | 100% | 99.30% | 0.997 |
| (85.2%; 100%) | (98.3%; 99.8%) | (0.994; 1.000) | |
| C17≥0.092 or C3/C16 ≥2.338 | 91.30% | 98.50% | 0.949 |
| (72%; 98.9%) | (97.2%; 99.3%) | (0.890; 1.000) | |
| C17≥0.092 or C3≥5.633 or C3/C16≥2.338 | 100% | 98.50% | 0.993 |
| (85.2%; 100%) | (97.2%; 99.3%) | (0.988; 0.997) |
Discussion
In this paper we have demonstrated that a new metabolite, C17 acylcarnitine, is suggestive for the diagnosis of propionate metabolism defects (MMA and PA) and should be considered an important biomarker. The presence of large amounts of odd-numbered long-chain fatty acids in adipose tissue (from 8% to 10% of total fatty acids vs 1% in normal controls) during fetal life in newborns suffering of propionate metabolism defects has been largely reported from Wendel and collaborators (15–17). During periods of acute catabolism, such as in the early days of life, the mobilization and the oxidation of odd-chain fatty acids from adipose tissue leads to the production of extensive amounts of toxic propionyl-CoA in the mitochondria. Fatty acid synthase catalyzes the synthesis of palmitate from acetyl-CoA and malonyl-CoA, in the presence of NADPH, into long-chain saturated fatty acids. The excess of propionyl-CoA in the mitochondria could be considered to be the likely cause of the production of a small percentage of heptadecanoic fatty acid. This acid, like most other carboxylic acids, can be activated by acyl-CoA synthases to form heptadecanoyl-CoA, which cannot be oxidized to carbon dioxide in mammalian cells. As a result, heptadecanoyl-CoA accumulates in cells and becomes a substrate for several acyl-CoA transferases. These reactions generate heptadecanoic-conjugates and among them C17 detectable by newborn screening tests. The Tuscany region’s newborn screening program began in 2004; out of more than 500000 newborns screened, 23 affected by inborn errors of propionate metabolism have been identified. The study of C17 neonatal concentration in this group showed 21 measurements higher than cutoff values, confirming the high prognostic significance of the biomarker. It should be noted that this biomarker would have failed in the identification of two newborns with MMA mut (−) phenotype (and one newborn affected by transcobalamin II deficiency; disorder not included in our NBS panel). In our experience the sensitivity of C17 can be considered comparable to or higher than C3/C2 ratio (20/23 correct identifications, plus transcobalamin II deficiency). Considering our cut-off levels, other ratios presented less sensitivity (Table 1).
In 2007, after one CblC false negative result, we developed a 2nd TT for the detection of 3-OH propionic acid and methylmalonic acid on DBS (8), reducing the upper C3 cutoff value to 98th percentile of our population, corresponding to 3.3 µmol/L. The previous C3 cutoff 5.65 µmol/L corresponded to 99.95th percentile. Eight affected newborns (three mut (−) and five CblC) tested after 2007 presented with a C3 concentration lower than 5.65 µmol/L. Furthermore, one CblC patient was subjected to 2nd TT during this study because he presented with a high C17 level (0.14 µmol/L) even with normal C3 concentration (3.05 µmol/L).
Statistical analysis demonstrated that C17 is able to distinguish negative newborn specimen results (controls + false negative) from positive ones better than other parameters previously used in our NBS program (Figure S2A). It should be noted that C17 alone seems to be the biomarker with the best correct classification as compared to other biomarkers. For NBS laboratories that are still not performing 2nd TT, the inclusion of C17 as specific biomarker could significantly improve the sensitivity of the first tier test.
Otherwise we consider the combination of first and second-tier testing for many metabolic disorders to be the best working practice for achieving the most benefit from NBS programs (18). In our daily practice we have modified our NBS protocol as reported in Figure S3.
Different considerations must be done for maternal vitamin B12 deficiencies. Poor nutrition, such as a vegetarian diet during pregnancy, can lead to vitamin B12 deficiency causing serious problems in newborn metabolism. Some babies whose mothers had vitamin B12 deficiency show a MMA and homocysteine accumulation (19). A severe maternal vitamin B12 deficiency in breast-fed infants can cause failure to thrive, developmental regression, anemia and nerve-related disorders. Prolonged metabolic decompensation can lead to severe lethargy progressing slowly to coma and irreversible neurological damage (20).
Early identification of these cases can make life-saving interventions prior to clinical manifestation possible (21). However, vitamin B12 supplementation should be always recommended in vegetarian/vegan mothers during pregnancy as well as while breastfeeding.
In our NBS program we were able to identify 19 maternal vitamin B12 deficiencies, one maternal homocystinuria and one maternal gastrin deficiency because our C3 cut-off. The power of C17 alone for the detection of maternal disorders (without 2nd TT) should be considered only moderately important, as it was capable of identifying only 10/23 cases (sensitivity 43.48%) (Table 2). On the other hand, two additional vitamin B12 deficiencies were discovered during this study through 2nd TT because they presented C17 at 0.1 µmol/L and 0.13 µmol/L upon NBS test even if the C3 values were normal (2.61 and 3.22 µmol/L, respectively). Moreover the possibility of detecting a maternal deficiency is consistently low if 2nd TT is not performed and the corresponding C3 cut-off is maintained high to reduce the C3-related false positive rate.
Conclusions
The results of this study strongly suggest the evaluation of C17 as primary biomarker (together with C3 and ratios) for propionate metabolism defects, especially in NBS laboratories where 2nd TT is still not available.
The use of 2nd TT is a worldwide NBS good practice parameter that greatly improves the specificity of the first screening results. However, 2nd TT depends on the results of the first tier test and the final sensitivity will be influenced by the cut off value of the primary marker. While this study has some limitations (exclusion of low birth weight infants <1800g and newborns receiving parenteral nutrition or a blood transfusion before newborn screening test), in our experience the evaluation of the new biomarker C17 improved sensitivity suggesting the utility of performing the 2nd TT even when the primary marker C3 falls within normal limits (two maternal vitamin B12 deficiencies and 1 CblC). However, a false-negative result cannot be excluded when both biomarkers (C3 and C17) are lower than the cut offs and 2nd TT is not performed.
Supplementary Material
Figure S1A: Extracted ion chromatogram of the transition 414>85 from a CblC patient DBS in the Orbitrap MS. Theoretically the peak 3 is the less polar undissociated C17 form where the carboxylic group R-COOH is protonated: pH<pKa. Vice versa the peak 2 corresponds to the more polar dissociated form.
Figure S1B and S1C: MS/MS high resolution fragmentation of peak 2 and 3. Both peaks were recognized with a formula C24H48O4N corresponding to the acylcarnitine C17. Moreover in both product ion scan profiles the fragment 253.2 m/z corresponding to C17H33O acyl group is present
Figure S2A. Distribution of biomarkers in cases (n=23), controls (n=530) and false positives (n=82). Y axis: C3, C17 and Met are expressed as µmol/L
Figure S2B. Distribution of biomarkers in maternal cases (n=23) and controls (n=115)
Y axis: C3, C17 and Met are expressed as µmol/L.
Figure S3. The new flowchart adopted by Tuscany NBS program to identify patients affected by MMA or PA acidemia
Acknowledgments
Use of trade names is for identification only and does not imply endorsement by the US Centers for Disease Control and Prevention, the Public Health Service, or the U.S. Department of Health and Human Services. The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the US Centers for Disease Control and Prevention.
Abbreviations
- C16:1-OH
3-hydroxypalmitoleoyl-carnitine
- PA
propionic acidemia
- MMA
methylmalonic acidemia
- LCHADD
long-chain 3-hydroxyacyl-CoA dehydrogenase deficiency
- DBS
dried blood spot
- C17
heptadecanoylcarnitine
- C3
propionylcarnitine
- C3/C2
propionylcarnitine/acetylcarnitine
- C3/C16
propionylcarnitine/palmitoylcarnitine
- C3/Met
propionylcarnitine/methionine
- 2nd TT
second-tier test
- NBS
newborn screening
- MRM
multiple reaction monitoring
- HCD
higher energy collisional dissociation
- ESI
electrospray ionization
- RT
retention time
- ROC
receiver operating characteristic
- AUC
area under the curve
- CI
confidence intervals
References
- 1.Thompson GN. Inborn errors of propionate metabolism: methylmalonic and propionic acidaemias. J Paediatr Child Health. 1992;28:134–5. doi: 10.1111/j.1440-1754.1992.tb02627.x. [DOI] [PubMed] [Google Scholar]
- 2.Fenton WA, Gravel RA, Rosenblatt DS. Disorders of propionate and methylmalonate metabolism. In: Scriver CR, Beaudet AL, Sly WS, Valle D, editors. The Metabolic and Molecular Bases of Inherited Disease. 8. New York: McGraw-Hill; 2001. pp. 2165–93. [Google Scholar]
- 3.Quadros EV, Lai SC, Nakayama Y, Sequeira JM, Hannibal L, Wang S, et al. Positive newborn screen for methylmalonic acidemia identifies the first mutation in TCblR/CD320, the gene for cellular uptake of transcobalamin-bound vitamin B(12) Hum Mutat. 2010;31:924–9. doi: 10.1002/humu.21297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dobson CM, Gradinger A, Longo N, Wu X, Leclerc D, Lerner-Ellis J, et al. Homozygous nonsense mutation in the MCEE gene and siRNA suppression of methylmalonyl-CoA epimerase expression: a novel cause of mild methylmalonic acidemia. Mol Genet Metab. 2006;88:327–33. doi: 10.1016/j.ymgme.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 5.Deodato F, Boenzi S, Santorelli FM, Dionisi-Vici C. Methylmalonic and propionic acidemia. Am J Med Genet C Semin Med Genet. 2006;142C:104–12. doi: 10.1002/ajmg.c.30090. [DOI] [PubMed] [Google Scholar]
- 6.la Marca G, Malvagia S, Casetta B, Pasquini E, Donati MA, Zammarchi E. Progress in expanded newborn screening for metabolic conditions by LC-MS/MS in Tuscany: update on methods to reduce false tests. J Inherit Metab Dis. 2008;31(Suppl2):S395–404. doi: 10.1007/s10545-008-0965-z. [DOI] [PubMed] [Google Scholar]
- 7.McHugh D, Cameron CA, Abdenur JE, Abdulrahman M, Adair O, Al Nuaimi SA, et al. Clinical validation of cutoff target ranges in newborn screening of metabolic disorders by tandem mass spectrometry: a worldwide collaborative project. Genet Med. 2011;13:230–54. doi: 10.1097/GIM.0b013e31820d5e67. [DOI] [PubMed] [Google Scholar]
- 8.la Marca G, Malvagia S, Pasquini E, Innocenti M, Donati MA, Zammarchi E. Rapid 2nd-tier test for measurement of 3-OH-propionic and methylmalonic acids on dried blood spots: reducing the false-positive rate for propionylcarnitine during expanded newborn screening by liquid chromatography-tandem mass spectrometry. Clin Chem. 2007;53:1364–9. doi: 10.1373/clinchem.2007.087775. [DOI] [PubMed] [Google Scholar]
- 9.Chace DH, DiPerna JC, Kalas TA, Johnson RW, Naylor EW. Rapid diagnosis of methylmalonic and propionic acidemias: quantitative tandem mass spectrometric analysis of propionylcarnitine in filter-paper blood specimens obtained from newborns. Clin Chem. 2001;47:2040–4. [PubMed] [Google Scholar]
- 10.Wilcken B, Wiley V, Hammond J, Carpenter K. Screening newborns for inborn errors of metabolism by tandem mass spectrometry. N Engl J Med. 2003;348:2304–12. doi: 10.1056/NEJMoa025225. [DOI] [PubMed] [Google Scholar]
- 11.Lindner M, Ho S, Kölker S, Abdoh G, Hoffmann GF, Burgard P. J Newborn screening for methylmalonic acidemias--optimization by statistical parameter combination. Inherit Metab Dis. 2008;31:379–85. doi: 10.1007/s10545-008-0892-z. [DOI] [PubMed] [Google Scholar]
- 12.la Marca G. Mass spectrometry in clinical chemistry: the case of newborn screening. J Pharm Biomed Anal. 2014;101:174–82. doi: 10.1016/j.jpba.2014.03.047. [DOI] [PubMed] [Google Scholar]
- 13.Van Belle G, Fisher LD, Heagerty PJ, Lumley T. Biostatistics: A Methodology for the health sciences. 2. Wiley; 2004. [Google Scholar]
- 14.Ho JK, Duclos RI, Hamilton JA. Interactions of acyl carnitines with model membranes: a (13)C-NMR study. J Lipid Res. 2002;43:1429–39. doi: 10.1194/jlr.m200137-jlr200. [DOI] [PubMed] [Google Scholar]
- 15.Wendel U. Abnormality of odd-numbered long-chain fatty acids in erythrocyte membrane lipids from patients with disorders of propionate metabolism. Pediatr Res. 1989;25:147–50. doi: 10.1203/00006450-198902000-00014. [DOI] [PubMed] [Google Scholar]
- 16.Wendel U, Baumgartner R, van der Meer SB, Spaapen LJ. Accumulation of odd-numbered long-chain fatty acids in fetuses and neonates with inherited disorders of propionate metabolism. Pediatr Res. 1991;29:403–5. doi: 10.1203/00006450-199104000-00015. [DOI] [PubMed] [Google Scholar]
- 17.Wendel U, Zass R, Leupold D. Contribution of odd-numbered fatty acid oxidation to propionate production in neonates with methylmalonic and propionic acidaemias. Eur J Pediatr. 1993;152:1021–3. doi: 10.1007/BF01957229. [DOI] [PubMed] [Google Scholar]
- 18.Ombrone D, Giocaliere E, Forni G, Malvagia S, la Marca G. Expanded newbornscreening by mass spectrometry: New tests, future perspectives. Mass Spectrom Rev. 2015 May 7; doi: 10.1002/mas.21463. [DOI] [PubMed] [Google Scholar]
- 19.Campbell CD, Ganesh J, Ficicioglu C. Two newborns with nutritional vitamin B12 deficiency: challenges in newborn screening for vitamin B12 deficiency. Haematologica. 2005;90(Suppl12):S45. [PubMed] [Google Scholar]
- 20.Stabler SP, Allen RH, Savage DG, Lindenbaum J. Clinical spectrum and diagnosis of cobalamin deficiency. Blood. 1990;76:871–81. [PubMed] [Google Scholar]
- 21.Demir N, Koc A, Üstyol L, Peker E, Abuhandan M. Clinical and neurological findings of severe vitamin B12 deficiency in infancy and importance of early diagnosis and treatment. J Paediatr Child Health. 2013;49:820–4. doi: 10.1111/jpc.12292. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1A: Extracted ion chromatogram of the transition 414>85 from a CblC patient DBS in the Orbitrap MS. Theoretically the peak 3 is the less polar undissociated C17 form where the carboxylic group R-COOH is protonated: pH<pKa. Vice versa the peak 2 corresponds to the more polar dissociated form.
Figure S1B and S1C: MS/MS high resolution fragmentation of peak 2 and 3. Both peaks were recognized with a formula C24H48O4N corresponding to the acylcarnitine C17. Moreover in both product ion scan profiles the fragment 253.2 m/z corresponding to C17H33O acyl group is present
Figure S2A. Distribution of biomarkers in cases (n=23), controls (n=530) and false positives (n=82). Y axis: C3, C17 and Met are expressed as µmol/L
Figure S2B. Distribution of biomarkers in maternal cases (n=23) and controls (n=115)
Y axis: C3, C17 and Met are expressed as µmol/L.
Figure S3. The new flowchart adopted by Tuscany NBS program to identify patients affected by MMA or PA acidemia