Summary
The Drosophila dNab2 protein is an ortholog of human ZC3H14, a poly(A) RNA-binding protein required for intellectual function. dNab2 supports memory and axon projection, but its molecular role in neurons is undefined. Here we present a network of interactions that links dNab2 to cytoplasmic control of neuronal mRNAs in conjunction with and the Fragile-X protein ortholog dFMRP. dNab2 and dfmr1 interact genetically in control of neurodevelopment and olfactory memory and their encoded proteins co-localize in puncta within neuronal processes. dNab2 regulates CaMKII but not futsch mRNA, implying a selective role in control of dFMRP-bound transcripts. Reciprocally, dFMRP and vertebrate FMRP restrict mRNA poly(A)-tail length similar to dNab2/ZC3H14. Parallel studies of murine hippocampal neurons indicate that ZC3H14 is also a cytoplasmic regulator of neuronal mRNAs. In sum these findings suggest that dNab2 represses expression of a subset of dFMRP-target mRNAs, which could underlie brain-specific defects in patients lacking ZC3H14.
Introduction
RNA binding proteins (RBPs) play important roles in the biogenesis and expression of virtually all types of eukaryotic RNAs including protein-coding mRNAs (Moore, 2005). Despite these broad roles, mutations in genes that encode RBPs often lead to tissue-specific disease pathology, particularly within the brain and nervous system (Castello et al., 2013). Examples of this link include the Fragile-X mental retardation protein FMRP and the spinal muscular atrophy protein SMN (Edens et al., 2015; Gross et al., 2012). The prevalence of neurological disorders caused by defects in RBPs likely reflects the enhanced role post-transcriptional mechanisms play in translational control within distal neuronal processes.
The ZC3H14 (zinc finger CCCH-type 14) gene encodes a ubiquitously expressed RBP that is lost in an inherited form of autosomal recessive, non-syndromic intellectual disability (Pak et al., 2011). Patients homozygous for nonsense mutations in ZC3H14 have reduced IQ but lack associated dysmorphic features. Loss of the ubiquitously expressed Drosophila ZC3H14 homolog, dNab2, produces defects in adult viability, motor function, and brain morphology that are fully rescued by neuronal dNab2 re-expression, and partially rescued by human ZC3H14 expression (Kelly et al., 2016; Kelly et al., 2014; Pak et al., 2011). These data reveal an important, and evidently conserved, role for human ZC3H14 and fly dNab2 in neurons.
ZC3H14 and dNab2 are predominantly localized to the nucleus, but are members of a conserved protein family whose founding member, S. cerevisiae Nab2, shuttles between the nucleus and cytoplasm (Green et al., 2002; Leung et al., 2009; Pak et al., 2011). ZC3H14 and dNab2 share a domain structure of an N-terminal PWI (proline/tryptophan/isoleucine)-like domain, a nuclear localization sequence, and five well conserved C-terminal CysCysCysHis (CCCH)-type zinc fingers (ZnFs) (Leung et al., 2009). These ZnF domains bind synthetic polyadenosine RNA probes in vitro (Kelly et al., 2010; Pak et al., 2011), implying that dNab2 and ZC3H14 interact with adenosine-rich tracts in vivo. In support of this hypothesis, ZC3H14 colocalizes with poly(A) mRNA speckles in rodent hippocampal neurons (Pak et al., 2011), and its loss increases bulk poly(A) tail PAT (PAT) length among RNAs in cultured N2a cells (Kelly et al., 2014). dNab2 also restricts PAT length in vivo and genetic interactions between dNab2 and components of the polyadenylation machinery (e.g. the PABP poly(A) binding protein and the hiiragi poly(A) polymerase) indicate that altered PAT length may underlie dNab2 mutant phenotypes (Pak et al., 2011). Altered PAT length can affect multiple steps in RNA metabolism including turnover and translational efficiency (Eichhorn et al., 2016; Subtelny et al., 2014).
dNab2 plays important roles within the central nervous system (CNS). Pan-neuron dNab2 depletion within the peripheral nervous system (PNS) and CNS replicates almost all phenotypes resulting from zygotic loss of dNab2, while dNab2 depletion from motor neurons does not (Pak et al., 2011). Moreover, pan-neuron dNab2 depletion impairs short-term memory and disrupts axon projection into the α/β lobes of the mushroom bodies (MBs) (Kelly et al., 2016), twin neuropil structures in the brain required for associative olfactory learning and memory (Heisenberg, 2003). In dNab2 mutants, β-axons misproject across the brain midline and α-axons show a high frequency of branching defects (Kelly et al., 2016). Selective depletion of dNab2 in Kenyon cells, which give rise to MB α/β axons (Armstrong et al., 1998), is sufficient to phenocopy these dNab2 zygotic defects, and dNab2 re-expression in these cells is sufficient to rescue them (Kelly et al., 2016). However, there is little evidence of how dNab2 regulates bound RNAs and whether this regulation occurs exclusively in the nucleus, as suggested by the nuclear steady-state localization of dNab2, Nab2 and ZC3H14 (Anderson et al., 1993; Leung et al., 2009), or involves a role for dNab2 in cytoplasm.
Here we describe a genetic screen for dNab2 interacting factors in the Drosophila eye that uncovers physical and functional interactions between dNab2 and the Drosophila ortholog of the Fragile X Mental Retardation Protein (FMRP). FMRP is an RBP and is lost in fragile X syndrome (FXS), the most common genetic cause of intellectual disability (Bassell and Warren, 2008). FMRP undergoes nucleocytoplsmic shuttling and is enriched in the cytoplasm at steady-state. Cytoplasmic FMRP regulates ~800 polyadenylated neuronal mRNAs, allowing for finely tuned pre- and post-synaptic translation of their encoded proteins (Darnell et al., 2011; Richter et al., 2015). Genetic interactions between dNab2 and the Drosophila FMRP gene (dfmr1) correspond at a molecular level to an RNAse-resistant physical association of dNab2 and dFMRP proteins in neurons. Within brain neurons, dNab2 and dFMRP co-localize in the soma but are also detected within discrete mRNP-like foci distributed along neuronal processes. A corresponding memory defect in dNab2/+,dfmr1/+ trans-heterozygotes indicates that dNab2 may co-regulate a subset of mRNAs bound by dFMRP. Indeed, dNab2 associates with the dFMRP-regulated mRNA encoding CaMKII (calmodulin-dependent kinase-II) and is required for repression of a CaMKII translational reporter in neurons. By contrast, dNab2 does not appear to regulate a second dFMRP-target mRNA encoding Futsch/Map1β, implying that the spectrum of dNab2-regulated mRNAs only partially overlaps with dFMRP. Moreover, we find evidence that dFMRP/FMRP restrict PAT length of neuronal mRNAs in a manner similar to dNab2/ZC3H14. Finally, we show that ZC3H14 is present in hippocampal axons and dendrites, where it is enriched in RNP and 80S ribosomal fractions. In sum these data represent a significant advance in understanding dNab2/ZC3H14 by defining a role for these disease-associated RBPs in translational control of neuronal mRNAs that, in Drosophila, occurs in conjunction with the dFMRP protein.
Results
dfmr1 alleles interact with a dNab2 transgene in the eye
To identify factors that interact with dNab2 in neurons, we exploited the finding that dNab2 expression in Drosophila retinal cells (GMR-Gal4,UAS-dNab2, hereafter referred to as ‘GMR>dNab2’) produces an rough-eye phenotype that is readily modified (Pak et al., 2011) (Fig. 1A). GMR>dNab2 eyes (“dNab2 o/e” in Fig. 1B) are reduced in size, lack full pigmentation and have disorganized ommatidia, presumably due to effects of excess dNab2 on endogenous retinal RNAs. A selected group of 200 alleles (loss-of-function, RNAi depletion, or EP-type overexpression), corresponding to 135 genes that encode factors with (i) established roles in neurodevelopment or neuronal function, (ii) RNA-binding activity, or (iii) roles in mushroom body (MB) development, were evaluated for modification of the GMR>dNab2 phenotype. This approach identified 15 enhancers corresponding to 10 genes, and 28 suppressors corresponding to 16 genes (Fig. 1A and Table S1). Among the modifiers are alleles of the previously defined dNab2-interacting genes poly(A) binding protein-2 (PABP2) and hiiragi (poly(A) polymerase) (Pak et al., 2011), in addition to previously undefined interactors like the fragile-X syndrome mental retardation ortholog (dfmr1), cytoplasmic poly(A) binding protein (PABC1), and the elongation factor-1α (EF-1 α) and eIF-4e translation factors.
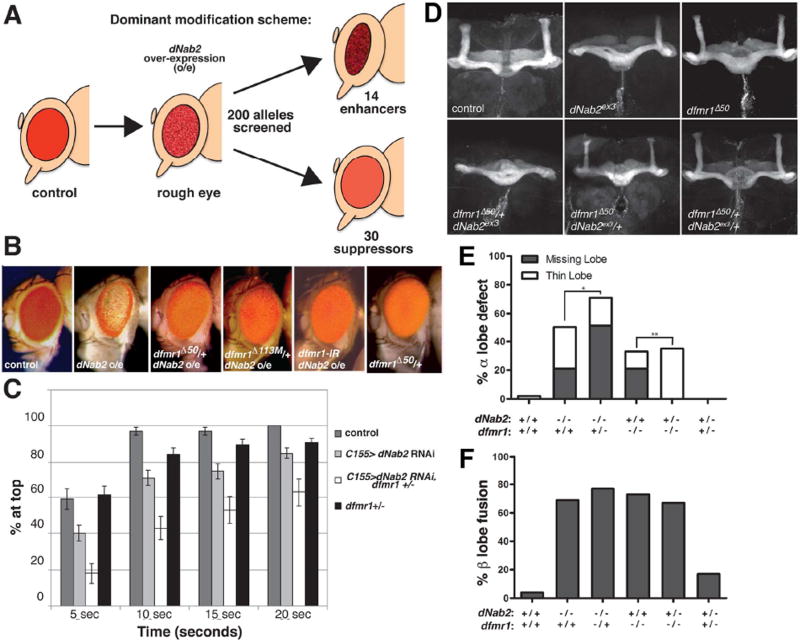
Figure 1. Genetic interactions between dNab2 and dfmr1.
(A) Schematic of the GMR>dNab2 screen. GMR-Gal4 overexpression (o/e) of dNab2 from the dNab2EP3716 allele leads to a rough eye phenotype that was enhanced by 14 and suppressed by 30 of the 200 candidate alleles. (B) Adult eyes from control (GMR-Gal4/+), dNab2 o/e (GMR-Gal4/+ ;dNab2EP3716/+), dNab2 o/e+dfmr1 heterozygote (GMR-Gal 4/+; dfmr1Δ50/dNab2EP3716 and GMR-Gal4/+;dfmr1Δ113/dNab2EP371), dNab2 o/e with dfmr1 RNAi (GMR-Gal 4/+;UAS-dfmr1RNAi/dNab2EP3716) and dfmr1 heterozygote (GMR-Gal4/+ ;dfmr1Δ50/+) adult females. (C) Negative geotaxis behavior of 5-day old control (elavC155-Gal4), pan-neuron dNab2 RNAi (elavC155-Gal4,UAS-dNab2RNAi), pan-neuron dNab2 RNAi+dfmr1 heterozygote (elavC155-Gal4,UAS-dNab2RNRi ,dfmr1Δ113M/+), or dfmr1 heterozygote (elavC155-Gal4,dfmrΔ113/+) flies. Data represent % of flies reaching the cylinder top at each time point. Each genotype represents ≥10 independent trials (10 flies/trial). Error bars=SD. (C) Anti-Fas2 stained wildtype (wt; isogenic precise excision pex41 of the element used to create dNab2ex3), dNab2 null (dNab2ex3/ex3 ), dfmr1 null (dfmr1Δ50/Δ50), dNab2 null with one copy of dmr1 (dNab2ex3 ,dfmr1Δ50/dNab2ex3, +), dfmr1 null lacking one copy of dNab2 (dNab2ex3 ,dfmr1Δ50/+,dfmr1Δ50), or trans-heterozygote (dNab2ex3,dfmr1Δ50/+,+) brains. Penetrance of (D) α-lobe or (C) β-lobe defects in the same genotypes as C with individual lobes counted as discrete events (≥24 brains per genotype). *p=4.8×10 and **p=1.5×10 (Chi square test).
Multiple GMR>dNab2 modifier alleles correspond to factors that act within a translational pathway centered on dfmr1 (bolded in Table S1). The dfmr1Δ50 and dfmr1Δ113 loss-of-function alleles each dominantly suppress GMR>dNab2, as does co-expression of a dfmr1 RNAi transgene, indicating that Drosophila FMRP (dFMRP), is required for excess dNab2 to disrupt eye morphology (Fig. 1B). Moreover, UAS-dNab2 and UAS-dfmr1 transgenes are individually viable but synthetically lethal when co-expressed with GMR-Gal4 in retinal neurons. The basis for this synthetic effect could be enhancement of dfmr1-induced apoptosis reported in earlier studies (Wan et al., 2000). dfmr1-interacting genes also modify the GMR>dNab2 phenotype (Table S1), including the miR components Ago1 and Gw182, the Rm62/dmp68 RNA helicase, the RBPs staufen and Ataxin-2, and Timp, a protease inhibitor implicated in synaptic FXS overgrowth in mice and flies (Barbee et al., 2006; Cziko et al., 2009; Jin et al., 2004; Siller and Broadie, 2011; Sudhakaran et al., 2014). This pattern of genetic links suggests that dNab2 may interact with the dFMRP pathway in retinal neurons.
dfmr1 alleles modify locomotor and mushroom body phenotypes caused by dNab2 loss
Interactions between dNab2 and dfmr1 alleles were examined in two additional neuronal contexts: locomotor behavior and MB development. Pan-neuronal RNAi of dNab2 (elavC155>dNab2RNAi) causes a locomotor defect in a negative geotaxis assay (Pak et al., 2011) that is dominantly enhanced by the dfmr1Δ113M null allele (Fig. 1C), which is consistent with its suppressive effect on gain-of-function GMR-dNab2. This dNab2RNAi locomotor defect is enhanced by overexpression of dfmr1, indicating that dFMRP and dNab2 are not redundant in this context (Fig. S1). Endogenous dNab2 and dFMRP are both expressed within Kenyon neurons whose axons branch to form the MB lobes (Bossie et al., 1992; Kelly et al., 2016; Michel et al., 2004). Null alleles of dNab2 and dfmr1 (dNab2ex3 and dfmr1Δ50) elicit similar MB defects, including missing or thinned α-lobes, that occur with similar severity and penetrance (Fig. 1D,E) (Kelly et al., 2016; Michel et al., 2004)) and are reciprocally sensitive to the genetic dose of the other factor: dfmr1Δ50 and the weaker dfmr1Δ113 allele (Michel et al., 2004) dominantly increase the frequency of α-lobe defects in dNab2ex3 mutants, while dNab2ex3 dominantly rescues α-lobe defects in dfmr1Δ50 and dfmr1Δ113 mutants (Fig. 1D–E and S2). These opposing effects imply that dNab2 is required for normal α-lobe development but supports aberrant α-lobe development in the absence of dFMRP. This dependence on dFMRP status could reflect linked or sequential roles for dNab2 and dFMRP on a shared cohort of RNAs. The lack of genetic interactions between dNab2 and dfmr1 in β-lobe axons (Fig. 1F and S2) could indicate that dNab2-dFMRP co-regulate α-lobe-specific RNAs. Expression of a UAS-dfmr1 transgene in dNab2ex3 neurons severely disrupts MBs (Fig. S2), again arguing that dFMRP and dNab2 may interact functionally but are not redundant.
dNab2 co-localizes with dFMRP in neurites
The genetic links between dNab2 and dfmr1 suggest that their encoded proteins might associate within neurons. At steady-state, dNab2 localizes to nuclei (Kelly et al., 2012; Pak et al., 2011) while dFMRP is cytoplasmic (Santos et al., 2014). However, homologs of both proteins undergo nucleocytoplasmic shuttling in association with bound RNAs (Feng et al., 1997; Green et al., 2002; Kim et al., 2009). As previously reported (Pak et al., 2011), dNab2 is enriched in neuronal nuclei of 3-day old cultured adult brain neurons co-stained with anti-dNab2 antibody and anti-HRP to visualize neuronal membranes (three examples in Fig. 2A–C). However, two-thirds of neurons also contain a punctate pool dNab2 distributed into the cytoplasm of neuronal processes (right panels in Figs. 2A–C are magnified views of single processes) that is absent in dNab2 null neurons (Fig. 2D). Approximately 80% of cultured, CD8:GFP-labelled Kenyon cells (OK107-Gal4,UAS-CD8:GFP) also contain dNab2 puncta in processes (Fig. 2E). The absence of cytoplasmic dNab2 in some Kenyon cells could reflect lobe-specific differences (e.g. α,β,γ-lobe) or developmental age (e.g. early vs. late born neurons) (Kunz et al., 2012). In aggregate, these data reveal that dNab2 localizes to the nuclei and distal processes of neurons.
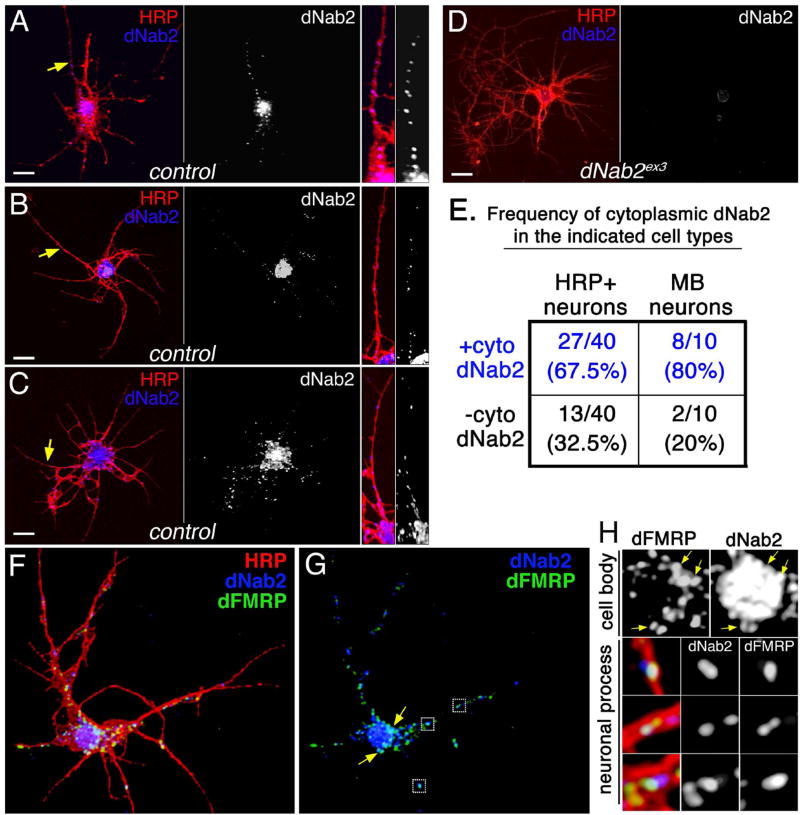
Figure 2. dNab2 colocalizes with dFMRP in mRNP-like puncta.
(A-C) Control (wt) or (D) dNab2 null (ex3) 24hr APF (after puparium formation) brain neurons cultured for 72hr and labeled with anti-HRP (red; neuronal membranes) and anti-dNab2 (blue). Scale bar=10µm. Rightmost panels in A-C are magnified views of dNab2 puncta in neurites (yellow arrows). (E) Frequency of cytoplasmic dNab2 in wt neurons (“brain neurons”; left) or Kenyon cells (“MB neurons”; right) labeled by CD8:GFP expression (CD8-GFP/+;;OK107>Gal4/+). (F-H) A single wt 24h APF brain neuron triple labeled with anti-HRP (red), anti-dFMRP (green), anti-dNab2 (blue). (G) Overlapping dNab2:dFMRP signals in the cell body (arrows) or neuronal process (boxes). (H) Magnified views of regions highlighted in G showing colocalization of dNab2 and dFMRP in the soma (“cell body”; see arrows) and processes (“neuronal process”).
Given the genetic interactions between dNab2 and dfmr1, antibodies to these two RBPs were used to assess their colocalization in the cytoplasm of cultured brain neurons. As described previously dFMRP is detected at low levels in the nucleus, higher levels in the cell body cytoplasm, and in puncta that distribute along the length of processes (Fig. 2F–H) (Barbee et al., 2006; Cziko et al., 2009; Feng et al., 1997; Wan et al., 2000). These puncta resemble reported dFMRP-containing mRNPs that contain other RNA processing factors such as PABC (Cziko et al., 2009), which is a genetic modifier of GMR>dNab2 (Table S1). Significantly, dNab2 colocalizes with dFMRP puncta in the cell body (yellow arrows in Fig. 2G and corresponding magnified views in Fig. 2H, cell body) and in neuronal processes (boxed regions in Fig. 2G, and corresponding magnified views in Fig. 2H, neuronal process). Quantification of this overlap within processes indicates that ~20% of dNab2 overlaps with dFMRP-positive puncta, while ~25% of dFMRP overlaps with dNab2-positive puncta (by Manders Overlap Coefficient, n=12 processes). These data suggest that dNab2 is a component of some dFMRP granules in brain neurons and provide a potential molecular context for the observed genetic interactions between dNab2 and dfmr1.
The dNab2 and dFMRP proteins associate and support olfactory memory
An adapted version of the RNA-tagging technique (Yang et al., 2005) was used to assess physical interaction of dNab2 and dFMRP in brain neurons. Briefly, head lysates of flies expressing either Flag-dNab2 or Flag-hPABP (human poly(A) RNA binding protein) in neurons (elavC155>UAS-Flag-Nab2 or Flag-hPABP) were precipitated with anti-Flag (Fig. 3A), then probed to detect recovery of Flag-dNab2 or Flag-hPABP (“anti-Flag” panel), or with anti-dFMRP (6A15, Morales et al., 2002) to detect co-purifying endogenous dFMRP (Fig. 3B) (“anti-dFMRP” panel). Notably, dFMRP is detected in Flag-dNab2 precipitates but not in Flag-hPABP or control (elavC155-Gal4 alone) precipitates. Addition of RNAse does not block recovery of dFMRP with Flag-dNab2, indicating that this association is RNAse-resistant (Fig. S3).
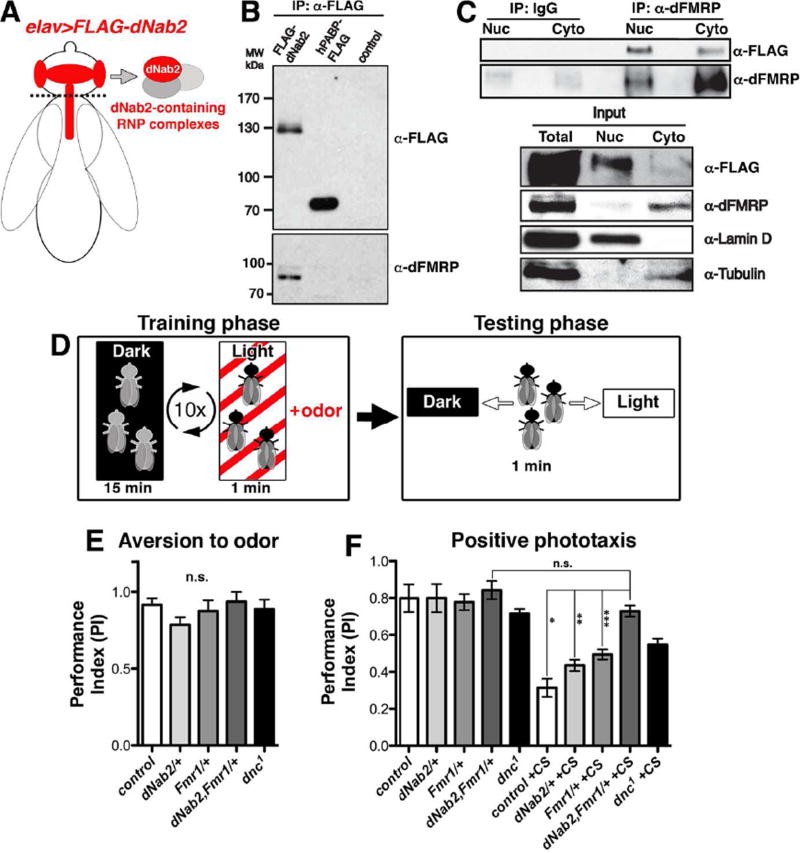
Figure 3. dNab2 and dFMRP associate and co-regulate olfactory memory.
(A) Schematic of the Flag-dNab2 transgenic system (elavC155-Gal4;;UAS-Flag-dNab2/+) used to recover Flag-dNab2-associated proteins in B and C. (B) Anti-Flag (top) or anti-dFMRP (bottom) immunoblots of anti-Flag immunoprecipitates (IPs) from elav>Flag-dNab2, hPABP-Flag (elavC155-Gal4;UAS-hPABP-Flag/+), or Gal4 only (“control”; elavC155-Gal4) adult heads. (C) Fractionated cytoplasmic (Cyto) and nuclear (Nuc) lysates from whole elav>Flag-dNab2 adults IPed with control IgG or anti-dFMRP (mAb 6A15) and immunoblotted for the Flag epitope or dFMRP. Input and fractionation controls (anti-Lamin and Tubulin) are indicated. (D) Scheme of the aversive olfactory conditioning system used in E and F. (E) Performance index (PI) of methylcyclohexanol (MCH) aversion in the indicated genotypes: control (white: w+,iso1), dNab2/+ (light grey: w+;;dNab2ex3/+), dfmr1/+ (grey: w+;;dfmr1Δ50/+), dNab2,dfmr1/+ (dark grey: w+;;dNab2ex3,dfmr1Δ50/+,+), or dnc1 (black) (n.s.=not significant). (F) PI indices of were untrained (left) or trained (+CS, right) genotypes in E. Error bars=SEM (n.s.=non-significant, p=0.15; *p=0.002, **p=.002, and ***p=0.005).
To confirm the dNab2-dFMRP association and biochemically text its localization, flies expressing neuronal Flag-dNab2 were separated into nuclear (Nuc) and cytoplasmic (Cyto) fractions and then subject to IP for endogenous dFMRP (Fig. 3C). Fractionation was confirmed with Lamin-D (nucleus) and β-Tubulin (cytoplasm) antibodies. Although dNab2 and dFMRP show inverse patterns of enrichment in the nucleus and cytoplasm, dNab2 is recovered in association with dFMRP from both compartments (Fig. 3C). These biochemical data support the microscopy data in Fig. 2A–C and provide additional evidence that dNab2 physically associates with dFMRP in multiple neuronal compartments.
dNab2 is required for courtship conditioning (Kelly et al., 2016), suggesting that it may regulate memory in conjunction with dFMRP. We therefore used an aversive olfactory conditioning assay (see Fig. 3D) to test whether heterozygosity for dNab2 could sensitize memory circuits to loss of a single copy of dfmr1 (i.e. trans-heterozygotes). Control adult flies (Fig. 3E–F, white bars) display a strong positive response to light (phototaxis) that can be suppressed by a training regimen of ten (10) rounds of light-exposure paired with the aversive odor methylcyclohexanol (MCH) (conditioned stimulus; +CS) followed by a period in darkness without MCH. Unconditioned dNab2ex3,dfmr1Δ50 trans-heterozygotes (Fig. 3E–F, dark grey bars) exhibit strong responses to light exposure and MCH when these stimuli are tested individually, but impaired MCH-induced suppression of phototaxis relative to control wildtype animals (white bars) or those carrying only the dNab2ex3 allele (light grey bar) or dfmr1Δ50 (grey bars) allele. Importantly, the memory defect in dNab2ex3,dfmr1Δ50 trans-heterozygotes is enhanced relative to the mild defect in dfmr1Δ50 heterozygotes (Fig. 3F) (see also Cziko et al., 2009; Sudhakaran et al., 2014), indicating that reduced dFMRP renders olfactory memory pathways sensitive to dNab2 dosage. The hypomorphic allele of dnc (dnc1), which encodes a cyclic AMP phosphodiesterase required for memory (Tully and Quinn, 1985), also shows a memory defect in phototaxis suppression (Fig. 3E–F, black bars), confirming the utility of the assay.
dNab2 interacts with the CaMKII mRNA and represses a CaMKII translational reporter
The data presented above suggest that dNab2 and dFMRP may co-regulate mRNAs encoding learning and memory factors. The CaMKII mRNA is among the most well-validated dFMRP/FMRP targets in Drosophila and mammals (Darnell et al., 2011; Zalfa et al., 2003), and encodes a kinase that plays a critical role in synaptic strengthening during learning and memory (Ashraf et al., 2006; Chen et al., 2012; Griffith et al., 1993; Malik et al., 2013; Malik and Hodge, 2014). The RNA-tagging technique was used to test association of Flag-dNab2 and CaMKII mRNA in neurons. Precipitation of Flag-dNab2 from elavC155>Flag-dNab2 head lysates strongly enriches for CaMKII mRNA, but not the abundant, polyadenylated rp49 mRNA (Chintapalli et al., 2013) (Fig. 4A). dNab2 thus appears to show in vivo specificity in its association with polyadenylated transcripts.
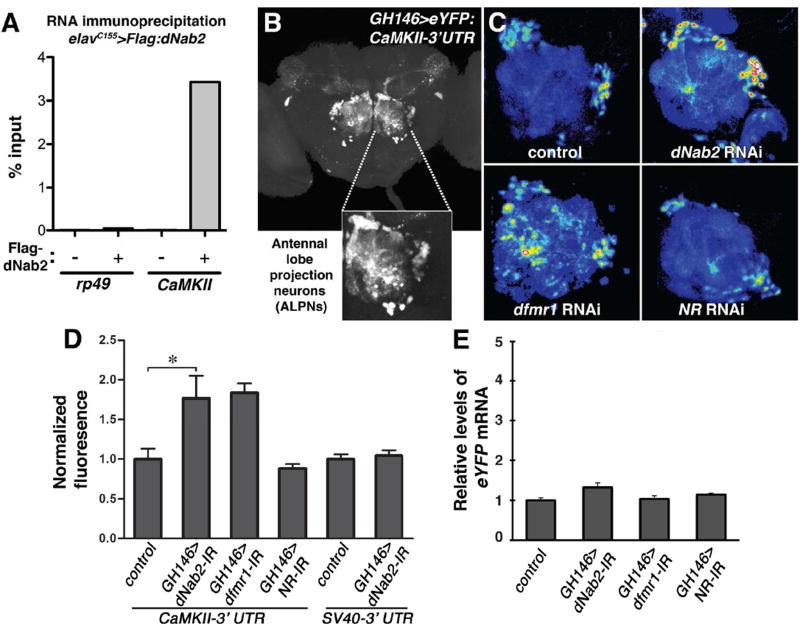
Figure 4. dNab2 regulates the CaMKII 3’UTR and associates with CaMKII mRNA.
(A) qPCR to detect rp49 and CaMKII transcripts in anti-Flag immunoprecipitates of elavC155>Flag-dNab2 heads. Percent of input mRNA recovered by IP is indicated. Note enrichment of CaMKII relative to rp49. (B) Confocal image of CaMKII-3’UTR reporter (GH146>eYFP:CaMKII-3’UTR) expression in a wt adult brain with magnified view of eYFP+ antennal lobe projection neurons (ALNPs). (C) Reporter expression in control, dNab2 RNAi (UAS-dNab2RNAi), dfmr1 RNAi (UAS-dfmr1RNAi), and NR1 RNAi (UAS-NMDAR-1RNAi) in GH146-Gal4 ALPNs. Expression represented as a 16-color intensity scale. (D) Mean eYFP fluorescence values of CaMKII-3’UTR and SV40-3’UTR reporters for indicated genotypes. Data are normalized to mean fluorescence of GH146-Gal4,UAS-eYFP:CaMKII-3’UTR or GH146-Gal4,UAS-eYFP:SV40-3’UTR ALPNs. Error bars=SEM (*p<0.05). (E) qPCR analysis of eYFP:CaMKII-3’UTR mRNA in brains of the indicated genotypes.
The evidence of physical association of dNab2 with CamKII was complemented by analysis of an in vivo reporter that detects regulatory inputs into 3’-sequences of the CamKII mRNA. This reporter contains the CaMKII 3’-untranslated region (UTR) fused to a Gal4-inducible eYFP coding-sequence, and is sensitive to dFMRP-mediated repression in antennal lobe projection neurons (ALPNs) (Ashraf et al., 2006; Sudhakaran et al., 2014). Expression of the CaMKII reporter (GH146-Gal4>UAS-eYFP:CaMKII-3’UTR) in ALPNs leads to eYFP fluoresence in the cell bodies and dendrites (Fig. 4B). As described previously (Sudhakaran et al., 2014), dFMRP RNAi increases eYFP fluorescence in ALNPs approximately two-fold, while RNAi of the NMDA receptor (NR1) has no effect (Fig. 4C–D). RNAi of dNab2 in ALPNs elevates eYFP expression to a similar extent as dfmr1 RNAi, but has no effect on an unrelated reporter comprised of eGFP fused to the SV40-3’UTR. qPCR confirms that the effects of dNab2 and dFMRP RNAi on eYFP fluorescence occur without a substantial effect on steady-state levels of the hybrid eYFP:CaMKII-3’UTR mRNA (Fig. 4E). dNab2 overexpression does not suppress the effect of dfmr1 RNAi on eYFP:CaMKII-3’UTR expression (Fig. S4), indicating that dNab2 is not redundant to dFMRP in translational effects mediated though the CaMKII 3’ UTR.
dNab2 plays a minor role in futsch regulation
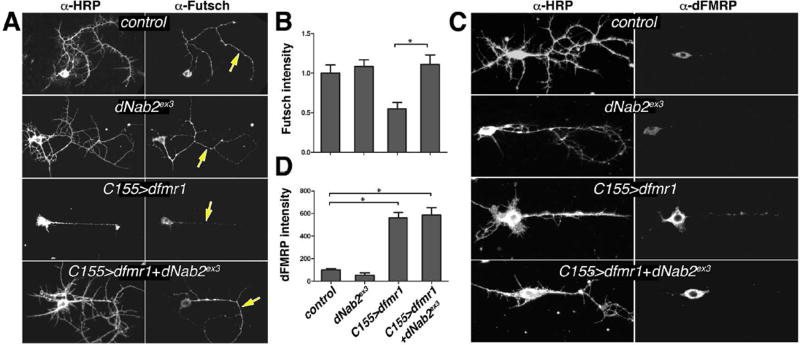
The interaction between dNab2 and CaMKII mRNA prompted analysis of futsch, a second dFMRP-target mRNA. Futsch is an ortholog of the microtubule-associated protein-1β (Map-1β) and its mRNA is a conserved target of dFMRP and FMRP (Hummel et al., 2000; Lu et al., 2004; Zhang et al., 2001). Excess Futsch promotes synaptic growth at the larval neuromuscular junction (NMJ) of dfmr1 mutant larvae (Roos et al., 2000; Zhang et al., 2001). Notably, dNab2 alleles have no effect on NMJ growth (Pak et al., 2011), suggesting that dNab2 may not regulate Futsch in vivo. Consistent with this hypothesis, the levels of Futsch protein are unaltered in dNab2-null brain neurons as assessed by anti-Futsch staining intensity (Fig. 5A–C), a technique used previously to assess the dFMRP regulation of Futsch at NMJs (Coyne et al., 2015). futsch mRNA is also not significantly enriched in IPs of neuronal Flag-dNab2 relative to control brains (Fig. S5), suggesting that futsch mRNA does not associate with dNab2 in brains. Consistent with its role as a repressor of Futsch translation (Zhang et al., 2001), dFMRP expression reduces Futsch levels in cultured brain neurons relative to controls (Fig. 5A,B), especially in shafts of major neuronal processes (yellow arrows). Notably, dNab2 loss suppresses this effect without effecting dfmr1 transgene expression (Fig. 5A–D), arguing that dNab2 may be ectopically recruited to regulate futsch when dFMRP is overexpresed. However, the lack of effect of dNab2 alleles on Futsch levels argues that dNab2 is not normally required to repress futsch mRNA in neurons.
Figure 5. dNab2 plays a more minor role in futsch regulation.
Paired images of (A) anti-Futsch or (C) anti-dFMRP labelled 24h APF brain neurons co-stained with anti-HRP. Genotypes: control (elavC155), dNab2ex3 (elavC155;dNab2ex3/ex3), C155>dfmr1 (elavC155>UAS-dfmr1), or C155>dfmr1+dNab2ex3 (elavC155>UAS-dfmr1;dNab2ex3/ex3). Yellow arrows highlight differences in Futsch staining in central processes. Quantitation of (B) Futsch (n=15 shafts) or (D) dFMRP (n=12 shafts) levels presented as mean fluorescence intensity from individual neuronal processes among the same genotypes as in A and C. Data are normalized to control (elavC155) in each graph. Error bars=SEM (*p<0.05).
dFMRP and FMRP restrict poly(A) tail (PAT) length
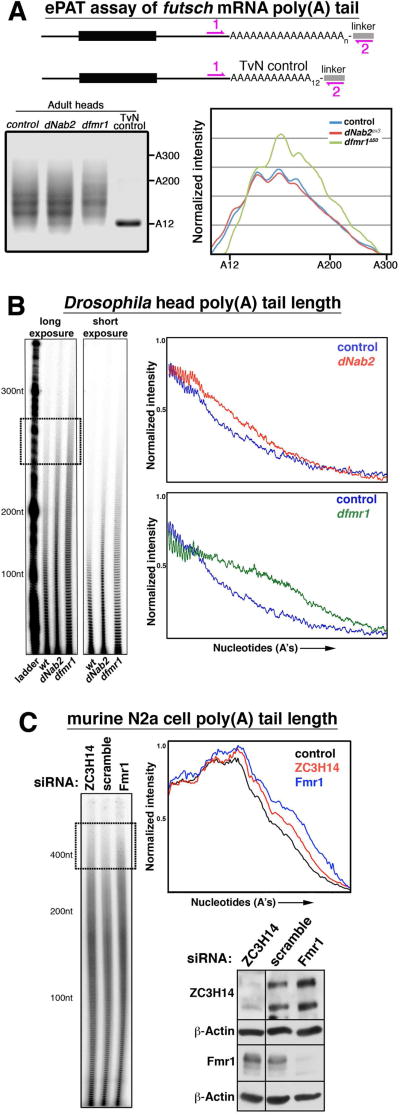
The differential requirement for dNab2 and dFMRP in Futsch regulation prompted analysis of dNab2 or dFMRP loss on futsch PAT length using the ePAT assay (Fig. 6A). Consistent the observation that dNab2 loss does not elevate Futsch protein levels in individual neurons, loss of dNab2 also had no effect on futsch PAT length relative to controls. futsch is thus the first mRNA identified whose poly(A) tail length is regulated independently of dNab2. By contrast, futsch PAT length is extended in dfmr1 mutants heads (Fig. 6A, gel lane 3 and graph). Given the large number of FMRP/dFMRP mRNA targets, we next tested the effect of dFMRP/FMRP loss on bulk PAT length in adult Drosophila heads and cultured mouse N2a neuroblastoma cells. Remarkably, dfmr1 mutant adult heads and FMRP-depleted N2a cells both show elongated PAT lengths to a degree that mirrors or exceeds the effect of dNab2/ZC3H14 loss (Fig. 6B–C). These data indicate that the role of dFMRP in control of futsch expression is paralleled by a role in limiting futsch PAT length in vivo that is not shared by dNab2, and that dFMRP/FMRP appears to be required to restrict bulk PAT length in neurons.
Figure 6. Effect of dFMRP/FMRP loss onPAT length.
(A) Schematic of extended poly(A) tail length (ePAT) assay using linker PCR amplification of futsch PAT and the TvN control fragment (12 adenosines, “A12”) from control, dNab2 null (dNab2ex3, dNab2ex3) or dfmr1 null (dfmr1Δ50/Δ50) heads. Size standards indicated (A200/A300). Right panel=densitometry trace of the PCR products. (B) Bulk PAT length among total RNAs harvested from adult heads of the same genotypes in A. Short and long exposures (with size “ladder”) are shown, along with densitometry traces of each lane normalized for band intensity. (C) Bulk PAT length in N2a cells treated with ZC3H14, Fmr1, or scramble siRNAs and accompanying densitometry trace. Sizes are indicated. Boxed region highlights elongated PATs in the ~400A size range in ZC3H14 and Fmr1 siRNA cells. Western confirmation of siRNA knockdown is shown.
ZC3H14 localizes to axons and dendrites and associates with RNPs
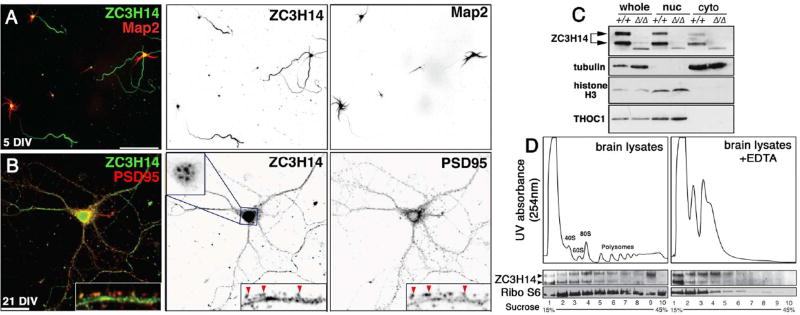
The finding that dNab2 localizes to neurons prompted analysis of the subcellular distribution of ZC3H14 in cultured hippocampal neurons. An anti-ZC3H14 antibody (Leung et al., 2009) detects ZC3H14 in hippocampal nuclei (as described in (Pak et al., 2011) (Fig. 7A–B) but also in cytoplasmic processes of differentiated hippocampal neurons after either 5 or 21 days in vitro culture (DIV). In 5-DIV neurons, cytoplasmic ZC3H14 is enriched in Tau-positive axons relative to Map2-positive dendrites (Fig. 7A). At 21-DIV, ZC3H14 is distributed into dendrites and PSD95-positive dendritic spines with well-elaborated dendritic arbors (Fig. 7B, arrowheads). ZC3H14 is also detected in the cytoplasmic fraction of murine brains (doublet in Fig. 7C). Recovery of THOC1, a nuclear RBP (Li et al., 2005), in the nuclear fraction confirms that biochemical evidence of cytoplasmic ZC3H14 is not a non-specific pattern common to all RBPs. Anti-ZC3H14 specificity was confirmed with lysates generated from ZC3H14Δex13/Δex13 knockout mouse brains (Rha et al., 2017).
Figure 7. ZC3H14 localizes to axons in primary hippocampal neurons and associates with polyribosomes in mouse cortical lysates.
(A,B) Confocal images of primary hippocampal neurons cultured 5 or 21 days in vitro (DIV) from P1 (post-natal day 1) mice and stained with anti-ZC3H14 (green) and (A) anti-Map2 (red) or (B) anti-PSD95 (red). Scale bars=50 µm. Individual channels are presented as inverted grayscale images. Magnified insets in B show distribution of ZC3H14 in dendritic shafts and PSD95-positive spines (red arrowheads). ZC3H14 in 21 DIV neurons is shown at reduced gain in order to resolve nuclear speckles in this cell type (Pak et al., 2011). (C) Immunoblot of fractionated Zc3h14+/+ and Zc3h14Δ13/Δ13 brains to detect ZC3H14, α-Tubulin (cytoplasmic marker), Histone H3 (nuclear marker), and THOC1, a nuclear RBP. (D) Cytoplasmic polysome profiles of wt P13 brain cortexes across a 15–45% linear sucrose gradient prepared +/− EDTA with 254nm absorption profiles (ribosomes and polysomes are indicated). Lower panels show immunoblot for ZC3H14 and S6 ribosomal protein (Ribo S6) across the indicated fractions.
The pool of ZC3H14 protein that distributes into distal hippocampal processes is likely to be part of larger mRNP complexes that modulate mRNA processing and translation (Donlin-Asp et al., 2017). This hypothesis was tested by linear sucrose density gradient fractionation of cytoplasmic P13 (postnatal day 13) mouse brain lysates generated in the presence or absence of the Ca+2 chelator EDTA, which disrupts mRNP complexes, including mono- and polyribosomes (Stefani et al., 2004) (Fig. 7D). In untreated cytoplasmic brain lysates, ZC3H14 co-sediments into multiple fractions across the sucrose density gradient, showing enrichment in fractions that contain 80S mono-ribosomes (Fig. 7D, left panel). Addition of EDTA results in a dramatic shift of ZC3H14 into lighter fractions and disruption of RNP complexes, as indicated by the loss of polyribosome peaks in the RNA absorption profile and a shift of the ribosomal S6 protein, a component of the 40S subunit (Roux et al., 2007) (Fig. 7D, right panel). A parallel analysis of cytoplasmic lysates generated from cultured cells confirms the effect of EDTA on ZC3H14-containing complexes and the P0 protein, a subunit of the 60S ribosomal subunit (Fig. S6). Addition of puromycin, which disrupts translating ribosomes (Franklin and Godfrey, 1966), also depletes a fraction of ZC3H14 that co-sediments with polyribosomes (Fig. S6; see asterisks, lanes 6–9). In aggregate, these data indicate that endogenous ZC3H14 localizes to nuclei, cell bodies, and distal neuronal compartments, including presynaptic axons and postsynaptic dendrites and spines, where it is principally found in RNPs and 80S ribosomal complexes with likely roles in regulating RNA translation.
Discussion
Here we report the results of a candidate-based screen for factors that interact genetically with the Drosophila dNab2 gene, which encodes an RBP whose human ortholog is lost in an inherited intellectual disability. Identified interators include components of the translation machinery (PABC1, EF-1α and eIF-4e) and elements of a pathway centered on the Drosophila ortholog of the FMRP translational repressor (dfmr1 itself, Argonaute-1, Gw182, Rm62, staufen, and Ataxin-2), suggesting that dNab2 functions within the dFMRP pathway. Additional genetic tests support this hypothesis. dfmr1 alleles suppress a rough-eye phenotype caused by transgenic expression of dNab2 in retinal neurons, while dfmr1 alleles enhance a locomotor defect caused by neuronal RNAi of dNab2. Genetic interactions also occur in the CNS, where dfmr1 heterozygosity enhances the frequency of MB α-lobe defects in dNab2 mutants. Notably dNab2 heterozygosity suppresses MB α-lobe defects in dfmr1 mutants, implying a functional hierarchy in which dNab2 effects are dependent on dFMRP status. The inability of either RBP to rescue phenotypes caused by loss of the other argues for a model in which dNab2 and dFMRP participate in common mechanism(s) but are not functionally redundant.
Genetic interactions between the dNab2 and dfmr1 genes are paralleled by a dNab2:dFMRP protein complex detected in neurons. This dNab2:dFMRP interaction, which could involve other factors, includes a cytoplasmic pool of dNab2 that partially co-localizes with dFMRP in mRNP-like granules in neuronal processes, suggesting that the two RBPs may associate with some of the same RNAs. Indeed, dNab2 can interact with and regulate the CaMKII mRNA, a dFMRP target, but is not required to regulate futsch, a second dFMRP target. The finding that trans-heterozygosity for dNab2 and dfmr1 impairs olfactory memory provides additional evidence that dNab2:dFMRP co-regulate some neuronal mRNAs. Finally, we find that murine ZC3H14 is also present in axons and dendrites of murine hippocampal neurons, and associates with mRNPs and elements of the translational machinery. FMRP also localizes to dendrites and axons, and regulates filopodial dynamics and motility of axonal growth cones (e.g. Antar et al., 2006). In aggregate, these data significantly advance our understanding of the role of dNab2/ZC3H14 proteins in neurons by defining a cytoplasmic pool of these proteins associated with translational control of mRNAs that, in Drosophila, occurs in conjunction with dFMRP.
This study highlights the dNab2:dFMRP association but also suggests that dNab2 can function independently of dFMRP. For example, dNab2 and dFMRP are each required for MB αβ-lobe structure (Kelly et al., 2016; Michel et al., 2004), yet dosage sensitive interactions between dNab2 and dfmr1 alleles are only evident in α-lobes, suggesting that dNab2 and dFMRP may co-regulate RNAs within specific axon branches. In addition, dNab2 selectively regulates CaMKII but not futsch, and that asymmetry is reflected at the level of the futsch PAT, which is unchanged in dNab2 mutant brains but extended in dfmr1 mutant brains. The failure of dNab2 alleles to alter Futsch protein levels is consistent with their lack of effect on the Futsch-dependent process of NMJ development (Pak et al., 2011). Together, these data suggest that the futsch mRNA is not a physiologic target of dNab2 and that dNab2 only regulates a subset of dFMRP-bound transcripts.
dFMRP protein is a well-established translational repressor, but the data reveal a previously unappreciated requirement for dFMRP/FMRP to inhibit mRNA PAT length, which in the case of futsch is likely to stem from a direct binding by dFMRP. These effects on PAT length could simply be a secondary consequence of enhanced futsch translation in dfmr1/Fmr1 mutant cells. However, loss of the cytoplasmic polyadenylation element binding protein (CPEB), which promotes cytoplasmic PAT extension in mammals and flies (Cziko et al., 2009; Keleman et al., 2007; Mastushita-Sakai et al., 2010; Udagawa et al., 2012), rescues FXS phenotypes in Fmr1 knockout mice (Udagawa et al., 2013). One interpretation of this result is that inappropriate PAT elongation contributes to excess translation in FXS, similar to the positive correlation between PAT length and translation observed among germline and embryonic mRNAs (Eichhorn et al., 2016; Subtelny et al., 2014). These data thus raise the possibility that altered mRNA polyadenylation may be an unappreciated feature of translational dysregulation in neurons lacking dfmr1/Fmr1.
The dNab2:dFMRP complex suggests that dNab2 may regulate gene expression through its interaction with dFMRP. FMRP inhibits translational initiation (Napoli et al., 2008; Schenck et al., 2003; Schenck et al., 2001), blocks ribosome movement along polyribosome-associated mRNAs (Darnell and Klann, 2013), and interacts with elements of the microRNA machinery (Bozzetti et al., 2015; Caudy et al., 2002; Ishizuka et al., 2002; Muddashetty et al., 2011). The dNab2-sensitive CaMKII 3’UTR GFP sensor is also regulated by the miRNA pathway (Ashraf et al., 2006; Sudhakaran et al., 2014), and multiple factors involved in microRNA-induced silencing interact genetically with dNab2 (see Table S1). The precise role dNab2 plays on bound mRNAs is not clear. PAT elongation induced by dNab2 loss could enhance recruitment of cytoplasmic PABPs that promote translation-coupled circularization of mRNAs (Preiss and Hentze, 1999). dNab2 and its ortholog ZC3H14 both repress PAT length and may thus indirectly limit cytoplasmic PABPs binding to key transcripts. Alternatively, they may directly compete with these PABPs for binding to polyadenosine tails, and thus occlude access of other factors involved in translation.
Consistent with the role of dNab2 in translational regulation, its ortholog ZC3H14 localizes to axons, dendrites, and dendritic spines in hippocampal neurons and co-sediments with 80S ribosomes. FMRP is primarily associated with polysomes, and can inhibit translation by ribosome stalling (e.g. Darnell et al., 2011). Intriguingly, the FMRP-target mRNA CamK11a mRNA is enriched in anti-ZC3H14 precipitates and CaMKIIa levels increase in the hippocampus of Zc3h14Δ13/Δ13 knockout mice compared to control mice (Rha et al., 2017), raising the possibility that Drosophila and vertebrate CaMKII mRNAs are conserved targets of dNab2/ZC3H14. Intriguingly, the FMRP-related protein Fxr1 (Morales et al., 2002; Stackpole et al., 2014) co-precipitates with the zinc-finger domain of ZC3H14 (Hu and Gao, 2014), suggesting that ZC3H14 may interact with FMRP family members in a manner analogous to dNab2 and dFMRP.
In sum, the data presented here provide evidence that the dNab2 localizes both to the nucleus and cytoplasm of Drosophila neuronal processes, and that it interacts physically and functionally with the dFMRP protein. Additional data provide evidence of an equivalent pool of cytoplasmic ZC3H14 that interacts with RNP complexes found in the axons and dendrites in the mouse brain. Given the link between FMRP and intellectual disability in humans (Santoro et al., 2011), these interactions raise the question of whether defects in translational silencing of mRNAs transported to distal sites within neuronal processes contribute to neurodevelopmental and cognitive defects in Drosophila lacking dNab2 or in humans lacking ZC3H14.
Experimental Procedures
Drosophila genetics
Crosses were maintained in 25°C humidified incubators with 12hr light-dark cycles. The ex3, pex41 (precise excision 41) and UAS-Flag-dNab2 alleles have been described previously (Pak et al., 2011). Modifier stocks are identified by source/stock in Table S1. Drivers: GMR (BL1350), elavC155 (BL458), OK107 (BL854), and GH146 (BL30026). Alleles: dNab2EP3716 (UAS-dNab2, BL17159), dfmr1Δ50 (BL6930), dfmr1Δ113M (BL6929), dnc1 (BL6020), UAS-NR1IR (BL25941), UAS-CD8-GFP (Lee and Luo, 1999), UAS-dNab2IR (VDRC 27487), UAS-dfmr1IR (BL35200), Pabp2EP2264 (gift of M. Simonelig), UAS-dfmr1 (gift of T. Jongens), UAS-eYFP-CaMKII-3’UTR (gift of S. Kunes), and UAS-GFP-SV40-3’UTR (gift of D. Bilder).
Behavioral assays
Negative geotaxis was tested as described previously (Pak et al., 2011). Aversive olfactory conditioning was performed essentially as described (Krashes and Waddell, 2011). Males were outcrossed to Oregon-R virgins to generate F1s with wildtype visual acuity (e.g. w+/w−;;dNab2ex3/+). The w+,dnc1 allele was tested directly. Groups of thirty (15 male:15 female) 3-day old adults were aged o/n in fresh vials then tested for light:dark preference in an optically sealed T-maze, or for odor avoidance in darkness with a 1cm square of Whatman with 30ul methocyclohexanol (MCH) (Sigma). Flies were trained by 10× cycles of 1min light+MCH/15min dark-MCH, then re-tested for light:dark preference in a fresh T-maze for 1min. Performance indices (PI=(attracted)-(avoided)/(attracted)+(avoided)) were calculated for each trial (≥4 trials per condition).
Drosophila brain dissection, immunohistochemistry and imaging
Brain dissections performed exactly as described previously (Kelly et al., 2016). Anti-FasII (1D4, DSHB) used at 1:20 dilution. Maximum intensity projections generated with Zeiss Zen™ software. Adult eyes imaged with a Leica DFC500 camera.
Drosophila neuronal culture
24APF pupal brains were disassociated in Liberase (Roche) and plated in Schenider’s Medium (10% FBS, 0.05 mg/ml insulin) on Laminin/ConA coated coverslips. 72hr cultures were fixed in 4% paraformaldehyde, dehydrated in EtOH @ −20°C, then rehydrated incubated in 1° antibody, washed in PBT, and incubated in 2° antibody. Rabbit anti-dNab2 was described previously (Pak et al., 2011) and used at 1:1000. Anti-dFMRP 6A15 (Abcam) was used at 1:400. Anti-HRP FITC (Jackson Laboratories) was used at 1:500.
Immunoprecipitation
The RNA-tagging technique was adapted from Yang et al. (Yang et al., 2005). Briefly, 5-day old adult heads were lysed (50mM Tris-HCl (pH 8.1), 10mM EDTA, 150mM NaCl, 1%SDS), diluted in 50mM Tris-HCl (pH 8.1), 10mM EDTA, 50mM NaCl, cleared by 12,000rpm, then IPed with anti-Flag-M2 agarose (Sigma). Fractionated lysates (below) were precipitated with the 6A15 anti-dFMRP mAb (Abcam). Precipitates were eluted with Elution buffer (EB: 50mM Tris-HCl (pH 7.0), 10 mM EDTA, 1.3% SDS). All buffers contain RNaseIN (Promega) and cOmplete protease inhibitor (Roche).
Fractionation
Five adults per genotype were homogenized in 250µl of ice-cold nuclear isolation buffer (NIB: 10 mM Tris-Cl (pH 7.4), 10 mM NaCl, 3 mM MgCl2, 0.5%NP-40, cOmplete protease inhibitor), incubated @ 4°C for 5min, then centrifuged at 500xg (“Cyto”). Pelleted nuclei were washed in NIB, collected by a 500xg spin, then sonicated in NIB (“Nuc”). Mouse brains were homogenized in CLB buffer (10mM HEPES, 10mM NaCl, 1mM KH2PO4, 5mM NaHCO3, 5mM EDTA, 1mM CaCl2, 0.5 mM MgCl2). One-tenth of the sample was retained (whole extract). Cyto and nuc fractions were then isolated as described (Guillemin et al., 2005). All fractions were sonicated and cleared at 13,000rpm.
Western Blotting
Samples were run on 5% SDS-PAGE gels, transferred to PVDF membrane (Bio-Rad), blocked and then probed with antibody: anti-Flag M2 (Sigma) at 1:1000, anti-dFMR1 monoclonal antibody 6A15 at 1:1500, anti-Lamin (DSHB) at 1:2000, anti-histone H3 at 1:100, anti-THOC1 at 1:100.
Hippocampal culture and imaging
Neuronal isolation and culture were performed as described (Kaech and Banker, 2006). P1 hippocampi were dissected, dissociated and plated on poly-d-Lysine-treated coverslips (EMD Millipore) in Neurobasal medium with B-27 and Glutamax (Invitrogen). Neurons were fixed with 4% paraformaldehyde, washed and permeabilized with 0.2% Triton X-100 then blocked with 4% BSA, 1% NGS and 0.1% TX-100. 1° antibodies: anti-ZC3H14 (1:500 (Leung et al., 2009)), Map2 (1:500; Sigma, M1406), Tau (Chemicon, MAB3420). Anti-rabbit or anti-mouse Alexa 488/546 antibodies were used as secondary antibodies. Cells were imaged between DIV4–6 or 20–22 using a NIKON TiE inverted microscope.
ePAT and poly(A) tail length assays
The ePAT assay was performed exactly as described (Chartier et al., 2017). Bulk PAT length analysis was performed as described (Apponi et al., 2010).
Polyribosome fractionation
Polysome analysis was performed as described (Muddashetty et al., 2007) with modifications. The cortex of P13 brains were dissected in ice-cold buffer (10mM HEPES, pH 7.3, 150mM KCl, 5mM MgCl2, 100ug/ml cycloheximide), then homogenized in 1ml of lysis buffer (10mM HEPES, pH 7.3, 150mM KCl, 5mM MgCl2, 100ug/ml cycloheximide, cOmplete protease inhibitor (Roche), 100U/ml SUPERase-In (LifeTechnologies)) with or without 30mM EDTA or 25uM puromycin. Homogenates were spun at 2000xg. Supe (S1) was transferred to new tubes, and supplemented with Igepal to 1%, incubated on ice, and spun 20,000xg. The resulting supe (S2) was loaded onto a 15–45% wt/wt linear density sucrose gradient in 10mM HEPES, pH 7.3, 150mM KCl, 5mM MgCl2, 100ug/ml cycloheximide, 100U/ml SUPERase-In. Gradients were spun 38,000rpm in a Beckman SW41 rotor and fractionated into 10×1.1-ml fractions with continuous monitoring at OD254.
Statistical Methods
Student’s Unpaired T-test and the Chi-square Test (GraphPad Prism™) were used as indicated to analyze significance between data points and between observed vs. expected data values. Sample sizes (n) and significance P-values (p) are denoted in the text. P-values are denoted by asterisks (e.g. *p<0.05). Manders Overlap Coefficient (MOC) analysis was carried out by R.S.B. according to (Dunn et al., 2011).
Supplementary Material
Highlights.
dNab2 is the fly ortholog of a human RBP lost in inherited intellectual disability
A cytoplasmic pool of dNab2 interacts with the Fragile-X homolog dFMRP
dNab2 regulates the CamKII mRNA and supports memory with dFMRP
dFMRP and dNab2 both restrict poly(A) length of neuronal mRNAs
Acknowledgments
We thank Bloomington Drosophila Stock Center (Indiana) and Developmental Studies Hybridoma Bank (Iowa) for stocks and antibodies. We are grateful to S. Kelly for assistance with brain dissection and neuronal culture, and to T. Jongens, S. Kunes, M. Metzstein, M. Simonelig and D. Bilder for providing stocks, and S. Sanyal for the T-mazes. Financial support: MH10730501 (K.H.M. and A.H.C.), MH109026 (G.J.B.), U54-NS091859 (S.T.W.), GM083889, MH104632, and MH108025 (J.Q.Z), F31-NS092437 (O.F.O) and F31-HD07922601 (R.S.B.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
Conceptualization, R.S.B., K.H.M. and A.H.C; Methodology, R.S.B., K.H.M., and A.H.C.; Investigation, R.S.B., A.B., J.C.R, J.O., J.R., S.L., C.G., C.P., K.J.M., S.K.J., and M.R.S.; Writing-Original Draft, R.S.B. and K.H.M.; Writing-Review and Editing, R.S.B., K.H.M., and A.H.C.; Resources, S.T.W., G.J.B. and J.Q.Z.; Supervision, K.H.M., A.H.C., S.T.W., G.J.B. and J.Q.Z.; Funding Acquisition, K.H.M., A.H.C., and R.S.B.
References
- Anderson JT, Wilson SM, Datar KV, Swanson MS. NAB2: a yeast nuclear polyadenylated RNA-binding protein essential for cell viability. Mol Cell Biol. 1993;13:2730–2741. doi: 10.1128/mcb.13.5.2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antar LN, Li C, Zhang H, Carroll RC, Bassell GJ. Local functions for FMRP in axon growth cone motility and activity-dependent regulation of filopodia and spine synapses. Mol Cell Neurosci. 2006;32:37–48. doi: 10.1016/j.mcn.2006.02.001. [DOI] [PubMed] [Google Scholar]
- Apponi LH, Leung SW, Williams KR, Valentini SR, Corbett AH, Pavlath GK. Loss of nuclear poly(A)-binding protein 1 causes defects in myogenesis and mRNA biogenesis. Hum Mol Genet. 2010;19:1058–1065. doi: 10.1093/hmg/ddp569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong JD, de Belle JS, Wang Z, Kaiser K. Metamorphosis of the mushroom bodies; large-scale rearrangements of the neural substrates for associative learning and memory in Drosophila. Learn Mem. 1998;5:102–114. [PMC free article] [PubMed] [Google Scholar]
- Ashraf SI, McLoon AL, Sclarsic SM, Kunes S. Synaptic protein synthesis associated with memory is regulated by the RISC pathway in Drosophila. Cell. 2006;124:191–205. doi: 10.1016/j.cell.2005.12.017. [DOI] [PubMed] [Google Scholar]
- Barbee SA, Estes PS, Cziko AM, Hillebrand J, Luedeman RA, Coller JM, Johnson N, Howlett IC, Geng C, Ueda R, et al. Staufen- and FMRP-containing neuronal RNPs are structurally and functionally related to somatic P bodies. Neuron. 2006;52:997–1009. doi: 10.1016/j.neuron.2006.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassell GJ, Warren ST. Fragile X syndrome: loss of local mRNA regulation alters synaptic development and function. Neuron. 2008;60:201–214. doi: 10.1016/j.neuron.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossie MA, DeHoratius C, Barcelo G, Silver P. A mutant nuclear protein with similarity to RNA binding proteins interferes with nuclear import in yeast. Mol Biol Cell. 1992;3:875–893. doi: 10.1091/mbc.3.8.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozzetti MP, Specchia V, Cattenoz PB, Laneve P, Geusa A, Sahin HB, Di Tommaso S, Friscini A, Massari S, Diebold C, et al. The Drosophila fragile X mental retardation protein participates in the piRNA pathway. J Cell Sci. 2015;128:2070–2084. doi: 10.1242/jcs.161810. [DOI] [PubMed] [Google Scholar]
- Castello A, Fischer B, Hentze MW, Preiss T. RNA-binding proteins in Mendelian disease. Trends Genet. 2013;29:318–327. doi: 10.1016/j.tig.2013.01.004. [DOI] [PubMed] [Google Scholar]
- Caudy AA, Myers M, Hannon GJ, Hammond SM. Fragile X-related protein and VIG associate with the RNA interference machinery. Genes Dev. 2002;16:2491–2496. doi: 10.1101/gad.1025202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chartier A, Joly W, Simonelig M. Measurement of mRNA Poly(A) Tail Lengths in Drosophila Female Germ Cells and Germ-Line Stem Cells. Methods Mol Biol. 2017;1463:93–102. doi: 10.1007/978-1-4939-4017-2_7. [DOI] [PubMed] [Google Scholar]
- Chen CC, Wu JK, Lin HW, Pai TP, Fu TF, Wu CL, Tully T, Chiang AS. Visualizing long-term memory formation in two neurons of the Drosophila brain. Science. 2012;335:678–685. doi: 10.1126/science.1212735. [DOI] [PubMed] [Google Scholar]
- Chintapalli VR, Al Bratty M, Korzekwa D, Watson DG, Dow JA. Mapping an atlas of tissue-specific Drosophila melanogaster metabolomes by high resolution mass spectrometry. PLoS One. 2013;8:e78066. doi: 10.1371/journal.pone.0078066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne AN, Yamada SB, Siddegowda BB, Estes PS, Zaepfel BL, Johannesmeyer JS, Lockwood DB, Pham LT, Hart MP, Cassel JA, et al. Fragile X protein mitigates TDP-43 toxicity by remodeling RNA granules and restoring translation. Hum Mol Genet. 2015;24:6886–6898. doi: 10.1093/hmg/ddv389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cziko AM, McCann CT, Howlett IC, Barbee SA, Duncan RP, Luedemann R, Zarnescu D, Zinsmaier KE, Parker RR, Ramaswami M. Genetic modifiers of dFMR1 encode RNA granule components in Drosophila. Genetics. 2009;182:1051–1060. doi: 10.1534/genetics.109.103234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JC, Klann E. The translation of translational control by FMRP: therapeutic targets for FXS. Nat Neurosci. 2013;16:1530–1536. doi: 10.1038/nn.3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JC, Van Driesche SJ, Zhang C, Hung KY, Mele A, Fraser CE, Stone EF, Chen C, Fak JJ, Chi SW, et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011;146:247–261. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donlin-Asp PG, Rossoll W, Bassell GJ. Spatially and temporally regulating translation via mRNA binding proteins in cellular and neuronal function. FEBS Lett. 2017 doi: 10.1002/1873-3468.12621. [DOI] [PubMed] [Google Scholar]
- Dunn KW, Kamocka MM, McDonald JH. A practical guide to evaluating colocalization in biological microscopy. Am J Physiol Cell Physiol. 2011;300:C723–742. doi: 10.1152/ajpcell.00462.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edens BM, Ajroud-Driss S, Ma L, Ma YC. Molecular mechanisms and animal models of spinal muscular atrophy. Biochim Biophys Acta. 2015;1852:685–692. doi: 10.1016/j.bbadis.2014.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichhorn SW, Subtelny AO, Kronja I, Kwasnieski JC, Orr-Weaver TL, Bartel DP. mRNA poly(A)-tail changes specified by deadenylation broadly reshape translation in Drosophila oocytes and early embryos. Elife. 2016;5 doi: 10.7554/eLife.16955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Gutekunst CA, Eberhart DE, Yi H, Warren ST, Hersch SM. Fragile X mental retardation protein: nucleocytoplasmic shuttling and association with somatodendritic ribosomes. J Neurosci. 1997;17:1539–1547. doi: 10.1523/JNEUROSCI.17-05-01539.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin TJ, Godfrey A. Polyribosomes in rat-liver preparations. The Biochemical journal. 1966;98:513–521. doi: 10.1042/bj0980513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green DM, Marfatia KA, Crafton EB, Zhang X, Cheng X, Corbett AH. Nab2p is required for poly(A) RNA export in Saccharomyces cerevisiae and is regulated by arginine methylation via Hmt1p. J Biol Chem. 2002;277:7752–7760. doi: 10.1074/jbc.M110053200. [DOI] [PubMed] [Google Scholar]
- Griffith LC, Verselis LM, Aitken KM, Kyriacou CP, Danho W, Greenspan RJ. Inhibition of calcium/calmodulin-dependent protein kinase in Drosophila disrupts behavioral plasticity. Neuron. 1993;10:501–509. doi: 10.1016/0896-6273(93)90337-q. [DOI] [PubMed] [Google Scholar]
- Gross C, Berry-Kravis EM, Bassell GJ. Therapeutic strategies in fragile X syndrome: dysregulated mGluR signaling and beyond. Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology. 2012;37:178–195. doi: 10.1038/npp.2011.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillemin I, Becker M, Ociepka K, Friauf E, Nothwang HG. A subcellular prefractionation protocol for minute amounts of mammalian cell cultures and tissue. Proteomics. 2005;5:35–45. doi: 10.1002/pmic.200400892. [DOI] [PubMed] [Google Scholar]
- Heisenberg M. Mushroom body memoir: from maps to models. Nature reviews Neuroscience. 2003;4:266–275. doi: 10.1038/nrn1074. [DOI] [PubMed] [Google Scholar]
- Hu J, Gao S. Mutant of RNA Binding Protein Zc3h14 Causes Cell Growth Delay/Arrest through Inducing Multinucleation and DNA Damage. JSM Biochem Mol Biol. 2014;2 [Google Scholar]
- Hummel T, Krukkert K, Roos J, Davis G, Klambt C. Drosophila Futsch/22C10 is a MAP1B–like protein required for dendritic and axonal development. Neuron. 2000;26:357–370. doi: 10.1016/s0896-6273(00)81169-1. [DOI] [PubMed] [Google Scholar]
- Ishizuka A, Siomi MC, Siomi H. A Drosophila fragile X protein interacts with components of RNAi and ribosomal proteins. Genes Dev. 2002;16:2497–2508. doi: 10.1101/gad.1022002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin P, Zarnescu DC, Ceman S, Nakamoto M, Mowrey J, Jongens TA, Nelson DL, Moses K, Warren ST. Biochemical and genetic interaction between the fragile X mental retardation protein and the microRNA pathway. Nat Neurosci. 2004;7:113–117. doi: 10.1038/nn1174. [DOI] [PubMed] [Google Scholar]
- Kaech S, Banker G. Culturing hippocampal neurons. Nat Protoc. 2006;1:2406–2415. doi: 10.1038/nprot.2006.356. [DOI] [PubMed] [Google Scholar]
- Keleman K, Kruttner S, Alenius M, Dickson BJ. Function of the Drosophila CPEB protein Orb2 in long-term courtship memory. Nat Neurosci. 2007;10:1587–1593. doi: 10.1038/nn1996. [DOI] [PubMed] [Google Scholar]
- Kelly S, Pak C, Garshasbi M, Kuss A, Corbett AH, Moberg K. New kid on the ID block: Neural functions of the Nab2/ZC3H14 class of Cys3His tandem zinc-finger polyadenosine RNA binding proteins. RNA Biol. 2012;9 doi: 10.4161/rna.20187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly SM, Bienkowski R, Banerjee A, Melicharek DJ, Brewer ZA, Marenda DR, Corbett AH, Moberg KH. The Drosophila ortholog of the Zc3h14 RNA binding protein acts within neurons to pattern axon projection in the developing brain. Dev Neurobiol. 2016;76:93–106. doi: 10.1002/dneu.22301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly SM, Leung SW, Apponi LH, Bramley AM, Tran EJ, Chekanova JA, Wente SR, Corbett AH. Recognition of polyadenosine RNA by the zinc finger domain of nuclear poly(A) RNA-binding protein 2 (Nab2) is required for correct mRNA 3’-end formation. J Biol Chem. 2010;285:26022–26032. doi: 10.1074/jbc.M110.141127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly SM, Leung SW, Pak C, Banerjee A, Moberg KH, Corbett AH. A conserved role for the zinc finger polyadenosine RNA binding protein, ZC3H14, in control of poly(A) tail length. RNA. 2014;20:681–688. doi: 10.1261/rna.043984.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Bellini M, Ceman S. Fragile X mental retardation protein FMRP binds mRNAs in the nucleus. Mol Cell Biol. 2009;29:214–228. doi: 10.1128/MCB.01377-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krashes MJ, Waddell S. Drosophila aversive olfactory conditioning. Cold Spring Harb Protoc. 2011;2011 doi: 10.1101/pdb.prot5608. pdb prot5608. [DOI] [PubMed] [Google Scholar]
- Kunz T, Kraft KF, Technau GM, Urbach R. Origin of Drosophila mushroom body neuroblasts and generation of divergent embryonic lineages. Development. 2012;139:2510–2522. doi: 10.1242/dev.077883. [DOI] [PubMed] [Google Scholar]
- Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- Leung SW, Apponi LH, Cornejo OE, Kitchen CM, Valentini SR, Pavlath GK, Dunham CM, Corbett AH. Splice variants of the human ZC3H14 gene generate multiple isoforms of a zinc finger polyadenosine RNA binding protein. Gene. 2009;439:71–78. doi: 10.1016/j.gene.2009.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Wang X, Zhang X, Goodrich DW. Human hHpr1/p84/Thoc1 regulates transcriptional elongation and physically links RNA polymerase II and RNA processing factors. Mol Cell Biol. 2005;25:4023–4033. doi: 10.1128/MCB.25.10.4023-4033.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R, Wang H, Liang Z, Ku L, O’Donnell WT, Li W, Warren ST, Feng Y. The fragile X protein controls microtubule-associated protein 1B translation and microtubule stability in brain neuron development. Proc Natl Acad Sci U S A. 2004;101:15201–15206. doi: 10.1073/pnas.0404995101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik BR, Gillespie JM, Hodge JJ. CASK and CaMKII function in the mushroom body alpha’/beta’ neurons during Drosophila memory formation. Frontiers in neural circuits. 2013;7:52. doi: 10.3389/fncir.2013.00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik BR, Hodge JJ. CASK and CaMKII function in Drosophila memory. Front Neurosci. 2014;8:178. doi: 10.3389/fnins.2014.00178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastushita-Sakai T, White-Grindley E, Samuelson J, Seidel C, Si K. Drosophila Orb2 targets genes involved in neuronal growth, synapse formation, and protein turnover. Proc Natl Acad Sci U S A. 2010;107:11987–11992. doi: 10.1073/pnas.1004433107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel CI, Kraft R, Restifo LL. Defective neuronal development in the mushroom bodies of Drosophila fragile X mental retardation 1 mutants. J Neurosci. 2004;24:5798–5809. doi: 10.1523/JNEUROSCI.1102-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MJ. From birth to death: the complex lives of eukaryotic mRNAs. Science. 2005;309:1514–1518. doi: 10.1126/science.1111443. [DOI] [PubMed] [Google Scholar]
- Morales J, Hiesinger PR, Schroeder AJ, Kume K, Verstreken P, Jackson FR, Nelson DL, Hassan BA. Drosophila fragile X protein, DFXR, regulates neuronal morphology and function in the brain. Neuron. 2002;34:961–972. doi: 10.1016/s0896-6273(02)00731-6. [DOI] [PubMed] [Google Scholar]
- Muddashetty RS, Kelic S, Gross C, Xu M, Bassell GJ. Dysregulated metabotropic glutamate receptor-dependent translation of AMPA receptor and postsynaptic density-95 mRNAs at synapses in a mouse model of fragile X syndrome. J Neurosci. 2007;27:5338–5348. doi: 10.1523/JNEUROSCI.0937-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muddashetty RS, Nalavadi VC, Gross C, Yao X, Xing L, Laur O, Warren ST, Bassell GJ. Reversible inhibition of PSD-95 mRNA translation by miR-125a, FMRP phosphorylation, and mGluR signaling. Mol Cell. 2011;42:673–688. doi: 10.1016/j.molcel.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napoli I, Mercaldo V, Boyl PP, Eleuteri B, Zalfa F, De Rubeis S, Di Marino D, Mohr E, Massimi M, Falconi M, et al. The fragile X syndrome protein represses activity-dependent translation through CYFIP1, a new 4E–BP. Cell. 2008;134:1042–1054. doi: 10.1016/j.cell.2008.07.031. [DOI] [PubMed] [Google Scholar]
- Pak C, Garshasbi M, Kahrizi K, Gross C, Apponi LH, Noto JJ, Kelly SM, Leung SW, Tzschach A, Behjati F, et al. Mutation of the conserved polyadenosine RNA binding protein, ZC3H14/dNab2, impairs neural function in Drosophila and humans. Proc Natl Acad Sci U S A. 2011;108:12390–12395. doi: 10.1073/pnas.1107103108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preiss T, Hentze MW. From factors to mechanisms: translation and translational control in eukaryotes. Curr Opin Genet Dev. 1999;9:515–521. doi: 10.1016/s0959-437x(99)00005-2. [DOI] [PubMed] [Google Scholar]
- Rha J, Jones SK, Fidler J, Banerjee A, Leung SW, Morris KJ, Wong JC, Inglis GAS, Shapiro L, Deng Q, et al. The RNA-binding Protein, ZC3H14, is Required for Proper Poly(A) Tail Length Control, Expression of Synaptic Proteins, and Brain Function in Mice. Hum Mol Genet. 2017 doi: 10.1093/hmg/ddx248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter JD, Bassell GJ, Klann E. Dysregulation and restoration of translational homeostasis in fragile X syndrome. Nat Rev Neurosci. 2015;16:595–605. doi: 10.1038/nrn4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roos J, Hummel T, Ng N, Klambt C, Davis GW. Drosophila Futsch regulates synaptic microtubule organization and is necessary for synaptic growth. Neuron. 2000;26:371–382. doi: 10.1016/s0896-6273(00)81170-8. [DOI] [PubMed] [Google Scholar]
- Roux PP, Shahbazian D, Vu H, Holz MK, Cohen MS, Taunton J, Sonenberg N, Blenis J. RAS/ERK signaling promotes site-specific ribosomal protein S6 phosphorylation via RSK and stimulates cap-dependent translation. J Biol Chem. 2007;282:14056–14064. doi: 10.1074/jbc.M700906200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro MR, Bray SM, Warren ST. Molecular Mechanisms of Fragile X Syndrome: A Twenty-Year Perspective. Annu Rev Pathol. 2011 doi: 10.1146/annurev-pathol-011811-132457. [DOI] [PubMed] [Google Scholar]
- Santos AR, Kanellopoulos AK, Bagni C. Learning and behavioral deficits associated with the absence of the fragile X mental retardation protein: what a fly and mouse model can teach us. Learn Mem. 2014;21:543–555. doi: 10.1101/lm.035956.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenck A, Bardoni B, Langmann C, Harden N, Mandel JL, Giangrande A. CYFIP/Sra-1 controls neuronal connectivity in Drosophila and links the Rac1 GTPase pathway to the fragile X protein. Neuron. 2003;38:887–898. doi: 10.1016/s0896-6273(03)00354-4. [DOI] [PubMed] [Google Scholar]
- Schenck A, Bardoni B, Moro A, Bagni C, Mandel JL. A highly conserved protein family interacting with the fragile X mental retardation protein (FMRP) and displaying selective interactions with FMRP-related proteins FXR1P and FXR2P. Proc Natl Acad Sci U S A. 2001;98:8844–8849. doi: 10.1073/pnas.151231598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siller SS, Broadie K. Neural circuit architecture defects in a Drosophila model of Fragile X syndrome are alleviated by minocycline treatment and genetic removal of matrix metalloproteinase. Dis Model Mech. 2011;4:673–685. doi: 10.1242/dmm.008045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stackpole EE, Akins MR, Fallon JR. N-myristoylation regulates the axonal distribution of the Fragile X-related protein FXR2P. Mol Cell Neurosci. 2014;62:42–50. doi: 10.1016/j.mcn.2014.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefani G, Fraser CE, Darnell JC, Darnell RB. Fragile X mental retardation protein is associated with translating polyribosomes in neuronal cells. J Neurosci. 2004;24:7272–7276. doi: 10.1523/JNEUROSCI.2306-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subtelny AO, Eichhorn SW, Chen GR, Sive H, Bartel DP. Poly(A)-tail profiling reveals an embryonic switch in translational control. Nature. 2014;508:66–71. doi: 10.1038/nature13007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudhakaran IP, Hillebrand J, Dervan A, Das S, Holohan EE, Hulsmeier J, Sarov M, Parker R, VijayRaghavan K, Ramaswami M. FMRP and Ataxin-2 function together in long-term olfactory habituation and neuronal translational control. Proc Natl Acad Sci U S A. 2014;111:E99–E108. doi: 10.1073/pnas.1309543111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tully T, Quinn WG. Classical conditioning and retention in normal and mutant Drosophila melanogaster. J Comp Physiol A. 1985;157:263–277. doi: 10.1007/BF01350033. [DOI] [PubMed] [Google Scholar]
- Udagawa T, Farny NG, Jakovcevski M, Kaphzan H, Alarcon JM, Anilkumar S, Ivshina M, Hurt JA, Nagaoka K, Nalavadi VC, et al. Genetic and acute CPEB1 depletion ameliorate fragile X pathophysiology. Nat Med. 2013;19:1473–1477. doi: 10.1038/nm.3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udagawa T, Swanger SA, Takeuchi K, Kim JH, Nalavadi V, Shin J, Lorenz LJ, Zukin RS, Bassell GJ, Richter JD. Bidirectional control of mRNA translation and synaptic plasticity by the cytoplasmic polyadenylation complex. Mol Cell. 2012;47:253–266. doi: 10.1016/j.molcel.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan L, Dockendorff TC, Jongens TA, Dreyfuss G. Characterization of dFMR1, a Drosophila melanogaster homolog of the fragile X mental retardation protein. Mol Cell Biol. 2000;20:8536–8547. doi: 10.1128/mcb.20.22.8536-8547.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Edenberg HJ, Davis RL. Isolation of mRNA from specific tissues of Drosophila by mRNA tagging. Nucleic Acids Res. 2005;33:e148. doi: 10.1093/nar/gni149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalfa F, Giorgi M, Primerano B, Moro A, Di Penta A, Reis S, Oostra B, Bagni C. The fragile X syndrome protein FMRP associates with BC1 RNA and regulates the translation of specific mRNAs at synapses. Cell. 2003;112:317–327. doi: 10.1016/s0092-8674(03)00079-5. [DOI] [PubMed] [Google Scholar]
- Zhang YQ, Bailey AM, Matthies HJ, Renden RB, Smith MA, Speese SD, Rubin GM, Broadie K. Drosophila fragile X-related gene regulates the MAP1B homolog Futsch to control synaptic structure and function. Cell. 2001;107:591–603. doi: 10.1016/s0092-8674(01)00589-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.