Abstract
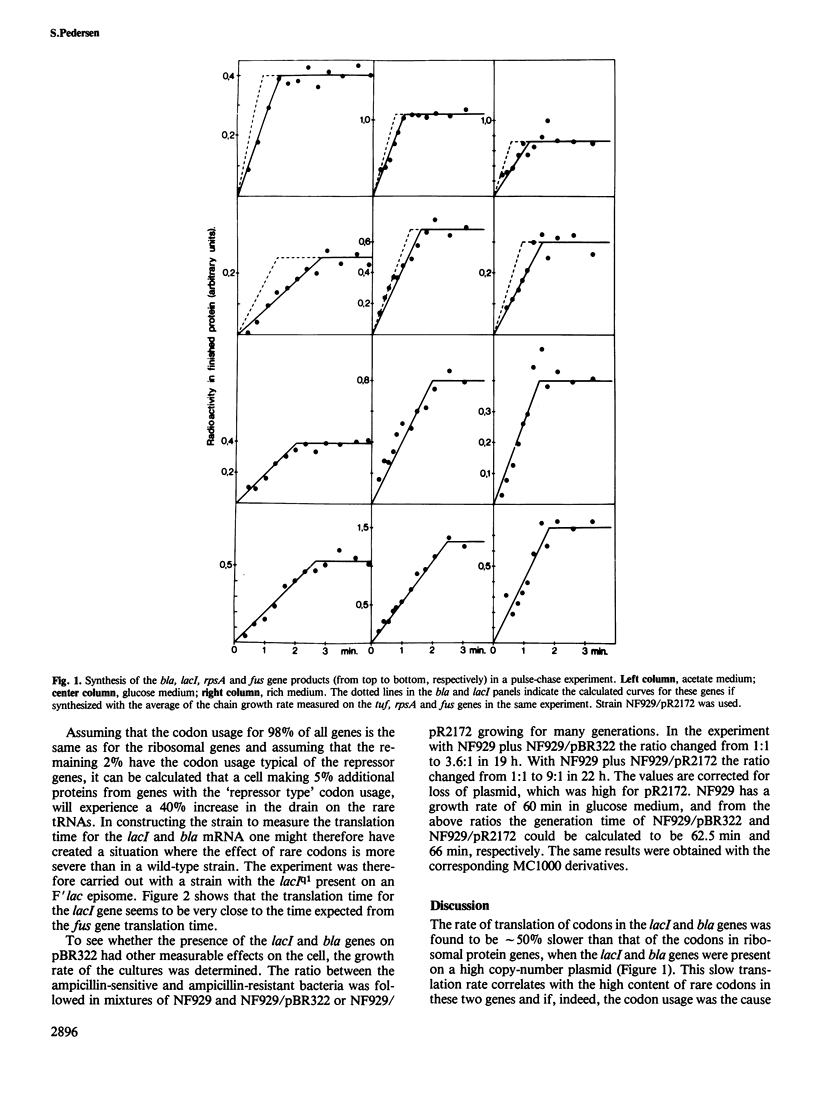
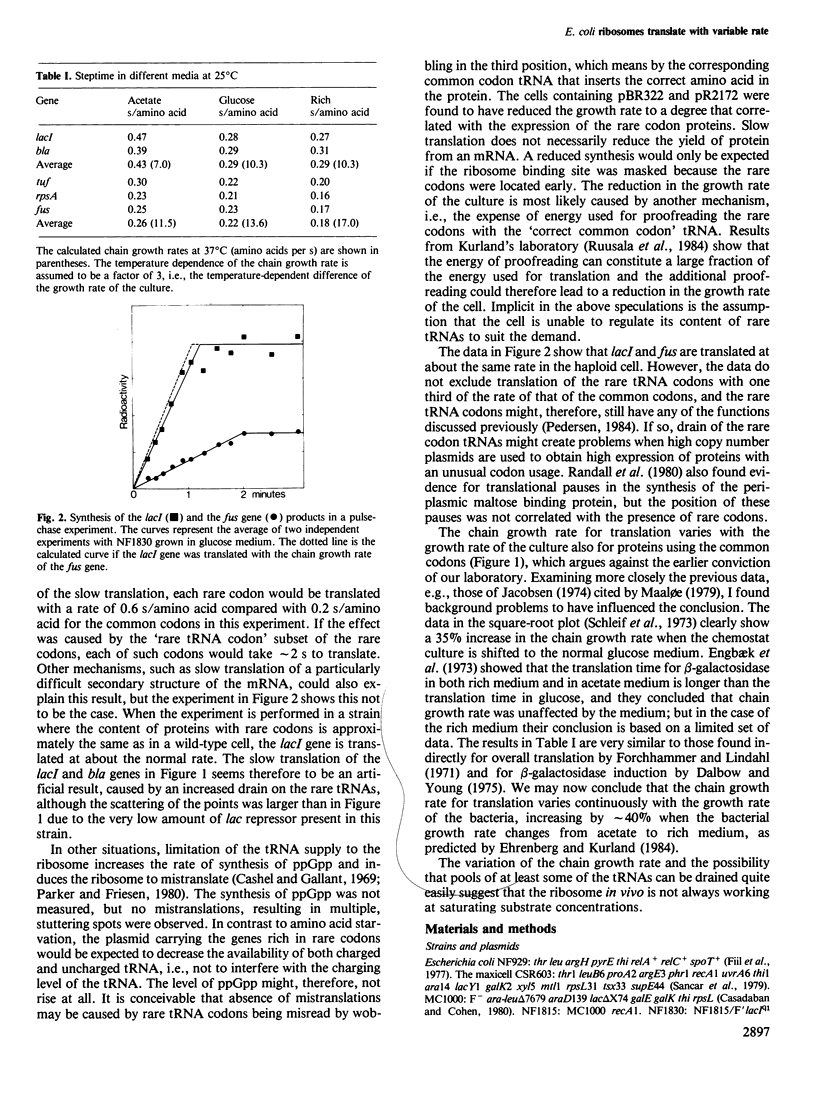
The question of whether or not 'rare' codons are translated with the same rate as 'common' codons was investigated by measuring the translation time for two genes, lacI and bla, rich in rare codons, and comparing the results with the translation times measured on fus, tsf, tuf and rpsA which have very few rare codons. The rate of synthesis of the lac repressor was first measured with the up-promoter mutation lacIq1 present on the high copy number plasmid pBR322. In such a strain the average translation times for lacI and bla were 50% slower than the rate calculated from the translation time for the four ribosomal proteins. In a strain having lacIq1 on an F'lac episome this difference was much smaller, thus slow translation of genes rich in rare codons is exaggerated in strains with increased drain on the rare codon tRNAs. The data do not exclude that only a subset of the rare codons is translated more slowly. Translation times were also measured in cells growing in different media, and the translation chain growth rate was found to increase by approximately 40% going from acetate medium to a fully supplemented medium.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arai K., Clark B. F., Duffy L., Jones M. D., Kaziro Y., Laursen R. A., L'Italien J., Miller D. L., Nagarkatti S., Nakamura S. Primary structure of elongation factor Tu from Escherichia coli. Proc Natl Acad Sci U S A. 1980 Mar;77(3):1326–1330. doi: 10.1073/pnas.77.3.1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Bremer H., Yuan D. Chain growth rate of messenger RNA in Escherichia coli infected with bacteriophage T4. J Mol Biol. 1968 Jun 28;34(3):527–540. doi: 10.1016/0022-2836(68)90178-2. [DOI] [PubMed] [Google Scholar]
- Casadaban M. J., Cohen S. N. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J Mol Biol. 1980 Apr;138(2):179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- Cashel M., Gallant J. Two compounds implicated in the function of the RC gene of Escherichia coli. Nature. 1969 Mar 1;221(5183):838–841. doi: 10.1038/221838a0. [DOI] [PubMed] [Google Scholar]
- Christiansen L., Pedersen S. Cloning, restriction endonuclease mapping and post-transcriptional regulation of rpsA, the structural gene for ribosomal protein S1. Mol Gen Genet. 1981;181(4):548–551. doi: 10.1007/BF00428751. [DOI] [PubMed] [Google Scholar]
- Dalbow D. G., Young R. Synthesis time of beta-galactosidase in Escherichia coli B/r as a function of growth rate. Biochem J. 1975 Jul;150(1):13–20. doi: 10.1042/bj1500013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engbaek F., Kjeldgaard N. O., Maaloe O. Chain growth rate of -galactosidase during exponential growth and amino acid starvation. J Mol Biol. 1973 Mar 25;75(1):109–118. doi: 10.1016/0022-2836(73)90532-9. [DOI] [PubMed] [Google Scholar]
- Farabaugh P. J. Sequence of the lacI gene. Nature. 1978 Aug 24;274(5673):765–769. doi: 10.1038/274765a0. [DOI] [PubMed] [Google Scholar]
- Fiers W., Contreras R., Duerinck F., Haegeman G., Iserentant D., Merregaert J., Min Jou W., Molemans F., Raeymaekers A., Van den Berghe A. Complete nucleotide sequence of bacteriophage MS2 RNA: primary and secondary structure of the replicase gene. Nature. 1976 Apr 8;260(5551):500–507. doi: 10.1038/260500a0. [DOI] [PubMed] [Google Scholar]
- Fiil N. P., Willumsen B. M., Friesen J. D., von Meyenburg K. Interaction of alleles of the relA, relC and spoT genes in Escherichia coli: analysis of the interconversion of GTP, ppGpp and pppGpp. Mol Gen Genet. 1977 Jan 7;150(1):87–101. doi: 10.1007/BF02425329. [DOI] [PubMed] [Google Scholar]
- Forchhammer J., Lindahl L. Growth rate of polypeptide chains as a function of the cell growth rate in a mutant of Escherichia coli 15. J Mol Biol. 1971 Feb 14;55(3):563–568. doi: 10.1016/0022-2836(71)90337-8. [DOI] [PubMed] [Google Scholar]
- Gouy M., Gautier C. Codon usage in bacteria: correlation with gene expressivity. Nucleic Acids Res. 1982 Nov 25;10(22):7055–7074. doi: 10.1093/nar/10.22.7055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosjean H., Fiers W. Preferential codon usage in prokaryotic genes: the optimal codon-anticodon interaction energy and the selective codon usage in efficiently expressed genes. Gene. 1982 Jun;18(3):199–209. doi: 10.1016/0378-1119(82)90157-3. [DOI] [PubMed] [Google Scholar]
- Hearst J. E., Isaacs S. T., Kanne D., Rapoport H., Straub K. The reaction of the psoralens with deoxyribonucleic acid. Q Rev Biophys. 1984 Feb;17(1):1–44. doi: 10.1017/s0033583500005242. [DOI] [PubMed] [Google Scholar]
- Ikemura T. Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes. J Mol Biol. 1981 Feb 15;146(1):1–21. doi: 10.1016/0022-2836(81)90363-6. [DOI] [PubMed] [Google Scholar]
- Neidhardt F. C., Bloch P. L., Pedersen S., Reeh S. Chemical measurement of steady-state levels of ten aminoacyl-transfer ribonucleic acid synthetases in Escherichia coli. J Bacteriol. 1977 Jan;129(1):378–387. doi: 10.1128/jb.129.1.378-387.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Farrell P. H. High resolution two-dimensional electrophoresis of proteins. J Biol Chem. 1975 May 25;250(10):4007–4021. [PMC free article] [PubMed] [Google Scholar]
- Ovchinnikov YuA, Alakhov YuB, Bundulis YuP, Bundule M. A., Dovgas N. V., Kozlov V. P., Motuz L. P., Vinokurov L. M. The primary structure of elongation factor G from Escherichia coli. A complete amino acid sequence. FEBS Lett. 1982 Mar 8;139(1):130–135. doi: 10.1016/0014-5793(82)80503-6. [DOI] [PubMed] [Google Scholar]
- Parker J., Friesen J. D. "Two out of three" codon reading leading to mistranslation in vivo. Mol Gen Genet. 1980 Feb;177(3):439–445. doi: 10.1007/BF00271482. [DOI] [PubMed] [Google Scholar]
- Pedersen S., Bloch P. L., Reeh S., Neidhardt F. C. Patterns of protein synthesis in E. coli: a catalog of the amount of 140 individual proteins at different growth rates. Cell. 1978 May;14(1):179–190. doi: 10.1016/0092-8674(78)90312-4. [DOI] [PubMed] [Google Scholar]
- Pedersen S., Reeh S. V. Analysis of the proteins synthesized in ultraviolet light-irradiated Escherichia coli following infection with the bacteriophages lambdadrifd 18 and lambdadfus-3. Mol Gen Genet. 1976 Mar 30;144(3):339–343. doi: 10.1007/BF00341733. [DOI] [PubMed] [Google Scholar]
- Post L. E., Strycharz G. D., Nomura M., Lewis H., Dennis P. P. Nucleotide sequence of the ribosomal protein gene cluster adjacent to the gene for RNA polymerase subunit beta in Escherichia coli. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1697–1701. doi: 10.1073/pnas.76.4.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall L. L., Josefsson L. G., Hardy S. J. Novel intermediates in the synthesis of maltose-binding protein in Escherichia coli. Eur J Biochem. 1980 Jun;107(2):375–379. doi: 10.1111/j.1432-1033.1980.tb06039.x. [DOI] [PubMed] [Google Scholar]
- Reeh S., Pedersen S., Friesen J. D. Biosynthetic regulation of individual proteins in relA+ and relA strains of Escherichia coli during amino acid starvation. Mol Gen Genet. 1976 Dec 22;149(3):279–289. doi: 10.1007/BF00268529. [DOI] [PubMed] [Google Scholar]
- SCHAECHTER M., MAALOE O., KJELDGAARD N. O. Dependency on medium and temperature of cell size and chemical composition during balanced grown of Salmonella typhimurium. J Gen Microbiol. 1958 Dec;19(3):592–606. doi: 10.1099/00221287-19-3-592. [DOI] [PubMed] [Google Scholar]
- Sancar A., Hack A. M., Rupp W. D. Simple method for identification of plasmid-coded proteins. J Bacteriol. 1979 Jan;137(1):692–693. doi: 10.1128/jb.137.1.692-693.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleif R., Hess W., Finkelstein S., Ellis D. Induction kinetics of the L-arabinose operon of Escherichia coli. J Bacteriol. 1973 Jul;115(1):9–14. doi: 10.1128/jb.115.1.9-14.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnier J., Kimura M., Foulaki K., Subramanian A. R., Isono K., Wittmann-Liebold B. Primary structure of Escherichia coli ribosomal protein S1 and of its gene rpsA. Proc Natl Acad Sci U S A. 1982 Feb;79(4):1008–1011. doi: 10.1073/pnas.79.4.1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutcliffe J. G. Nucleotide sequence of the ampicillin resistance gene of Escherichia coli plasmid pBR322. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3737–3741. doi: 10.1073/pnas.75.8.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota T., Sugisaki H., Takanami M., Kaziro Y. The nucleotide sequence of the cloned tufA gene of Escherichia coli. Gene. 1980 Dec;12(1-2):25–31. doi: 10.1016/0378-1119(80)90012-8. [DOI] [PubMed] [Google Scholar]
- Yokota T., Sugisaki H., Takanami M., Kaziro Y. The nucleotide sequence of the cloned tufA gene of Escherichia coli. Gene. 1980 Dec;12(1-2):25–31. doi: 10.1016/0378-1119(80)90012-8. [DOI] [PubMed] [Google Scholar]
- Zengel J. M., Archer R. H., Lindahl L. The nucleotide sequence of the Escherichia coli fus gene, coding for elongation factor G. Nucleic Acids Res. 1984 Feb 24;12(4):2181–2192. doi: 10.1093/nar/12.4.2181. [DOI] [PMC free article] [PubMed] [Google Scholar]



