Summary
Bone marrow (BM) fibrosis is associated with poor prognosis in patients with de novo myelodysplastic syndromes (MDS). TP53 mutations and TP53 (p53) overexpression in MDS are also associated with poor patient outcomes. The prevalence and significance of TP53 mutations and TP53 overexpression in MDS with fibrosis are unknown.
We studied 67 patients with de novo MDS demonstrating moderate to severe reticulin fibrosis (MDS-F). Expression of TP53 was evaluated in BM core biopsy specimens using dual-colour CD34/TP53 immunohistochemistry with computer-assisted image analysis. Mutation analysis was performed using next-generation sequencing, or Sanger sequencing methods.
TP53 mutations were present in 44.4% of cases. TP53 mutation was significantly associated with TP53 expression (p= 0.0294). High levels of TP53 expression (3+ in ≥10% cells) were associated with higher BM blast counts (p=0.0149); alterations of chromosomes 5 (p= 0.0009) or 7 (p= 0.0141); complex karyotype (p= 0.0002); high- and very-high risk IPSS-R groups (p =0.009); and TP53 mutations (p=0.0003). High TP53 expression independently predicted shorter overall survival (OS) by multivariate analysis (p=<0.001). Expression of TP53 by CD34-positive cells was associated with shorter OS and leukaemia-free survival (p=0.0428). TP53 overexpression is a predictor of poor outcome in patients with MDS-F.
Keywords: myelodysplastic syndrome, TP53, TP53 expression, next-generation sequencing, fibrosis
Introduction
Moderate to severe bone marrow (BM) fibrosis can be detected at presentation in a subset of patients with myelodysplastic syndromes (MDS). Several studies have demonstrated that fibrosis in MDS is associated with adverse features, including shorter overall survival (OS). (Buesche, et al 2008, Della Porta and Malcovati 2011, Della Porta, et al 2009, Fu, et al 2014, Lambertenghi-Deliliers, et al 1991, Verhoef, et al 1991) Notably, the adverse impact on OS seems to be independent of other factors, such as BM blast count and poor-risk cytogenetics in multivariate modelling. (Della Porta, et al 2009) As a result, widely applied prognostication schemes, including the Revised International Prognostic Scoring System (IPSS-R), that rely heavily on blast percentage and cytogenetics have been reported to be of limited utility in predicting outcomes in patients with MDS associated with fibrosis (MDS-F). (Fu, et al 2014, Machherndl-Spandl, et al 2014)
TP53 overexpression has been shown to be associated with a poor clinical outcome in a subset of de novo MDS cases. (Cleven, et al 2015, Honkaniemi, et al 2014, Saft, et al 2014) TP53 overexpression correlates with TP53 mutations, which can be observed in up to 7% of de novo MDS cases and have been shown to be associated with adverse clinical features and poor patient outcomes.(Bejar, et al 2011) However, the frequency of TP53 protein overexpression in de novo MDS-F remains unknown.
In this study we demonstrate a relatively high frequency of TP53 protein overexpression in MDS-F and demonstrate the utility of TP53 immunohistochemistry as a prognostic marker in this disease subset.
Methods
Patients
This study was approved by the Institutional Review Board of The University of Texas MD Anderson Cancer Center (MDACC) and carried out in accordance with the Declaration of Helsinki. All patients who presented to our institution with de novo MDS between 1 September 2002 and 22 October 2014 were identified. Selection criteria for the study group were as follows: 1) 18 years of age or older at time of diagnosis; 2) diagnosis of MDS as defined by the 2008 World Health Organization (WHO) classification (Brunning, et al 2008, Vardiman, et al 2009); and, 3) moderate (MF2) or severe (MF3) BM fibrosis at the time of initial diagnosis, graded according to the European Bone Marrow Fibrosis Network (EUMNET) criteria(Thiele, et al 2005). We excluded patients with myeloproliferative neoplasms, myelodysplastic/ myeloproliferative neoplasms, therapy-related myeloid neoplasms, and those who had received therapy for MDS (other than supportive therapy) prior to the onset of fibrosis. Clinical and laboratory data were obtained by chart review with emphasis on variables demonstrated to have prognostic utility in MDS patients.(Greenberg, et al 2012)
Bone marrow morphology and fibrosis assessment
All diagnostic BM samples were reviewed and the diagnosis of MDS was confirmed. Cellularity was assessed relative to age according to the EUMNET criteria.(Thiele, et al 2005) The presence or absence of morphological dysplasia in the haematopoietic cell lineages was recorded.(Della Porta, et al 2015) BM blast percentage was enumerated by a 500-cell count using Wright-Giemsa-stained aspirate smears and/or touch imprints. All cases were classified into the appropriate MDS category according to the WHO 2008 scheme. (Vardiman, et al 2009) Reticulin and Masson Trichrome stains performed on tissue sections prepared from paraffin-embedded trephine biopsy specimens and assessed for extent of reticulin and collagen fibrosis, respectively.
Immunohistochemistry and digital image analysis
Dual-colour immunohistochemistry (IHC) was performed using paraffin-embedded BM biopsy sections on a Leica Bond platform using anti-TP53 (DO-7, Dako, Carpinteria, CA) and anti-CD34 (My10, BD Biosciences, San Jose, CA) antibodies.
Immunohistochemical results were reviewed independently by two haematopathologists blinded to the clinical and laboratory features. The percentage of CD34+ cells was assessed by manual count in 5 or more randomly selected high-power fields at 400× magnification on an Olympus microscope (Olympus, Waltham, MA) and recorded as a fraction of total BM nucleated cells in 5% increments. The intensity of TP53 staining was graded on a scale of 0–3+ (0: no staining; 1+: weak staining; 2+: moderate staining; 3+ strong staining). The presence of strong co-expression of TP53 by CD34+ cells (3+ TP53 expression in CD34-positive cells) and the presence of CD34+ clusters (≥3 CD34+ cells, as described previously(Della Porta, et al 2009)) were recorded.
All dual-stained immunohistochemistry slides were scanned using the Aperio ScanScope (Aperio Technologies, Vista, CA, USA). Representative areas of the BM core biopsy specimen were selected for digital image analysis using the nuclear algorithm of the Aperio ImageScope software. All digital analysis data were confirmed by manual review. The percentage of cells with TP53 expression and the intensity (on a scale of 1–3+) were recorded as a fraction of total BM nucleated cells. A previously established cut-off of ≥1% 3+ TP53 positive cells was used to consider a case TP53 positive. (Saft, et al 2014)
Cytogenetic analysis
Conventional cytogenetic analysis was performed as part of routine clinical workup using previously described techniques.(Khoury, et al 2003) Patients were assigned to cytogenetic risk groups according to the Revised International Prognostic Scoring System for Myelodysplastic Syndromes (IPSS-R). (Greenberg, et al 2012) In addition, complementary or confirmatory fluorescence in situ hybridization (FISH) analysis for abnormalities of chromosomes 5, 7, 8, 17 and 20 was performed in a subset of patients.
Molecular analysis
Mutation analysis was performed using DNA extracted from BM aspirate samples in a subset of patients using the following techniques: Next-generation sequencing-based mutation analysis of hotspot genomic loci (EZH2, DNMT3A, GNAS, IDH1, IDH2, JAK2, KIT, KMT2A [MLL], MPL, NPM1, NOTCH1, NRAS, KRAS, and TP53) or exonic regions (ABL1, EGFR, GATA2, IKZF2, MDM2, NOTCH1, RUNX1, ASXL1, EZH2, HRAS, JAK2, KMT2A, NPM1, TET2, BRAF, IDH1, KIT, NRAS, TP53, DNMT3A, GATA1, IDH2, KRAS, MYD88, PTPN11, WT1) was performed using the Illumina MiSeq sequencer (Illumina, San Diego, CA) as described previously.(Zhang, et al 2014) FLT3 (internal tandem duplication and D835) and NPM1 (exon12, codons 956–971) mutations were assessed by polymerase chain reaction (PCR) followed by capillary electrophoresis on Genetic Analyser (Applied Biosystems, Foster City, CA), as described previously.(Warren, et al 2012) In a subset of patients, mutations in TP53 (exons 4–9), IDH1 (exon 4) and IDH2 (exon 4) were assessed using Sanger sequencing (Applied Biosystems), and mutations in NRAS and KRAS (codons 12, 13, and 61), MPL (codon 515) and JAK2 (codon 617) were analysed using pyrosequencing (Biotage, Uppsala, Sweden).
Statistical analysis
Statistical analysis was performed using GraphPad Prism 6 (GraphPad Software, Inc., La Jolla, CA) and IBM SPSS Statistics 22.0 (IBM, Armonk, NY). Fisher’s exact test and Mann-Whitney U test were used to assess categorical and continuous variables, respectively. The Spearman rank method was used to asses correlations. The Kaplan-Meier method was used to asses overall and acute myeloid leukaemia (AML)-free survival using the log-rank (Mantel-Cox) test. Overall survival (OS) was calculated as the time period from the date of MDS diagnosis to the date of last follow-up or death of any cause. Leukaemia-free survival was defined as the time period from the date of diagnosis to the date of progression to AML. Patients who received allogeneic stem cell transplant (SCT) were censored at the time of transplant. Multivariate analysis was conducted using backward stepwise (Wald) elimination of prognostic factors to determine those that are independently impactful. A p< 0.05 was considered statistically significant.
Results
Clinical and laboratory features of patients with MDS-F
The study group included 67 patients, 42 (62.7%) men and 25 (37.3%) women with a median age of 64.6 years at the time of diagnosis (range, 18.1–91.3 years). These cases of MDS were classified as follows: 27 (40.3%) refractory cytopenia with multilineage dysplasia (RCMD); 21 (31.3%) refractory anaemia with excess blasts-1 (RAEB-1); 16 (23.9%) refractory anaemia with excess blasts-2 (RAEB-2); and 3 (4.5%) refractory cytopenia with unilineage dysplasia (RCUD). Constitutional symptoms and palpable organomegaly were seen in 32 (47.8%) and 7 of 66 (10.6%) patients, respectively. The median haemoglobin (Hb), absolute neutrophil count (ANC) and platelet count at the time of presentation were 95 g/l (range, 65–137), 1.52 × 109/l (range, 0.1–7.8) and 53 × 109/l (range, 2–403), respectively. Thirty-nine patients (58.2%) were deemed to be transfusion-dependent. Sixty-three patients were stratified into IPSS-R risk groups as follows: 3 (4.8%) very low; 14 (22.2%) low; 18 (28.6%) intermediate; 8 (12.7%) high; and 20 (31.7%) as very high-risk.
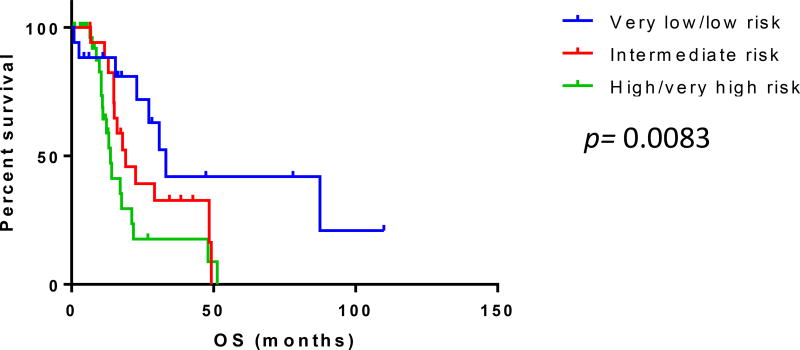
Clinical follow-up was available for all patients (median 17.1 months; range, 0.85–113.0). Overall, 22 (32.8%) patients progressed to AML within a median of 13.2 months and 45 (67.2%) patients died within a median of 19.1 months. Patients within the very low and low-risk IPSS-R groups had significantly longer OS compared with patients within other risk categories (p=0.0083). [Fig 1]
Figure 1.
Overall survival (OS) of patients with myelodysplastic syndrome with fibrosis grouped according to the Revised International Prognostics Stratification System. Patients within the very low and low risk groups according had significantly longer overall survival compared to those within other risk groups (p=0.0083).
Cytogenetic findings
Sufficient cytogenetic data were available for 66 patients (median number of metaphases analysed=20; range, 11–30). Of these, 26 (39.3%) patients had a diploid karyotype and 17 (25.8%) had a complex karyotype (≥3 alterations). In one patient, only 2 metaphases were available for evaluation that showed 45,XY,−7; monosomy 7 was further confirmed by interphase FISH. Monosomy 5/del(5q) was detected in 14 (21.5%) patients; none had isolated del 5q. Monosomy 7/del(7q) was detected in 11 of 66 (16.6%) patients and other, less frequent abnormalities included trisomy 8 (15.4%), monosomy 17/del(17p) (13.8%) and del(20q) (9.2%). IPSS-R cytogenetic risk categories were available for 66 patients and were as follows: 30 (45.5%) good; 15 (22.7%) intermediate; 5 (7.5%) poor and 16 (24.2%) very poor.
Molecular analysis
The most frequently observed mutation in this patient group was in TP53, in 8 of 17 (44.4%) patients, including two TP53 mutations in three of these patients. Other gene mutations identified included ASXL1 (1/3; 33.3%), GNAS (1/15; 6.7%), JAK2 (2/37; 5.4%), IDH1 (1/22; 4.5%); IDH2 (1/22; 4.5%); KRAS (2/58; 3.4%), and FLT3 (1/59; 1.7 %) (Fig S1). No mutations in KIT (n=37), NPM1 (n=29), MPL (n=19), EZH2 (n=15), NOTCH1 (n=15), DNMT3A (n=16), RUNX1 (n=3), TET2 (n=3), KMT2A (N=3), or KMT2A (n=3) were detected.
Of the 11 TP53 mutations identified, nine were missense mutations, one was a nonsense mutation resulting in early termination and one was a deletion resulting in early termination of the gene. The latter two mutations were both present in conjunction with additional missense TP53 mutations (Supplementary Table 1).
Clinical correlates of TP53 overexpression in MDS-F
As no data on TP53 expression in MDS-F exists, we initially determined a meaningful cut-off for TP53 expression by immunohistochemistry using TP53 mutation status with an allelic frequency >5% as a standard, in line with previous reports(Lambertenghi-Deliliers, et al 1991) and our clinical reporting procedures. Starting with a previously established cut-off of 3+ expression in ≥1% of cells(Saft, et al 2014) (overexpression), 41/67 (61.2%) MDS-F cases were positive for TP53 overexpression (range, 1–65%). Using this cut-off, TP53 overexpression was significantly associated with TP53 mutations (p=0.0294). Strong TP53 overexpression was observed mostly in early granulocytic (40/41) and erythroid (39/41) precursors, and less often in megakaryocytes (26/41). In cases with wild-type TP53, the median number of 3+ TP53-positive cells was 0.76% (range, 0.22–2.59%). However, when we compared TP53 expression to TP53 mutation status, the extent of TP53 overexpression in TP53-mutated cases ranged from 4.6–65.0% (median 21.0%), with all but one case (6/7) having 3+ expression in ≥10%. [Fig 2] Notably, in the one case with 3+ TP53 expression in only 4.6% of cells, the estimated allelic frequency of the TP53 mutation was commensurately low (<5%). On the basis of these findings, we defined 3+ TP53 expression in ≥10% of cellularity as a clinically relevant cut-off, thereafter designated TP53high. [Fig S2] Eleven (16.4%) patients had TP53high MDS-F. The remaining patients (n=56) (TP53neg-low) included 24 who had <1% 3+ TP53 expression and 32 those who had 1–9% 3+ TP53 expression. The clinical and pathological features of the TP53high and TP53neg-low groups are summarized in Tables I and II, respectively.
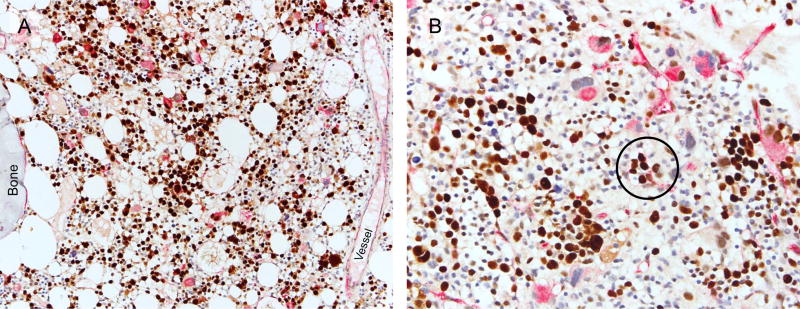
Figure 2.
Dual-colour immunohistochemistry for assessment of TP53 (brown) and CD34 (red) expression in myelodysplastic syndrome with fibrosis. (A) Strong TP53 expression is detected in 65% of cells in this sample within the erythroid, granulocytic, and megakaryocytic lineages (200×). (B) Another case with strong TP53 expression in 21% of cells. Note strong TP53 expression in CD34-positive cells in clusters (circled) (400×).
Table I.
Clinical and laboratory features of MDS-F patients by TP53 expression status.
| Variable | MDS-F with TP53neg-low | MDS-F with TP53high | p value | |
|---|---|---|---|---|
| n=56 | n=11 | |||
| Age at diagnosis, median years (range) | 65.3 (27.3–91.3) | 60.4 (18.1–76.3) | 0.4145 | |
| Sex | ||||
| Male | 35 (62.5) | 7 (63.6) | 1.0000 | |
| Female | 21 (37.5) | 4 (36.4) | ||
| Transfusion dependence | 36 (64.3) | 3 (27.3) | 0.0417 | |
| Palpable organomegaly | 18 (32.1) | 1 (9.1) | 0.1592 | |
| Complete blood count | ||||
| ANC (× 109/l), median (range) | 1.74 (0.1–7.8) | 0.85 (0.12–5.6) | 0.0985 | |
| Haemoglobin (g/l), median (range) | 96 (62–137) | 93 (76–116) | 0.4641 | |
| Platelet count (× 109/l), median (range) | 57 (2–409) | 33 (7–236) | 0.1460 | |
| Circulating blast %, median (range) | 0 (0–9) | 0 (0–6) | 0.2767 | |
| Cytogenetics categories* | n=55 | |||
| Diploid | 18 (32.7) | 2 (18.2) | 0.4816 | |
| Del 20q | 4 (7.3) | 2 (18.2) | 0.2598 | |
| −5/ del 5q | 7 (12.7) | 7 (63.6) | 0.0009 | |
| −7/ del 7q | 6 (10.9) | 5 (45.5) | 0.0141 | |
| −17/ del 17p | 6 (10.9) | 3 (27.3) | 0.1648 | |
| +8 | 9 (16.3) | 1 (9.1) | 1.0000 | |
| Complex karyotype* | 8 (14.5) | 8 (72.7) | 0.0002 | |
| IPSS-R “poor”/“very poor” cytogenetics | 13 (23.6) | 8 (72.7) | 0.0030 | |
| IPSS-R category | n=52 | 0.0090† | ||
| Very low | 2 (3.8) | 0 | ||
| Low | 15 (28.8) | 0 | ||
| Intermediate | 16 (30.8) | 2 (18.2) | ||
| High | 6 (11.5) | 2 (18.2) | ||
| Very high | 13 (25) | 7 (63.6) | ||
Abbreviations: MDS-F: myelodysplastic syndrome with fibrosis; ANC: absolute neutrophil count; IPSS-R: Revised International Prognostic Scoring System for Myelodysplastic Syndromes.
Defined as >3 karyotypic abnormalities.
“Very low” and “low-risk” vs. “high and very high-risk” groups.
Table II.
Pathological features of MDS-F patients by TP53 expression status.
| Variable | MDS-F with TP53neg- low |
MDS-F with TP53high |
p value | |
|---|---|---|---|---|
| BM cellularity relative to age | 0.4097 | |||
| Low | 3 (5.4) | 0 | ||
| Normal | 5 (8.9) | 0 | ||
| High | 48 (85.7) | 11 (100) | ||
| BM blast % median (range) | 4 (0–19) | 8 (0–16) | 0.0149 | |
| MDS categories | 0.0985 | |||
| RCUD | 3 (5.4) | 0 | ||
| RCMD | 26 (46.4) | 1 (9.1) | ||
| RAEB-1 | 12 (21.4) | 4 (36.4) | ||
| RAEB-2 | 15 (26.8) | 6 (54.5) | ||
| % CD34-positive cells | 0.0762 | |||
| <5 | 38 (67.9) | 4(36.4) | ||
| 5–9 | 2 (3.6) | 6(54.5) | ||
| 10–14 | 4 (7.1) | 0 | ||
| 15–19 | 3 (5.3) | 1(9.1) | ||
| CD34-positive clusters | 25 (44.6) | 6(54.5) | 0.7425 | |
| TP53 mutation | 1/10 (10) | 7/7 (100) | 0.0003 | |
Abbreviations: BM: Bone marrow; MDS-F: myelodysplastic syndrome with fibrosis; RCUD: refractory cytopenia with unilineage dysplasia; RCMD: refractory cytopenia with multilineage dysplasia; RAEB-1: refractory anaemia with excess blasts type 1; RAEB-2: refractory anaemia with excess blasts type 2
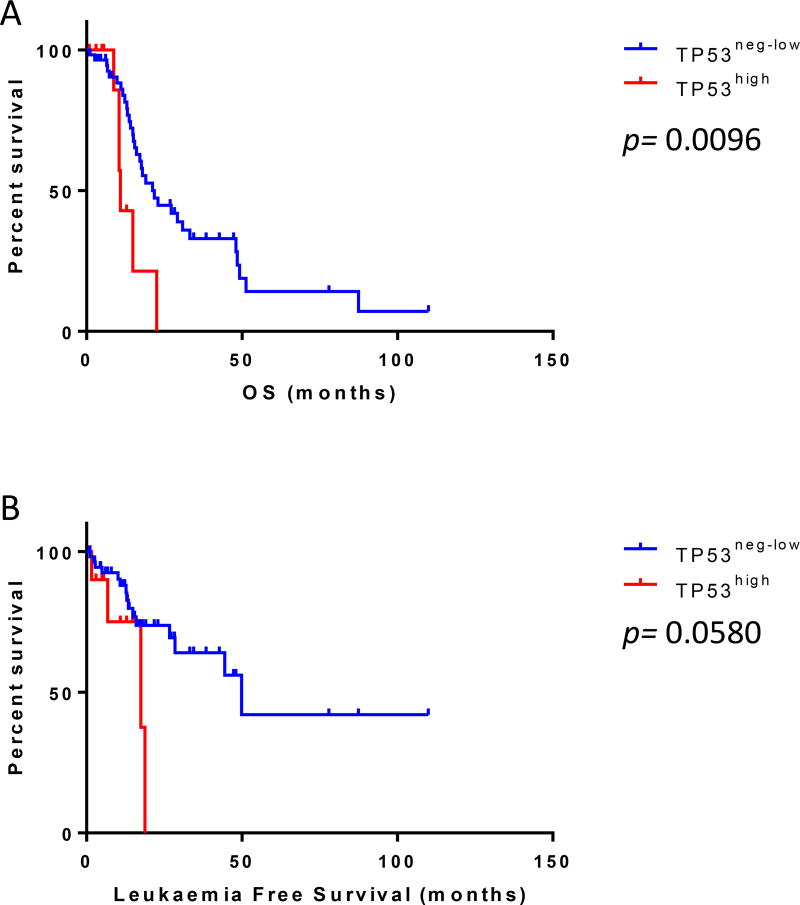
Patients in the TP53high group had higher BM blast counts (p=0.0149); more frequently had high-risk cytogenetics (p=0.0030) - including alterations of chromosome 7 (p=0.0141) and complex karyotype (p=0.0002); a higher frequency of high and very-high IPSS-R risk categories; and alterations of chromosome 5 (p=0.0009) compared with patients in the TP53neg-low group. Patients in the TP53high group were less often transfusion-dependent compared to those in the TP53neg-low group (27.3% vs. 64.3%, p=0.0417). There was no significant difference between the two groups with respect to age, sex, constitutional symptoms, organomegaly, complete blood count indices, BM cellularity, MDS diagnostic categories, dysplasia, or the presence of CD34+ cell clusters. Notably, patients in the TP53high group had a significantly shorter OS compared with patients in the TP53neg-low group (median 10.9 vs. 21.8 months, p=0.0096; Hazard ratio [HR]: 2.91, 95% confidence interval [CI]: 1.60–23.13) [Fig 3A] and a trend towards shorter leukaemia-free survival (p=0.0580) [Fig 3B]
Figure 3.
TP53 expression and survival. Patients with myelodysplastic syndrome with fibrosis in the TP53high group had (A) a significantly shorter overall survival (OS) compared to patients in the TP53neg-low group (p=0.0096) and (B) tended to have a shorter leukaemia-free survival (p=0.058).
Patients were stratified into five groups according to the most aggressive type of therapy administered during the course of disease. Twenty-eight (41.8%) patients received hypomethylating therapy, 14 (20.9%) were treated with supportive measures, 13 (19.4%) received allogeneic SCT, 7 (10.4%) were treated with chemotherapy and 5 (7.5%) were treated with other therapeutic measures, including immunotherapy. Analysing outcomes of patients in these clinical groups, those who received SCT had a significantly longer OS compared with patients in the other groups (p=0.0166). Three of the patients treated with SCT had TP53high disease (range, 40–70%) and TP53 mutation; these patients were alive and free of disease at latest follow-up (range 8.2–18.2 months).
Clinical correlates of CD34/TP53 co-expression and CD34-positive clusters in MDS-F
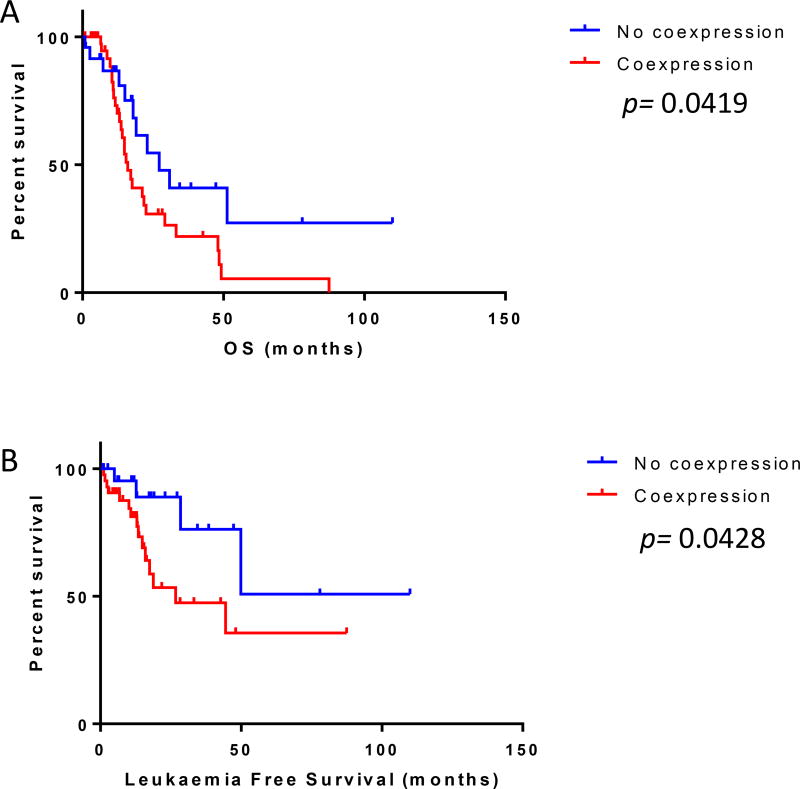
In view of previous reports suggesting a prognostic role for CD34-positive clusters in MDS (Della Porta et al, 2009), and to assess the prevalence of TP53 overexpression in blasts versus maturing elements, we performed dual-colour immunohistochemistry to colocalize CD34-positive and TP53-positive cells in MDS-F. Forty-three (64.2%) cases showed strong (3+) TP53 overexpression in CD34+ cells. The presence of CD34+ cells with strong TP53 coexpression was associated with increased (>5%) BM blasts (p= 0.0019). Patients with CD34/TP53 coexpression had a significantly shorter OS (median 16.01 vs. 27.22 months, p=0.0419; HR= 2.014, 95% CI: 1.033–3.657) and leukaemia-free survival (p=0.0428; HR= 2.909; 95% CI: 1.049–6.427). (Fig 4A–B)
Figure 4.
TP53 expression by CD34-positive cells and survival. Patients with myelodysplastic syndrome with fibrosis and strong (3+) TP53 expression by CD34-positive cells had (A) significantly shorter overall survival (OS, p=0.0419) and (B) leukaemia-free survival (p=0.0428) in comparison to those without p53 expression in CD34-positive cells.
We then assessed the impact of CD34-positive clusters, independent of TP53 expression, in this cohort of patients with MDS-F. CD34-positive cell clusters were identified in 31 (46.2%) patient BM samples. The presence of CD34-positive cells clusters was associated with increased (>5%) BM blasts (p=<0.0001). However, there was no association between CD34-positive clusters and OS or leukaemia-free survival.
Multivariate analysis
We performed multivariate analysis using backward stepwise elimination of prognostic factors to determine the independent impact of each variable. We included age at diagnosis, sex, Hb, white blood cell count, ANC, platelet count, BM blast count, transfusion dependence, alterations of chromosome 5, alterations of chromosome 7, presence of complex karyotypic abnormalities, IPSS-R cytogenetic risk group (Poor and very poor risk groups vs. others), expression in ≥10% of cells (TP53high), presence of CD34 clusters and TP53/CD34 coexpression in our initial multivariate analysis model and found that age (p=<0.0001), Hb (p=0.002) and TP53high status (p=0.007) were independently associated with shorter OS while other prognostic factors were not statistically significant. We then removed IPSS-R cytogenetic risk group as a potential confounding factor to assess the effects of each karyotypic abnormality individually and found that age (p=<0.0001), Hb (p=0.001), TP53high status (p=0.003), presence of CD34 clusters (p=0.049) and complex karyotype emerged as significantly associated with shorter OS while all other factors included were not. We further removed the possible confounding factor of complex karyotype to assess the potentially independent influence of chromosome 5 and/or 7 alterations and found that age (p=0.001), Hb (p=0.002), TP53high status (p=0.008), and alterations of chromosome 5 (p=0.016) were associated with shorter OS while other factors were not. We then reintroduced IPSS-R cytogenetic risk assignment, representing the overall contribution of karyotype and removed Hb as a potential confounding factor to assess the effect of transfusion dependence. At this stage we also removed all other cytogenetic-related prognostic factors, as well as those that were previously insignificant, including sex, CD34 clusters, CD34/TP53 coexpression, WBC, ANC and platelet count, and found that age (p=<0.0001), IPSS-R cytogenetic risk group (p=0.002) and TP53high status (p=<0.001) were associated with shorter OS while transfusion dependence and BM blasts were not. The combination of analyses used in this stepwise elimination method resulted in a final multivariate analysis model that includes age (p=<0.001), TP53high status (p=<0.001), Hb (p=0.012) and IPSS-R cytogenetic risk group (p=0.008). The details of the multivariate analysis are provided in Table III.
Table III.
Multivariate analysis of prognostic variables in MDS-F.
| Variable | p Value | HR | 95% CI | |
|---|---|---|---|---|
| Lower | Upper | |||
| Age | <0.0001 | 1.072 | 1.041 | 1.103 |
| Haemoglobin | 0.012 | 0.751 | 0.601 | 0.940 |
| IPSS-R cytogenetics* | 0.008 | 2.444 | 1.265 | 4.719 |
| P53 ≥10% | <0.001 | 5.754 | 2.416 | 13.705 |
Abbreviations: MDS-F: myelodysplastic syndrome with fibrosis; HR: Hazard ratio; 95% CI: 95% confidence interval; ANC: absolute neutrophil count; BM: Bone marrow; IPSS-R: Revised International Prognostic Scoring System for Myelodysplastic Syndromes.
Poor and very poor risk groups vs. others.
Discussion
In the current study we show a relatively high frequency of TP53 expression in de novo MDS-F and demonstrate the utility of dual-colour immunohistochemistry for TP53 and CD34 as an effective tool to enhance risk-stratification for patients with MDS-F.
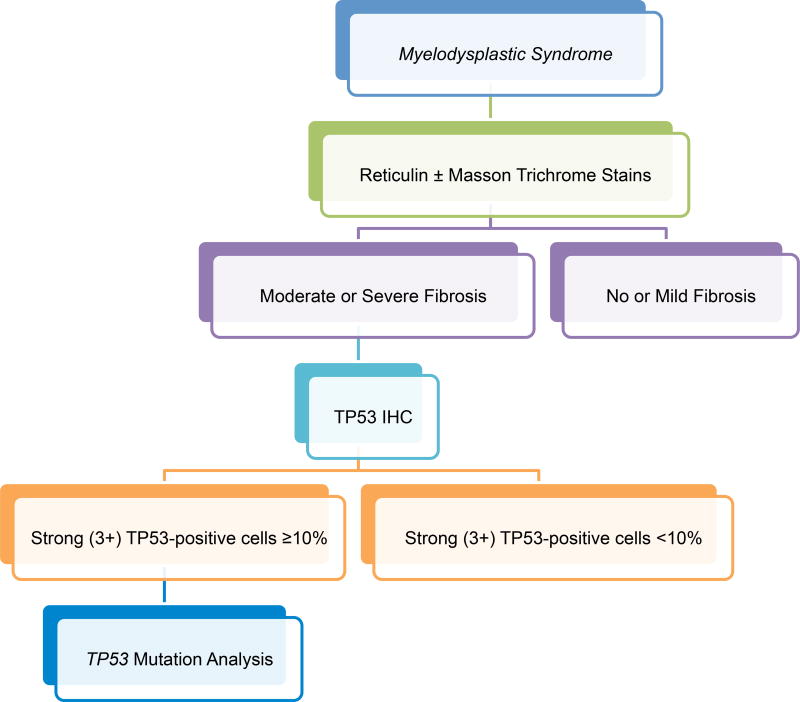
To our knowledge, this study is the first to examine the prognostic impact of TP53 protein expression in MDS-F and demonstrate the independent association of TP53 overexpression with shorter OS in this group of patients. We acknowledge that a cut-off of ≥10% 3+ TP53-positive cells is arbitrary; however, in contrast with earlier studies that used a cut-off of ≥1% 3+ TP53 expression in low-risk and therapy-related MDS (Cleven, et al 2015, Saft, et al 2014), we found that using a cut-off of ≥10% 3+ TP53 expression in MDS-F was associated with patient outcome whereas lower cut-offs [e.g. 1% and 5%; although 5% was predictive of shorter OS by univariate analysis, it was not independently associated with outcome when co-analysed with other prognostic features (data not shown)]. This observation is supported by the fact that all TP53 mutated cases, except one, showed ≥10% 3+ TP53 protein expression by immunohistochemistry. Although not encountered in our study, it should be noted that one drawback to using immunohistochemistry for determining TP53 mutation status and patient prognostication is that assessment of TP53 overexpression by IHC may render false negative results in cases with mutations that result in protein truncation. Two cases with early termination mutations were included in our study; however, both of these cases had additional missense TP53 mutations resulting in protein overexpression and therefore showed ≥10% 3+ TP53 -positive cells by IHC. Similar to previous studies (Cleven et al 2015; Saft et al 2014), we found that the presence of cells with weak (1+) to moderate (2+) TP53 staining had no association with clinical features or patient outcomes (data not shown). Based on our findings, we propose a working algorithm (Fig 5) that incorporates reticulin staining and TP53 immunohistochemistry using BM core biopsy specimens in cases of de novo MDS that will help in identifying this high-risk group of patients and enhance risk assessment.
Figure 5.
Proposed bone marrow evaluation algorithm for myelodysplastic syndromes. Proposed bone marrow evaluation algorithm for patients with de novo myelodysplastic syndromes incorporating fibrosis assessment, TP53 immunohistochemistry (IHC), and TP53 mutation analysis.
Although only assessed in a subset of cases, TP53 was the most frequently mutated gene in this study with a frequency of 44%. This frequency is higher than the reported 7–10% reported frequency of TP53 mutations in de novo MDS patients in general, (Bejar, et al 2014, Kulasekararaj, et al 2013) and more similar to the frequency of TP53 mutations in therapy-related MDS. (Andersen, et al 2005, Christiansen, et al 2001) The unfavourable effect of TP53 mutations in MDS has been well established. (Bally, et al 2014, Jadersten, et al 2011, Kulasekararaj, et al 2013, Volkert, et al 2014) Although the exact pathogenic role of TP53 mutations in MDS and AML is unknown, recent studies have demonstrated that TP53 mutant haematopoietic stem cells have a competitive survival advantage compared to their TP53 wild type counterparts during the course of chemotherapy, even in patients where the TP53 mutant clone represents a minor subclone. (Bondar and Medzhitov 2010, Marusyk, et al 2010, Wong, et al 2015) The observation of frequent TP53 mutations in MDS-F may, in part, explain the frequent association of this disease subtype with adverse clinical features and the dismal prognosis of this subset of patients. (Buesche, et al 2008, Della Porta and Malcovati 2011, Fu, et al 2014, Marisavljevic, et al 2004)
The association of TP53 protein overexpression with alterations of chromosome 5 (−5, del 5q) has been shown in de novo and therapy-related MDS by others. (Cleven, et al 2015, Kulasekararaj, et al 2013, Saft, et al 2014) We also observed a strong association between TP53 protein expression by immunohistochemistry and abnormalities of chromosome 5 in this cohort of patients with MDS-F and there was also a significant t association between TP53 protein expression and high-risk karyotypic features including alterations of chromosome 7 (monosomy 7, del 7q) and a complex karyotype.
Earlier studies have shown that patients with MDS and severe BM fibrosis have unfavourable outcomes when treated with SCT. (Kroger, et al 2011) In a recent study of TP53 protein expression in therapy-related MDS (Cleven, et al 2015), the authors showed that TP53-positive cases treated with SCT had very poor outcomes. In contrast, three patients with TP53 protein overexpression and TP53 mutation in this study who underwent allogeneic SCT had favourable outcomes and were alive and free of disease. This is obviously a small number of patients; however, the potential role of SCT in patients with MDS-F and TP53 protein expression/TP53 mutation should be systematically analysed in a larger cohort of patients.
We found that the presence of CD34+ cells showing 3+ TP53 expression was associated with increased BM blasts and shorter OS and leukaemia-free survival by univariate analysis. Previous studies have established the prognostic impact of increased BM blast counts in MDS-F. (Machherndl-Spandl, et al 2014) Incorporation of anti-CD34 in this immunohistochemistry cocktail is helpful in the enumeration of BM blast counts in this group of patients where the presence of moderate to severe BM fibrosis often leads to suboptimal BM aspirate smears, hampering accurate assessment of BM blast cell counts.
In summary, we show a high frequency of TP53 overexpression in > 10% BM cells in de novo MDS-F. TP53 overexpression correlates with TP53 mutations and is an independent adverse prognostic factor that may aid in risk-stratification for MDS-F patients.
Supplementary Material
Acknowledgments
The authors thank Leiloni Gilbert for assistance with manuscript preparation, Sherry Pierce for assistance with database queries, and Dr. Jaime Rodriguez-Canales and Lynda Corley for assistance with image analysis. This work was supported in part by the MD Anderson Cancer Center Support Grant (CA016672) from the National Cancer Institute.
Footnotes
Author contributions
SL: Data collection, data analysis and manuscript preparation; ZZ: Statistical analysis; AA-I, SAW, GGM, MY, SW, HMK, CCY, RNM, RL, LJM and CEB-R: Data analysis and manuscript preparation; JDK: Conception and design of study, data analysis and manuscript preparation. All authors read and approved the final manuscript.
References
- Andersen MK, Christiansen DH, Pedersen-Bjergaard J. Centromeric breakage and highly rearranged chromosome derivatives associated with mutations of TP53 are common in therapy-related MDS and AML after therapy with alkylating agents: an M-FISH study. Genes Chromosomes Cancer. 2005;42:358–371. doi: 10.1002/gcc.20145. [DOI] [PubMed] [Google Scholar]
- Bally C, Ades L, Renneville A, Sebert M, Eclache V, Preudhomme C, Mozziconacci MJ, de The H, Lehmann-Che J, Fenaux P. Prognostic value of TP53 gene mutations in myelodysplastic syndromes and acute myeloid leukemia treated with azacitidine. Leuk Res. 2014;38:751–755. doi: 10.1016/j.leukres.2014.03.012. [DOI] [PubMed] [Google Scholar]
- Bejar R, Stevenson K, Abdel-Wahab O, Galili N, Nilsson B, Garcia-Manero G, Kantarjian H, Raza A, Levine RL, Neuberg D, Ebert BL. Clinical effect of point mutations in myelodysplastic syndromes. N Engl J Med. 2011;364:2496–2506. doi: 10.1056/NEJMoa1013343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejar R, Stevenson KE, Caughey B, Lindsley RC, Mar BG, Stojanov P, Getz G, Steensma DP, Ritz J, Soiffer R, Antin JH, Alyea E, Armand P, Ho V, Koreth J, Neuberg D, Cutler CS, Ebert BL. Somatic mutations predict poor outcome in patients with myelodysplastic syndrome after hematopoietic stem-cell transplantation. J Clin Oncol. 2014;32:2691–2698. doi: 10.1200/JCO.2013.52.3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondar T, Medzhitov R. p53-mediated hematopoietic stem and progenitor cell competition. Cell Stem Cell. 2010;6:309–322. doi: 10.1016/j.stem.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunning RD, Orazi A, Germing U, Le Beau MM, Porwit A, Baumann I, Vardiman JW, Hellstrom-Lindberg E. Myelodysplastic syndromes/neoplasms, overview. In: Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, Thiele J, Vardiman JW, editors. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. IARC; Lyon: 2008. pp. 88–93. [Google Scholar]
- Buesche G, Teoman H, Wilczak W, Ganser A, Hecker H, Wilkens L, Gohring G, Schlegelberger B, Bock O, Georgii A, Kreipe H. Marrow fibrosis predicts early fatal marrow failure in patients with myelodysplastic syndromes. Leukemia. 2008;22:313–322. doi: 10.1038/sj.leu.2405030. [DOI] [PubMed] [Google Scholar]
- Christiansen DH, Andersen MK, Pedersen-Bjergaard J. Mutations with loss of heterozygosity of p53 are common in therapy-related myelodysplasia and acute myeloid leukemia after exposure to alkylating agents and significantly associated with deletion or loss of 5q, a complex karyotype, and a poor prognosis. J Clin Oncol. 2001;19:1405–1413. doi: 10.1200/JCO.2001.19.5.1405. [DOI] [PubMed] [Google Scholar]
- Cleven AH, Nardi V, Ok CY, Goswami M, Dal Cin P, Zheng Z, Iafrate AJ, Abdul Hamid MA, Wang SA, Hasserjian RP. High p53 protein expression in therapy-related myeloid neoplasms is associated with adverse karyotype and poor outcome. Mod Pathol. 2015;28:552–563. doi: 10.1038/modpathol.2014.153. [DOI] [PubMed] [Google Scholar]
- Della Porta MG, Malcovati L. Myelodysplastic syndromes with bone marrow fibrosis. Haematologica. 2011;96:180–183. doi: 10.3324/haematol.2010.039875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Della Porta MG, Malcovati L, Boveri E, Travaglino E, Pietra D, Pascutto C, Passamonti F, Invernizzi R, Castello A, Magrini U, Lazzarino M, Cazzola M. Clinical relevance of bone marrow fibrosis and CD34-positive cell clusters in primary myelodysplastic syndromes. J Clin Oncol. 2009;27:754–762. doi: 10.1200/JCO.2008.18.2246. [DOI] [PubMed] [Google Scholar]
- Della Porta MG, Travaglino E, Boveri E, Ponzoni M, Malcovati L, Papaemmanuil E, Rigolin GM, Pascutto C, Croci G, Gianelli U, Milani R, Ambaglio I, Elena C, Ubezio M, Da Via MC, Bono E, Pietra D, Quaglia F, Bastia R, Ferretti V, Cuneo A, Morra E, Campbell PJ, Orazi A, Invernizzi R, Cazzola M. Minimal morphological criteria for defining bone marrow dysplasia: a basis for clinical implementation of WHO classification of myelodysplastic syndromes. Leukemia. 2015;29:66–75. doi: 10.1038/leu.2014.161. [DOI] [PubMed] [Google Scholar]
- Fu B, Jaso JM, Sargent RL, Goswami M, Verstovsek S, Medeiros LJ, Wang SA. Bone marrow fibrosis in patients with primary myelodysplastic syndromes has prognostic value using current therapies and new risk stratification systems. Mod Pathol. 2014;27:681–689. doi: 10.1038/modpathol.2013.187. [DOI] [PubMed] [Google Scholar]
- Greenberg PL, Tuechler H, Schanz J, Sanz G, Garcia-Manero G, Sole F, Bennett JM, Bowen D, Fenaux P, Dreyfus F, Kantarjian H, Kuendgen A, Levis A, Malcovati L, Cazzola M, Cermak J, Fonatsch C, Le Beau MM, Slovak ML, Krieger O, Luebbert M, Maciejewski J, Magalhaes SM, Miyazaki Y, Pfeilstocker M, Sekeres M, Sperr WR, Stauder R, Tauro S, Valent P, Vallespi T, van de Loosdrecht AA, Germing U, Haase D. Revised international prognostic scoring system for myelodysplastic syndromes. Blood. 2012;120:2454–2465. doi: 10.1182/blood-2012-03-420489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honkaniemi E, Mattsson K, Barbany G, Sander B, Gustafsson B. Elevated p53 protein expression; a predictor of relapse in rare chronic myeloid malignancies in children? Pediatr Hematol Oncol. 2014;31:327–339. doi: 10.3109/08880018.2014.898723. [DOI] [PubMed] [Google Scholar]
- Jadersten M, Saft L, Smith A, Kulasekararaj A, Pomplun S, Gohring G, Hedlund A, Hast R, Schlegelberger B, Porwit A, Hellstrom-Lindberg E, Mufti GJ. TP53 mutations in low-risk myelodysplastic syndromes with del(5q) predict disease progression. J Clin Oncol. 2011;29:1971–1979. doi: 10.1200/JCO.2010.31.8576. [DOI] [PubMed] [Google Scholar]
- Khoury JD, Sen F, Abruzzo LV, Hayes K, Glassman A, Medeiros LJ. Cytogenetic findings in blastoid mantle cell lymphoma. Hum Pathol. 2003;34:1022–1029. doi: 10.1053/s0046-8177(03)00412-x. [DOI] [PubMed] [Google Scholar]
- Kroger N, Zabelina T, van Biezen A, Brand R, Niederwieser D, Martino R, Lim ZY, Onida F, Schmid C, Garderet L, Robin M, van Gelder M, Marks R, Symeonidis A, Kobbe G, de Witte T. Allogeneic stem cell transplantation for myelodysplastic syndromes with bone marrow fibrosis. Haematologica. 2011;96:291–297. doi: 10.3324/haematol.2010.031229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulasekararaj AG, Smith AE, Mian SA, Mohamedali AM, Krishnamurthy P, Lea NC, Gaken J, Pennaneach C, Ireland R, Czepulkowski B, Pomplun S, Marsh JC, Mufti GJ. TP53 mutations in myelodysplastic syndrome are strongly correlated with aberrations of chromosome 5, and correlate with adverse prognosis. Br J Haematol. 2013;160:660–672. doi: 10.1111/bjh.12203. [DOI] [PubMed] [Google Scholar]
- Lambertenghi-Deliliers G, Orazi A, Luksch R, Annaloro C, Soligo D. Myelodysplastic syndrome with increased marrow fibrosis: a distinct clinico-pathological entity. Br J Haematol. 1991;78:161–166. doi: 10.1111/j.1365-2141.1991.tb04411.x. [DOI] [PubMed] [Google Scholar]
- Machherndl-Spandl S, Sega W, Bosmuller H, Germing U, Gruber C, Nachtkamp K, Reinecke P, Sperr WR, Wimazal F, Mullauer L, Sotlar K, Horny HP, Tuchler H, Valent P, Krieger O. Prognostic impact of blast cell counts in dysplastic bone marrow disorders (MDS and CMML I) with concomitant fibrosis. Ann Hematol. 2014;93:57–64. doi: 10.1007/s00277-013-1945-4. [DOI] [PubMed] [Google Scholar]
- Marisavljevic D, Rolovic Z, Cemerikic V, Boskovic D, Colovic M. Myelofibrosis in primary myelodysplastic syndromes: clinical and biological significance. Med Oncol. 2004;21:325–331. doi: 10.1385/MO:21:4:325. [DOI] [PubMed] [Google Scholar]
- Marusyk A, Porter CC, Zaberezhnyy V, DeGregori J. Irradiation selects for p53-deficient hematopoietic progenitors. PLoS Biol. 2010;8:e1000324. doi: 10.1371/journal.pbio.1000324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saft L, Karimi M, Ghaderi M, Matolcsy A, Mufti GJ, Kulasekararaj A, Gohring G, Giagounidis A, Selleslag D, Muus P, Sanz G, Mittelman M, Bowen D, Porwit A, Fu T, Backstrom J, Fenaux P, MacBeth KJ, Hellstrom-Lindberg E. p53 protein expression independently predicts outcome in patients with lower-risk myelodysplastic syndromes with del(5q) Haematologica. 2014;99:1041–1049. doi: 10.3324/haematol.2013.098103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiele J, Kvasnicka HM, Facchetti F, Franco V, van der Walt J, Orazi A. European consensus on grading bone marrow fibrosis and assessment of cellularity. Haematologica. 2005;90:1128–1132. [PubMed] [Google Scholar]
- Vardiman JW, Thiele J, Arber DA, Brunning RD, Borowitz MJ, Porwit A, Harris NL, Le Beau MM, Hellstrom-Lindberg E, Tefferi A, Bloomfield CD. The 2008 revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia: rationale and important changes. Blood. 2009;114:937–951. doi: 10.1182/blood-2009-03-209262. [DOI] [PubMed] [Google Scholar]
- Verhoef GE, De Wolf-Peeters C, Ferrant A, Deprez S, Meeus P, Stul M, Zachee P, Cassiman JJ, Van den Berghe H, Boogaerts MA. Myelodysplastic syndromes with bone marrow fibrosis: a myelodysplastic disorder with proliferative features. Ann Hematol. 1991;63:235–241. doi: 10.1007/BF01698371. [DOI] [PubMed] [Google Scholar]
- Volkert S, Kohlmann A, Schnittger S, Kern W, Haferlach T, Haferlach C. Association of the type of 5q loss with complex karyotype, clonal evolution, TP53 mutation status, and prognosis in acute myeloid leukemia and myelodysplastic syndrome. Genes Chromosomes Cancer. 2014;53:402–410. doi: 10.1002/gcc.22151. [DOI] [PubMed] [Google Scholar]
- Warren M, Luthra R, Yin CC, Ravandi F, Cortes JE, Kantarjian HM, Medeiros LJ, Zuo Z. Clinical impact of change of FLT3 mutation status in acute myeloid leukemia patients. Mod Pathol. 2012;25:1405–1412. doi: 10.1038/modpathol.2012.88. [DOI] [PubMed] [Google Scholar]
- Wong TN, Ramsingh G, Young AL, Miller CA, Touma W, Welch JS, Lamprecht TL, Shen D, Hundal J, Fulton RS, Heath S, Baty JD, Klco JM, Ding L, Mardis ER, Westervelt P, DiPersio JF, Walter MJ, Graubert TA, Ley TJ, Druley TE, Link DC, Wilson RK. Role of TP53 mutations in the origin and evolution of therapy-related acute myeloid leukaemia. Nature. 2015;26:552–555. doi: 10.1038/nature13968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Singh RR, Patel KP, Stingo F, Routbort M, You MJ, Miranda RN, Garcia-Manero G, Kantarjian HM, Medeiros LJ, Luthra R, Khoury JD. BRAF kinase domain mutations are present in a subset of chronic myelomonocytic leukemia with wild-type RAS. Am J Hematol. 2014;89:499–504. doi: 10.1002/ajh.23652. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.