Abstract
The functions of the Bloom syndrome helicase (BLM) and its orthologs are well characterized in mitotic DNA damage repair, but their roles within the context of meiotic recombination are less clear. In meiotic recombination, multiple repair pathways are used to repair meiotic DSBs, and current studies suggest that BLM may regulate the use of these pathways. Based on literature from S. cerevisiae, A. thaliana, M. musculus, D. melanogaster, and C. elegans, we present a unified model for a critical meiotic role of BLM and its orthologs. In this model, BLM and its orthologs utilize helicase activity to regulate the use of various pathways in meiotic recombination by continuously disassembling recombination intermediates. This unwinding activity provides the meiotic program with a steady pool of early recombination substrates, increasing the probability for a DSB to be processed by the appropriate pathway. As a result of BLM activity, crossovers are properly placed throughout the genome, promoting proper chromosomal disjunction at the end of meiosis. This unified model can be used to further refine the complex role of BLM and its orthologs in meiotic recombination.
Keywords: Bloom syndrome helicase, crossover patterning, meiosis, meiotic recombination, double-strand break repair, resolvases, model organism genetics
Graphical Abstract
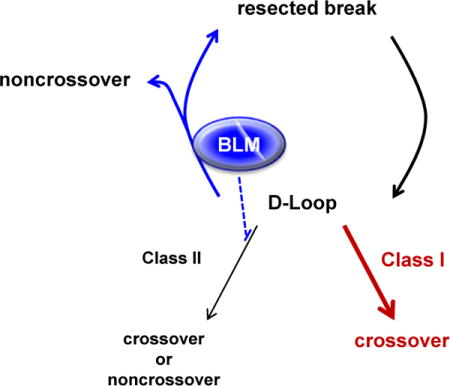
In meiotic recombination, Blm utilizes its helicase activity to disassemble early recombination intermediates, thereby retaining a pool of recombination sites from which some can be selected to enter the Class I crossover pathway while simultaneously promoting SDSA and preventing the use of the Class II pathway.
Introduction
Double-strand breaks (DSBs) can be detrimental to mitotically dividing cells if repaired as crossovers reviewed in [1]). During repair by homologous recombination (HR), cells employ a high-fidelity DSB repair mechanism that uses a homologous template (Figure 1). The proposed steps within the mitotic HR model for DSB repair are well defined (Figure 1). First, the ends of the DSB are resected to form 3′ tails (Figure 1B). One 3′ tail invades a homologous template to form a key intermediate, the D-loop, which can be enlarged by DNA synthesis (Figure 1C). With aid from helicases, the D-loop can be unwound from the invaded template either prior to synthesis to reverse the strand exchange, or after synthesis to promote synthesis-dependent strand annealing (SDSA) to produce a noncrossover repair product (Figure 1D). Alternatively, the other resected end can anneal to the displaced template strand and prime synthesis across the break (Figure 1E). Ligation of this intermediate generates a double-Holliday junction (dHJ) structure (Figure 1F). Helicases and associated topoisomerases can dissolve the dHJ, again generating noncrossover products (Figure 1G). In the absence of dissolution, the toxic dHJ must be removed; specialized structure-specific nucleases (SSNs) cleave each HJ in an unbiased manner (either orientation of cleavage; Figure 1H), generating either crossover (Figure 1I) or noncrossover (Figure 1J) products with equal probability. Because noncrossovers are the favorable outcome in DSB repair in mitotic cells, the helicase-dependent pathways SDSA and dissolution are considered to be the primary mitotic HR mechanisms, and as a result, anti-crossover helicases play a large role in maintaining genomic stability.
Figure 1.
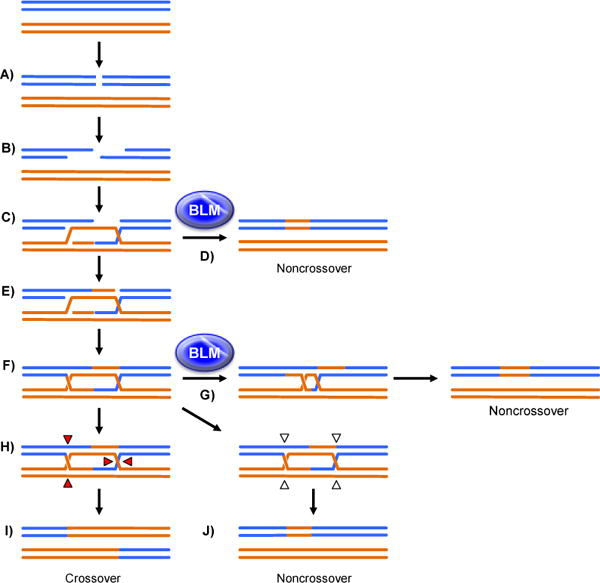
Mitotic Homologous Recombination (HR). A: Mitotic HR begins with a spontaneous DSB. B: The DSB is resected to yield 3′ single-stranded tails. C: A 3′ tail invades a homologous template, displacing a strand to create a D-loop. D: After synthesis off of the template, the nascent strand can be unwound and annealed to the other resected end of the DSB strand through synthesis-dependent strand annealing (SDSA), which is facilitated by BLM, to create a noncrossover. Here and in subsequent figures E: Alternatively, the displaced strand can anneal to the other side of the DSB to prime synthesis. F: After synthesis and ligation, a double-Holliday junction (dHJ) intermediate is formed. G: The dHJ can be unwound in a process called dissolution, facilitated by BLM, to generate a noncrossover. H: If not dissolved, the dHJ can be cleaved by endonucleases at the red triangles (or at the opposite strands at each HJ) to create a crossover. I: dHJs can also be cleaved by endonucleases at the open arrows (or at the opposite strands at each HJ) to generate a noncrossover (J). Dotted lines indicated nascent synthesis. Final products are represented after repair of any mismatches or other heterologies between parental molecules so as to emphasize transfer of sequence information from the template.
Bloom syndrome helicase (BLM), a member of the conserved RecQ helicase family, has been implicated in anti-crossover activity in mitotic cells. Bi-allelic mutations in BLM give rise to Bloom syndrome, a recessive human genetic disorder characterized by short stature, sterility, and cancer predisposition [2]. Cells from Bloom syndrome patients exhibit elevated mitotic recombination, suggesting that BLM prevents mitotic crossovers [3]. In vitro studies have demonstrated that BLM can disassemble D-loops and, with its partners topoisomerase 3α, RMI1, and RMI2 (the BTR complex), catalyze dHJ dissolution [4–7]. In vivo studies in Drosophila showed that in the absence of Blm, mitotic SDSA is severely diminished and mitotic crossovers are increased [8, 9] (Figure 1D). In S. cerevisiae, the Blm ortholog Sgs1 promotes dissolution of dHJs during mitotic-like repair [10] (Figure 1G). Collectively, these in vitro and in vivo studies establish that BLM and its orthologs have multiple conserved roles in generating noncrossovers within the mitotic HR pathway.
In contrast to mitotically dividing cells, meiotic cells employ a specialized version of HR to repair a subset of DSBs as crossovers between homologs, ultimately promoting proper chromosomal segregation at the first meiotic division. The meiotic DSBs that are not selected to become crossovers are repaired into noncrossovers, primarily via SDSA [11, 12], a feature of HR shared between meiotic and mitotic cells. Since SDSA may be facilitated largely by BLM in mitotic HR, it is intuitive to propose that the role of BLM in meiotic HR is to generate noncrossovers via SDSA. Indeed, studies in multiple organisms have found BLM orthologs to be essential for normal meiosis [13–21]. However, it has become increasingly clear that the roles of BLM orthologs in meiotic HR extend beyond promoting noncrossover recombination and that these roles may include, perhaps surprisingly, promoting crossover formation.
In this review, we discuss the many meiotic roles of BLM orthologs in the model organisms Saccharomyces cerevisiae (budding yeast; Sgs1), Arabidopsis thaliana (thale cress; RECQ4A and RECQ4B), Mus musculus (mouse; BLM), Drosophila melanogaster (fruit fly; Blm), and Caenorhabditis elegans (nematode; HIM-6). We review the proposal of two types of crossovers (Class I and Class II) and expand this to consider two classes of meiotic HR crossover pathways, the Class I and Class II pathways, uniting ideas from several sources [16,22–25]. Finally, we review and propose hypotheses for the involvement of BLM orthologs in regulating the use and execution of these crossover pathways.
Pathways for generating Class I and Class II crossovers: An expanded model for meiotic HR
Meiotic HR is initiated by the formation of programmed DSBs (Figure 2A) [reviewed in 26]), which are repaired using the homolog to yield crossover or noncrossover chromosomes. While the majority of meiotic DSBs are repaired as noncrossovers via SDSA [16] (Figure 2B), crossover formation between homologs is required for accurate chromosomal disjunction at the end of meiosis I. To ensure proper segregation, crossovers must be properly placed along the chromosome and among chromosomes [27, 28]. Thus, several crossover patterning phenomena are enforced upon meiotic HR to designate which DSBs (or repair intermediates; for simplicity, we will refer to these as DSBs) will be repaired as crossovers versus noncrossovers.
Figure 2.

Meiotic Homologous Recombination. A: Meiotic HR is initiated by a programmed DSB. B: After resection and D-loop formation, intermediates can undergo SDSA to yield noncrossovers (NCOs). C: Crossover-designated intermediates in the Class I pathway are stabilized by pro-crossover complexes. D: Meiosis-specific endonucleases resolve dHJs, E: exclusively generating crossovers that are patterned along and among chromosomes and poised to promote disjunction at the end of meiosis. F: Alternatively, Class II intermediates are processed independent of pro-crossover complexes. G: Ligated dHJs can either be dissolved (not pictured) or cleaved in an unbiased manner, H: resulting in a 1:1 ratio of crossovers: noncrossovers. Brackets: Crossovers created exclusively through the Class II pathway can yield nondisjunction due to lack of crossover patterning [21]. Note: For simplicity, coordination of dHJ resolution between multiple nuclease subunits is not depicted.
Two important crossover patterning phenomena are interference and assurance. Crossover interference reduces the probability of one DSB being repaired into a crossover if a nearby DSB has already been designated to become a crossover, resulting in crossovers being widely spaced along a chromosome arm (reviewed in [29]). Crossover assurance is the guarantee that each chromosome pair will receive at least one crossover regardless of chromosome size (reviewed in [30]). Together, these henomena give rise to interfering crossovers that are distributed so as to ensure accurate chromosome segregation at the end of meiosis.
Interfering crossovers in wild-type meiosis are formed through a pathway that employs meiosis-specific “pro-crossover” protein complexes (Figure 2C). In Arabidopsis, S. cerevisiae, C. elegans, and M. musculus, a central component of this complex consists of the Msh4 and Msh5 orthologs, which form the MutSɣ heterodimer (reviewed in [31]). Drosophila and other higher flies are unique among animals in lacking Msh4 and Msh5 orthologs, and instead appear to use a protein complex termed mei‐MCM [32]. Incorporation or stabilization of these complexes is thought to designate an intermediate to become a crossover. The final crossover-designated intermediate is cleaved by an endonuclease (Figure 2D) to produce mostly or exclusively crossover products (Figure 2E) [11, 33]. In Arabidopsis, S. cerevisiae, and M. musculus, this endonuclease complex includes MutLɣ (Mlh1 and Mlh3) (reviewed in [31]), but in Drosophila the catalytically active subunit is Mei-9 (the ortholog of the nucleotide excision repair protein Rad1/XPF) [34] and in C. elegans several partially redundant nucleases, including XPF‐1 and MUS-81, are used [14, 17, 35].
In many organisms, a minor fraction of meiotic crossovers and crossovers that occur in mutants that lack MutSɣ do not exhibit interference, leading Zalevsky et al. [22] to propose that there are two different meiotic crossover pathways that produce crossovers with different properties. Crossovers that exhibit interference are referred to as Class I crossovers, whereas those that do not participate in interference are called Class II crossovers [23, 24].
The Class I and Class II labels are generally used to differentiate the crossovers, but we find it helpful to expand this usage to the pathways that generate these crossovers. The pathway described above, which uses pro-crossover protein complexes and produces interfering crossovers, is the Class I pathway (Figure 2, green box). On the other hand, crossover patterning and biased resolution do not occur in the Class II pathway (Figure 2, red box) [16, 21, 24, 25]. Rather, meiotic intermediates that enter the Class II pathway are repaired similarly to DSBs in the mitotic HR pathway. Some dHJs may be produced, with at least some being independent of stabilization by the pro-crossover complexes (Figure 2F). These joint molecules can either be disassembled into noncrossovers through dissolution, or can be resolved through cleavage by an SSN, such as Mus81 [16, 25], in an unbiased manner (Figure 2G), generating crossovers and noncrossovers in a 1:1 ratio (Figure 2H).
Although crossovers generated by the Class II pathway are competent to form chiasmata (physical connections between homologs due to a crossover), in the absence of Class I crossovers, they may not be sufficient to guarantee proper disjunction due to their unpatterned placement throughout the genome [21]. Thus, crossover formation by the Class I pathway is favored over the Class II pathway. This crossover pathway favoritism suggests that there is a regulatory mechanism promoting DSB processing through the Class I pathway or prohibiting the use of the Class II pathway. Bearing in mind that the majority of the DSBs are processed via SDSA to generate noncrossovers, regulation of meiotic DSB processing through three pathways – SDSA, Class I, and Class II – is complex and not well understood.
S. cerevisiae Sgs1: The pioneering meiotic pathway regulator
Crossover patterning enforced in the Class I pathway leads to optimal crossover placement throughout the genome for promoting proper disjunction of homologs. Thus, it is reasonable to hypothesize a regulatory mechanism that promotes crossover formation through the Class I pathway. Perhaps surprisingly, this mechanism appears to involve the ortholog of BLM in Saccharomyces cerevisiae, Sgs1, which is generally considered an anti-crossover helicase.
Historically, Sgs1 is one of the first anti-crossover helicases to be examined in meiosis, initially being demonstrated to be required for efficient sporulation in 1996 [36]. Since then, myriad studies have shown that Sgs1 is an integral regulator of S. cerevisiae recombination. In S. cerevisiae, most noncrossovers appear before the dHJs and crossovers [11]. In sgs1 mutants, this early noncrossover formation is abolished [16], and dHJs are subsequently resolved by mitotic SSNs rather than the meiosis-specific resolvase MutLɣ [16, 25]. Further, most, if not all, crossovers generated in sgs1 mutants appear to be independent of the Class I pro-crossover stabilization complex MutSɣ [16] and exhibit reduced interference [13], suggesting these crossovers are created through the Class II pathway. Additionally, an increase in sister-chromatid recombination is observed when Sgs1 is absent [13, 37]. Together, these results suggest that Sgs1 has multiple roles in meiotic recombination, facilitating both early noncrossover formation and formation of Class I crossovers between homologous chromosomes.
From data obtained from the studies described above, an elegant model has been proposed for the roles of Sgs1 in meiosis [12, 16]. We present a modification of this model that incorporates the Class I and Class II pathway concept (Figure 3). After the formation of a DSB, 5′ resection occurs, yielding a break with two 3′ overhangs (Figure 3A). These 3′ ends can stochastically invade any homologous template (though there is a mechanism that discourages use of the sister chromatid, e.g. [38]), and it is the job of Sgs1 to continuously disassemble the resulting D-loops to maintain a constant pool of intermediates at very early stages of recombination, not yet committed to go down any particular repair pathway. In some instances, the two 3′ overhangs can invade different chromatids (e.g. one into each sister chromatid of the homologous chromosome) to generate aberrant joint molecules; Sgs1 and its interacting partners must dismantle these intermediates [12] (Figure 3B). In cases where one end has invaded a chromatid of the homolog (Figure 3C), the resulting D-loop can be captured by MutSɣ pro-crossover complex, thereby blocking its disassembly by Blm (Figure 3D). These stabilized intermediates are then processed through the Class I crossover pathway. D‐loops that have undergone synthesis but have not been stabilized by MutSɣ can undergo Sgs1-faciliatated SDSA to form noncrossovers (Figure 3E). Lastly, intermediates that are not unwound by Blm and that are not stabilized by pro-crossover complexes are processed by the Class II pathway (called ALT by Kaur et al. [12]) (Figure 3F).
Figure 3.
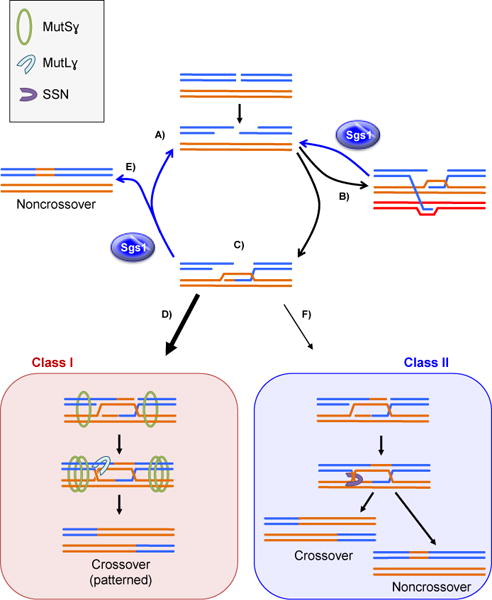
Model of Sgs1 in S. cerevisiae within meiotic recombination. A: After formation of a DSB, 5′ ends are resected to reveal 3′ tails. B: 3′ tails can inappropriately invade multiple homologs or sister chromatids (not pictured) to create aberrant joint molecules. These joint molecules are unwound by Sgs1 to regenerate the 3′ resected ends. C: The 3′ tail can invade the homologous chromosome to create a D-loop. D: A D-loop can be processed by the Class I pathway, to be subsequently stabilized by the MutSɣ complex and resolved by MutLɣ complex to generate a patterned crossover. E: D-loops that have undergone synthesis can be unwound by Sgs1, creating noncrossovers via SDSA. Sgs1 can unwind the D-loop if synthesis did not occur, or if there is too little synthesis for complementarity, Sgs1unwinds to regenerate the resected intermediate with some synthesis. F: D-loops can be processed by the Class II pathway, where they are eventually cleaved into equal ratios of crossovers to noncrossovers.
This model parsimoniously incorporates extensive data that have been obtained from sgs1 mutants in S. cerevisiae meiosis while considering the biochemical activities of BLM and Sgs1. Sgs1 promotes formation of crossovers through the Class I pathway by constantly regenerating early recombination intermediates, and at the same time, prevents Class II processing by unwinding inappropriate D-loops. Yet, there are limitations to this model. In particular, a subset of crossovers produced in the absence of Sgs1 may still be dependent on MutSɣ [39], suggesting that either the Class I pathway is still active sgs1 mutants, though at lower levels, or that some Class II crossovers are MutSɣ-dependent. We favor the former interpretation (which is asserted throughout this review), as MutSɣ-dependence is a defining criterion of Class I crossovers [22, 23]. Indeed, if the Class I pathway is partially active in sgs1 mutants, then Sgs1 may have a more direct role in promoting the full use of the Class I pathway in wild type meiotic HR than what is proposed above.
While Sgs1 has proven to be critical for proper meiotic HR, it is important to note that Sgs1 is not acting alone within meiotic HR; instead, its meiotic roles are dependent upon its interacting partners, topoisomerase 3α (Top 3α) and RMI1 – a complex referred to as the STR complex (Table 1). In S. cerevisiae, recent studies by Kaur et al. [12] and Tang et al. [40] demonstrate that all of the described meiotic functions of Sgs1 are dependent upon Top3 and Rmi1. Unexpectedly, these studies revealed that Rmi1 and Top3 act independently of Sgs1 to disentangle chromosomes in late meiotic prophase, allowing for proper chromosomal segregation [12,40]. Similar BLM-independent roles for RMI1 and TOP3α have been described in Arabidopsis [20, 41–43] and C. elegans [44], but it is likely that functions described below for BLM orthologs also require these interacting proteins.
Table 1.
STR/BTR orthologs in model organisms.
| Organism | Protein
|
BLM-independent meiotic functions? | |||
|---|---|---|---|---|---|
| BLM | TOP3α | RMI1 | RMI2 | ||
| S. cerevisiae | Sgs1 | Top3 | Rmi1 | N/A | Yes [12,40] |
| A. thaliana | RECQ4A/B | TOP3α | RMI1 | RMI2 | Yes [20,41–43] |
| M. musculus | Blm | Top3α | Rmi1 | Rmi2 | unknown |
| D. melanogaster | Blm | Top3α | N/A | N/A | unknown |
| C. elegans | HIM-6 | TOP-3 | RMH-1 | N/A | Yes [44] |
N/A: No ortholog in this species.
Arabidopsis RECQ4A/B: Paralogs that promote and prevent crossovers
As discussed above, in S. cerevisiae, it is posited that Sgs1 acts as both a promoter of Class I crossover formation and an inhibitor of Class II crossover formation through maintaining a steady flux of early recombination intermediates (Figure 3). As in S. cerevisiae, Arabidopsis thaliana meiosis is dependent upon the presence of BLM orthologs. The paralogs RECQ4A and RECQ4B (herein referred to as RECQ4A/B), are BLM orthologs that appear to be redundant in meiosis [20]. The meiotic redundancy between RECQ4A/B is in stark contrast with these paralogs’ mitotic roles: RECQ4A mutants display hypersensitivity to DNA damaging agents and increased recombination, but RECQ4B mutants do not exhibit these defects [45]. The apparent discordance in redundancy of RECQ4A and RECQ4B in meiosis and mitosis suggests that these paralogs have different, perhaps specialized, roles in meiotic recombination.
During Arabidopsis meiotic HR, RECQ4A/B appear in abundance on chromosome axes as shown through immunofluorescence (IF), presumably soon after the onset of programmed DSBs, suggesting RECQ4A/B act early in HR [46]. In the absence of RECQ4A/B, aberrant connections between homologs are observed [20], suggesting that RECQ4A/B, like S. cerevisiae Sgs1, dismantles multi-chromatid recombination intermediates. Additionally, in RECQ4A/B null mutants, the dependence of crossover formation on MutSɣ is eliminated [20], suggesting that RECQ4A/B has some role in guiding DSBs into the Class I pathway. Interestingly, in the absence of RECQ4A/B, crossovers are increased 6-fold without a change in DSB number. Although there is an increase in crossovers, the number of MutSɣ-dependent interactions is not changed when compared to wildtype, but crossover interference is abolished [20], suggesting that Class I crossovers are still being generated in RECQ4A/B null mutants. Seguela-Arnaurd and colleagues [20] suggest that the dramatic increase in observed crossovers is due to a combination of the elimination of RECQ4A/B-dependent SDSA in the Class I pathway and unbiased cleavage of intermediates in the Class II pathway.
In light of the Sgs1 model in S. cerevisiae (Figure 3), and considering the in vitro biochemical properties of BLM [5, 6, 47, 48], we propose that RECQ4A/B acts similarly to Sgs1 in meiotic recombination. In this model, RECQ4A/B uses its helicase activity to dismantle aberrant multi-chromatid recombination molecules and unwind D-loops to create a steady flux of early recombination intermediates that have the potential to be designated as interfering crossovers and subsequently stabilized by MutSɣ. D-loops that have not been designated to go down the Class I crossover pathway, but have undergone synthesis, are also unwound by RECQ4A/B, resulting in noncrossovers via SDSA. In the absence of RECQ4A/B, both the Class I and Class II pathways are active; only the SDSA pathway is inactive, giving rise to a dramatic increase in crossovers.
Although we propose that RECQ4A/B and Sgs1 act similarly in a meiotic context, the phenotypes of RECQ4A/B and sgs1 mutants are not identical. Without RECQ4A/B, Class I crossovers are presumed to be near or above wild type levels [20], a result that is not observed in sgs1 mutants [37]. Furthermore, the dramatic 6-fold increase in crossovers seen in RECQ4A/B null mutants is not observed in sgs1 mutants [16, 25]. These differences in mutant phenotypes suggest that RECQ4A/B and Sgs1 may have subtle differences in regulating meiotic recombination intermediates, or that another helicase in S. cerevisiae, such as Mph1,) may be able to promote SDSA [49, 50].
Mammalian BLM: Inhibitor of inappropriate interactions
As in Arabidopsis and S. cerevisiae, it is widely accepted that the Class I pathway generates the majority of crossovers in mouse meiosis. Although the Class II pathway in mouse meiosis has not been defined by the presence of non-interfering crossovers, Holloway et al. showed that Mus81, a SSN implicated in the Class II pathway in budding yeast and Arabidopsis [23, 25, 51], generates a subset of the crossovers that are independent of the Class I endonuclease MutLɣ [52]. This result suggests that the Class II pathway is indeed active in mouse meiosis.
In addition to utilizing alternative SSNs, for a crossover pathway to be considered truly Class II it must also be independent of pro-crossover complexes and produce unpatterned crossovers. To this point, Holloway and colleagues show that in Mus81 mutants, the number of MutSɣ foci does not change, but MutLɣ foci – used also as a proxy for crossovers – increase, resulting in a decrease in interference [52]. These results suggest two models for this second crossover pathway in mammalian meiosis: 1) Mus81-dependent recombination intermediates are not stabilized by MutSɣ but can be resolved by the Class I nuclease MutLɣ when Mus81 is absent (Figure 4A); or 2) Mus81-dependent recombination intermediates are stabilized by MutSɣ and are resolved by MutLɣ in Mus81 mutants (Figure 4B). Additional experiments, including determining whether Mus81 crossovers are MutSɣ-independent and examining interference in Mus81-dependent crossovers, are required to determine if this Class II mammalian pathway is similar to the Class II pathway proposed for yeast, worms, and plants.
Figure 4.
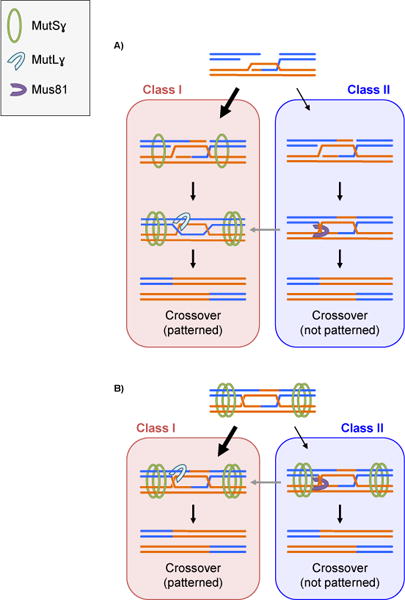
Models of Class I/Class II crossover pathways in mammals. A: D-loop formation can either be processed by the Class I pathway, whereby MutSɣ stabilization and MutLɣ resolution can occur. If processed by the Class II pathway, the intermediate will be cleaved by Mus81 to generate a crossover. Gray arrow: In the absence of Mus81, Class II intermediates are stabilized by MutSɣ and resolved by MutLɣ. B: Alternatively, MutSɣ stabilizes the recombination intermediate prior to Class I/Class II processing. Gray arrow: In the absence of Mus81, MutLɣ resolves the intermediate. Blm is not illustrated but it could have a disassembly function similar to that of orthologs in other model organisms.
There have been few investigations of the roles of Blm in mammalian meiosis. Cytological studies demonstrate that Blm is present throughout the progression of prophase I in mouse meiosis, from the time of early DSB processing (as inferred from Dmc1 co-localization) to later recombination processing (as inferred by Mlh1 staining) [15, 53, 54]. As in Arabidopsis, progression of the Class I pathway appears to be normal in mouse Blm mutants, as determined by dynamics of Msh4 and Mlh1/3 foci. However, altered meiotic chromatid structures that are suggestive of promiscuous homolog or sister interactions are observed [52], as in Arabidopsis [20].
The data presented above suggest that in the absence of Blm the Class I crossover pathway may still be active and aberrant chromatid interactions may be occurring. Assuming the second pathway in mammalian meiosis is equivalent to the Class II pathway defined for other model organisms (Figure 4A), and noting the similarities of Blm mutant cytological phenotypes to those seen in Arabidopsis, we suggest a model in which Blm acts similarly to Sgs1 in S. cerevisiae (Figure 3) and RECQ4A/B in Arabidopsis. In this model, mammalian Blm controls the flux of early recombination intermediates by 1) unwinding inappropriate chromatid interactions, 2) disassembling D-loops that are not stabilized by MutSɣ, and 3) generating noncrossovers from D-loops that have undergone synthesis and are not stabilized for entry into the Class II pathway.
Drosophila Blm: Promoter of patterning through pathway selection
In Drosophila meiosis, most – if not all – crossovers are formed through the Class I pathway. Typical of Class I products, these crossovers exhibit crossover patterning phenomena such as interference and the centromere effect (the inhibition of proximal crossovers), both of which were initially discovered in Drosophila [55, 56], are dependent on the pro-crossover mei-MCM complex [32], and are presumably produced through biased resolution by MEI-9 [34, 57]. In the absence of MEI-9, residual crossovers occur, providing the first hint that a second crossover pathway may be active in Drosophila meiosis [34, 58]. However, a more complete description of properties of Class II crossovers came out of studies of the meiotic roles of Blm [21, 32].
In Blm mutants, crossovers are no longer dependent on the mei-MCM complex [32] or MEI-9 [21], and crossover patterning phenomena are either reduced or completely abolished [21]. A mutation that allow production of Blm protein but abolishes ATPase activity results in the same defects [21]. Collectively, these data indicate that Blm utilizes its helicase function to promote crossover formation through the Class I pathway, while inhibiting the use of the Class II pathway.
With the model for S. cerevisiae Sgs1 in mind, we propose that Drosophila Blm also maintains the flux of early recombination intermediate, thereby regulating the use of the Class I, Class II, and SDSA pathways (Figure 5). After resection, there is invasion of a 3′ end into the homolog. The synaptonemal complex, which in Drosophila is built prior to the introduction of DSBs [59], may provide a barrier to sister chromatid invasion. After D-loop formation, either 1) Blm unwinds the structure to regenerate a resected end ready to invade a homologous template once again; 2) synthesis occurs, followed by unwinding by Blm to promote noncrossover generation via SDSA (Figure 5A); or 3) the mei-MCM complex stabilizes the intermediate so that disassembly by Blm is blocked and it can enter the Class I crossover pathway (Figure 5B).
Figure 5.

Model of Blm in Drosophila meiotic recombination. A: After D-loop formation, Blm can unwind intermediate that have undergone synthesis to create a noncrossover, or if no synthesis occurred, Blm can unwind to regenerate the 3′ resected end intermediate. B: Crossover-designated D-loop intermediates are processed through the Class I pathway to be resolved into patterned crossovers. C: Alternatively, D-loop intermediates can be processed by the Class II pathway to generate both crossovers and noncrossovers.
In this model, Blm is protecting early recombination intermediates from being processed by the Class II pathway as well as promoting Class I crossover designation by maintaining a pool of substrates upon which Class I machinery can act. This model predicts that in the absence of Blm, the Class I pathway is still active (perhaps at lower levels than in wild type) and that the SDSA pathway is abolished or impaired, suggesting that most of the DSB intermediates are processed via the Class II pathway (Figure 5C).
Notably, it is yet to be determined cytologically whether the Class I pathway remains active in Blm mutants, as predicted by the model in Figure 5, but genetically this assertion is not supported. In mei-9; Blm double mutants the crossover frequency is similar to that of Blm single mutants, which is about six-fold higher than in mei-9 single mutants, suggesting that MEI-9 is not actively generating Class I crossovers in Blm mutants [21]. Additional studies must be done to determine how much (if any) Class I pathway activity remains in Blm mutants.
C. elegans HIM-6: The Class I facilitator
In S. cerevisiae, Arabidopsis, and possibly M. musculus, the Class II pathway generates a subset of meiotic crossovers [23, 51, 52]. During wildtype meiotic HR in C. elegans, however, only the Class I pathway is active, producing exactly one crossover per chromosome pair [22, 60]. Because the Class II pathway is not utilized in wildtype C. elegans meiosis, the need for a Class I pathway regulatory role of HIM-6, the C. elegans ortholog of BLM, should be eliminated. Consistent with this prediction, gross meiotic progression and crossover designation are normal in HIM-6 mutants, as shown by MutSɣ cytology and other meiotic markers [18]. However, HIM-6 is not dispensable for meiosis; rather it is required for wild type levels of both noncrossovers and crossovers, placing HIM-6 in two important steps in meiotic HR [18].
RAD-51 foci, which mark early recombination intermediates, persist in HIM-6 mutants, perhaps at sites of intermediates that would have become noncrossovers through SDSA [35]. This suggests that noncrossover formation via SDSA is dependent upon HIM-6, similar to what is proposed in yeast, plants, and mice [15, 16, 20]. Further, an increase in MutSɣ-independent homolog interactions is observed in him-6 mutants [18], suggesting that aberrant interactions may occur in lieu of noncrossover formation.
In C. elegans, crossover-designated intermediates are resolved into crossovers by two parallel endonuclease pathways, one requiring XPF-1 and the other using SLX-1 and MUS-81 [14, 17, 35]. Epistasis experiments place HIM-6 in the XPF‐1 crossover pathway [17], and in the absence of HIM-6 a subset of crossover-designated intermediates fail to mature into crossovers [18]. This observation leads to the hypothesis that HIM-6 is acting directly on late recombination intermediates to bias their resolution into crossovers by XPF-1.
Together, these data suggest a model in which HIM-6 has two important functions: to generate noncrossovers via SDSA (Figure 5A) and to confer crossover resolution bias with XPF-1 (Figure 5B). These roles can be incorporated into the model posited for yeast (Figure 3). After resection, the 3′ ends of the DSB invade the homologous chromosome. As in Drosophila, the synaptonemal complex is built prior to the onset of DSBs [61], perhaps eliminating sister chromatid invasion. D-loops that are not stabilized by MutSɣ are disassembled by HIM-6 only to stochastically invade again, and D-loops that have undergone synthesis are unwound by HIM-6 to generate noncrossovers via SDSA. Intermediates that are stabilized by MutSɣ are resolved by the two parallel pathways, one in which HIM-6 is used to confer resolution bias.
Models speculating how crossover-biased resolution occurs through two parallel, partially redundant endonuclease pathways have been proposed [17, 18]. In one pathway, SLX-1 is proposed to nick a Holliday junction and MUS-81 is then responsible for the counter-nick on the opposing strand. In the parallel endonuclease pathway, HIM-6 acts as a dimer to unwind the intermediate to create a molecule that the XPF-1 homodimer can access and cleave in a biased manner, also resulting in a crossover (Figure 5C). There is not yet any evidence that XPF-1 and HIM-6 interact physically, but previous studies have revealed physical interactions between BLM and MUS-81 in human mitotic cells [62]. Both resolution mechanisms have been proposed to occur in the crossover-biased orientation, with HIM-6 imparting bias to the XPF-1 nuclease [17, 18]. However, to achieve crossover bias in dHJ resolution, cleavage of the two Holliday junctions must be coordinated with one another (Figure 1H) and it is unclear how either resolvase pathway might achieve that.
Meiotic crossovers are historically classified as Class I based on three different criteria: crossovers display interference, are dependent on pro-crossover complexes, and are generated in a biased manner by a meiosis-specific endonuclease complex. Class I crossovers in C. elegans experience perfect interference (only one crossover per chromosome pair), and are dependent on the pro-crossover complex MutSɣ. Yet opposed to other BLM orthologs, HIM-6 is proposed to have only a later function in meiotic HR by facilitating crossover resolution bias in the Class I pathway. Although HIM-6 has a later pro-crossover role in C. elegans while other Blm orthologs have an earlier pro-crossover role in meiotic HR, the alleged bias-conferring role of HIM-6 provides HIM-6 with a function in meiosis that echoes that of BLM orthologs in yeast, plants, mice, and flies – to promote the formation of Class I crossovers.
Meiotic BLM: A united model to promote Class I crossover formation
Through examination of the body of BLM meiosis literature, a unified model for the role of meiotic BLM can be proposed. In the model organisms discussed above, BLM and its orthologs use its helicase function to continuously generate substrates that can be selected for stabilization by the Class I pathway, thereby promoting the formation of Class I crossovers. While doing so, BLM orthologs are preventing the processing of these intermediates by the Class II pathway (if such a pathway exists in the organism). If sister chromatid or multi-chromatid associations occur, BLM orthologs disassemble these inappropriate interactions. Lastly, intermediates that have undergone synthesis but are not selected to enter the Class I crossover pathway are unwound by BLM orthologs to promote formation of noncrossovers through the SDSA pathway. Thus, using helicase activity, the meiotic roles of BLM across species may be summed up into three conserved roles: (1) promote Class I crossover formation; (2) protect against the use of the Class II pathway; and (3) enable the SDSA noncrossover pathway.
Conclusion
Although the role of BLM orthologs in mitotic HR has been extensively characterized, roles in meiotic HR have been less clear. In this review, we have discussed roles of Bloom syndrome helicase orthologs during meiotic recombination. Among different model organisms, these orthologs are proposed to utilize their helicase activity to continuously disassemble recombination intermediates, thereby regulating meiotic recombination in multiple ways. By constantly unwinding intermediates, BLM and its orthologs prevent inter-sister recombination, multi-chromatid interactions, and processing by the Class II pathway, while promoting both SDSA noncrossover formation and Class I crossover formation. While the models proposed here are subject to change – and the delineation between Class I/Class II/SDSA pathways is certainly more ambiguous than what is presented – this review provides a nearly unified model for BLM in meiosis. With this model as a starting point, the characterization of BLM and its homologs in meiosis can be further defined, providing both the meiosis community and the BLM community with a deeper understanding of the complex roles of BLM in meiotic recombination.
Figure 6.
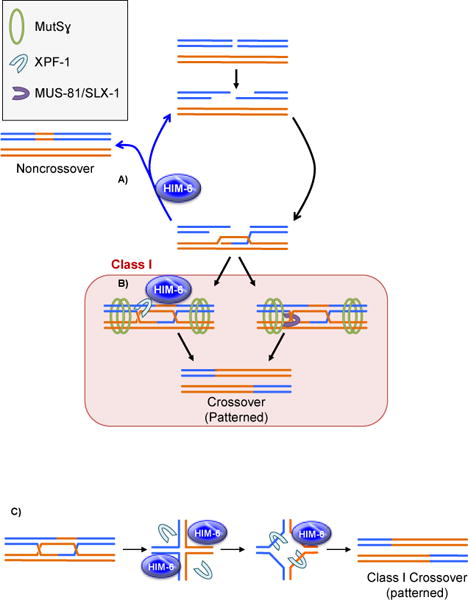
Model of HIM-6 in C. elegans meiotic recombination. A: HIM-6 can disassemble D-loops to regenerate 3′ resected end intermediates. If the D-loop underwent synthesis, HIM-6 can unwind the intermediate to generate a noncrossover via SDSA. B: Crossover-designated intermediates are resolved into crossovers through one of two parallel pathways: the HIM-6–XPF-1 pathway and the MUS-81–SLX‐1 pathway. C: Speculative mechanism of HIM-6 facilitating biased resolution of recombination intermediates: Two HIM-6 molecules unwind an unstable ligated intermediate, forming a structure available for a dimer of XPF-1 to cut and resolve into a crossover.
Acknowledgments
We thank the Sekelsky lab, especially Michaelyn Hartmann, for critical review of this manuscript. We appreciate the helpful and insightful comments from the reviewers. TH is supported in part by NIH grants 5T32GM007092 and 1F31AG055157. Research in the laboratory of JS is supported by a grant from the NIGMS (1R35GM118127).
Abbreviations
- BTR
BLM-Top3α-RMI1-RMI2 complex
- dHJ
double-Holliday junction
- DSB
double-strand break
- HR
homologous recombination
- SDSA
synthesis-dependent strand annealing
- SSN
structure-selective nuclease
Footnotes
Conflict of Interest
The authors declare that they have no conflicts of interest.
References
- 1.Andersen SL, Sekelsky J. Meiotic versus mitotic recombination: two different routes for double-strand break repair. BioEssays. 2010;32:1058–66. doi: 10.1002/bies.201000087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.German J. Bloom syndrome: a Mendelian prototype of somatic mutational disease. Medicine. 1993;72:393–406. [PubMed] [Google Scholar]
- 3.Chaganti RS, Schonberg S, German J. A manyfold increase in sister chromatid exchanges in Bloom’s syndrome lymphocytes. Proc Natl Acad Sci USA. 1974;71:4508–12. doi: 10.1073/pnas.71.11.4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bachrati CZ, Borts RH, Hickson ID. Mobile D-loops are a preferred substrate for the Bloom’s syndrome helicase. Nucleic Acids Res. 2006;34:2269–79. doi: 10.1093/nar/gkl258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Brabant AJ, Ye T, Sanz M, German IJ, et al. Binding and melting of D-loops by the Bloom syndrome helicase. Biochemistry. 2000;39:14617–25. doi: 10.1021/bi0018640. [DOI] [PubMed] [Google Scholar]
- 6.Wu L, Hickson ID. The Bloom’s syndrome helicase suppresses crossing over during homologous recombination. Nature. 2003;426:870–4. doi: 10.1038/nature02253. [DOI] [PubMed] [Google Scholar]
- 7.Wu L, Bachrati CZ, Ou J, Xu C, et al. BLAP75/RMI1 promotes the BLM-dependent dissolution of homologous recombination intermediates. Proc Natl Acad Sci USA. 2006;103:4068–73. doi: 10.1073/pnas.0508295103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adams MD, McVey M, Sekelsky J. Drosophila BLM in double-strand break repair by synthesis-dependent strand annealing. Science. 2003;299:265–7. doi: 10.1126/science.1077198. [DOI] [PubMed] [Google Scholar]
- 9.McVey M, Andersen SL, Broze Y, Sekelsky J. Multiple functions of Drosophila BLM helicase in maintenance of genome stability. Genetics. 2007;176:1979–92. doi: 10.1534/genetics.106.070052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dayani Y, Simchen G, Lichten M. Meiotic recombination intermediates are resolved with minimal crossover formation during return-to-growth, an analogue of the mitotic cell cycle. PLoS Genet. 2011;7:e1002083. doi: 10.1371/journal.pgen.1002083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Allers T, Lichten M. Differential timing and control of noncrossover and crossover recombination during meiosis. Cell. 2001;106:47–57. doi: 10.1016/s0092-8674(01)00416-0. [DOI] [PubMed] [Google Scholar]
- 12.Kaur H, De Muyt A, Lichten M. Top3-Rmi1 DNA single-strand decatenase is integral to the formation and resolution of meiotic recombination intermediates. Mol Cell. 2015;57:583–94. doi: 10.1016/j.molcel.2015.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oh SD, Lao JP, Hwang PY, Taylor AF, et al. BLM ortholog, Sgs1, prevents aberrant crossing-over by suppressing formation of multichromatid joint molecules. Cell. 2007;130:259–72. doi: 10.1016/j.cell.2007.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saito TT, Youds JL, Boulton SJ, Colaiácovo MP. Caenorhabditis elegans HIM-18/SLX-4 interacts with SLX-1 and XPF-1 and maintains genomic integrity in the germline by processing recombination intermediates. PLoS Genet. 2009;5:e1000735. doi: 10.1371/journal.pgen.1000735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holloway JK, Morelli MA, Borst PL, Cohen PE. Mammalian BLM helicase is critical for integrating multiple pathways of meiotic recombination. J Cell Biol. 2010;188:779–89. doi: 10.1083/jcb.200909048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Muyt A, Jessop L, Kolar E, Sourirajan A, et al. BLM helicase ortholog Sgs1 is a central regulator of meiotic recombination intermediate metabolism. Mol Cell. 2012;46:43–53. doi: 10.1016/j.molcel.2012.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Agostinho A, Meier B, Sonneville R, Jagut M, et al. Combinatorial regulation of meiotic Holliday junction resolution in C. elegans by HIM-6 (BLM) helicase, SLX-4, and the SLX-1, MUS-81 and XPF-1 nucleases. PLoS Genet. 2013;9:e1003591. doi: 10.1371/journal.pgen.1003591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schvarzstein M, Pattabiraman D, Libuda DE, Ramadugu A, et al. DNA helicase HIM-6/BLM both promotes MutSgamma-dependent crossovers and antagonizes MutSgamma-independent interhomolog associations during Caenorhabditis elegans meiosis. Genetics. 2014;198:193–207. doi: 10.1534/genetics.114.161513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rockmill B, Fung JC, Branda SS, Roeder GS. The Sgs1 helicase regulates chromosome synapsis and meiotic crossing over. Curr Biol. 2003;13:1954–62. doi: 10.1016/j.cub.2003.10.059. [DOI] [PubMed] [Google Scholar]
- 20.Seguela-Arnaud M, Crismani W, Larcheveque C, Mazel J, et al. Multiple mechanisms limit meiotic crossovers: TOP3alpha and two BLM homologs antagonize crossovers in parallel to FANCM. Proc Natl Acad Sci USA. 2015;112:4713–8. doi: 10.1073/pnas.1423107112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hatkevich T, Kohl KP, McMahan S, Hartmann MA, et al. Bloom syndrome helicase promotes meiotic crossover patterning and homolog disjunction. Curr Biol. 2017;27:1–5. doi: 10.1016/j.cub.2016.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zalevsky J, MacQueen AJ, Duffy JB, Kemphues KJ, et al. Crossing over during Caenorhabditis elegans meiosis requires a conserved MutS-based pathway that is partially dispensable in budding yeast. Genetics. 1999;153:1271–83. doi: 10.1093/genetics/153.3.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de los Santos T, Hunter N, Lee C, Larkin B, et al. The Mus81/Mms4 endonuclease acts independently of double-Holliday junction resolution to promote a distinct subset of crossovers during meiosis in budding yeast. Genetics. 2003;164:81–94. doi: 10.1093/genetics/164.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kohl KP, Sekelsky J. Meiotic and mitotic recombination in meiosis. Genetics. 2013;194:327–34. doi: 10.1534/genetics.113.150581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zakharyevich K, Tang S, Ma Y, Hunter N. Delineation of joint molecule resolution pathways in meiosis identifies a crossover-specific resolvase. Cell. 2012;149:334–47. doi: 10.1016/j.cell.2012.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lam I, Keeney S. Mechanism and regulation of meiotic recombination initiation. Cold Spring Harb Perspect Biol. 2014;7:a016634. doi: 10.1101/cshperspect.a016634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koehler KE, Boulton CL, Collins HE, French RL, et al. Spontaneous X chromosome MI and MII nondisjunction events in Drosophila melanogaster oocytes have different recombinational histories. Nat Genet. 1996;14:406–14. doi: 10.1038/ng1296-406. [DOI] [PubMed] [Google Scholar]
- 28.Lamb NE, Freeman SB, Savage-Austin A, Pettay D, et al. Susceptible chiasmate configurations of chromosome 21 predispose to non-disjunction in both maternal meiosis I and meiosis II. Nat Genet. 1996;14:400–5. doi: 10.1038/ng1296-400. [DOI] [PubMed] [Google Scholar]
- 29.Berchowitz LE, Copenhaver GP. Genetic interference: don’t stand so close to me. Curr Genomics. 2010;11:91–102. doi: 10.2174/138920210790886835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jones GH. The control of chiasma distribution. Symp Soc Exp Biol. 1984;38:293–320. [PubMed] [Google Scholar]
- 31.Manhart CM, Alani E. Roles for mismatch repair family proteins in promoting meiotic crossing over. DNA Repair (Amst) 2016;38:84–93. doi: 10.1016/j.dnarep.2015.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kohl KP, Jones CD, Sekelsky J. Evolution of an MCM complex in flies that promotes meiotic crossovers by blocking BLM helicase. Science. 2012;338:1363–5. doi: 10.1126/science.1228190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yokoo R, Zawadzki KA, Nabeshima K, Drake M, et al. COSA-1 reveals robust homeostasis and separable licensing and reinforcement steps governing meiotic crossovers. Cell. 2012;149:75–87. doi: 10.1016/j.cell.2012.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sekelsky JJ, McKim KS, Chin GM, Hawley RS. The Drosophila meiotic recombination gene mei-9 encodes a homologue of the yeast excision repair protein Rad1. Genetics. 1995;141:619–27. doi: 10.1093/genetics/141.2.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O’Neil NJ, Martin JS, Youds JL, Ward JD, et al. Joint molecule resolution requires the redundant activities of MUS-81 and XPF-1 during C. elegans meiosis. PLoS Genet. 2013;9:e1003582. doi: 10.1371/journal.pgen.1003582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Watt PM, Hickson ID, Borts RH, Louis EJ. SGS1, a homologue of the Bloom’s and Werner’s syndrome genes, is required for maintenance of genome stability in Saccharomyces cerevisiae. Genetics. 1996;144:935–45. doi: 10.1093/genetics/144.3.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jessop L, Lichten M. Mus81/Mms4 endonuclease and Sgs1 helicase collaborate to ensure proper recombination intermediate metabolism during meiosis. Mol Cell. 2008;31:313–23. doi: 10.1016/j.molcel.2008.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Niu H, Wan L, Baumgartner B, Schaefer D, et al. Partner choice during meiosis is regulated by Hop1-promoted dimerization of Mek1. Mol Biol Cell. 2005;16:5804–18. doi: 10.1091/mbc.E05-05-0465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jessop L, Rockmill B, Roeder GS, Lichten M. Meiotic chromosome synapsis-promoting proteins antagonize the anti-crossover activity of Sgs1. PLoS Genet. 2006;2:e155. doi: 10.1371/journal.pgen.0020155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tang S, Wu MK, Zhang R, Hunter N. Pervasive and essential roles of the Top3-Rmi1 decatenase orchestrate recombination and facilitate chromosome segregation in meiosis. Mol Cell. 2015;57:607–21. doi: 10.1016/j.molcel.2015.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chelysheva L, Vezon D, Belcram K, Gendrot G, et al. The Arabidopsis BLAP75/Rmi1 homologue plays crucial roles in meiotic double-strand break repair. PLoS Genet. 2008;4:e1000309. doi: 10.1371/journal.pgen.1000309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hartung F, Suer S, Knoll A, Wurz-Wildersinn R, et al. Topoisomerase 3alpha and RMI1 suppress somatic crossovers and are essential for resolution of meiotic recombination intermediates in Arabidopsis thaliana. PLoS Genet. 2008;4:e1000285. doi: 10.1371/journal.pgen.1000285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Seguela-Arnaud M, Choinard S, Larcheveque C, Girard C, et al. RMI1 and TOP3alpha limit meiotic CO formation through their C-terminal domains. Nucleic Acids Res. 2017;45:1860–71. doi: 10.1093/nar/gkw1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jagut M, Hamminger P, Woglar A, Millonigg S, et al. Separable roles for a Caenorhabditis elegans RMI1 homolog in promoting and antagonizing meiotic crossovers ensure faithful chromosome inheritance. PLoS Biol. 2016;14:e1002412. doi: 10.1371/journal.pbio.1002412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hartung F, Suer S, Puchta H. Two closely related RecQ helicases have antagonistic roles in homologous recombination and DNA repair in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2007;104:18836–41. doi: 10.1073/pnas.0705998104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Higgins JD, Ferdous M, Osman K, Franklin FC. The RecQ helicase AtRECQ4A is required to remove inter-chromosomal telomeric connections that arise during meiotic recombination in Arabidopsis. Plant J. 2011;65:492–502. doi: 10.1111/j.1365-313X.2010.04438.x. [DOI] [PubMed] [Google Scholar]
- 47.Karow JK, Chakraverty RK, Hickson ID. The Bloom’s syndrome gene product is a 3′–5′ DNA helicase. J Biol Chem. 1997;272:30611–4. doi: 10.1074/jbc.272.49.30611. [DOI] [PubMed] [Google Scholar]
- 48.Karow JK, Constantinou A, Li JL, West SC, et al. The Bloom’s syndrome gene product promotes branch migration of holliday junctions. Proc Natl Acad Sci USA. 2000;97:6504–8. doi: 10.1073/pnas.100448097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mitchel K, Lehner K, Jinks-Robertson S. Heteroduplex DNA position defines the roles of the Sgs1, Srs2, and Mph1 helicases in promoting distinct recombination outcomes. PLoS Genet. 2013;9:e1003340. doi: 10.1371/journal.pgen.1003340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scheller J, Schurer A, Rudolph C, Hettwer S, et al. MPH1, a yeast gene encoding a DEAH protein, plays a role in protection of the genome from spontaneous and chemically induced damage. Genetics. 2000;155:1069–81. doi: 10.1093/genetics/155.3.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Berchowitz LE, Francis KE, Bey AL, Copenhaver GP. The role of AtMUS81 in interference-insensitive crossovers in A. thaliana. PLoS Genet. 2007;3:e132. doi: 10.1371/journal.pgen.0030132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Holloway JK, Booth J, Edelmann W, McGowan CH, et al. MUS81 generates a subset of MLH1-MLH3-independent crossovers in mammalian meiosis. PLoS Genet. 2008;4:e1000186. doi: 10.1371/journal.pgen.1000186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moens PB, Freire R, Tarsounas M, Spyropoulos B, et al. Expression and nuclear localization of BLM, a chromosome stability protein mutated in Bloom’s syndrome, suggest a role in recombination during meiotic prophase. J Cell Sci. 2000;113(Pt 4):663–72. doi: 10.1242/jcs.113.4.663. [DOI] [PubMed] [Google Scholar]
- 54.Walpita D, Plug AW, Neff NF, German J, et al. Bloom’s syndrome protein, BLM, colocalizes with replication protein A in meiotic prophase nuclei of mammalian spermatocytes. Proc Natl Acad Sci USA. 1999;96:5622–7. doi: 10.1073/pnas.96.10.5622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Beadle GW. A possible influence of the spindle fibre on crossing-over in Drosophila. Proc Natl Acad Sci USA. 1932;18:160–5. doi: 10.1073/pnas.18.2.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sturtevant AH. The linear arrangement of six sex-linked factors in Drosophila, as shown by their mode of association. J Exp Biol. 1913;14:43–59. [Google Scholar]
- 57.Crown KN, McMahan S, Sekelsky J. Eliminating both canonical and short-patch mismatch repair in Drosophila melanogaster suggests a new meiotic recombination model. PLoS Genet. 2014;10:e1004583. doi: 10.1371/journal.pgen.1004583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Baker BS, Carpenter ATC. Genetic analysis of sex chromosomal meiotic mutants in Drosophila melanogaster. Genetics. 1972;71:255–86. doi: 10.1093/genetics/71.2.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McKim KS, Green-Marroquin BL, Sekelsky JJ, Chin G, et al. Meiotic synapsis in the absence of recombination. Science. 1998;279:876–8. doi: 10.1126/science.279.5352.876. [DOI] [PubMed] [Google Scholar]
- 60.Kelly KO, Dernburg AF, Stanfield GM, Villeneuve AM. Caenorhabditis elegans msh-5 is required for both normal and radiation-induced meiotic crossing over but not for completion of meiosis. Genetics. 2000;156:617–30. doi: 10.1093/genetics/156.2.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dernburg AF, McDonald K, Moulder G, Barstead R, et al. Meiotic recombination in C. elegans initiates by a conserved mechanism and is dispensable for homologous chromosome synapsis. Cell. 1998;94:387–98. doi: 10.1016/s0092-8674(00)81481-6. [DOI] [PubMed] [Google Scholar]
- 62.Zhang R, Sengupta S, Yang Q, Linke SP, et al. BLM helicase facilitates Mus81 endonuclease activity in human cells. Cancer Res. 2005;65:2526–31. doi: 10.1158/0008-5472.CAN-04-2421. [DOI] [PubMed] [Google Scholar]


