Abstract
Simultaneous delivery of multiple genes and proteins (e.g., transcription factors, TFs) is an emerging issue surrounding therapeutic research due to their ability to regulate cellular circuitry. Current gene and protein delivery strategies, however, are based on slow batch synthesis, which is ineffective, poorly controlled, and incapable of simultaneous delivery of both genes and proteins with synergistic functions. Consequently, advances in this field have been limited to in vitro studies. Here, by integrating microfluidic technologies with a supramolecular synthetic strategy, we present a high-throughput approach for formulating and screening multifunctional supramolecular nanoparticles (MFSNPs) self-assembled from a collection of functional modules to achieve simultaneous delivery of one gene and TF with unprecedented efficiency both in vitro and in vivo. We envision that this new approach could open a new avenue for immunotherapy, stem cell reprogramming, and other therapeutic applications.
Keywords: supramolecular assembly, nanoparticles, microfluidic technology, biomolecular delivery, transcription factor
Graphical abstract
Presently, research interests have been directed toward transcription factors for the purpose of investigating disease therapy due to their ability to control cell behavior.[1–3] Transcription factors are proteins that contain DNA-binding domains to recognize matching DNA sequences adjacent to the genes they regulate. Researchers have attempted to control transcription factors’ function by delivering the related DNA[1] or the transcription factor itself[2,3] into the cell to steer the cell’s fate towards a desired direction (e.g. apoptosis or cell differentiation, etc.). The transportation of transcription factors to a target location is especially challenging because most are easily decomposed by various enzymes in the body. Therefore, researchers have a wide variety of techniques for the delivery of genes and proteins.[4–12] However, the conventional approaches, including viral vectors,[4] nanocapsules,[5] nanoparticles,[6,7] liposomes,[7–9] copolymer,[10,11] and polypeptide[12] employed for optimization of delivery systems require empirical and multiple optimization cycles to repeat design/synthesis/assays processes. Given the enormous complexity of the biological system, it is unlikely that such a time- and cost-consuming developmental pipeline could lead to a crucial breakthrough in the search for optimal delivery vectors. As a result, most research has been limited to in vitro systems, apart from the recently reported multifunctional oligonucleotides.[13] Their advances in an in vivo system were highlighted[14] but still suffer from limitations on fast screening of the optimal combination due to their complex chemical synthesis. In our previous research, we proposed a convenient, flexible, and modular supramolecular synthetic strategy for delivery of intact (unmodified) TF in a highly efficient manner.[15] Yet, continuous efforts remain to be devoted to achieving in vivo applications.
Combining integrated microfluidic technologies[16–25] with a supramolecular synthetic strategy,[15,26–30] we demonstrate herein a high-throughput approach for formulating and screening multifunctional supramolecular nanoparticles (MFSNPs) that are capable of simultaneous delivery of gene and functional proteins with superb efficiency and controllable stoichiometry among individual payloads (Figure 1). As illustrated in Figure 1a, this approach utilized two microfluidic systems including a digital droplet generator (DDG; Supporting Information, Section 1.2–1.5, video available in Supporting Information)[20,22] and a microfluidic cell culture array[31,32]. To enable the capability of on-chip synthesis of MFSNPs using the DDG, we developed a modular assembly system based on the supramolecular synthetic strategy[15,26–30] (Figure 1b). By utilizing the molecular recognition between adamantane (Ad) and β-cyclodextrin (CD), a combinatorial library of DNA and proteins-encapsulated MFSNPs was generated by the DDG via systematically altering the ratios among the four functional modules. By coupling the DDG and the cell culture array, all the MFSNPs in the library could be exposed to cells for screening of the synthetic parameters that facilitated optimal delivery performance. By applying the synthetic parameters identified by the optimization studies (Figure 1b), the MFSNPs, which consist of a pair of a functionally complementary TF and gene[15,33] (GAL4-VP16 and pG5E4T-Fluc, respectively), were synthesized, and subsequently facilitated successful co-delivery of the protein and gene allowing them to function synergistically both in vitro and in vivo.
Figure 1. A high-throughput approach for formulating and screening MFSNPs for simultaneous delivery of genes and proteins.
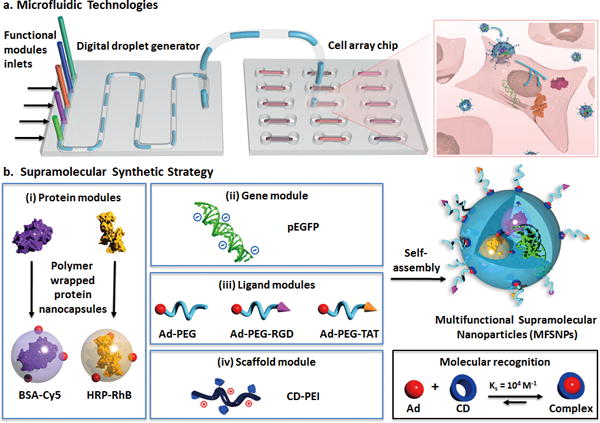
a) Two microfluidic systems, i.e., a digital droplet generator and a microfluidic cell array chip, were employed for formulating and screening of MFSNPs capable of simultaneous delivery of genes and functional proteins with superb efficiency and controllable stoichiometry among individual payloads. b) Our supramolecular synthetic strategy, based on an Ad and CD molecular recognition system, allows a convenient, flexible, and modular generation of a combinatorial library of genes and proteins-encapsulated MFSNPs by systematically altering the mixing ratio among the four functional modules, including (i) protein modules, (ii) gene module, (iii) ligand modules (Ad-PEG, Ad-PEG-RGD and Ad-PEG-TAT), and (iv) a scaffold module (CD-PEI).
One of the key features of this platform is its capacity to rapidly determine the optimal formulation of MFSNPs with the desired delivery efficiency. As illustrated in Figure 1b, the optimization studies using the model system consist of the following modules: (i) the protein modules include two different protein nanocapsules[5] (i.e., Cy5 labeled bovine serum albumin, BSA-Cy5, and rhodamin B-labeled horseradish peroxidase, HRP-RhB; Supporting Information, Section 1.8), which were made by encapsulating the protein within a thin layer of degradable polymer of which the surface was linked with Ad groups; (ii) the gene module can be any DNA of interest (i.e., EGFP-encode DNA plasmid, pEGFP); (iii) the function modules are a series of poly(ethylene glycol) (PEG) derivatives,[20,26,27] of which one end-group is terminated with an Ad group and the other end is terminated with a methyl group (Ad-PEG[26]; Supporting Information, Section 1.9) or a functional peptide, such as RGD (Ad-PEG-RGD[34]; Supporting Information, Section 1.10) and TAT (Ad-PEG-TAT[35]; Supporting Information, Section 1.11); and (iv) the scaffold module is CD-linked poly(ethylenimine) (CD-PEI[26]; Supporting Information, Section 1.12). This strategy yields generally applicable protein modules whose assembly properties are not affected by the core protein. Driven by the supramolecular synthetic strategy,[15,26–30] genes and proteins can be effectively incorporated together to form MFSNPs grafted with various functional molecules (Ad-PEG, Ad-PEG-RGD, and Ad-PEG-TAT) on their surface to achieve desired structural stability, delivery specificity, and cell transfusion capability, respectively. Moreover, because the gene and protein payloads are encapsulated inside the self-assembled MFSNPs, potential degradation[36] over the delivery process can be avoided.
By modulating the inlet of the functional module, three categories of MFSNPs with different surface ligands were synthesized (Supporting Information, Figure S1). The 1st category of MFSNPs was decorated with only Ad-PEG; the 2nd category (RGD-MFSNPs) contained Ad-PEG and Ad-PEG-RGD; and the 3rd category (RGD/TAT-MFSNPs) bears Ad-PEG, Ad-PEG-RGD, and Ad-PEG-TAT with pre-identified ratios (Supporting Information, Table S1)[15,28]. Each category contains 125 formulations of MFSNPs that were prepared by systematically altering the concentration of CD-PEI, BSA-Cy5, and HRP-RhB (Supporting Information, Section 1.4). As a result, 375 formulations of MFSNPs were prepared automatically with 1h (Supporting Information, Table S2). After 30-min incubation, each MFSNP droplet (0.2 μL) was then automatically diluted with Opti-MEM media to 2 μL, and introduced into an individually addressed cell culture chamber (containing NIH 3T3 cells, ca. 5 × 103 cells/chamber) in the cell culture array chip. After incubating the MFSNPs-treated cells in the cell array chip at 37 °C (5% CO2) for 24 h, the transfection and transduction efficiencies of each MFSNPs formulation were quantified by fluorescence microscopy-based single-cell image cytometry. A 3D plot (Figure 2a) summarizes the transfection and transduction efficiencies of the MFSNPs library, revealing that an optimal transfection/transduction performance was achieved using RGD/TAT-MFSNPs (3rd category) with a specific composition, i.e., CD-PEI = 20 ng, Ad-PEG = 100 ng, Ad-PEG-RGD = 5 ng, Ad-PEG-TAT = 9 ng, pEGFP = 10 ng, BSA-Cy5 = 39 ng and HRP-RhB = 27 ng in 200-nL PBS. Figure 2b shows the fluorescence images of cells treated with optimal RGD/TAT-MFSNPs, indicating the successful transfection of pEGFP and the successful transduction of both of the two proteins.
Figure 2. Optimization of MFSNPs for simultaneous delivery of genes and proteins.
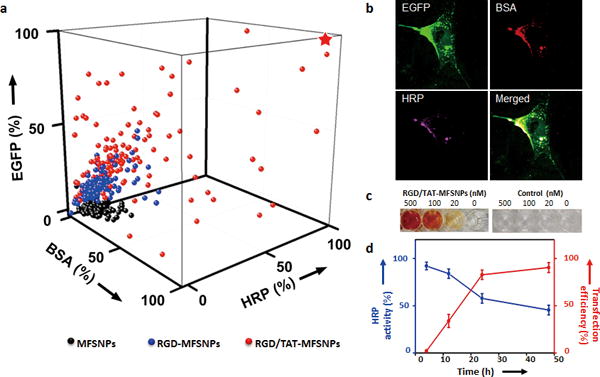
a) A 3D profile of gene transfection (pEGFP) and transduction performance (BSA-Cy5 and HRP-RhB) of three categories of MFSNPs produced by DDG. Black dots represent 125 data points observed for 1st category: MFSNPs; Blue dots (125 data points) for 2nd category: RGD-MFSNPs; Red dots (125 data points) for 3rd category: RGD/TAT-MFSNPs. b) Confocal micrographs that reflect transfection/transduction outcomes with RGD/TAT-MPSNP of high (★) delivery performance. c) A colorimetric assay using o-Dianisidine (3,3′-dimethoxybenzidine) (50 mM) as chromogenic substrates and H2O2 as an oxidizer to quantify intracellular enzymatic activity of HRP delivered by the RGD/TAT-MFSNPs. d) The time-dependent in vitro co-delivery performance of the optimal RGD/TAT-MFSNPs based on fluorescent signal quantification associated with EGFP expression and colorimetric readouts correlated to HRP activity, respectively.
To gain insight on how the structural property of RGD/TAT-MFSNPs affects the observed co-delivery performance, the optimal synthetic parameters (Figure 2a and b, ★) were chosen for scale-up preparation (200 droplets, 40 μL). The RGD/TAT-MFSNPs are of spherical morphology with diameters of 100 ± 4 nm and surface charges of 7.2 ± 2.5 mV, which were confirmed by transmission electron microscopy (TEM) and dynamic light scattering measurements (DLS; Supporting Information, Figure S5 – S7). Furthermore, a colorimetric assay was employed for quantifying the enzymatic activity of RGD/TAT-MFSNP-encapsulated HRP intracellular by using o-Dianisidine (3,3′-dimethoxybenzidine) as chromogenic substrates (Figure 2c). The time-dependent in vitro transfection and transduction performance of 100-nm RGD/TAT-MFSNPs was monitored in parallel by quantifying fluorescent signals associated with EGFP expression and colorimetric readouts correlated to HRP activity, respectively. As shown in Figure 2d, the highest co-delivery performance was achieved 24 to 48 h after RGD/TAT-MFSNP-treatment. Moreover, the other two formulations that led to sub-optimal delivery performances of RGD/TAT-MFSNPs were also carefully examined (Supporting Information, Figure S5 and Table S1). TEM images indicated that the size of those two sub-optimal RGD/TAT-MFSNPs were larger than 200 nm (Supporting Information, Figure S5d), which is unfavorable for high-performance delivery[7]. The stability of the RGD/TAT-MFSNPs in various conditions was also confirmed by a series of studies (Supporting Information, Figure S8–S14). In addition, no toxic effect was observed with RGD/TAT-MFSNPs (Supporting Information, Figure S15).
The uniqueness of the high-efficient co-delivery vector[15] lies in the fact that it can simultaneously introduce a functionally complementary protein and gene that will lead to synergistic outcomes for regulating cellular circuitry. To exploit the use of RGD/TAT-MFSNP vector for delivery of functionally complementary gene and TF, pG5E4T-Fluc (a plasmid vector that contains GAL4-VP16 matching recognition sequences and a luciferase reporter), and GAL4-VP16 (a transcription factor fusion protein) were chosen for conducting in vitro and in vivo studies (Figure 3a). Again, GAL4-VP16 was first coated with a degradable polymer of which the surface was linked with Ad groups. After formulation and intracellular delivery of pG5E4T-Fluc and GAL4-VP16, the luciferase reporter in pG5E4T-Fluc was specifically activated by GAL4-VP16, generating a real-time readout (bioluminescence) reflecting a synergistic outcome of the co-delivered payloads. By using the previously optimized synthetic parameters, 10 droplets (2 μL in total volume) containing desired RGD/TAT-MFSNPs were generated from CD-PEI (200 ng), Ad-PEG (1000 ng), Ad-PEG-RGD (50 ng), Ad-PEG-TAT (90 ng), pG5E4T-Fluc (100 ng), and GAL4-VP16 (660 ng) in the DDG. The resulting RGD/TAT-MFSNPs were introduced into a 96-well plate (containing NIH 3T3 cells, ca. 1 × 104 cells/well) along with control systems, including pG5E4T-Fluc plasmid and the protein module containing GAL4-VP16. After incubating (5% CO2) at 37 °C for 24, 48, and 72 h, the cells were lysed for quantification of bioluminescence using either a plate reader or a cooled charge-coupled device (CCD) camera (IVIS, Xenogen) (Figure 3b and c). Compared to background-level bioluminescence intensities observed from the control experiments, the bioluminescence intensity of the RGD/TAT-MFSNPs-treated cells was significantly higher, suggesting that the RGD/TAT-MFSNPs are capable of delivering both pG54ET-Fluc and GAL4-VP16 simultaneously into the cells in a highly efficient manner as well as allowing both the protein and gene functioning synergistically.
Figure 3. In vitro and in vivo co-delivery of functionally complementary gene and protein.
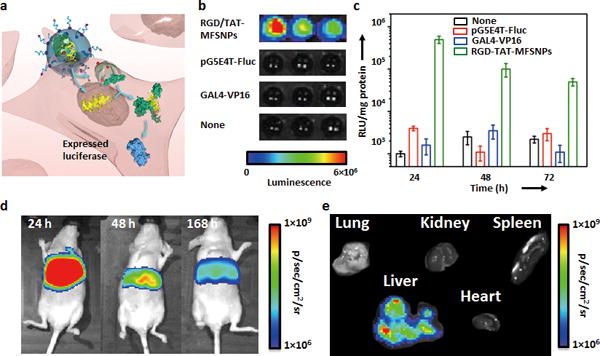
a) Simultaneous delivery of gene (pG5E4T-Fluc) and functionally complementary protein (GAL4-VP16) with optimal 100-nm RGD/TAT-MFSNP. b) Bioluminescence imaging and c) time-dependent luciferase expression of the 100-nm RGD/TAT-MFSNPs-treated cells along with the control experiments based on pG5E4T-Fluc plasmid and GAL4-VP16. Error bars were obtained from three independent experiments. d) Monitoring in vivo synergetic effects of the co-delivered gene and protein after i.v. injection of 100-nm RGD/TAT-MFSNPs (containing 1 μg pG5E4T-Fluc and 6.6 μg Ad-n-GAL4-VP16) into nu/nu nude mice (n = 3) via tail veins. e) Ex vivo experiments of 100-nm RGD/TAT-MFSNPs-treated mice were performed after 168 h post injection.
Finally, the in vivo experiments were performed by injection of the RGD/TAT-MFSNPs that encapsulated pG5E4T-Fluc (1 μg) and GAL4-VP16 (6.6 μg) into nu/nu nude mice (n = 3) via tail veins. As illustrated in Figure 3d and S16 (Supporting Information), a strong bioluminescent signal was observed from the liver of the treated animal, indicating efficient delivery of the RGD/TAT-MFSNPs into the hepatocytes. The intensity of the signal at 168 h post injection was decreased in magnitude two-fold compared to that at 48 h post injection (Supporting Information, Figure S16 and S17). Ex vivo results were consistent with bioluminescent imaging data (Figure 3e). Furthermore, negligible in vivo cytotoxicity of the optimal co-delivery system was observed with both histological study and blood chemistry assay (Supporting Information, Figure S18 and S19).
In conclusion, by integrating microfluidic technologies with a supramolecular synthetic strategy, we have demonstrated a new high-throughput approach for formulating and screening MFSNPs. Using our DDG, hundreds of formulations with variable delivery performance were programmed into the MFSNPs combinatorial library. Subsequently, in vitro screening of the MFSNPs library in cell array chips identified specific formulations that lead to desired delivery performance. The optimal formulations were then employed to encapsulate functionally complementary TF and gene into MFSNP-based vectors, and the respective synergistic outcomes were achieved both in vitro and in vivo. This approach could open up a new avenue[14] for immunotherapy, stem cell reprogramming and other therapeutic applications.
Supplementary Material
Acknowledgments
This research was supported by National Institutes of Health (R21 487GM098982 and R21 EB016270) and California Institute of Regenerative Medicine (RT1-01022).
Footnotes
Supporting information for this article is available on the WWW under http://www.angewandte.org or from the author
Contributor Information
Dr., Prof. Yang Liu, Key Laboratory of Functional Polymer Materials of Ministry of Education, Institute of Polymer Chemistry, State Key Laboratory of Medicinal Chemical Biology, Nankai University, Tianjin (China) California NanoSystems Institute, Department of Chemical and Bimolecular Engineering, UCLA (USA); Laboratory of Biological Effects of Nanomaterials and Nanosafety, National Center for Nanoscience and Technology (NCNST), Chinese Academy of Science, No. 11 Beiyitiao, Zhongguancun, Beijing (China).
Juanjuan Du, California NanoSystems Institute, Department of Chemical and Bimolecular Engineering, UCLA (USA).
Dr. Jin-sil Choi, Crump Institute for Molecular Imaging, California NanoSystems Institute, Department of Molecular and Medical Pharmacology, UCLA, CA 90095 (USA)
Dr. Kuan-Ju Chen, Crump Institute for Molecular Imaging, California NanoSystems Institute, Department of Molecular and Medical Pharmacology, UCLA, CA 90095 (USA)
Dr. Shuang Hou, Crump Institute for Molecular Imaging, California NanoSystems Institute, Department of Molecular and Medical Pharmacology, UCLA, CA 90095 (USA)
Ming Yan, California NanoSystems Institute, Department of Chemical and Bimolecular Engineering, UCLA (USA).
Prof. Wei-Yu Lin, Department of Medicinal and Applied Chemistry, Kaohsiung Medical University, 100, Shih-Chuan 1st Road, Kaohsiung, 80708 (Taiwan)
Kevin Sean Chen, Crump Institute for Molecular Imaging, California NanoSystems Institute, Department of Molecular and Medical Pharmacology, UCLA, CA 90095 (USA).
Tracy Ro, Crump Institute for Molecular Imaging, California NanoSystems Institute, Department of Molecular and Medical Pharmacology, UCLA, CA 90095 (USA).
Prof. Gerald S. Lipshutz, Division of Liver and Pancreas Transplantation, Department of Surgery, David Geffen School of Medicine, UCLA (USA)
Prof. Lily Wu, Crump Institute for Molecular Imaging, California NanoSystems Institute, Department of Molecular and Medical Pharmacology, UCLA, CA 90095 (USA)
Prof. Linqi Shi, Key Laboratory of Functional Polymer Materials of Ministry of Education, Institute of Polymer Chemistry, State Key Laboratory of Medicinal Chemical Biology, Nankai University, Tianjin (China)
Prof. Yunfeng Lu, California NanoSystems Institute, Department of Chemical and Bimolecular Engineering, UCLA (USA)
Prof. Hsian-Rong Tseng, Crump Institute for Molecular Imaging, California NanoSystems Institute, Department of Molecular and Medical Pharmacology, UCLA, CA 90095 (USA)
Prof. Hao Wang, Laboratory of Biological Effects of Nanomaterials and Nanosafety, National Center for Nanoscience and Technology (NCNST), Chinese Academy of Science, No. 11 Beiyitiao, Zhongguancun, Beijing (China)
References
- 1.Darnell JE., Jr Nat Rev Cancer. 2002;2:740–749. doi: 10.1038/nrc906. [DOI] [PubMed] [Google Scholar]
- 2.Thomas M. Reg Med. 2010;5:441–450. doi: 10.2217/rme.10.17. [DOI] [PubMed] [Google Scholar]
- 3.Maherali N, Hochedlinger K. Adv Drug Deliv Rev. 2008;3:595–605. [Google Scholar]
- 4.Takahashi K, Yamanaka S. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 5.Yan M, et al. Nat Nanotechnol. 2010;5:48–53. doi: 10.1038/nnano.2009.341. [DOI] [PubMed] [Google Scholar]
- 6.Ghosh P, Yang XC, Arvizo R, Zhu ZJ, Agasti SS, Mo ZH, Rotello VM. J Am Chem Soc. 2010;132:2642–2645. doi: 10.1021/ja907887z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davis ME, Chen Z, Shin DM. Nat Rev Drug Discov. 2008;7:771–782. doi: 10.1038/nrd2614. [DOI] [PubMed] [Google Scholar]
- 8.Nie SM, Xing Y, Kim GJ, Simons JW. Annu Rev Biomed Eng. 2007;9:257–288. doi: 10.1146/annurev.bioeng.9.060906.152025. [DOI] [PubMed] [Google Scholar]
- 9.Zuris JA, et al. Nat Biology. 2015;33:73–80. [Google Scholar]
- 10.Kakizawa Y, Kataoka K. Adv Drug Deliv Rev. 2002;54:203–222. doi: 10.1016/s0169-409x(02)00017-0. [DOI] [PubMed] [Google Scholar]
- 11.Wang Y, Gao S, Ye W-H, Yoon HS, Yang Y-Y. Nat Mater. 2006;5:791–796. doi: 10.1038/nmat1737. [DOI] [PubMed] [Google Scholar]
- 12.Gabrielson NP, Lu H, Yin L, Li D, Wang F, Cheng J. Angew Chem Int Ed. 2012;51:1143–1147. doi: 10.1002/anie.201104262. [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew Chem. 2012;124:1169–1173. [Google Scholar]
- 13.Lee K, Rafi M, Wang X, Aran K, Feng X, Sterzo Carlo Lo, Tang R, Lingampalli N, Kim HJ, Murthy N. Nat Mater. 2015;14:701–706. doi: 10.1038/nmat4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Servick K. Science News. 2015 doi: 10.1126/science.aab2549. [DOI] [Google Scholar]
- 15.Liu Y, Wang H, Kamei K, Yan M, Chen KJ, Yuan QH, Shi LQ, Lu YF, Tseng HR. Angew Chem Int Ed. 2011;50:3058–3062. doi: 10.1002/anie.201005740. [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew Chem. 2011;123:3114–3118. [Google Scholar]
- 16.Lee CC, et al. Science. 2005;310:1793–1796. doi: 10.1126/science.1118919. [DOI] [PubMed] [Google Scholar]
- 17.Wang Y, et al. Lab Chip. 2009;9:2281–2285. doi: 10.1039/b907430a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang JY, Sui GD, Mocharla VP, Lin RJ, Phelps ME, Kolb HC, Tseng HR. Angew Chem Int Ed. 2006;45:5276–5281. doi: 10.1002/anie.200601677. [DOI] [PubMed] [Google Scholar]; Angew Chem. 2006;118:5402–5407. [Google Scholar]
- 19.Wang YJ, et al. Lab Chip. 2009;9:2281–2285. doi: 10.1039/b907430a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang H, et al. ACS Nano. 2010;4:6235–6243. doi: 10.1021/nn101908e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin WY, Wang YJ, Wang ST, Tseng HR. Nano Today. 2009;4:470–481. doi: 10.1016/j.nantod.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu K, Chen YC, Tseng HR, Shen CKF, van Dam RM. Microfluid Nanofluid. 2010;9:933–943. doi: 10.1007/s10404-010-0617-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yoshida J-i, Nagaki A, Yamada T. Chem- Eur J. 2008;14:7450–7459. doi: 10.1002/chem.200800582. [DOI] [PubMed] [Google Scholar]
- 24.Marre S, Jensen KF. Chem Soc Rev. 2010;39:1183–1202. doi: 10.1039/b821324k. [DOI] [PubMed] [Google Scholar]
- 25.deMello AJ. Nature. 2006;442:394–402. doi: 10.1038/nature05062. [DOI] [PubMed] [Google Scholar]
- 26.Wang H, et al. Angew Chem Int Ed. 2009;48:4344–4348. doi: 10.1002/anie.200900063. [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew Chem. 2009;121:4408–4412. [Google Scholar]
- 27.Wang H, Chen KJ, Wang ST, Ohashi M, Kamei K, Sun J, Ha JH, Liu K, Tseng HR. Chem Commun. 2010;46:1851–1853. doi: 10.1039/b923711a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang ST, Chen KJ, Wu TH, Wang H, Lin WY, Ohashi M, Chiou PY, Tseng HR. Angew Chem Int Ed. 2010;49:3777–3781. doi: 10.1002/anie.201000062. [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew Chem. 2010;122:3865–3869. [Google Scholar]
- 29.Chen KJ, et al. Biomaterials. 2011;32:2160–2165. doi: 10.1016/j.biomaterials.2010.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stoffelen C, Huskens J. Chem Commun. 2013;49:6740–6742. doi: 10.1039/c3cc43045f. [DOI] [PubMed] [Google Scholar]
- 31.Sun J, et al. Cancer Res. 2010;70:6128–6138. doi: 10.1158/0008-5472.CAN-10-0076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kamei K, et al. Lab Chip. 2010;10:1113–1119. doi: 10.1039/b922884e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sadowski I, Ma J, Triezenberg S, Ptashne M. Nature. 1988;335:563–564. doi: 10.1038/335563a0. [DOI] [PubMed] [Google Scholar]
- 34.Merkel OM, Germershaus O, Wada CK, Tarcha PJ, Merdan T, Kissel T. Bioconjugate Chem. 2009;20:1270–1280. doi: 10.1021/bc9001695. [DOI] [PubMed] [Google Scholar]
- 35.Torchilin VP. Peptide Science. 2008;90:604–610. doi: 10.1002/bip.20989. [DOI] [PubMed] [Google Scholar]
- 36.Fagain CO. Biochimica et Biophysical Acta. 1995;1252:1–14. doi: 10.1016/0167-4838(95)00133-f. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


