Abstract
We have identified 6201 alternative splice relationships in human genes, through a genome-wide analysis of expressed sequence tags (ESTs). Starting with ∼2.1 million human mRNA and EST sequences, we mapped expressed sequences onto the draft human genome sequence and only accepted splices that obeyed the standard splice site consensus. A large fraction (47%) of these were observed multiple times, indicating that they comprise a substantial fraction of the mRNA species. The vast majority of the detected alternative forms appear to be novel, and produce highly specific, biologically meaningful control of function in both known and novel human genes, e.g. specific removal of the lysosomal targeting signal from HLA-DM β chain, replacement of the C-terminal transmembrane domain and cytoplasmic tail in an FC receptor β chain homolog with a different transmembrane domain and cytoplasmic tail, likely modulating its signal transduction activity. Our data indicate that a large proportion of human genes, probably 42% or more, are alternatively spliced, but that this appears to be observed mainly in certain types of molecules (e.g. cell surface receptors) and systemic functions, particularly the immune system and nervous system. These results provide a comprehensive dataset for understanding the role of alternative splicing in the human genome, accessible at http://www.bioinformatics.ucla.edu/HASDB.
INTRODUCTION
Alternative splicing is an important mechanism for modulating gene function. It can change how a gene acts in different tissues and developmental states by generating distinct mRNA isoforms composed of different selections of exons. Alternative splicing has been implicated in many processes, including sex determination (1), apoptosis (2) and acoustic tuning in the ear (3). Recently, it has been suggested that if alternative splicing is widespread in the human genome, it could represent a relatively efficient expansion of the genome’s ‘vocabulary’ of variant genes, by producing multiple functional forms of many genes. Its functional implications can be simple, generating a single alternative form, or can produce remarkable diversity. In the Drosophila gene Dscam, combinatorial alternative splicing of ‘cassettes’ of exons reminiscent of the combinatorial generation of immunoglobulin diversity, produces thousands of distinct functional isoforms (4). This gene, homologous to the human gene for Down’s syndrome cell adhesion molecule (DSCAM), appears to be involved in neuronal guidance, where such diversity could be useful as a molecular ‘address’.
Alternative splicing has been studied intensively in hundreds of human genes (1,5), and it appears to be widespread, occurring in 5–30% of human genes (6,7) or perhaps as many as 35–40% (8,9). Recently, it has been reported that alternative splicing can be detected in expressed sequence tag (EST) sequencing (9) and has been analyzed in a collection of full-length mRNAs (8). Based upon estimates of the total number of human genes (10,11), it is likely that at least 10 000–20 000 human genes are alternatively spliced. However, currently only 899 alternatively spliced human genes are catalogued in the Alternative Splicing Database of Mammals (AsMamDB) (12).
We have performed a genome-wide analysis of alternative splicing based on human expressed sequence data, which greatly expands our knowledge of this central function in human molecular biology (Table 1). We have identified tens of thousands of splices, and thousands of alternative splices, in several thousand human genes. We have mapped all of these onto the draft human genome sequence, and verified that the putative splice junctions detected in the expressed sequences map onto genomic exon–intron junctions that match the known splice site consensus. Based on this genome-wide analysis of gene structure and alternative splicing, we have constructed a Human Alternative Splicing Database, at http://www.bioinformatics.ucla.edu/HASDB. In this paper we also show how our database can be used to study the impact of alternative splicing on protein function. We present an initial analysis of the patterns and functional role of alternative splicing across the human genome.
Table 1. Alternative splicing in mRNA and EST sequence data.
| All genes | Genes with mRNA | On chromosome 22 | |||||||
| |
No. splices |
No. clusters |
|
No. splices |
No. clusters |
|
No. splices |
No. clusters |
|
| Initial unigene clusters | 86 244 | 16 240 | 548 | ||||||
| Mapped to draft genome (10/00) | 47 422 | 55% | 6603 | 41% | 421 | 77% | |||
| Detected splices | 39 862 | 8429 | 18% | 30 495 | 4024 | 61% | 1797 | 220 | 52% |
| Alternative splice relationships | 6201 | 2272 | 27% | 5009 | 1687 | 42% | 313 | 94 | 43% |
| (with multiple evidence) | 2892 | 1306 | 2505 | 1089 | 141 | 56 | |||
Tabulation of the number of splices and number of distinct gene clusters in which they were observed, from the total dataset (All genes), clusters containing partial and/or full-length mRNAs (Genes with mRNA) and a control set of 548 clusters that have been STS-mapped to chromosome 22 (On chromosome 22). Percentages are given for the fraction of gene clusters successfully mapped by our procedure to the October 2000 draft human genome sequence (14) (Mapped to draft genome 10/00); the fraction of mapped gene clusters observed to contain at least one splice (Detected splices); the fraction of gene clusters containing alternative splices, out of the total observed to contain at least one splice (Alternative splice relationships); and the subset of alternative splices that were observed in multiple ESTs or a human-verified mRNA, as opposed to being observed in just a single EST (with multiple evidence).
As we seek to show with examples in this paper, our database could be a useful resource to researchers who have found a new cDNA or human gene and wish to find additional information. It can help answer a wide range of questions, e.g. ‘Are the two bands on a western blot due to alternative splicing?’ or ‘Do the genes in protein family X all use alternative splicing as a mechanism to modulate function?’ The database integrates a variety of data for each gene, ranging from genomic map location to gene structure, with links to external resources such as GenBank, OMIM, SWISS-PROT etc. It provides a detailed alignment of the ESTs, mRNAs, genomic DNA and protein sequence, showing single nucleotide polymorphisms (SNPs) (13), exons and introns, splice site junctions, alternative splices and, most importantly, the raw experimental evidence for all of these features, including chromatogram traces from public EST sequencing projects.
MATERIALS AND METHODS
Data sources
Our analysis is based on two major types of data: human genomic sequence assemblies and human EST sequences. Human genomic assembly sequences (accession no. NT_XXXX) and ‘draft’ BAC clone sequences [accession nos ACXXXX, ALXXXXX, etc. (14)] were downloaded from NCBI (ftp://ncbi.nlm.nih.gov/genome/seq and ftp://ncbi.nlm.nih.gov/genbank/gbhtgXXX.seq.gz). For partially sequenced clones, ‘draft’ fragments of 4 kb or longer continuous sequence were included in our analysis. All the work described in this paper is based on the October 2000 release of these data. Human EST sequences were downloaded from UNIGENE (ftp://ncbi.nlm.nih.gov/repository/UniGene). We used the EST clustering provided by UNIGENE, and did not perform our own re-clustering of the EST sequences. All the work described in this paper is based on the December 2000 release of UNIGENE.
Genomic mapping of expressed sequence clusters
Consensus sequences from our previous analysis of human expressed sequences (13) were searched against a database of human genomic assembly sequences using BLAST (15). We used consensus sequences to eliminate non-consensus features of each UNIGENE cluster, such as EST sequencing errors, chimeric ESTs or contamination by a minority of similar but paralogous sequences. The consensus sequence excludes these features, and should prevent them from affecting the genomic search. Our assembly and consensus analysis of UNIGENE was previously described as part of SNP discovery from human expressed sequences (13), and the consensus sequences are available at http://www.bioinformatics.ucla.edu/snp/. Briefly, after assembly, the maximum likelihood traversal of the EST–mRNA alignment is generated using dynamic programming, producing a consensus sequence that excludes minority features such as sequencing errors, sequence differences due to paralog contamination, unaligned ends and inserts due to chimeric sequences or unspliced introns.
The search for the genomic location of each UNIGENE cluster was performed in two stages. First, we identify the candidate gene regions in the genomic sequence for a given consensus using a BLAST threshold of E < 10–50 and a nucleotide mismatch penalty of 11. Secondly, to check the candidate gene location, we searched for radiation hybrid mapping data for sequence tagged sites (STS) linked with this gene. Candidate regions that did not agree with the STS mapping information for the cluster were discarded. Thirdly, we identified the putative exons, by using a lower threshold (E < 10–10) that will report shorter exons. The resulting BLAST hits must span the entire consensus, allowing only up to 100 bp of unmatched sequence at the consensus ends. Allowing BLAST this short unmatched region at the ends is necessary, since it may not identify very small exons reliably. Genomic candidates are assessed in order of ascending expectation value, until a candidate passes our second BLAST stage. The matching genomic region, plus 2 kb on either end to allow for short or fragmentary exons at the ends that BLAST may have missed, is aligned with the complete set of ESTs and mRNAs for the UNIGENE cluster using dynamic programming (16,17), truncating the gap extension penalty beyond 16 bp to allow for introns. The full EST and mRNA sequence must match the genomic sequence to be kept for the alternative splicing analysis. If an EST has 6 bp or more of insert relative to genomic, it is excluded. Using this procedure we mapped 47 422 of the 86 244 UNIGENE clusters onto the genomic sequence. Based on our analysis of chromosome 22, and comparison with NCBI’s Acembly gene mapping, we estimate a false negative rate for our mapping procedure of 20%, and an upper bound for its false positive rate of 3–5% (see Results).
Alternative splicing analysis
Splicing is detected by a computational procedure that analyzes the genomic–EST–mRNA multiple sequence alignment. Briefly, the gene structure is marked on the genomic sequence, based on its alignment with ESTs and mRNAs, by drawing a connection between each pair of genomic letters aligned to a pair of letters in an expressed sequence that are adjacent (i.e. nucleotide i and i+1). Thus, an exon is identified by a contiguous segment of connected letters, an intron by a contiguous segment of unconnected letters and a splice by a connection that jumps from one genomic letter to a distant genomic letter. Thus, a candidate splice is detected as a gap between two exons that match a single contiguous region of one or more ESTs. We report splices only for connections that skip >10 bp in the genomic sequence (representing an effective minimum intron length) to screen out sequencing error or alignment heterogeneity artefacts. Individual splice observations from different ESTs are joined together when their 5′ splice sites match within 6 bp in the genomic sequence, and their 3′ splice sites also match within 6 bp. This level of variation is permitted to screen out sequencing error and alignment artefacts that could give spurious alternative splices. All candidate splices were checked against the standard consensus splicing sequences, and all candidates with mismatches were discarded. It is possible that some of these mismatches may be viable deviations from the consensus sequence and represent real splices. However, we have excluded them from consideration in the results presented in this paper. This procedure was designed to be robust, and even in cases with a mis-assembled genomic sequence should not report spurious splices. Instead, genomic mis-assembly would likely cause mismatch with the ESTs, and complete exclusion of the cluster from our analysis. It should be noted that EST–mRNA versus genomic sequence alignments occasionally contain degenerate alignment positions, in which one or more nucleotides are identical in the genomic sequence on either side of a gap (intron). In this case our software checks each of the equivalent alignments to identify the correct splice junction.
Alternative splices are reported when two detected splices overlap in the genomic sequence (and thus are mutually exclusive events). One important consequence of this definition is that alternative splicing always requires positive evidence (i.e. strong match of EST to genomic) on both sides of each compared splice. An alternative splice will never be reported simply because one EST is longer or shorter than others, or even if vector sequence was attached at one end. [Vector sequences are screened out of UNIGENE (18) data. However, it is still important to note that heterogeneity at EST ends will not give rise to reported alternative splices.] All splices, alternative splices, individual splice observations in specific sequences, source library information, gene information, genomic mapping information, etc. are stored within a relational database (MySQL), and are accessible for query via the web (http://www.bioinformatics.ucla.edu/HASDB). To assess the fraction of alternative splices detected based on mRNA, EST versus mRNA or EST versus EST evidence (Fig. 5D), we used a database query to compute these numbers for all the alternative splices in our database.
Figure 5.
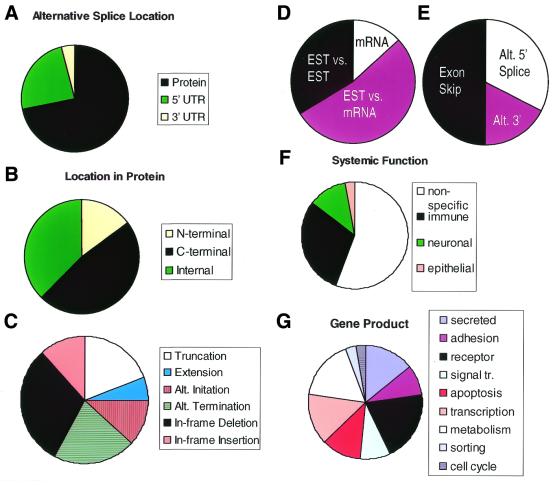
Analysis of a random sample of alternative spliced genes. (A) Fraction of the observed alternative splice in protein coding region, 5′ and 3′ UTR. (B) Fraction replacing the protein N-terminus, C-terminus or internal regions. (C) Fraction causing truncation of protein product due to frame-shift; extension of the protein product due to frame-shift; switch to a new initiator codon while preserving protein reading frame; switch to a new terminator codon from an alternative exon; in-frame deletion of codons, preserving reading frame; in-frame insertion of codons, preserving frame. (D) Origin of alternative splicing evidence: detected in mRNA (presumed not novel); detected in EST (by comparison with an mRNA); detected in EST (by comparison with other ESTs). (E) Type of alternative splice. (F) Categorization of alternatively spliced genes by systemic function (see text). (G) Categorization by gene product (see text).
Functional analysis of alternative splicing
We have performed extensive visual analysis and verification of our results, for hundreds of different genes. We used the GeneMine software system (19) to validate all aspects of the genomic mapping of our clusters, the exons, introns, splice sites, alternative splicing analysis and impact on protein structure and function, by thoroughly examining each of these features in the genomic–EST–mRNA multiple sequence alignments. The GeneMine software is freely available to academic researchers (http://www.bioinformatics.ucla.edu/genemine).
To characterize the functional impacts of alternative splicing, a random sample of 50 clusters with alternative splicing and at least one full-length mRNA was generated (Table 2). The mRNA requirement was imposed to ensure that the cluster would contain as complete a set of the gene’s exons as possible, to cover the full coding region and untranslated regions (UTRs). Without such coverage in many cases it is not even possible to define what the actual bounds of the coding region are, let alone get unbiased sampling of the coding region versus UTRs. To characterize the function of each gene product at both the cellular and systemic level required careful manual evaluation and study (i.e. not only sequence analysis but also digging into the available literature and information on the web). We did not feel that the twin objectives of accurate classification of the functional effects of alternative splicing and lack of selection bias could be provided reliably by electronic annotations at this time, although this is a very interesting area for further work. The effect of each alternative splice was evaluated manually, by careful examination of the complete alignment and available information using the GeneMine software. Most importantly, we considered all possible changes in the boundaries of the coding region (alternative initiation, alternative termination, truncation, extension, in-frame deletion and insertion). Since an alternative splice can change where the coding region starts and ends, it is incorrect to classify it as the UTR simply because it is upstream of the translation start site given by the GenBank annotation for the gene. We have adopted the policy that any alternative splice that alters the protein product will be classified as a ‘coding region’, regardless of its location relative to the GenBank CDS annotation. In the process, the alternative splices affecting the coding region were identified as changing the N-terminal, C-terminal or internal region of the protein.
Table 2. Random gene sample used for functional analysis.
| Cluster ID |
Gene |
Title |
| Hs.104519 | PLD2 | Phospholipase D2 |
| Hs.84190 | SLC19A1 | Solute carrier family 19 (folate transport) |
| Hs.366 | PTS | 6-Pyruvoyltetrahydropterin synthase |
| Hs.43812 | STX10 | Syntaxin 10 |
| Hs.6483 | CXORF5 | Chromosome X open reading frame 5 |
| Hs.52166 | LOC51275 | Apoptosis-related protein PNAS-1 |
| Hs.172894 | BID | BH3 interacting domain death agonist |
| Hs.20887 | FLJ10392 | Hypothetical protein |
| Hs.26994 | FLJ20477 | Hypothetical protein |
| Hs.76873 | HYAL2 | Hyaluronoglucosaminidase 2 |
| Hs.81337 | LGALS9 | Lectin, galactoside-binding, soluble, 9 (galectin 9) |
| Hs.198246 | GC | Group-specific component (vitamin D binding protein) |
| Hs.155247 | ALDOC | Aldolase C, fructose-bisphosphate |
| Hs.125139 | FLJ11004 | Hypothetical protein |
| Hs.89575 | CD79B | CD79B antigen (immunoglobulin-associated β) |
| Hs.49427 | LOC51291 | Gem-interacting protein |
| Hs.7100 | CL25084 | Hypothetical protein |
| Hs.11042 | LOC51248 | Hypothetical protein |
| Hs.82359 | TNFRSF6 | Tumor necrosis factor receptor superfamily, member 6 |
| Hs.2839 | NDP | Norrie disease (pseudoglioma) |
| Hs.94498 | LILRA2 | Leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 2 |
| Hs.169294 | TCF7 | Transcription factor 7 (T-cell specific, HMG-box) |
| Hs.75562 | DDR1 | Discoidin domain receptor family, member 1 |
| Hs.3657 | KIAA0784 | KIAA0784 protein |
| Hs.99855 | FPRL1 | Formyl peptide receptor-like 1 |
| Hs.1252 | APOH | Apolipoprotein H (β-2-glycoprotein I) |
| Hs.171595 | HTATSF1 | HIV TAT specific factor I |
| Hs.278522 | PSG6 | Pregnancy specific β-1-glycoprotein 6 |
| Hs.55847 | LOC51258 | Hypothetical protein |
| Hs.76285 | DKFZP564B167 | DKFZP564B167 protein |
| Hs.89506 | PAX6 | Paired box gene 6 (aniridia, keratitis) |
| Hs.1334 | MYB | v-myb avian myeloblastosis viral oncogene homolog |
| Hs.7768 | FIBP | Fibroblast growth factor (acidic) intracellular binding protein |
| Hs.3280 | CASP6 | Caspase 6, apoptosis-related cysteine protease |
| Hs.6710 | MPDU1 | Mannose-P-dolichol utilization defect 1 |
| Hs.78921 | AKAP1 | A kinase (PRKA) anchor protein 1 |
| Hs.96038 | RIT | Ric (Drosophila)-like |
| Hs.73851 | ATP5J | ATP synthase, H+ transporting, mitos F0 complex, subunitF6 |
| Hs.167031 | DKFZP566D133 | DKFZP566D133 protein |
| Hs.49767 | NDUFS6 | NADH dehydrogenase (ubq) Fe-S protein 6 (13 kDa) (NADH CoQ reductase) |
| Hs.151761 | KIAA0100 | KIAA0100 gene product |
| Hs.83937 | FLJ20323 | Hypothetical protein |
| Hs.1162 | HLA-DMB | Major histocompatibility complex, class II, DM β |
| Hs.38044 | DKFZP564M082 | DKFZP564M082 protein |
| Hs.99526 | OBP2B | Odorant-binding protein 2B |
| Hs.15159 | HSPC224 | Transmembrane proteolipid |
| Hs.69285 | NRP1 | Neuropilin 1 |
| Hs.10028 | CG1I | Putative cyclin G1 interacting protein |
| Hs.198272 | NDUFB2 | NADH dehydrogenase (ubq) 1 β subcomplex, 2(8 kDa, AGGG) |
| Hs.75486 | HSF4 | Heat shock transcription factor 4 |
A random sample of 50 UNIGENE clusters containing at least one full-length mRNA was generated. The UNIGENE cluster ID, gene symbol and title are described.
RESULTS
Detection of alternative splicing
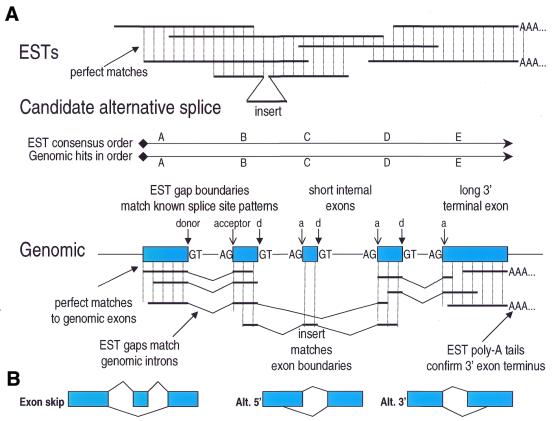
Our analysis of alternative splicing is based strictly on experimental data, not theoretical models. Rather than seeking to predict alternative splices, we directly detect them as large inserts in EST data from the publicly available dbEST (20) and UNIGENE (18) databases. We measure the evidence for a genuine alternative splice via a series of criteria (Fig. 1). First, a set of ESTs must match over their full lengths, on both sides of a putative alternative splice (allowing for sequence error). A large insert in the middle of such a perfect match is a candidate alternative splice. Unlike many other types of genomics results such as SNPs and variations in expression level, alternative splicing does not resemble common experimental noise (such as sequencing error).
Figure 1.
Detection and validation of alternative splicing. (A) Types of evidence for alternative splicing (see text). (B) Types of alternative splicing detected in this study include exon skipping, alternative 5′ splice donor sites and alternative 3′ splice acceptor sites.
Next, the EST consensus sequence is mapped to the draft human genome sequence by homology search. Because human genes are broken into short exons, a genomic hit typically consists of many short matches. To be valid, these matches must be perfect (again allowing only for sequencing error), must all be in the same orientation (strand) and form a complete, correctly ordered walk through the EST consensus sequence. We require that each genomic–EST match region (putative exon) be bounded by consensus splice donor site and acceptor site sequences in the neighboring genomic (intron) sequence. Our results give an average internal exon size of 144 bp, with only 4% of internal exons >300 bp in length, similar to results obtained for known genes (21). Only 0.2% (79/39 862) of our introns were <60 bp, and the median intron length was 935 bp. The typical gene pattern of short internal exons ending in a single, long 3′ exon can usually be verified because 3′-end sequences are highly represented in the EST data, and because 3′ ESTs can be identified by their conspicuous poly(A) tails, which directly indicate the end of the 3′ exon.
To assess the accuracy of our gene mapping and exon/intron structure, we have compared against the completely independent data produced by NCBI’s Acembly, a human curated gene annotation effort (data downloaded from ftp://ncbi.nlm.nih.gov/genomes/H_sapiens). LocusLink provides an independent linkage between individual RefSeq genes and UNIGENE clusters (22). For genes mapped independently to the genomic sequence by RefSeq and our procedure, 97.3% mapped to the same genomic contig. Moreover, of those genes, 95% were mapped to the same nucleotides of the contig. While Acembly’s mapping should not be assumed to be perfect, this high level of agreement between independent efforts is encouraging. Our exon details (derived in our procedure from our splice detection) match the NCBI Acembly exons in 97% of cases at the 5′ splice site, and 96% at the 3′ splice site (overall, 94% of the exons were identical). Our splice details matched the NCBI Acembly introns in 93% of cases at the 5′-end, and 92% at the 3′-end (86% matched exactly at both ends). Because of alternative splicing, a 100% correspondence is not expected.
A candidate alternative splice insert (from the EST) must pass a series of tests. First, it must also be found in the genomic sequence, matching an exonic region in the genomic sequence whose boundaries correspond to known splice site sequences. Since these splice site sequences are mostly intronic, this provides an independent validation of the alternative splice. It should be emphasized that differences in where ESTs begin and end in a gene (e.g. a shorter EST might give the appearance of a truncated gene product) will never be interpreted as an alternative splice by our procedure. We focus exclusively on detecting splicing, i.e. a contiguous region of the transcript that has been removed during mRNA processing. Detecting a splice in an EST requires extensive matches to both upstream and downstream exons. Our analysis identified 39 862 splices in 8429 clusters. Our analysis only reports alternative splices, i.e. pairs of validated splices that are mutually exclusive. Thus unspliced introns or other genomic contaminants will never be reported, since they result in the absence of a splice, not the creation of a new, mutually exclusive splice. To call an alternative splice, our procedure requires a pair of splices that match exactly at one splice site, and differ at the second splice site. This procedure can detect exon skipping, alternative 5′ donor sites, and alternative 3′ acceptor sites (Fig. 1B). 6201 such alternative splice relationships were identified in 2272 clusters. These diverse forms of evidence produce strong log odds scores for each detected alternative splice. A detailed statistical analysis of this evidence will be presented elsewhere (D.Miller, J.Aten, C.Grasso, B.Modrek and C.Lee, manuscript in preparation).
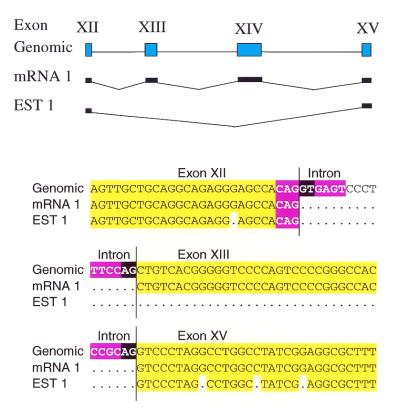
As a typical validation example from our database, we illustrate the dystrophia myotonica protein kinase (DMPK) gene (Fig. 2), whose alternative splicing has previously been studied extensively. In DMPK, we identified three alternative splices in the EST data, all of which are verified by independent experimental results in the existing literature (23). Of the three alternative splices, one deletes the last 15 bp of exon 8, another skips exon 12 and exon 13, and the last deletes just 4 bp in exon 14. Figure 2 shows one of these alternative splice forms including junction and quality of match of the EST evidence versus the genomic sequence.
Figure 2.
Alternative splicing of DMPK. (A) Gene structure for exons XII–XV of the dystrophia myotonica protein kinase gene (DMPK), in contig NT000991 of chromosome 19. Two splice forms are shown, one observed in an mRNA (mRNA1) and one in an EST (EST1). (B) Example sequence evidence for the two splice forms. Sequence EST1 skips directly from exon XII to exon XV. We detected three alternative splice forms in DMPK; all are confirmed by the experimental literature (23).
Novel alternative splice forms of a known gene
Figure 3 shows several novel alternative splices detected in a well-studied gene, HLA-DM β. Eighty ESTs from UNIGENE cluster Hs.1162 align to form a consensus sequence, which in turn matches an ordered series of segments on one strand of chromosome 6. The EST sequences match the genomic sequence closely, consistent with sequencing error. The EST sequences mark out a long 3′ exon (359 bp) plus a series of five short exons, whose sizes (36–288 bp) match the range expected for internal exons. This matches the known gene location and structure for HLA-DM β (24,25). Eight splices are observed in these ESTs, where sequence matching one exonic region skips directly to a downstream exonic region as indicated in Figure 3A. The 16 putative exon boundaries implied by the ESTs map precisely to strong consensus splice acceptor and donor sites in the genomic sequence (Fig. 3C).
Figure 3.
Alternative splicing of HLA-DMB. (A) Genomic structure of the HLA-DM β gene, in contig NT001520 of chromosome 6. Exons are shown as filled boxes, and the observed splices are shown on top of the genomic sequence. (B) The four alternative forms of HLA-DM β mRNA inferred from the expressed sequence data, colored to show the exons. The protein reading frame is indicated by an arrow beneath each form, showing the transmembrane domain (TM) and lysosomal targeting signal (LT). (C) The splice donor and acceptor sites for the eight putative splices observed in HLA-DM β. The primary consensus site sequences are highlighted in black and secondary consensus sequences (5) are marked in magenta.
Four different alternatively spliced forms of HLA-DM β are observed: splices 3+4+5 (including exons IV and V in the mRNA product); splices 6+5 (skipping exon IV); splices 3+7 (excluding exon V); splice 8 (skipping exons IV and V). Exons IV and V are 117 and 36 bp in length, and thus these alternative splices are all in-frame. The protein coding region begins in exon I and ends in exon VI, so these splices produce four different forms of the HLA-DM β chain that differ at their C-terminus.
Analysis of these forms reveals a remarkably simple and intriguing functional effect. HLA-DM is essential for the loading of class II MHC molecules with exogenous peptide antigens, a key step in antigen presentation and activation of the humoral immune response. This is thought to occur in early lysosomal compartments. HLA-DM is normally targeted to lysosomes, and its β chain contains a transmembrane domain anchoring its C-terminus (26,27). Exon IV is short, and corresponds precisely to the transmembrane domain. Exon V is very short, and encodes the lysosomal targeting signal YTPL, whose first residue begins at the start of the exon. Thus, the alternative splice regulates HLA-DM’s targeting to endosomal compartments (by including or excluding the YTPL signal), as well as its anchoring to the membrane. Given HLA-DM’s importance in antigen processing and presentation by class II MHC, this regulation is functionally interesting. Removing its targeting signal would likely redirect HLA-DM first to the plasma membrane, so that it would travel to lysosomes via endocytic pathways, altering the kinetics and conditions in which it first encounters class II MHC. It appears that the gene structure of the HLA-DM β gene has been carefully ‘designed’ to enable control of HLA-DM function, by pulling out both the transmembrane helix and the lysosomal targeting signal into separate short exons (IV, V) that can be alternatively spliced in-frame (exon VI supplies the last 4 amino acids of the protein, identical in all forms). The alternatively spliced forms were detected in uterus (two ESTs), placenta, lymph, stomach and colon. Despite the fact that HLA-DM is the subject of intense research, we have not been able to find any report of such alternative splicing in the published literature, and it is thought to be novel by an expert on HLA-DM (E.Mellins, personal communication).
Scope of alternative splicing in human genes
Our genome-wide analysis detected thousands of alternative splices in the current, publicly available human genome data (Table 1). 6201 alternative splice relationships were detected in which two splices shared a common donor or acceptor site, but spliced to a different site on their other end (i.e. exon skipping, alternative 5′ splice donor site or alternative 3′ splice acceptor site; Fig. 1B). We found alternative splices in 27% of genes for which we had enough expressed sequence to cover more than a single exon. However, this estimate, based on analysis of all EST clusters, likely underestimates the real occurrence of alternative splicing, because the available EST data typically cover only a small part of the complete gene. To test this hypothesis, we analyzed the alternative splicing rate in genes for which mRNA sequence was available (representing all or part of the full gene). We detected one or more alternative splice forms in 42% of these genes, significantly higher than the rate observed in EST-only clusters. This is in close agreement with a previous study of mRNA-based expressed sequence clusters (8). Since fragmentation of the genomic sequence can also block complete coverage of a gene, we assessed the rate of alternative splice detection in genes mapped to chromosome 22. Of these, 43% contained alternative splices, including both mRNA and EST-only clusters.
The current EST data appears to be incomplete. Our procedure identified splices (i.e. multiple exons) in only 18% of the mapped EST clusters. However, for clusters that we mapped to chromosome 22 (full genomic) that also had an mRNA sequence, 88% contained at least one splice. A variety of factors such as the fragmentation of the draft human genome sequence, the large size of introns and the tendency of the ESTs to cluster at the 3′-end bias the current dataset against finding full-length genes, and probably underestimate the true level of alternative splicing. Moreover, since the current EST data for each gene represent only a subset of the tissues and cell types in which that gene is expressed, it is likely that the total occurrence of alternative splicing is much greater than what our analyses can detect. A large fraction of the EST alternative splice forms were observed multiple times (from different clones and different libraries), indicating that they constitute a relatively high fraction of total mRNA. Of our alternative splices, 2892 (47%) were observed in two or more EST sequences. These data represent a ‘high confidence’ subset of the detected alternative splices.
Our analysis indicates that the vast majority of our database represents novel findings (Fig. 5D). Only 13% of our alternative splices were detected in mRNA sequences from GenBank, which presumably have been thoroughly studied. The remaining 87% could be detected only with ESTs. Our procedure also detected large numbers of alternative splicing events in completely novel genes. Approximately 1200 alternative splices were detected in clusters containing ESTs only.
Alternative splicing in a novel human gene
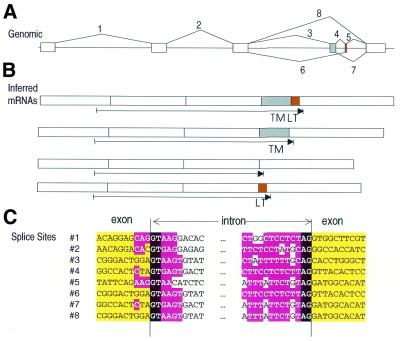
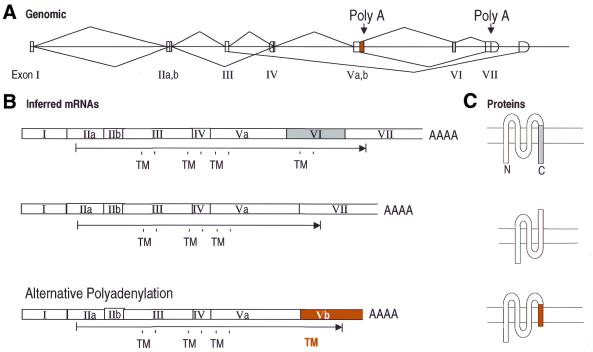
Figure 4 illustrates an example of alternative splice detection in a novel gene mapped in the human genome by our procedure. This gene has 33% identity to rat FCɛ receptor I β chain, and 25% identity to CD20, and has a pattern of four predicted transmembrane domains characteristic of both proteins. At least seven different forms are detectable, all of which affect the protein product. In a pattern strikingly reminiscent of HLA-DM β, the C-terminal transmembrane region and cytoplasmic tail of the major form (form 1) are placed on a single, short exon (exon VI), that can be included or excluded to create different forms. One particularly interesting form is created by ignoring the normal splice from exon V to exon VI, extending the coding region from exon Va for 142 bp (which we have designated exon Vb). A polyadenylation site is predicted at the end of this sequence, and the ESTs are observed to terminate in poly(A) at this point. This alternative termination replaces the coding region of exons VI and VII with 40 amino acids encoded by exon Vb [terminated by a STOP codon 23 bp before the poly(A) site]. Intriguingly, this replacement C-terminal sequence also contains a predicted transmembrane sequence, and thus neatly substitutes a new C-terminal transmembrane domain and cytoplasmic tail. The cytoplasmic tail in equivalent FC receptor chains plays a key role in activating cytoplasmic signal transduction molecules (28,29), so this alternative form likely modulates the signal transduction activity of this receptor. This form is detected in placenta and kidney, while the majority form was detected in many different libraries.
Figure 4.
Alternative splicing of Hs.11090, a putative FCɛ receptor β chain homolog. (A) Genomic structure of exons and splices, as in Figure 3. Potential polyadenylation sites important for the alternative gene forms are indicated. (B) Three alternative forms inferred from the expressed sequences. Predicted transmembrane domains (TM) are indicated (see text). (C) The corresponding protein forms, indicating topology across the membrane.
DISCUSSION
Our results provide a comprehensive dataset for understanding the role of alternative splicing in the human genome. First of all, what is the function of alternative splicing—modification of the protein product, or of the untranslated regions that could affect mRNA localization and stability? Analysis of a random sample from our database (Table 2) indicates that 74% of alternative splices modified the protein product, whereas 22% were confined to the 5′ UTR versus just 4% in the 3′ UTR (Fig. 5A). This may simply reflect the larger fraction of exons in human genes that are protein-coding as opposed to UTR. This result fits expectations from molecular biology studies (1), but disagrees strongly with a bioinformatics analysis of a small set of ESTs (9), which reported 80% of genes with alternative splicing had an alternative splice in 5′ UTR versus only 20% in coding regions. Our observation of little alternative splicing in 3′ UTR is striking in view of the strong bias in the EST data towards the 3′ exon. One possible explanation is that mRNA species with alternatively spliced 3′ UTR sequence could contain sequences that destabilize the mRNA, resulting in fewer observations of these forms. In contrast, the effect on the protein product is seen much more frequently at the C-terminal end (3′ in the mRNA) (Fig. 5B). We observed a tendency to replace the C-terminus (46%), as opposed to making an internal deletion, insertion or substitution (37%), or a replacement of the N-terminus (17%). In this respect, the examples we have shown (HLA-DM β and FCɛ receptor I β homolog) are representative. Alternative splicing appears to be strongly biased to preserve the protein coding frame (Fig. 5C). Only 19% of alternative splices resulted in a truncation of the protein product due to frame-shift; occasionally alternative splicing was observed to add a new, extended C-terminal sequence through frame-shift (6%). Alternative splicing resulted in a switch to a new AUG start site on an alternative exon in 15% of cases. In contrast, replacing the C-terminus by switching to a different exon containing an alternative STOP codon occurred in 20% of cases. In-frame deletion or insertion of a new sequence in the middle of the protein accounted for 29 and 11% of cases, respectively.
In what types of molecules is alternative splicing commonly observed? Figure 5G shows a molecular classification of a random sample of alternatively spliced genes. The most abundant category is cell surface functions/receptors (29%), which includes membrane-anchored receptors (e.g. CD79B), integral membrane proteins (e.g. folate transporter SLC19A1) and proteins involved in cell surface adhesion (e.g. lectin, hyaluronoglucosaminidase 2). In two related categories, an additional 14% of alternatively spliced genes encode secreted proteins (e.g. Norrie disease protein; group-specific component) and 9% encode signal transduction molecules (e.g. phospholipase D2; RIT). The next two major categories are transcriptional regulation (14%; e.g. MYB, PAX6) and apoptosis (11%; e.g. BID, PNAS-1). Together, these functions of transmission, reception and response to cellular signals comprise >75% of the observed alternatively spliced genes. Proteins involved in metabolism (e.g. aldolase C), and organelle-specific sorting proteins were also observed. This sample is by no means comprehensive or exact, but indicates a trend towards cell surface interactions and signaling.
What types of systemic functions are most often affected by alternative splicing? Twenty-nine percent of the alternatively spliced genes encoded functions specific to the immune system (Fig. 5F; e.g. T-cell specific transcription factor 7, TNF receptor superfamily member 6). In particular, alternative splicing of immune system cell surface receptors was very prevalent. Neuronal functions (e.g. neuropilin, brain-specific aldolase C) comprised 12% of the total. The remaining genes possessed no clearly specific systemic function. These data suggest alternative splicing may play a large role in immune system and nervous system functions which require precise control of cellular differentiation and activation, to process large amounts of information. Controlling how each cell responds to a diverse array of signals can be achieved through alternative splicing of its receptors and signal transduction molecules.
How often is alternative splicing clearly associated with a specific tissue? Based on a sample of 50 genes, ∼14% of alternatively spliced genes in our dataset showed evidence of tissue specificity for the minor isoform. This estimate is based on a conservative definition requiring that the minor isoform be observed multiple times in a specific tissue in which the major form was not observed. Since in many known cases of tissue-specific alternative splicing both minor and major forms are observed in the same tissue, this probably misses many cases of real tissue-specificity. Examples include DDR1, discoidin domain receptor, which has a minor form observed in muscle; and CG1I, a putative cyclin G1 interacting protein, which has isoforms observed specifically in ovary and brain. Within the small sample, tissue-specific minor isoforms were observed in novel, uncharacterized genes in brain, colon, testis and prostate.
How comprehensive is our dataset, and what are its prospects for growth? We have noted two causes of failure by our procedure to detect alternatively spliced forms that are known in the literature. First, a given gene may not map yet to the draft genome, a prerequisite in our procedure for analyzing its splicing. Secondly, some alternatively spliced mRNA forms are miscategorized as genomic DNA in GenBank, causing them to be excluded by our procedure. The former seems to be the most important cause of failure. Despite >90% completeness by total nucleotides sequenced, the draft genome used in this study (October 2000) only enabled mapping of 55% of UNIGENE expressed sequence clusters, because we require a full-length match versus the expressed gene sequence consensus (Table 1). The draft (i.e. incomplete) BAC clone sequences which constituted the majority of this dataset, consisted in large part of short sequence fragments (4–10 kb) separated by unsequenced regions. Such fragments are too small to map a typical human gene (10–30 kb) by our conservative procedure. This trend is even stronger for the subset of genes that have full-length mRNAs. Of these clusters, only 41% could be mapped over their full length to an available genomic contig. To check whether this is due to the draft genome’s fragmentation, we analyzed a subset of gene clusters that have been mapped by STS to chromosome 22, which has been almost completely sequenced. For these clusters, 77% could be mapped. Thus, given unbroken genomic sequence, our mapping procedure has a false negative rate of ∼20%. These data suggest that completion of the human genome sequence, along with improvements in our algorithms, will at least double the number of alternative splices detected. Our detection of alternative splicing should also grow with increasing EST data. In our current EST dataset (December 2000), splices were detected in only 18% of clusters, reflecting the fact that the average cluster consists of too few ESTs (one or two) and is too short (a few hundred base pairs) to cover more than a single exon. This is exaggerated by the strong bias of the ESTs to be from the 3′-end, since 3′ exons tend to be much longer than typical internal exons. In contrast, in genes for which a full-length or partial mRNA sequence was available and which were mapped to a region of full-length genomic sequence (e.g. chromosome 22), 88% contained at least one splice (and typically many more).
Acknowledgments
ACKNOWLEDGEMENTS
We wish to thank D. Black, D. Miller, S. Galbraith and D. Eisenberg for their helpful discussions and comments on this work, and K. Ke for assistance in constructing the HASDB web site. This work was supported by Department of Energy grant DEFG0387ER60615 and a grant from the Searle Scholars Program to C.L. B.M. is a predoctoral trainee supported by NSF IGERT Award #DGE-9987641.
References
- 1.Lopez A.J. (1998) Alternative splicing of pre-mRNA: developmental consequences and mechanisms of regulation. Annu. Rev. Genet., 32, 279–305. [DOI] [PubMed] [Google Scholar]
- 2.Boise L.H., Gonzalez-Garcia,M., Postema,C.E., Ding,L., Lindsten,T., Turka,L.A., Mao,X., Nunez,G. and Thompson,C.B. (1993) bcl-x, a bcl-2-related gene that functions as a dominant regulator of apoptotic cell death. Cell, 74, 597–608. [DOI] [PubMed] [Google Scholar]
- 3.Fettiplace R. and Fuchs,P.A. (1999) Mechanisms of hair cell tuning. Annu. Rev. Physiol., 61, 809–834. [DOI] [PubMed] [Google Scholar]
- 4.Schmucker D., Clemens,J.C., Shu,H., Worby,C.A., Xiao,J., Muda,M., Dixon,J.E. and Zipursky,S.L. (2000) Drosophila Dscam is an axon guidance receptor exhibiting extraordinary molecular diversity. Cell, 101, 671–684. [DOI] [PubMed] [Google Scholar]
- 5.Smith C.W.J. and Valcarcel,J. (2000) Alternative pre-mRNA splicing: the logic of combinatorial control. Trends Biochem. Sci., 25, 381–388. [DOI] [PubMed] [Google Scholar]
- 6.Sharp P.A. (1994) Split genes and RNA splicing. Cell, 77, 805–815. [DOI] [PubMed] [Google Scholar]
- 7.Sutcliffe J.G. and Milner,R.J. (1988) Alternative mRNA splicing: the Shaker gene. Trends Genet., 4, 297–299. [DOI] [PubMed] [Google Scholar]
- 8.Brett D., Hanke,J., Lehmann,G., Haase,S., Delbruck,S., Krueger,S., Reich,J. and Bork,P. (2000) EST comparison indicates 38% of human mRNAs contain possible alternative splice forms. FEBS Lett., 474, 83–86. [DOI] [PubMed] [Google Scholar]
- 9.Mironov A.A., Fickett,J.W. and Gelfand,M.S. (1999) Frequent alternative splicing of human genes. Genome Res., 9, 1288–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liang F., Holt,I., Pertea,G., Karamycheva,S., Salzberg,S.L. and Quackenbush,J. (2000) Gene Index analysis of the human genome estimates approximately 120,000 genes. Nat. Genet., 25, 239–240. [DOI] [PubMed] [Google Scholar]
- 11.Ewing B. and Green,P. (2000) Analysis of expressed sequence tags indicates 35,000 human genes. Nat. Genet., 25, 232–234. [DOI] [PubMed] [Google Scholar]
- 12.Ji H., Zhou,Q., Wen,F., Xia,H., Lu,X. and Li,Y. (2001) AsMamDB: an alternative splice database of mammals. Nucleic Acids Res., 29, 260–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Irizarry K., Kustanovich,V., Li,C., Brown,N., Nelson,S., Wong,W. and Lee,C. (2000) Genome-wide analysis of single-nucleotide polymorphisms in human expressed sequences. Nat. Genet., 26, 233–236. [DOI] [PubMed] [Google Scholar]
- 14.Jang W., Chen,W.C., Sicotte,H. and Schuler,G.D. (1999) Making effective use of human genomic sequence data. Trends Genet., 15, 284–286. [DOI] [PubMed] [Google Scholar]
- 15.Altschul S.F., Gish,W., Miller,W., Myers,E.W. and Lipman,D.J. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 16.Needleman S.B. and Wunsch,C.D. (1970) A general method applicable to the search for similarities in the amino acid sequence of two proteins. J. Mol. Biol., 48, 443–453. [DOI] [PubMed] [Google Scholar]
- 17.Smith T.F. and Waterman,M.S. (1981) Identification of common molecular subsequences. J. Mol. Biol., 147, 195–197. [DOI] [PubMed] [Google Scholar]
- 18.Schuler G. (1997) Pieces of the puzzle: expressed sequence tags and the catalog of human genes. J. Mol. Med., 75, 694–698. [DOI] [PubMed] [Google Scholar]
- 19.Lee C. and Irizarry,K. (2001) The GeneMine system for genome/proteome annotation and collaborative data-mining. IBM Syst. J., 40, in press. [Google Scholar]
- 20.Boguski M.S., Lowe,T.M. and Tolstoshev,C.M. (1993) dbEST—database for ‘expressed sequence tags’. Nat. Genet., 4, 332–333. [DOI] [PubMed] [Google Scholar]
- 21.Hawkins J.D. (1988) A survey on intron and exon lengths. Nucleic Acids Res., 16, 9893–9905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pruitt K.D. and Maglott,D.R. (2001) RefSeq and LocusLink: NCBI gene-centered resources. Nucleic Acids Res., 29, 137–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Groenen P.J., Wansink,D.G., Coerwinkel,M., van den Broek,W., Jansen,G. and Wieringa,B. (2000) Constitutive and regulated modes of splicing produce six major myotonic dystrophy protein kinase (DMPK) isoforms with distinct properties. Hum. Mol. Genet., 9, 605–616. [DOI] [PubMed] [Google Scholar]
- 24.Kelly A.P., Monaco,J.J., Cho,S.G. and Trowsdale,J. (1991) A new human HLA class II-related locus, DM. Nature, 353, 571–573. [DOI] [PubMed] [Google Scholar]
- 25.Shaman J., von Scheven,E., Morris,P., Chang,M.Y. and Mellins,E. (1995) Analysis of HLA-DMB mutants and -DMB genomic structure. Immunogenetics, 41, 117–124. [DOI] [PubMed] [Google Scholar]
- 26.Sanderson F., Kleijmeer,M.J., Kelly,A.P., Verwoerd,D., Tulp,A., Neefjes,J., Geueze,H.J. and Trowsdale,J. (1994) Accumulation of HLA-DM, a regulator of antigen presentation, in MHC class II compartments. Science, 266, 1566–1569. [DOI] [PubMed] [Google Scholar]
- 27.Potter P.K., Copier,J., Sacks,S.H., Calafat,J., Janssen,H., Neefjes,J. and Kelly,A.P. (1999) Accurate intracellular localization of HLA-DM requires correct spacing of a cytoplasmic YTPL targeting motif relative to the transmembrane domain. Eur. J. Immunol., 29, 3936–3944. [DOI] [PubMed] [Google Scholar]
- 28.Daeron M. (1997) Fc receptor biology. Annu. Rev. Immunol., 15, 203–234. [DOI] [PubMed] [Google Scholar]
- 29.Kinet J.P. (1999) The high affinity IgE receptor (FCɛRI): from physiology to pathology. Annu. Rev. Immunol., 17, 931–972. [DOI] [PubMed] [Google Scholar]