Abstract
(1) Runt-related transcription factor 1 (RUNX1) mutations in acute myeloid leukemia (AML) are often associated with worse prognosis. We assessed co-occurring mutations, response to therapy, and clinical outcomes in patients with and without mutant RUNX1 (mRUNX1); (2) We analyzed 328 AML patients, including 177 patients younger than 65 years who received intensive chemotherapy and 151 patients >65 years who received hypomethylating agents. RUNX1 and co-existing mutations were identified using next-generation sequencing; (3) RUNX1 mutations were identified in 5.1% of younger patients and 15.9% of older patients, and were significantly associated with increasing age (p = 0.01) as well as intermediate-risk cytogenetics including normal karyotype (p = 0.02) in the elderly cohort, and with lower lactate dehydrogenase (LDH; p = 0.02) and higher platelet count (p = 0.012) overall. Identified co-occurring mutations were primarily ASXL1 mutations in older patients and RAS mutations in younger patients; FLT3-ITD and IDH1/2 co-mutations were also frequent. Younger mRUNX1 AML patients treated with intensive chemotherapy experienced inferior treatment outcomes. In older patients with AML treated with hypomethylating agent (HMA) therapy, response and survival was independent of RUNX1 status. Older mRUNX1 patients with prior myelodysplastic syndrome or myeloproliferative neoplasms (MDS/MPN) had particularly dismal outcome. Future studies should focus on the prognostic implications of RUNX1 mutations relative to other co-occurring mutations, and the potential role of hypomethylating agents for this molecularly-defined group.
Keywords: RUNX1, mutations, acute myeloid leukemia, hypomethylating agents, chemotherapy, prognosis
1. Introduction
The genetic diversity of acute myeloid leukemia (AML) and the complexity of its multi-step pathogenesis now allow the classification of AML based on molecular events. The evolution of AML has traditionally been proposed to follow the “two-hit model” [1], in which two classes of mutations are required for cancer development. Class I mutations are activating mutations that stimulate cell survival and proliferation, while class II mutations are inactivating mutations that interfere with hematopoietic differentiation [2]. Runt-related transcription factor 1 (RUNX1) is a key hematopoietic transcription factor that regulates genes involved in myeloid differentiation, and is generally considered to be a classical tumor suppressor (class II) mutation [2]. Mutations of RUNX1 are reported in approximately 10–16% of AML patients [3,4] and 12–24% of myelodysplastic syndrome (MDS) patients [5,6]. RUNX1 alterations predominate in the morphologically undifferentiated French-American-British (FAB) M0 subtype. Clinical features associated with this mutation include older age, male sex, and absence of cytogenetic abnormalities [4].
In patients with AML, mutant RUNX1 often associates with certain class I mutations. Gene expression profiling has identified the co-occurrence of RUNX1 with mutations of the chromatin remodeling gene ASXL1 or partial tandem duplication of the transcription regulator MLL in all major cohorts to date, while RUNX1 and NPM1 (nucleophosmin) mutations are consistently negatively correlated [2,7]. ASXL1 mutations appear to be the most frequent co-mutation with RUNX1, and ASXL1/RUNX1 double mutants are associated with lower rates of therapeutic response [8,9]. Other significant associations, such as with alterations in the tyrosine kinase FLT3, splicing factor mutations, or isocitrate dehydrogenase (IDH1 or IDH2) has not been firmly established [3,4,7,10]; however, concomitant FLT3 mutations are thought to play a synergistic role with RUNX1 mutations in the development of AML [11]. Upregulation of genes normally expressed in hematopoietic progenitor cells or lymphoid cells, and downregulation of promoters of myelopoeisis also ascribe a unique gene expression signature to RUNX1-mutated AML [3,12], implicating upregulation of oncogenic pathways such as BCR, TLR-4 and NOTCH1 to the pathogenicity of mutant RUNX1.
Although standard intensive chemotherapy has yielded high remission rates and superior long-term survival in younger AML patients [13], elderly patients continue to experience lower response rates and poor long-term outcomes [14], and hypomethylating agents (HMAs) are typically utilized in the older patient population [14]. Clinical outcomes with respect to the influence of RUNX1 mutations based on age and treatment regimen have not been previously explored [4].
RUNX1 mutations are correlated with poor clinical outcomes. Gaidzik et al. [10] compared 53 mutant RUNX1 and 831 wild-type RUNX1 newly-diagnosed AML patients, and found inferior rates of event-free survival (EFS), relapse-free survival (RFS), and overall survival (OS) in RUNX1-mutated patients in patients 60 years of age or younger and treated with intensive chemotherapy (EFS, 8% vs. 30%; RFS, 26% vs. 44%; OS, 32% vs. 45%). In an analysis of AML patients treated with intensive chemotherapy by Tang et al. [7], the complete remission rate was lower in 62 newly-diagnosed patients with RUNX1-mutated AML compared with 408 without the mutation (56.8% vs. 77.5%). A statistically higher incidence of induction-related death in patients with mutant-RUNX1 AML (10.8% vs. 6.5%) was also identified. Other studies have also correlated RUNX1 mutations with resistance to chemotherapy and higher rates of refractory disease [2,7,10]. It is important to expand our understanding of the clinical outcomes of mutant and wild-type RUNX1 in relation to specific treatment modalities, with attention to the older patient population treated with hypomethylating or lower-intensity approaches where RUNX1 mutations are more often identified.
This study examined the frequency of RUNX1 mutations in newly diagnosed patients with AML, and their effect on clinical outcomes and treatment response rates, including younger patients receiving chemotherapy and elderly patients receiving HMAs. Co-existing mutations and their effect on the clinical course of AML in conjunction with RUNX1 were also examined.
2. Results
2.1. Frequency and Characteristics of RUNX1 Mutations
Overall, mutant RUNX1 was identified in 33 (10.1%) patients with newly diagnosed AML. These rates were significantly higher in patients 65 years of age or older, 24 (15.9%), compared with the younger adult cohort, 9 (5.1%). None of the patients were identified as having more than one pathogenic RUNX1 mutation at diagnosis. The majority (>90%) of RUNX1 mutations described were either frameshift or nonsense in nature, resulting in early amino acid chain termination and truncated proteins. The specific characteristics of these mutations, including their allelic frequencies, types, and exon locations, have been elaborated in Appendix A Table A1.
2.2. Association of RUNX1 with Clinical Characteristics
Table 1 summarizes the clinicopathologic variables evaluated with respect to the impact of RUNX1 mutational status. In the elderly cohort, a significant correlation was observed between the presence of mutated RUNX1 and increasing age (p = 0.01); mRUNX1 was rare in the younger patient cohort (5%). Along with an increasing age, RUNX1 mutations occurred more frequently in older patients with intermediate-risk cytogenetics (particularly those with a normal karyotype), as compared to those with complex cytogenetic abnormalities (p = 0.02). Lower lactate dehydrogenase (LDH) levels and higher platelet counts were also found to be significant in RUNX1-mutated patients within the entire cohort overall (p = 0.02 and p = 0.012 respectively). RUNX1 mutational status did not significantly correlate with WBC count, hemoglobin, or peripheral blood and bone marrow blasts, or history of prior myelodysplastic syndrome or myeloproliferative neoplasms (MDS/MPN) or therapy-related AML (t-AML). Among the mutant RUNX1 patients, 15.2% (n = 20) of the patients classified as FAB M0 subtype.
Table 1.
Clinical characteristics in the younger cohort receiving chemotherapy and the elderly cohort receiving hypomethylating agents according to RUNX1 mutation status.
| Clinical Characteristics | Younger Cohort (Chemotherapy) | p-Value | Elderly Cohort (HMA) | p-Value | ||
|---|---|---|---|---|---|---|
| Median (Range) | Median (Range) | |||||
| RUNX1mut | RUNX1WT | RUNX1mut | RUNX1WT | |||
| (n = 9) | (n = 168) | (n = 24) | (n = 127) | |||
| Age, year | 56 (31–63) | 51 (17–64) | 0.14 | 77 (65–92) | 73 (65–91) | 0.01 * |
| WBC, ×109/L | 4.95 (1.3–17.2) | 6.9 (0.5–378.4) | 0.43 | 3.2 (0.6–36.5) | 3.2 (0.3–164.5) | 0.98 |
| Platelet count, ×109/L | 57.5 (28–213) | 30 (1–584) | 0.05 | 61 (7–416) | 42 (3–324) | 0.20 |
| Hemoglobin, g/dL | 8.95 (7.4–12.1) | 9.3 (5.1–13.0) | 0.38 | 9.4 (5.8–13.1) | 9.4 (7.5–13.2) | 0.37 |
| Peripheral blood blasts, % | 28 (1–86) | 25.5 (0–97) | 0.61 | 7 (0–90) | 11 (0–95) | 0.51 |
| Neutrophils, % | 14.5 (0–45) | 12 (0–98) | 0.93 | 21 (0–81) | 21 (0–73.3) | 0.95 |
| Bone marrow blasts, % | 51 (28–93) | 54 (2–96) | 0.66 | 50 (12–84) | 44 (1–90) | 0.80 |
| Cytogenetic | 0.94 | 0.02 | ||||
| Complex | 2 | 37 | 2 | 47 | ||
| Diploid | 3 | 65 | 14 | 50 | ||
| ^ Intermediate | 4 | 66 | 8 | 30 | ||
| LDH, U/L | 668.5 | 888 | 0.10 | 532 | 650 | 0.44 |
| (526–1649) | (310–12,489) | (276–2314) | (210–3921) | |||
| t-AML | 0 | 10 | 0.45 | 4 | 18 | 0.75 |
| Prior MDS/MPD | 1 | 6 | 0.26 | 8 | 25 | 0.14 |
Abbreviations: LDH, lactate dehydrogenase; HMA, hypomethylating agent; Prior MDS/MPD: Prior myelodysplastic syndrome or myeloproliferative neoplasms; t-AML, therapy-related acute myeloid leukemia; WBC, white blood cell; * Boldface indicates statistical significance. ^ Intermediate also includes insufficient metaphases and not done.
2.3. Association of RUNX1 with Co-Occurring Mutations
In the older cohort, mutational analysis identified ASXL1 mutations as the most frequent aberration in the presence of mutant RUNX1, co-occurring at a frequency of 37.5%—significantly higher compared to ASXL1 occurrence in wild-type RUNX1 (14.2%) (p = 0.007). In comparison, the incidence of co-occurring RAS mutation was notable within the younger mRUNX1 cohort, occurring in 4 of 9 patients (44.4%) (p = 0.184). The other most frequent co-occurring mutations in the younger cohort were those in ASXL1, FLT3-ITD, and DNMT3A genes at 33% each; compared to wild-type RUNX1 (ASXL1, 2.4% p < 0.005; FLT3-ITD, 20.8% p = 0.379; DNMT3A, 20.2% p = 0.384). IDH1 and IDH2 mutations were also frequently identified in association with mutant RUNX1, with a frequency of mutant IDH1 as 11.1% in mRUNX1 compared to 4.8% wild-type (p = 0.407) in the younger cohort and 16.7% in mRUNX1 compared to 12.6% in wild-type RUNX1 in the elderly cohort (p = 0.603). IDH2 mutations occurred at 22.2% (mRUNX1) and 15.5% (wild-type) (p = 0.617) and 20.8% (mRUNX1) compared to 16.5% (wild-type) (p = 0.624) in the younger and the elderly cohorts, respectively.
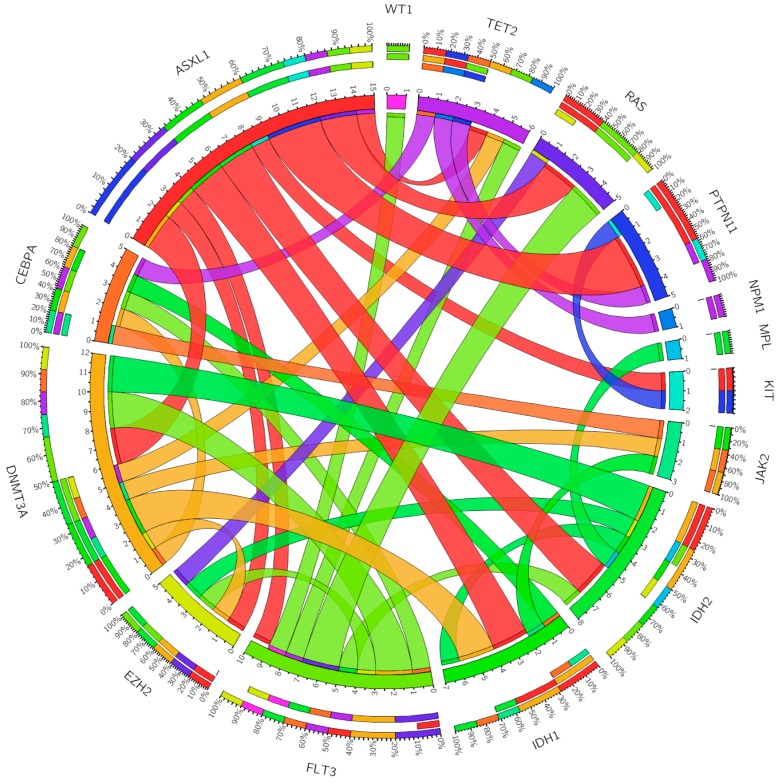
A number of other gene mutations were analyzed and showed varying association with mutated RUNX1 (Figure 1) [15]. The frequencies of these co-mutations with mutant RUNX1 in the younger and the elderly cohorts were, respectively, as follows: FLT3-ITD, 33.3% and 16.7%; NPM1, 0% and 4.2%; RAS, 44.4% and 8.3%; IDH1, 11.1% and 16.7%; IDH2, 22.2% and 20.8%; TET2, 0% and 12.5%; PTPN11, 11.1% and 8.3%; JAK2, 0% and 8.3%; CEBPA, 0% and 12.5%; DNMT3A, 33.3% and 16.7%; EZH2, 22.2% and 0%; KIT, 11.1% and 0%; MPL, 0% and 4.2%; WT1, 0% and 4.2%. Additionally, MLL translocation 11q23 was detected in four patients (1.2%) out of the total cohort of 328.
Figure 1.
Co-occurring mutations among patients with RUNX1 acute myeloid leukemia. The thickness of the connecting lines indicates the frequency with which the two mutations co-occur.
We additionally identified a number of mutations that were negatively associated with mutant RUNX1. Mutations in the NPM1 gene were significantly lower in both the older (4.2%) and younger cohorts (0%) with RUNX1 mutations, compared to those with wild-type RUNX1 (older, 15.0%; younger, 26.2%). A similar negative correlation was identified between RUNX1 and CEBPα mutations—particularly among the younger cohort, wherein CEBPα incidence with wild-type RUNX1 was 16.1%, and was not identified (0%) in the setting of mutated RUNX1.
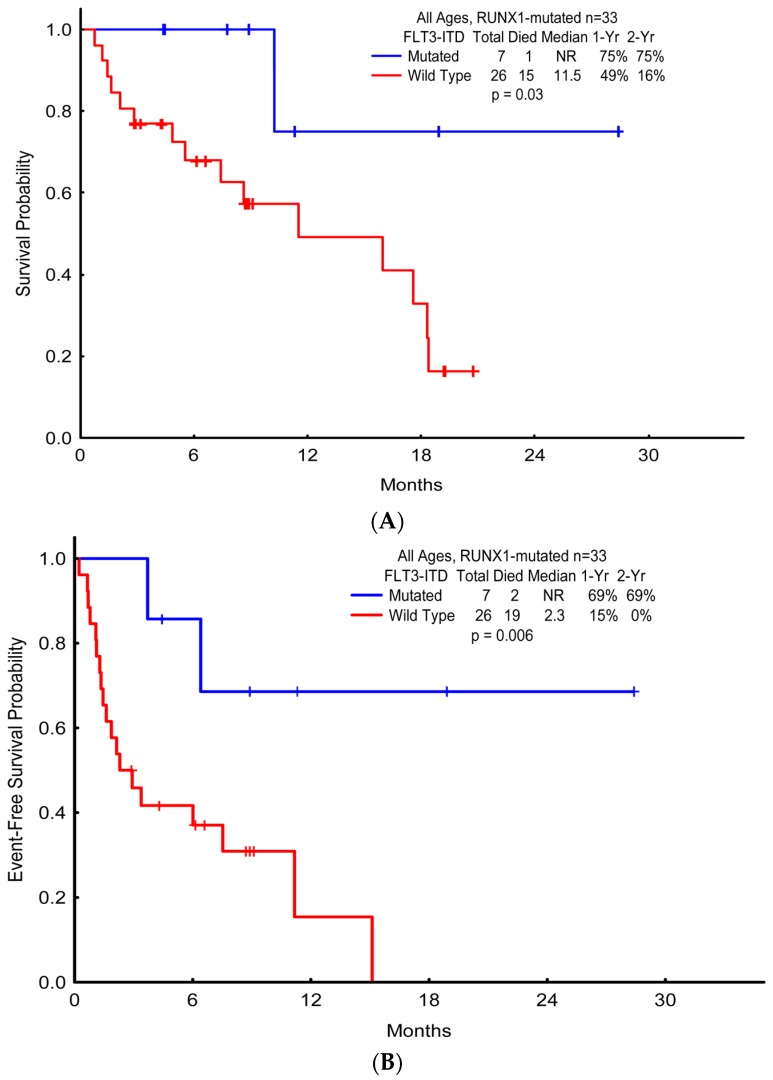
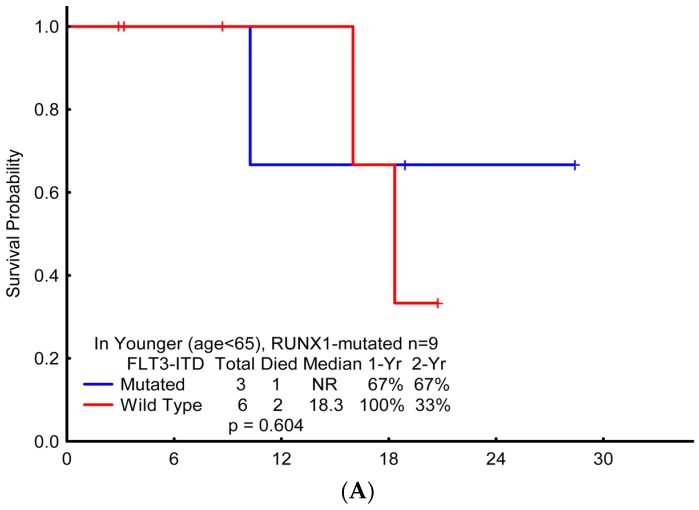
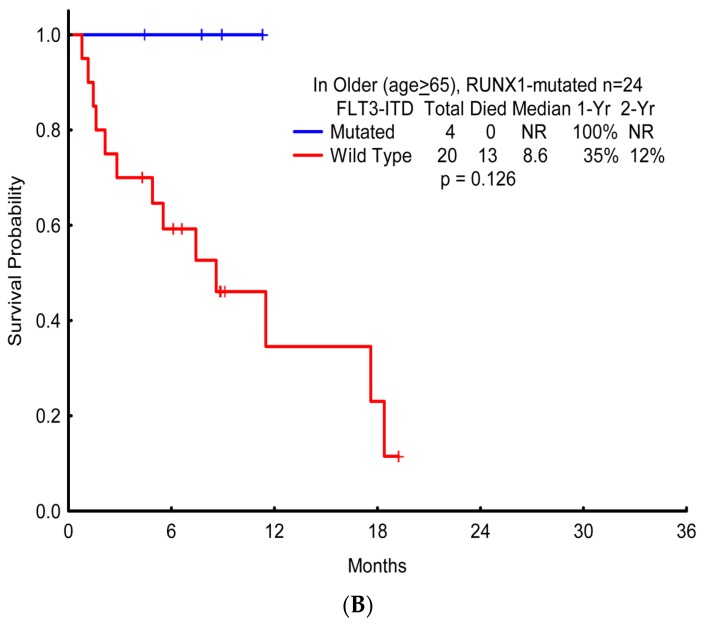
FLT3-ITD mutations were frequently identified in both cohorts, irrespective of mRUNX1: frequencies in the presence of mutated RUNX1 versus wild-type RUNX1 (older, 16.7% vs. 15.7%; younger, 30.0% vs. 20.8%). Of interest, co-occurring FLT3-ITD in the setting of mRUNX1 appeared to confer a more favorable prognosis (median OS with mutated FLT3-ITD vs. wild-type FLT3-ITD, NR vs. 11.5 months; p = 0.034) with a notable improved OS in FLT3-ITD and RUNX1 dual-mutated patients (Figure 2), although these differences were not significant when separated by age (younger, p = 0.604; older, p = 0.126) (Figure A1). This overall prognostic effect was additionally observed with improved EFS in dual-mutated patients (median EFS with mutated FLT-ITD3 vs. wild-type FLT3-ITD, NR vs. 2.3 months; p = 0.006).
Figure 2.
Survival probabilities of patients with RUNX1 and FLT3-ITD mutations. (A) Overall survival; (B) Event-free survival.
2.4. Association of RUNX1 with Response to Frontline AML Therapy
In the elderly AML cohort, response to frontline therapy was generally unaffected by RUNX1 mutational status (Table 2). There were no significant differences between patients with mutated and wild-type RUNX1 for complete remission (CR) rates, overall response rate (ORR);(CR + complete remission with incomplete platelet recovery (CRp) + hematological improvement (HI) + partial remission (PR), HI, and/or early death (ED; death within 28 days of initiating treatment) in the cohort as a whole. Notably, a significant difference was observed for the subset of older AML patients with prior MDS/MPN (Table 3). Overall, ORR was obtained in 80% of MDS/MPN patients in the older cohort with wild-type RUNX1, but was achieved in only 25% of the patients identified with mutant RUNX1 (p = 0.004).
Table 2.
Clinical outcomes of all patients according to age, treatment, and RUNX1 mutation status.
| Response | Younger Cohort (Chemotherapy) | Elderly Cohort (HMA) | ||||
|---|---|---|---|---|---|---|
| n (%) | n (%) | |||||
| RUNX1WT | RUNX1mut | p-Value | RUNX1WT | RUNX1mut | p-Value | |
| (n = 168) | (n = 9) | (n = 127) | (n = 24) | |||
| Complete remission | 128 (76.2) | 3 (33.3) | 0.004 * | 48 (37.8) | 9 (37.5) | 0.976 |
| Complete remission with incomplete platelet recovery | 9 (5.4) | 3 (33.3) | 0.271 | 11 (8.7) | 1 (4.2) | 0.667 |
| Hematological improvement | 4 (2.4) | 1 (11.1) | 0.631 | 17 (13.4) | 3 (12.5) | 0.603 |
| Partial remission | 2 (1.2) | 0 (0.0) | 4 (3.1) | 0 (0.0) | ||
| Overall response rate | 143 (85.0) | 7 (78.0) | 0.549 | 80 (63.0) | 13 (54.0) | 0.418 |
| No remission | 19 (11.3) | 2 (22.2) | 22 (17.3) | 7 (29.2) | ||
| Death | 5 (3.0) | 0 (0.0) | 22 (17.3) | 3 (12.5) | ||
| Early death ** | 1 (0.6) | 0 (0.0) | 0.818 | 3 (2.4) | 1 (4.2) | 0.617 |
* Boldface indicates statistical significance. Early death **: Death within 28 days of initiating treatment.
Table 3.
Clinical outcomes of patients with prior MDS or MPD according to age, treatment, and RUNX1 mutation status.
| Response | Younger Cohort (Chemotherapy) | Elderly Cohort (HMA) | ||||
|---|---|---|---|---|---|---|
| n (%) | n (%) | |||||
| RUNX1WT | RUNX1mut | p-Value | RUNX1WT | RUNX1mut | p-Value | |
| (n = 6) | (n = 1) | (n = 25) | (n = 8) | |||
| Complete remission | 2 (33.3) | 1 (100.0) | 0.211 | 11 (44.0) | 1 (12.5) | 0.107 |
| Complete remission with incomplete platelet recovery | 1 (16.7) | 0 (0.0) | 4 (16.0) | 0 (0.0) | ||
| Hematological improvement | 0 (0.0) | 0 (0.0) | 4 (16.0) | 1 (12.5) | ||
| Partial remission | 1 (16.7) | 0 (0.0) | 1 (4.0) | 0 (0.0) | ||
| Overall response rate | 4 (66.7) | 1 (100.0) | 0.497 | 20 (80.0) | 2 (25.0) | 0.004 * |
| No remission | 1 (16.7) | 0 (0.0) | 1 (4.0) | 3 (37.5) | ||
| Death | 1 (16.7) | 0 (0.0) | 4 (16.0) | 2 (25.0) | ||
| Early death ** | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (12.5) | ||
* Boldface indicates statistical significance. Early death **: Death within 28 days of initiating treatment.
In the younger cohort (Table 3), while CR rates were significantly reduced in patients with mutated versus wild-type RUNX1 (33.3% vs. 76.2%; p = 0.004), no significant difference in overall response rate was observed.
2.5. Association of RUNX1 with Clinical Outcomes
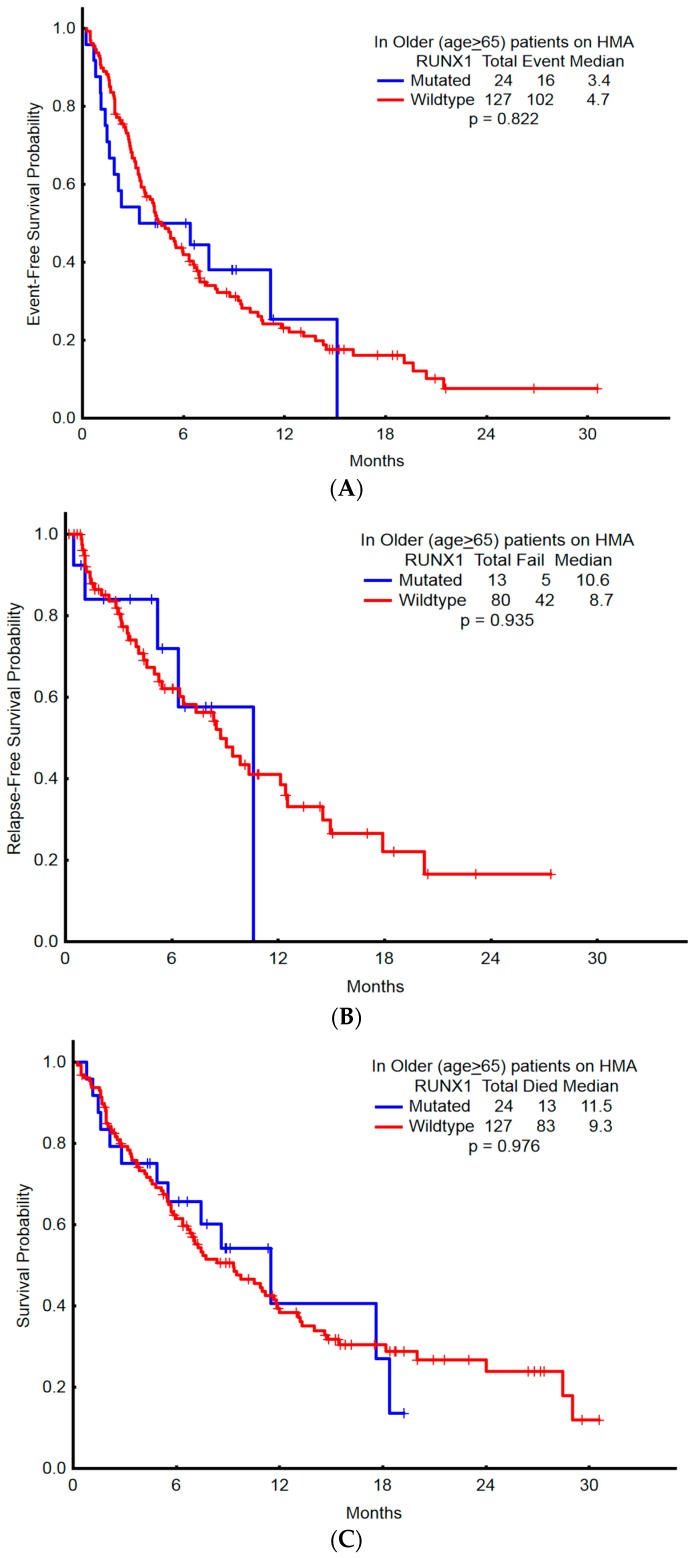
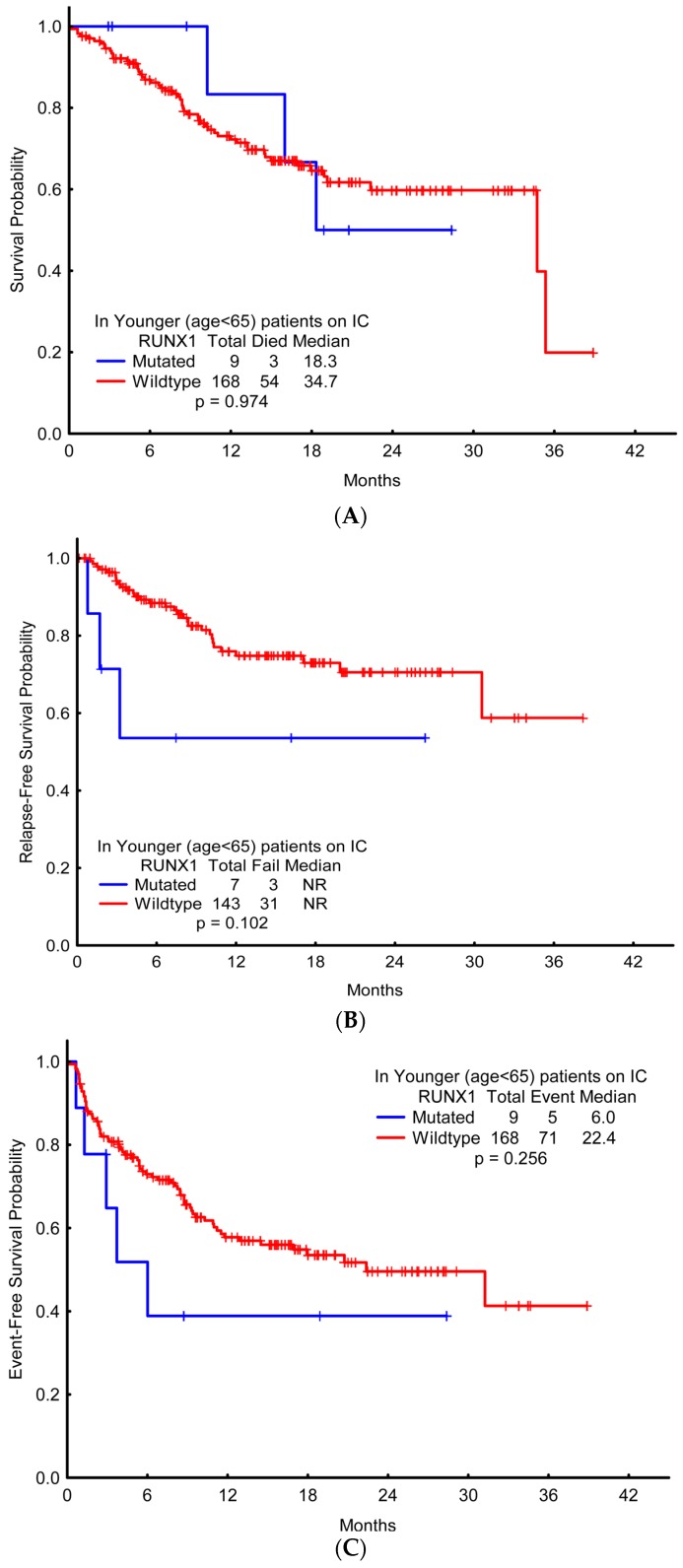
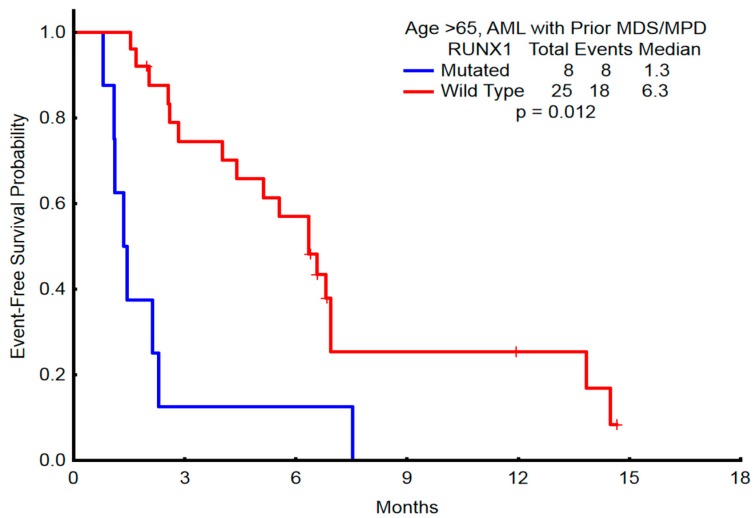
OS, EFS, and RFS were not significantly different between mutant RUNX1 compared with wild-type (Figure 3 and Figure 4). In the younger cohort, the two-year OS with wild-type RUNX1 and mutant RUNX1, respectively, was 60% and 50% (95% CI: ± 13% vs. ±98%); two-year RFS was 71% and 54% (95% CI: ±15% vs. ±85%); two-year EFS was 50% and 39% (95% CI: ±13% vs. ±85%). The clinical outcomes of the older cohort showed similar results, with no significant difference observed between the OS, EFS, and RFS of mutated-RUNX1 patients versus that of wild-type-RUNX1 patients. In a subgroup analysis of patients with a history of prior MDS/MPN (n = 40), those with mutant RUNX1 had significantly shorter EFS compared with wild-type RUNX1 (1.35 vs. 6.34 months; p = 0.012) (Figure 5) and trend to inferior OS (p = 0.079) in the older patient cohort.
Figure 3.
Survival probabilities among patients older than 65 years who received induction with hypomethylating agents, compared with the presence or absence of RUNX1 mutation. (A) Overall survival; (B) Relapse-free survival; (C) Event-free survival.
Figure 4.
Survival probabilities among patients aged below 65 years who received induction with chemotherapy, compared with the presence or absence of RUNX1 mutation. (A) Overall survival; (B) Relapse-free survival; (C) Event-free survival.
Figure 5.
Event-free survival in patients with prior MDS/MPN who were older than 65 years and received HMA induction therapy.
3. Discussion
In our study, RUNX1 mutations were detected at an overall incidence of 10.1%. This is consistent with previous studies that reported mutated RUNX1 alleles in 5.6% to 16.1% of patients with AML [3,4,7,10,12].
Mutations in RUNX1 associate with increasing age, also consistent with previous studies examining RUNX1 mutation status and age [4,7,12,16]. Accumulation of cellular stress and impaired repair of double-stranded DNA breaks (e.g., after radiation exposure) may contribute to genomic instability [17] and increase the frequency of genetic mutations, including RUNX1 [18,19,20]. Furthermore, RUNX1 mutations in our elderly cohort clustered among patients identified with intermediate cytogenetic risk including cytogenetically-normal AML, consistent with previous studies [7,10]. Other clinical characteristics (white blood cell (WBC) count, hemoglobin, peripheral blood and bone marrow blasts, t-AML, and AML secondary to MDS/MPN) were not associated with RUNX1 mutational status. Our analysis additionally confirmed a relationship between RUNX1 mutation and platelet count and LDH level, as has been previously reported [4,7].
The most commonly co-occurring mutation overall and particularly prevalent in the older cohort was ASXL1. Of interest, clonal expansion of ASXL1 is one of the most frequently identified genes in age-related clonal hematopoiesis, and subsequent RUNX1 mutations may be implicated in malignant progression in these individuals [21,22]. Among younger patients, co-occurring RAS mutations (44%) and FLT3-ITD (33%) were also frequent. Their classification as class I mutations represents their capacity to promote stem cell proliferation and thereby cooperate with RUNX1 mutations in AML pathogenesis [2,23].
Co-occurring FLT3 and RUNX1 mutations have been previously described, with a hypothesis that loss of function RUNX1 mutations may predispose hematopoietic cells to overexpress activating mutations of FLT3 [24]. When examining the clinical implications of this hypothesis in patients with mutant RUNX1, our analysis interestingly found superior OS and EFS in mRUNX1 associated with FLT3 mutations. Previous studies have identified that the adverse effect of RUNX1 is independent of FLT3-ITD [25]. Evaluation in future studies will be important to see if this is a reproducible finding. It is also important to note that NPM1 and biallelic CEBPα mutations confer a favorable prognosis in terms of disease-free survival and OS in the absence of FLT3 mutations [2], and both are inversely related to the incidence of RUNX1 mutations [7]. In our cohort, the incidence of NPM1 mutations declined from 26.2% to 0% in younger patients and from 15.0% to 4.2% in older patients in the absence and presence of RUNX1 mutations, respectively. Conversely, although partial tandem duplications in the MLL gene have been previously associated with mutated RUNX1 and implicated in shorter survival [2,7], this association was not identified in our patient cohort, as only four patients were identified with the MLL rearrangement 11q23. An association of co-occurring IDH1/IDH2 mutations with RUNX1 mutations was additionally identified, consistent with Gaidzik et al. [10]. Furthermore, mutations in RUNX1 and IDH1/2 have been collectively described as the most frequent genetic lesions in M0-AML [26], the prognostic impact of the latter remaining controversial [27].
While RUNX1 mutations are frequently associated with inferior responses to AML therapy, our analysis suggests that the poor outcomes seen in RUNX1 mutations occur primarily within younger patients treated with intensive therapeutic strategies, as well as the subgroup of RUNX1 mutant AML with prior MDS/MPN. The similar responses and outcome of the larger cohort of older mRUNX1 treated with HMA approaches was a notable finding of this analysis.
A previous study in patients with MDS identified that treatment with HMAs may partially mitigate the adverse prognostic impact of gene mutations in patients with RUNX1, ASXL1, EZH2, or ETV6 mutations [28]. Differences in outcomes between patient populations may also be linked with the distribution of co-existing mutations and cytogenetic abnormalities that may exacerbate or mitigate the effect of mutated RUNX1, [2] and our study is limited by a lack of complete co-mutational data, most notably for splicing factor mutations, as whole exome sequencing was not possible. The importance of RUNX1 as a marker of MRD in responding patients was additionally not able to be assessed in our study.
4. Materials and Methods
4.1. Patients
A total of 328 patients with treatment-naïve AML treated at the University of Texas MD Anderson Cancer Center between August 2013 and July 2016 were analyzed in this study. The presence of de novo AML, or history of prior myelodysplastic syndrome (MDS) or myeloproliferative neoplasms (MPNs) was collected. As treatment intensity at our institution during this time period was strongly and near-universally correlated to patient age at diagnosis, patients were categorized into two cohorts according to their age and therapeutic regimen: patients younger than 65 years who received intensive chemotherapy (younger cohort; n = 177) and patients ≥65 years who received HMA therapy (elderly cohort; n = 151).
Variables in our study included patient demographics (age and sex), relevant diagnostic tests including white blood cell (WBC) count, hemoglobin (Hb) levels, platelet count, lactate dehydrogenase (LDH) levels, and percentage of bone marrow blasts, cytogenetics, FAB classification, RUNX1 mutation, and co-occurrence of other mutations. Clinical outcomes were assessed using Kaplan–Meier estimates of OS, EFS, and RFS [29]. Response to therapy was evaluated using revised IWG criteria for AML [30].
All participating patients provided informed consent in compliance with the Declaration of Helsinki, and the study received ethical approval by the Institutional Review Board as part of a retrospective chart review protocol, PA11.0788 on 10 October 2011 within the Department of Leukemia at University of Texas MD Anderson Cancer Center.
4.2. Molecular Analysis
Diagnostic bone marrow samples were obtained for mutational analysis. Total genomic DNA was extracted from unenriched peripheral blood (PB) or bone marrow (BM) samples using ReliaPrep genomic DNA isolation kit (Promegal Corp, Madison, WI, USA) [31]. FLT3 (internal tandem duplication and D835) was assessed by PCR followed by capillary electrophoresis on a Genetic Analyzer (Applied Biosystems, Foster City, CA, USA) [32]. Briefly, a total of 250 ng DNA was utilized to prepare sequencing libraries using Agilent HaloPlex custom Kit (Agilent Technologies, Santa Clara, CA, USA) [33]. The entire coding sequences of 28 genes (ABL1, ASXL1, BRAF, DNMT3A, EZH2, FLT3, GATA1, GATA2, HRAS, IDH1, IDH2, IKZF2, JAK2, KIT, KRAS, MDM2, MLL, MPL, NPM1, NRAS, PTPN11, RUNX1, TET2, TP53, WT1) were interrogated on a custom-designed next-generation sequencing approach using the IlluminaMiSeq platform (Illumina; San Diego, CA, USA) [34]. The genomic reference sequence used was genome GRch37/hg19. The following software tools were utilized in the experimental setup and data analysis: Illumina Experiment Manager 1.6.0 (Illumina; San Diego, CA, USA), MiSeq Control Software 2.4 (Illumina; San Diego, CA, USA), Real Time Analysis 1.18.54 (Illumina; San Diego, CA, USA), Sequence Analysis Viewer 1.8.37 (Illumina; San Diego, CA, USA), MiSeq Reporter 2.5.1 (Illumina; San Diego, CA, USA), and SureCall 3.0.1.4 (Agilent Technologies; Santa Clara, CA, USA). A minimum of 80% reads at quality scores of AQ30 or higher were required to pass quality control. NM_001754 was utilized as the RUNX1 reference sequence.
The lower limit of detection of this assay (analytical sensitivity) for single nucleotide variations was determined to be 5% (one mutant allele in the background of nineteen wild type alleles) to 10% (one mutant allele in the background of nine wild type alleles). Testing of patients with active hematologic malignancies was limited to somatic mutations only. Known germline polymorphisms identified in >20% of our in-house patient population were excluded. Additionally, germline polymorphisms previously reported in population databases such as dbSNP and ExAC were classified as “variants of probable germline origin” and were excluded from this analysis.
4.3. Statistical Analysis
Continuous variables were described as means, medians, standard deviations, and ranges, whereas the categorical variables were presented as frequencies and percentages. The categorical variables in patients with mutated RUNX1 were compared with the wild-type using the Chi-squared test. Kaplan–Meier curves [26] were used to estimate unadjusted OS durations. OS was calculated from the start of treatment to the last follow up or death (censor = alive/dead). EFS was measured from the start of treatment to any adverse event or the last follow-up. Adverse events included either relapse (i.e., NR, ED) or death. RFS was determined from leukemia-free response (i.e., at the time of documented CR, CRp, CRi, PR) to relapse (loss), death, or last follow-up. For all analyses, p < 0.05 was considered statistically significant. All computations were carried out in Statistica 12.0 (StatSoftInc. Tulsa, OK, USA; 1984–2013).
5. Conclusions
In conclusion, RUNX1 mutations are among the most frequent recurring genetic abnormalities in patients with AML, often associated with specific clinical features and poor outcomes. ASXL1, DNMT3A, IDH1/2, FLT3-ITD, and RAS mutations frequently co-occur with RUNX1 mutations. FLT3-ITD mutations did not impart an inferior prognosis in mRUNX1 AML, and of note appeared to confer an improved OS in this molecularly defined cohort. While younger mRUNX1 patients treated with intensive chemotherapy and a subgroup of older mRUNX1 patients with prior MDS/MPN exhibited inferior treatment responses, the majority of mRUNX1 AML patients included older patients treated with HMA therapy, in whom treatment responses and clinical outcomes were not inferior compared to RUNX1 wild-type. Further studies investigating the treatment response to HMAs—in particular in younger patient populations with RUNX1-mutated AML—are required to fully establish their therapeutic role in the individualized treatment of these patients.
Acknowledgments
This work was supported in part by the MD Anderson Cancer Center Support Grant P30 CA016672, Leukemia SPORE CA100632 and Charif Souki Cancer Research Fund.
Appendix A
Table A1.
RUNX1 mutations according to their minor allele frequency, exon location, and mutation type.
| Mutation HGVS (MAF%) | Exon | Type |
|---|---|---|
| NM_001754.4 ( RUNX1):c.497G>A p.R166Q [46.4] | 5 | Missense |
| NM_001754.4 ( RUNX1):c.423_424insAAAGGGATTCT p.A142fs [5.9] | 5 | Frameshift |
| NM_001754.4 ( RUNX1):c.891_894dupCCCA p.A299fs [8.7] | 8 | Frameshift |
| NM_001754.4 ( RUNX1):c.238dupG p.E80fs [50.3] | 4 | Frameshift |
| NM_001754.4 ( RUNX1):c.900_901insTCCG p.P301fs [17.0], NM_001754.4 (RUNX1):c.987_988dupGT p.F330fs [32.3], NM_001754.4(RUNX1):c.1281_1285del p.I428fs [13.6] | “8,9” | Frameshift |
| NM_001754.4 ( RUNX1):c.292delG p.L98fs,NM_001754.4(RUNX1):c.492_493insAT p.G165fs [36.2] | “4,5” | Frameshift |
| NM_001754.4 ( RUNX1):c.368A>C p.D123A [9.2], NM_001754.4(RUNX1):c.367G>C p.D123H [9.3], NM_001754.4 (RUNX1):c.361_364del p.L121fs [10.1] | 5 | Frameshift |
| NM_001754.4( RUNX1):c.422_423insA p.A142fs [26.2] | 5 | Frameshift |
| NM_001754.4 ( RUNX1):c.418_422dupTACTC p.A142fs [21.0] | 5 | Frameshift |
|
NM_001754.4 (
RUNX1):c.422dupC p.A142fs [9.5], NM_001754.4(RUNX1):c.424_425insGGTGG p.A142fs [4.4] |
5 | Frameshift |
| NM_001754.4 ( RUNX1):c.939_942dupCTCT p.A315fs [43.5] | 8 | Frameshift |
| NM_001754.4 ( RUNX1):c.593A>G p.D198G [83.0] | 6 | Missense |
| NM_001754.4 ( RUNX1):c.592G>A p.D198N [31.7] | 6 | Missense |
| NM_001754.4 ( RUNX1):c.993_1005del p.D332fs | 9 | Frameshift |
| NM_001754.4 ( RUNX1):c.402dupT p.G135fs [41.1] | 5 | Frameshift |
| NM_001754.4 ( RUNX1):c.1023_1024insC p.I342fs | 9 | Frameshift |
| NM_001754.4 ( RUNX1):c.1251_1252insC p.M418fs | 9 | Frameshift |
| NM_001754.4 ( RUNX1):c.1244_1250dupAGTTCTC p.M418fs [19.5] | 9 | Frameshift |
| NM_001754.4 ( RUNX1):c.548dupC p.P184fs [45.3] | 6 | Frameshift |
| NM_001754.4 ( RUNX1):c.256_259delinsACCCGA p.P86fs | 4 | Missense |
| NM_001754.4 ( RUNX1):c.484A>G p.R162G [70.4] | 5 | Missense |
| NM_001754.4 ( RUNX1):c.485G>A p.R162K [42.2],NM_001754.4(RUNX1):c.388G>T p.V130F [44.7] | 5 | Missense |
| NM_001754.4 ( RUNX1):c.496C>T p.R166* [39.1] | 5 | Nonsense |
| NM_001754.4 ( RUNX1):c.601C>T p.R201* [40.7] | 6 | Nonsense |
| NM_001754.4 ( RUNX1):c.955_958dupAGTC p.R320fs [49.7] | 8 | Frameshift |
| NM_001754.4 ( RUNX1):c.952del p.S318fs [29.7] | 8 | Frameshift |
|
NM_001754.4 (
RUNX1):c.1352_1356dupACGTG p.V453fs [22.9], NM_001754.4 (RUNX1):c.611G>A p.R204Q [49.7] |
“9,6” | Frameshift |
| NM_001754.4 ( RUNX1):c.952T>G p.S318A [14.9], NM_001754.4 (RUNX1):c.954dupC p.S319fs [14.4], NM_001754.4(RUNX1) :c.950T>G p.L317R [15.0] | 8 | Frameshift |
| NM_001754.4 ( RUNX1):c.977del p.D326fs [17.1], NM_001754.4 (RUNX1):c.352-1_352insTTTAG p.? | 9 | Frameshift, splice |
| NM_001754.4 ( RUNX1):c.492_507dupCGGTCGAAGTGGAAGA p.G170fs [30.7], NM_001754.4 (RUNX1):c.484dupA p.R162fs [3.0] | 5 | Frameshift |
| NM_001754.4 ( RUNX1):c.486dupG p.F163fs [51.2] | 5 | Frameshift |
| NM_001754.4 ( RUNX1):c.1036dupC p.R346fs [2.8] | 9 | Frameshift |
| NM_001754.4 ( RUNX1):c.167T>C p.L56S [51.5] | 4 | Missense |
Figure A1.
Survival probabilities of FLT3-ITD and RUNX1 dual-mutated patients. (A) Patients aged below 65 years. (B) Patients older than 65 years.
Author Contributions
Maliha Khan Keyur P. Patel designed the study, collected and analyzed data, wrote the manuscript. Mark Brandt and Sherry Pierce provided statistical analysis. Jorge Cortes, Farhad Ravandi, Kapil Bhalla, Steven Kornblau, Gautam Borthakur, Marina Konopleva, Tapan Kadia, Kiran Naqvi, Keyur P. Patel and Hagop Kantarjian edited the manuscript. Courtney D. DiNardo conceived the study, designed the study, analyzed data, and wrote the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Gilliland D.G., Griffin J.D. The roles of FLT3 in hematopoiesis and leukemia. Blood. 2002;100:1532–1542. doi: 10.1182/blood-2002-02-0492. [DOI] [PubMed] [Google Scholar]
- 2.Takahashi S. Current findings for recurring mutations in acute myeloid leukemia. J. Hematol. Oncol. 2011;4:36. doi: 10.1186/1756-8722-4-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Greif P.A., Konstandin N.P., Metzeler K.H., Herold T., Pasalic Z., Ksienzyk B., Dufour A., Schneider F., Schneider S., Kakadia P.M., et al. RUNX1 mutations in cytogenetically normal acute myeloid leukemia are associated with a poor prognosis and up-regulation of lymphoid genes. Haematologica. 2012;97:1909–1915. doi: 10.3324/haematol.2012.064667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gaidzik V.I., Teleanu V., Papaemmanuil E., Weber D., Paschka P., Hahn J., Wallrabenstein T., Kolbinger B., Köhne C.H., Horst H.A., et al. RUNX1 mutations in acute myeloid leukemia are associated with distinct clinico-pathologic and genetic features. Leukemia. 2016;30:2282. doi: 10.1038/leu.2016.207. [DOI] [PubMed] [Google Scholar]
- 5.Harada H., Harada Y., Niimi H., Kyo T., Kimura A., Inaba T. High incidence of somatic mutations in the AML1/RUNX1 gene in myelodysplastic syndrome and low blast percentage myeloid leukemia with myelodysplasia. Blood. 2004;103:2316–2324. doi: 10.1182/blood-2003-09-3074. [DOI] [PubMed] [Google Scholar]
- 6.Chen C.Y., Lin L.I., Tang J.L., Ko B.S., Tsay W., Chou W.C., Yao M., Wu S.J., Tseng M.H., Tien H.F., et al. RUNX1 gene mutation in primary myelodysplastic syndrome—The mutation can be detected early at diagnosis or acquired during disease progression and is associated with poor outcome. Br. J. Haematol. 2007;139:405–414. doi: 10.1111/j.1365-2141.2007.06811.x. [DOI] [PubMed] [Google Scholar]
- 7.Tang J.L., Hou H.A., Chen C.Y., Liu C.Y., Chou W.C., Tseng M.H., Huang C.F., Lee F.Y., Liu M.C., Yao M., et al. AML1/RUNX1 mutations in 470 adult patients with de novo acute myeloid leukemia: Prognostic implication and interaction with other gene alterations. Blood. 2009;114:5352–5361. doi: 10.1182/blood-2009-05-223784. [DOI] [PubMed] [Google Scholar]
- 8.Chou W.C., Huang H.H., Hou H.A., Chen C.Y., Tang J.L., Yao M., Tsay W., Ko B.S., Wu S.J., Huang S.Y., et al. Distinct clinical and biological features of de novo acute myeloid leukemia with additional sex comb-like 1 (ASXL1) mutations. Blood. 2010;116:4086–4094. doi: 10.1182/blood-2010-05-283291. [DOI] [PubMed] [Google Scholar]
- 9.Schnittger S., Eder C., Alpermann T., Fasan A., Grossmann V., Kohlmann A., Kern W., Haferlach C., Haferlach T. ASXL1 exon 12 mutations are frequent in AML with intermediate risk karyotype and are independently associated with an adverse outcome. Leukemia. 2013;27:82–91. doi: 10.1038/leu.2012.262. [DOI] [PubMed] [Google Scholar]
- 10.Gaidzik V.I., Bullinger L., Schlenk R.F., Zimmermann A.S., Rock J., Paschka P., Corbacioglu A., Krauter J., Schlegelberger B., Ganser A., et al. RUNX1 mutations in acute myeloid leukemia: Results from a comprehensive genetic and clinical analysis from the AML study group. J. Clin. Oncol. 2011;29:364–372. doi: 10.1200/JCO.2010.30.7926. [DOI] [PubMed] [Google Scholar]
- 11.Osato M. Point mutations in the RUNX1/AML1 gene: Another actor in RUNX leukemia. Oncogene. 2004;23:4284–4296. doi: 10.1038/sj.onc.1207779. [DOI] [PubMed] [Google Scholar]
- 12.Mendler J.H., Maharry K., Radmacher M.D., Mrozek K., Becker H., Metzeler K.H., Schwind S., Whitman S.P., Khalife J., Kohlschmidt J., et al. RUNX1 mutations are associated with poor outcome in younger and older patients with cytogenetically normal acute myeloid leukemia and with distinct gene and MicroRNA expression signatures. J. Clin. Oncol. 2012;30:3109–3118. doi: 10.1200/JCO.2011.40.6652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hann I.M., Stevens R.F., Goldstone A.H., Rees J.K., Wheatley K., Gray R.G., Burnett A.K. Randomized comparison of DAT versus ADE as induction chemotherapy in children and younger adults with acute myeloid leukemia. Results of the Medical Research Council’s 10th AML trial (MRC AML10). Adult and Childhood Leukaemia Working Parties of the Medical Research Council. Blood. 1997;89:2311–2318. [PubMed] [Google Scholar]
- 14.Al-Ali H.K., Jaekel N., Niederwieser D. The role of hypomethylating agents in the treatment of elderly patients with AML. J. Geriatr. Oncol. 2014;5:89–105. doi: 10.1016/j.jgo.2013.08.004. [DOI] [PubMed] [Google Scholar]
- 15.Krzywinski M., Schein J., Birol I., Connors J., Gascoyne R., Horsman D., Jones S.J., Marra M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schneider F., Hoster E., Schneider S., Dufour A., Benthaus T., Kakadia P.M., Bohlander S.K., Braess J., Heinecke A., Sauerland M.C., et al. Age-dependent frequencies of NPM1 mutations and FLT3-ITD in patients with normal karyotype AML (NK-AML) Ann Hematol. 2012;91:9–18. doi: 10.1007/s00277-011-1280-6. [DOI] [PubMed] [Google Scholar]
- 17.Li Z., Zhang W., Chen Y., Guo W., Zhang J., Tang H., Xu Z., Zhang H., Tao Y., Wang F., et al. Impaired DNA double-strand break repair contributes to the age-associated rise of genomic instability in humans. Cell Death Differ. 2016;23:1765–1777. doi: 10.1038/cdd.2016.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Papamichos-Chronakis M., Peterson C.L. Chromatin and the genome integrity network. Nat. Rev. Genet. 2013;14:62–75. doi: 10.1038/nrg3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harada H., Harada Y., Tanaka H., Kimura A., Inaba T. Implications of somatic mutations in the AML1 gene in radiation-associated and therapy-related myelodysplastic syndrome/acute myeloid leukemia. Blood. 2003;101:673–680. doi: 10.1182/blood-2002-04-1010. [DOI] [PubMed] [Google Scholar]
- 20.Zharlyganova D., Harada H., Harada Y., Shinkarev S., Zhumadilov Z., Zhunusova A., Tchaizhunusova N.J., Apsalikov K.N., Kemaikin V., Zhumadilov K., et al. High frequency of AML1/RUNX1 point mutations in radiation-associated myelodysplastic syndrome around Semipalatinsk nuclear test site. J. Radiat. Res. 2008;49:549–555. doi: 10.1269/jrr.08040. [DOI] [PubMed] [Google Scholar]
- 21.Genovese G., Kähler A.K., Handsaker R.E., Lindberg J., Rose S.A., Bakhoum S.F., Chambert K., Mick E., Neale B.M., Fromer M., et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. NEJM. 2014;371:2477–2487. doi: 10.1056/NEJMoa1409405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xie M., Lu C., Wang J., McLellan M.D., Johnson K.J., Wendl M.C., McMichael J.F., Schmidt H.K., Yellapantula V., Miller C.A., et al. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat. Med. 2014;20:1472–1478. doi: 10.1038/nm.3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klein K., Kaspers G., Harrison C.J., Beverloo H.B., Reedijk A., Bongers M., Cloos J., Pession A., Reinhardt D., Zimmerman M., et al. Clinical Impact of Additional Cytogenetic Aberrations, cKIT and RAS Mutations, and Treatment Elements in Pediatric t(8;21)-AML: Results From an International Retrospective Study by the International Berlin-Frankfurt-Münster Study Group. J. Clin. Oncol. 2015;33:4247–4258. doi: 10.1200/JCO.2015.61.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsuno N., Osato M., Nanri T., Shigesada K., Fukushima T., Motoji T., Kusumoto S., Naoe T., Ohno R., Mitsuya H., et al. Dual mutations in the AML1 and FLT3 genes are associated with leukemogenesis in acute myeloblastic leukemia of the M0 type. Leukemia. 2003;17:2492–2499. doi: 10.1038/sj.leu.2403160. [DOI] [PubMed] [Google Scholar]
- 25.Schnittger S., Dicker F., Kern W., Wendland N., Sundermann J., Alpermann T., Haferlach C., Haferlach T. RUNX1 mutations are frequent in de novo AML with noncomplex karyotype and confer an unfavorable prognosis. Blood. 2011;117:2348–2357. doi: 10.1182/blood-2009-11-255976. [DOI] [PubMed] [Google Scholar]
- 26.Kao H.W., Liang D.C., Wu J.H., Kuo M.C., Wang P.N., Yang C.P., Shih Y.S., Lin T.H., Huang Y.H., Shih L.Y., et al. Gene mutation patterns in patients with minimally differentiated acute myeloid leukemia. Neoplasia. 2014;16:481–488. doi: 10.1016/j.neo.2014.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Green C.L., Evans C.M., Zhao L., Hills R.K., Burnett A.K., Linch D.C., Gale R.E. The prognostic significance of IDH2 mutations in AML depends on the location of the mutation. Blood. 2011;118:409–412. doi: 10.1182/blood-2010-12-322479. [DOI] [PubMed] [Google Scholar]
- 28.Bejar R., Lord A., Stevenson K., Bar-Natan M., Pérez-Ladaga A., Zaneveld J., Wang H., Caughey B., Stojanov P., Getz G., et al. TET2 mutations predict response to hypomethylating agents in myelodysplastic syndrome patients. Blood. 2014;124:2705–2712. doi: 10.1182/blood-2014-06-582809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaplan E.L., Meier P. Nonparametric estimation from incomplete observations. J. Am. Stat. Assoc. 1958;53:457–481. doi: 10.1080/01621459.1958.10501452. [DOI] [Google Scholar]
- 30.Cheson B.D., Bennett J.M., Kopecky K.J., Büchner T., Willman C.L., Estey E.H., Schiffer C.A., Doehner H., Tallman M.S., Lister T.A., et al. Revised recommendations of the International Working Group for Diagnosis, Standardization of Response Criteria, Treatment Outcomes, and Reporting Standards for Therapeutic Trials in Acute Myeloid Leukemia. J. Clin. Oncol. 2003;21:4642–4649. doi: 10.1200/JCO.2003.04.036. [DOI] [PubMed] [Google Scholar]
- 31.Casaril A.E., de Oliveira L.P., Alonso D.P., de Oliveira E.F., Barrios S.P.G., Infran J.D.O.M., de Souza Fernandes W., Oshiro E.T., Ferreira A.M.T., Ribolla P.E.M., et al. Standardization of DNA extraction from sand flies: Application to genotyping by next generation sequencing. Exp. Parasitol. 2017;177:66–72. doi: 10.1016/j.exppara.2017.04.010. [DOI] [PubMed] [Google Scholar]
- 32.Warren M., Luthra R., Yin C.C., Ravandi F., Cortes J.E., Kantarjian H.M., Medeiros L.J., Zuo Z. Clinical impact of change of FLT3 mutation status in acute myeloid leukemia patients. Mod. Pathol. 2012;25:1405–1412. doi: 10.1038/modpathol.2012.88. [DOI] [PubMed] [Google Scholar]
- 33.Wendt F.R., Zeng X., Churchill J.D., King J.L., Budowle B. Analysis of Short Tandem Repeat and Single Nucleotide Polymorphism Loci From Single-Source Samples Using a Custom HaloPlex Target Enrichment System Panel. Am. J. Forensic Med. Pathol. 2016;37:99–107. doi: 10.1097/PAF.0000000000000228. [DOI] [PubMed] [Google Scholar]
- 34.Patel K.P., Newberry K.J., Luthra R., Jabbour E., Pierce S., Cortes J., Singh R., Mehrotra M., Routbort M.J., Luthra M., et al. Correlation of mutation profile and response in patients with myelofibrosis treated with ruxolitinib. Blood. 2015;126:790–797. doi: 10.1182/blood-2015-03-633404. [DOI] [PMC free article] [PubMed] [Google Scholar]