Abstract
The ABCC1 gene is structurally and functionally related to the cystic fibrosis transmembrane conductance regulator gene (CFTR). Upregulation of ABCC1 is thought to improve lung function in patients with cystic fibrosis (CF); the mechanism underlying this effect is unknown. We analyzed the ABCC1 promoter single nucleotide polymorphism (SNP rs504348), plasma-induced ABCC1 mRNA expression levels, and ABCC1 methylation status and their correlation with clinical variables among CF subjects with differing CFTR mutations. We assigned 93 CF subjects into disease severity groups and genotyped SNP rs504348. For 23 CF subjects and 7 healthy controls, donor peripheral blood mononuclear cells (PBMCs) stimulated with plasma underwent gene expression analysis via qRT-PCR. ABCC1 promoter methylation was analyzed in the same 23 CF subjects. No significant correlation was observed between rs504348 genotypes and CF disease severity, but pancreatic insufficient CF subjects showed increased colonization with any form of Pseudomonas aeruginosa (OR = 3.125, 95% CI: 1.192–8.190) and mucoid P. aeruginosa (OR = 5.075, 95% CI: 1.307–28.620) compared to the pancreatic sufficient group. A significantly higher expression of ABCC1 mRNA was induced by CF plasma compared to healthy control plasma (p < 0.001). CF subjects with rs504348 (CC/CG) also had higher mRNA expression compared to those with the ancestral GG genotype (p < 0.005). ABCC1 promoter was completely unmethylated; therefore, we did not detect any association between methylation and CF disease severity. In silico predictions suggested that histone modifications are crucial for regulating ABCC1 expression in PBMCs. Our results suggest that ABCC1 expression has a role in CFTR activity thereby increasing our understanding of the molecular underpinnings of the clinical heterogeneity in CF.
Keywords: ABCC1, cystic fibrosis, DNA methylation, polymorphism
1. Introduction
Cystic fibrosis (CF) is a heritable multisystem disorder caused by mutations in the cystic fibrosis transmembrane conductance regulator (CFTR) gene [1,2]. Despite advances in newborn screening and treatments for CF, clinical heterogeneity remains a major challenge [3]. Modifier genes that may impact CF disease severity are emerging keys to deciphering clinical heterogeneity [4,5,6,7].
The gene encoding the multidrug resistance-associated protein 1, ABCC1/MRP1, is a candidate for further molecular investigation based on its structural and functional association with CFTR [8,9]. ABCC1, as well as CFTR (ABCC7) and 11 other genes associated with multidrug resistance, are subfamily C members of the ATP-binding cassette (ABC) transporter genes [10]. The diverse activities of subfamily C transporters include transportation of chemotherapeutic agents, amino acids, glutathione conjugates, and small peptides, as well as excretion of fungal and bacteria toxins [10,11,12]. CFTR is unique among the ABC subfamily C members of transporters due to its intrinsic ability to conduct chloride ions at a fast rate [12,13,14], but it shares its closest homology with ABCC1 [9,15,16].
Since the cloning of ABCC1 in the early 90s [17,18,19], progress has been made towards establishing a functional relationship between ABCC1 and CFTR. For example, functional complementation of dysfunctional CFTR by ABCC1 following chemotherapy that resulted in increased expression of ABCC1 was associated with improved lung function in a CF patient [15]. A study analyzing nasal epithelial cells in CF patients also showed that low ABCC1 transcript levels were associated with more severe disease [8]. Additionally, increased expression of ABCC1 in nasal cells of CF patients following treatment with azithromycin was associated with restoration of chloride channel conduction and improved lung function [20].
Polymorphisms or epigenetic modifications that alter the expression ABCC1 may underlie the clinical heterogeneity in lung function observed among CF patients. A functional single nucleotide polymorphism (SNP) in the promoter region of ABCC1 (rs504348) has been shown to modulate gene expression and impact lung function [21,22]. Mafficini, et al. [23] recently reported that this polymorphism is associated with disease severity in CF patients homozygous for the F508del mutation. However, this study has not been replicated using a cohort of CF patients with differing genotypes, and it is unclear whether this SNP impacts lung function by modulating ABCC1 expression.
DNA methylation is a predominant epigenetic mechanism that modulates gene expression [24]. Methylation of cytosine-phosphate-guanosine (CpG) dinucleotides present in CpG islands within gene promoters can inactivate and silence gene expression [25,26,27]. The ABCC1 promoter contains CpG islands that are potential targets for methylation analysis. Previous research in cancer cells has shown that the ABCC1 promoter is hypomethylated but that the methylation status is not correlated with ABCC1 gene expression [28]. Whether this epigenetic mechanism modulates ABCC1 expression in CF is unknown.
Analysis of peripheral blood cell gene expression signatures provides us with a non-invasive approach to diagnose many diseases, including CF. This may involve direct profiling of transcripts of peripheral blood mononuclear cells (PBMCs) from affected individuals [29,30]. Alternatively, analysis of gene expression changes after exposure of healthy donor PBMCs to patient serum or plasma has been used as a model in diabetes, Crohn’s disease, ulcerative colitis and juvenile rheumatoid arthritis [31,32,33,34]. To examine the association between ABCC1 expression and lung function in CF, we analyzed the functional ABCC1 promoter polymorphism (rs504348), plasma-induced ABCC1 expression in PBMCs, and ABCC1 promoter methylation status among CF subjects with differing CFTR genotypes and then examined the correlation of these molecular measures with clinical status.
2. Results
2.1. Baseline Characteristics of Subjects
A total of 93 CF subjects were recruited for this study. The median (interquartile range (IQR)) subject age was 10 (6, 19) years and 47.3% were males. The majority of CF subjects (74.2%) were pancreatic insufficient. More than half of the CF subjects (58.1%) were positive for Pseudomonas aeruginosa, of which 59.2% were positive for the mucoid form of P. aeruginosa. The median (IQR) sweat chloride level was 103 (87, 114) mmol/L, and the forced expiratory volume in 1 second (FEV1) percent predicted was 97 (72, 111). Spirometry information and culture results were recorded during the same clinical visit and the collection date was used to deduce sample age. Based on the adopted Epidemiologic Study of Cystic Fibrosis (ESCF) disease severity equation, most CF subjects (47.3%) had normal/very mild disease, while 19 CF subjects (20.4%) had severe disease. Fourteen percent of subjects were not assigned a severity group because they had no recorded FEV1 to estimate FEV1 percent predicted (Table 1).
Table 1.
Demographics of study subjects.
| Parameter | CF Subjects |
|---|---|
| Samples (n) | 93 |
| Age in years, median (IQR) | 10 (6,19) |
| Gender: Male, n (%) | 44 (47.3%) |
| Pancreatic insufficient, n (%) | 69 (74.2%) |
| P. aeruginosa, n (%) | 54 (58.1%) |
| Mucoid P. aeruginosa, n (%) | 32 (34.4%) |
| Sweat chloride (mmol/L), median (IQR) | 103 (87,114) |
| FEV1 (%) predicted, median (IQR) | 97 (72,111) |
| ESCF disease severity classification | n (%) |
| Normal/very mild | 44 (47.3%) |
| Mild | 10 (10.8%) |
| Moderate | 7 (7.5%) |
| Severe | 19 (20.4%) |
| Uncharacterized 1 | 13 (14.0%) |
| CFTR genotypes | n (%) |
| F508del homozygotes | 45 (48.4%) |
| F508del heterozygotes | 40 (43.0%) |
| Other 2 | 8 (8.6%) |
FEV1: Forced expiratory volume in 1 second; ESCF: Epidemiologic Study of Cystic Fibrosis. 1 Uncharacterized group consists of CF subjects less than 6 years old or no FEV1 (%) predicted values. 2 Other combinations of CFTR genotypes include: 394delTT/3272-26A>G, 711+5G>A/c.438_440delTCA, G551D/ R1070W, G542X/W1282X, G551D/R117H-7T, N1303K/R117H-7T, R117H-7T/R117H-7T and one unknown. CFTR mutations were defined according to the CFTR2 database or the Human Genome Variation Society (conventional name) nomenclature (www.hgvs.org) where available.
2.2. Association between Disease Severity Categories and Clinical Status
Pancreatic insufficient CF subjects had an increased risk of colonization with any form of P. aeruginosa (OR = 3.125, 95% CI: 1.192–8.190, p < 0.02) and colonization with mucoid P. aeruginosa (OR = 5.075, 95% CI: 1.307–28.620, p < 0.012) compared to the pancreatic sufficient group. Association between disease severity (defined by ESCF categories: Moderate and Severe; Mild and Normal/very mild) and pancreatic status, colonization with P. aeruginosa, or with mucoid P. aeruginosa was assessed using a Chi-Square or Fisher’s exact test. All 3 groups were associated with disease severity (p < 0.01). Pancreatic sufficient CF subjects were more likely to have Mild or Normal/mild disease severity than the pancreatic insufficient group (p < 0.01). CF subjects in the normal/very mild disease severity group were less likely to have mucoid P. aeruginosa colonization compared to CF subjects in the Moderate or Severe ESCF categories (p < 0.002) (Table 2).
Table 2.
Relationship of ESCF disease severity and clinical outcomes.
| Parameter | Status | ESCF Disease Severity Classification n (%) | Pancreatic Status n (%) | ||||
|---|---|---|---|---|---|---|---|
| Severe | Moderate | Mild | Normal/Very Mild | PI | PS | ||
| P. aeruginosa status *,§ | Positive | 15 (79) | 7 (100) | 5 (50) | 23 (52) | 45 (65) | 9 (38) |
| Mucoid P. aeruginosa status *,§ | Positive | 12 (63) | 5 (71) | 3 (30) | 12 (27) | 29 (42) | 3 (13) |
| Pancreatic status * | Pancreatic insufficient | 19 (100) | 6 (86) | 10 (100) | 28 (64) | -- | -- |
ESCF: Epidemiologic Study of Cystic Fibrosis; PI: Pancreatic insufficient; PS: Pancreatic sufficient; --: not applicable. * p < 0.01, Moderate or Severe versus Mild or Normal/very mild; § p < 0.05, PI versus PS.
2.3. Frequency Distribution of ABCC1 SNP rs504348 and Association with Clinical Status
Among the 93 CF probands, the observed frequencies for the rs504348 CC, CG, and GG genotypes were 8.6%, 25.8%, and 65.6%, respectively. The distribution was similar (p > 0.25) to those reported for the global population and Utah residents (rs504348 genotype distribution from the 1000 genome project can be retrieved from the Ensemble genomic browser at http://www.ensembl.org). Significant deviation from Hardy–Weinberg equilibrium (HWE) was observed in the global population (p < 0.0001), Utah residents (p < 0.03), and our study cohort (p < 0.03). The GG genotype was the most prevalent regardless of ESCF severity classification, pancreatic status, and colonization with P. aeruginosa; however, there was no association between any of the subject groups and rs504348 (C/G) genotypes (p > 0.05) (Table 3).
Table 3.
Frequency distribution of rs504348
| Parameter | Genotype | HWE p-Value | p-Value3 | ||
|---|---|---|---|---|---|
| CC | CG | GG | |||
| Populations 1 | n (%) | ||||
| Global population | 360 (14.4%) | 658 (26.3%) | 1486 (59.3%) | <0.0001 | -- |
| CEU | 6 (6.1%) | 22 (22.2%) | 71 (71.7%) | <0.03 | -- |
| Study cohort | 8 (8.6%) | 24 (25.8%) | 61 (65.6%) | <0.03 | -- |
| ESCF Disease Severity Classification | n (%) | >0.17 | |||
| Normal/very mild | 5 (5.4%) | 9 (9.7%) | 30 (32.3%) | -- | |
| Mild | 1 (1.1%) | 3 (3.2%) | 6 (6.5%) | -- | |
| Moderate | 0 (0%) | 1 (1.1%) | 6 (6.5%) | -- | |
| Severe | 2 (2.2%) | 3 (3.2%) | 14 (15.1%) | -- | |
| Uncharacterized 2 | 0 (0%) | 8 (8.6%) | 5 (5.4%) | -- | |
| Pancreatic Status | n (%) | >0.14 | |||
| Pancreatic insufficient | 5 (5.4%) | 15 (16.1%) | 49 (52.7%) | -- | |
| Pancreatic sufficient | 3 (3.2%) | 9 (9.7%) | 12 (12.9%) | -- | |
| P. aeruginosa | n (%) | >0.89 | |||
| Positive | 4 (4.3%) | 14 (15.1%) | 36 (38.7%) | -- | |
| Negative | 4 (4.3%) | 10 (10.8%) | 25 (26.9%) | -- | |
| Mucoid P. aeruginosa | n (%) | >0.31 | |||
| Positive | 4 (4.3%) | 10 (10.8%) | 18 (19.4%) | -- | |
| Negative | 4 (4.3%) | 14 (15.1%) | 43 (46.2%) | -- | |
HWE: Hardy–Weinberg equilibrium; ESCF: Epidemiologic Study of Cystic Fibrosis; --: not applicable. 1 Global population and CEU (Utah residents with Northern and Western European ancestry) adopted from 1000 Genomes Project (http://www.ensembl.org). 2 The uncharacterized group consisted of CF patients less than 6 years old or have no FEV1 (%) predicted values. 3 p-value estimated using Fisher’s Exact tests.
2.4. Analysis of Plasma-Induced ABCC1 mRNA Expression in CF
Induced ABCC1 mRNA expression in PBMC exposed to plasma from 23 CF subjects and 7 healthy controls were measured by RT-qPCR. The median and IQR age for CF subjects and healthy controls were 9.4 (6.7, 17.3) and 26.9 (7.0, 27.1) years, respectively. Just over half (56.5%) of the CF subjects were pancreatic insufficient. The ABCC1 SNP rs504348 was recorded in 78.3% of the CF subjects while 21.7% harbored the ancestral GG genotype (Table 4).
Table 4.
Clinical and demographic information for subjects included in expression analysis.
| Parameter | CF Subjects | Healthy Controls 1 |
|---|---|---|
| Samples, n | 23 | 7 |
| Age in years, median (IQR) | 9.4 (6.7, 17.3) | 26.9 (7.0, 27.1) |
| Pancreatic insufficient, n (%) | 13 (56.5%) | -- |
| ABCC1 genotypes | n (%) | |
| Ancestral GG | 5 (21.7%) | -- |
| rs504348 (CC/CG) | 18 (78.3%) | -- |
| CFTR genotypes 2 | n (%) | |
| F508del homozygotes | 11 (48.0%) | -- |
| F508del heterozygotes | 11 (48.0%) | -- |
| G542X/W1282X | 1 (4.0%) | -- |
1 All healthy control volunteers were unrelated and included for comparison with CF subjects. 2 CFTR mutations were defined according to the CFTR2 database or the Human Genome Variation Society (conventional name) nomenclature (www.hgvs.org) where available; --: not applicable.
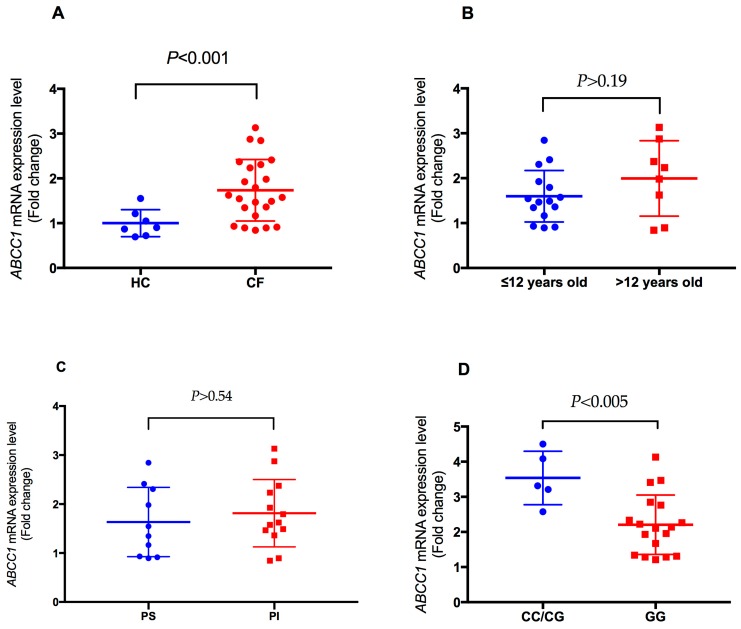
Plasma from CF subjects induced a significantly higher ABCC1 mRNA expression level compared to plasma from healthy controls (p < 0.001) (Figure 1A). To rule out any age effect on plasma-induced expression levels by CF subjects, we compared the expression levels between young (≤12 years of age) and old (>12 years of age) CF subjects. We found no significant difference in plasma-induced ABCC1 expression between the two groups (p > 0.19) (Figure 1B). Although plasma-induced expression was higher for pancreatic insufficient CF subjects than pancreatic sufficient CF subjects, the difference in expression levels was not significant (p > 0.54) (Figure 1C). However, plasma from CF subjects with rs504348 (CC/CG) induced significantly higher ABCC1 expression compared to plasma from subjects with the ancestral GG genotype (p < 0.005) (Figure 1D), which suggests a cis-expression quantitative trait loci (cis-eQTL) effect of rs504348 (Figure 1).
Figure 1.
Comparison of plasma-induced ABCC1 mRNA expression levels in PBMC. (A) Expression levels were higher for cystic fibrosis (CF) subjects than healthy controls (HC). No difference was observed between young (≤12 years of age) and old (>12 years of age) CF subjects (B) or between pancreatic insufficient (PI) and pancreatic sufficient (PS) CF patients (C). Significantly higher ABCC1 expression was induced with plasma from CF subjects with rs504348 (CC/CG) compared to homozygotes with the ancestral G allele (D). Significance estimated using t-tests.
2.5. ABCC1 Promoter Methylation
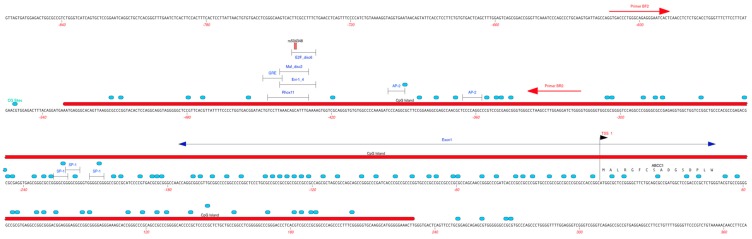
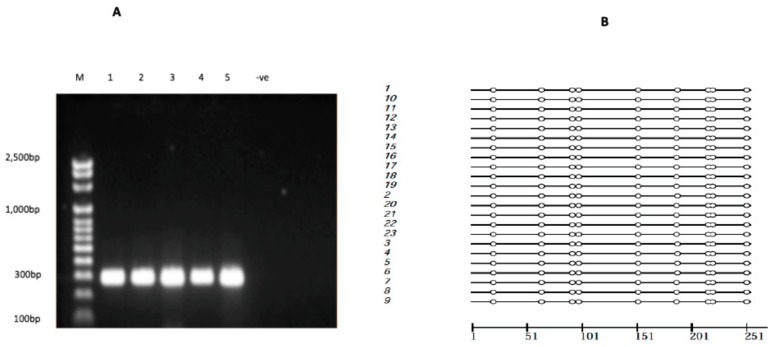
A 295-bp promoter region of ABCC1 (NG_028268.1) spanning −612 to −317 bp, containing a predicted CpG island (Figure 2), was amplified from 23 CF subjects using bisulfite PCR (Figure 3A) to determine the methylation status in peripheral blood tissue. Analysis of bisulfite sequence data indicated that the ABCC1 promoter was completely unmethylated in all CF subject samples analyzed (Figure 3B), indicating that promoter methylation may not be involved in regulating ABCC1 expression in peripheral blood of CF patients.
Figure 2.
Annotation of the ABCC1 promoter region targeted for methylation analysis. Red line, CpG island spanning −531 to +231 bp. BF2 and BR2 primers targeting CpG sites (blue circles) and regulatory factors (delimited lines), including the rs504348 (−435 bp) SNP were used for bisulfite PCR.
Figure 3.
ABCC1 promoter methylation analysis. (A) ABCC1 promoter bisulfite PCR products. Clear bands of ~295 bp shown in lanes 1–5 indicate successful bisulfite PCR. −ve represents unmodified human genomic DNA used as a negative control. (B) White cycles reported by Methtools represent unmethylated CpG sites [35].
2.6. Regulatory Effects of rs504348
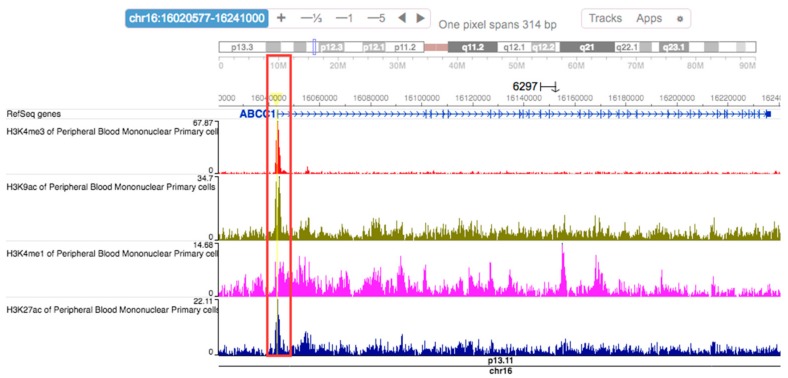
Computational predictions using HaploReg v4.1 [36] showed that the rs504348 SNP overlaps regulatory features of the ABCC1 promoter, including promoter histone marks, enhancer histone marks, DNAse 1 hypersensitive sites, transcription factor binding sites, and altered regulatory motifs in many tissues (Table 5). In PBMCs, a high cluster of ABCC1 promoter activity in the region spanning rs504348 was reported by HaploReg v4.1. Histone marks associated with active promoters (H3K4me3 and H3K9ac) and active enhancers (H3K4me1 and H3K27ac) were enriched in the region spanning rs504348, but no enrichment of repressive histone marks was observed in this region (Figure 4). Analysis using the portal for genotype-tissue expression project (GTEx Portal) and expression quantitative trait locus (eQTL) browser demonstrated that rs504348 (p = 4.2 × 10−9 and 2.1 × 10−4, respectively) modulates the expression of ABCC1 in whole blood, which supports the cis-eQTL effect of rs504348 (Table 5).
Table 5.
In silico predictions for rs504348 and variant (rs762775) in high linkage disequilibrium (LD).
| hg38 Chromosome (Position) | SNP | SNPs High LD (r2) | Promoter Histone Marks | Enhancer Histone Marks | DNAse | Proteins Bound | Motifs Changed | eQTL Hits |
|---|---|---|---|---|---|---|---|---|
| 16 (15949317) | rs504348 | 1 | 24 tissues, including PBMCs | Many tissues including PBMCs | 30 tissues | 8 bound proteins | 4 altered motifs | 2 hits 2 |
| 16 (15954300) | rs762775 1 | 0.62 | 13 tissues | 1 tissue | 2 altered motifs | 2 hits |
1 rs762775 was the only SNP detected to be in high (r2 > 0.6) linkage disequilibrium (LD) with rs504348. 2 The 2 eQTL hits are results from GTEx Portal and Blood eQTL. Both results show rs504348 significantly (p < 0.05) impacts ABCC1 expression.
Figure 4.
WashU Epigenome Browser schematic of histone marks enriched at the ABCC1 promoter SNP rs504348. Peaks at H3K4me3, H3K9ac, H3K4me1, and H3K27ac (highlighted in red box) indicate that these histone modifications contribute to the chromatin state assignment at rs504348 in PBMCs and validated results from computational predictions using HaploReg v4.1.
3. Discussion
In this study, we investigated the ABCC1 promoter SNP rs504348 by measuring plasma-induced ABCC1 mRNA expression levels, and rs504348 ABCC1 promoter methylation in CF subjects with differing CFTR genotypes. We correlated these molecular measures with clinical status to better understand the molecular mechanisms linking increased expression of ABCC1 and improved lung function observed in CF patients. Previous studies reported that the rs504348 polymorphism in the ABCC1 promoter has been reported to impact lung function [22,23], ABCC1 mRNA expression [37,38], and CF disease severity [23]. We found higher plasma-induced ABCC1 mRNA expression in CF subjects (Figure 1), but no significant association between rs504348 and clinical markers of CF disease severity (Table 3).
A prior study found that the rare CC genotype in CF is associated with a younger age at which FEV1 <60% predicted was first observed and a younger age of chronic infection with P. aeruginosa [23]. We analyzed the rs504348 genotypes and various markers of CF disease severity, such as ESCF classification [39,40], pancreatic status, and colonization with P. aeruginosa (Table 3), but found no significant association. Although we reported a similar rs504348 frequency distribution as the previous study, we also observed a significant deviation from HWE. We analyzed data from the 1000 Genome Project [41] but the results again showed a significant deviation from HWE in both the global population and Utah residents with Northern and Western European ancestry. One possible source for the observed disequilibrium could be the population stratification [42,43]. Our study cohort was not case-matched by genetic ancestry and although our study had a relatively small sample size, which may influence HWE estimates [44], even the larger sample sizes used for the 1000 Genome Project estimates showed significant deviations from HWE (Table 3).
Despite the limitation of small sample size in our study, we showed that CF subjects with CFTR genotypes associated with pancreatic insufficiency were more likely to be colonized with P. aeruginosa or the mucoid form of P. aeruginosa than those with genotypes associated with pancreatic sufficiency (Table 2). Pancreatic insufficiency or sufficiency is genetically determined [45,46], and the correlation between CFTR genotype and pancreatic phenotype is well established [47,48,49,50]. Pancreatic insufficient CF subjects harboring two severe mutations were more likely to have severe disease than pancreatic sufficient CF subjects harboring either two mild mutations or a combination of a severe and a mild mutation [45,46]. Although the association between CFTR genotypes and pancreatic disease is well characterized, the association between CFTR genotype and pulmonary phenotypes is less understood, perhaps due to variable fluctuations in lung function in CF patients [3,51].
FEV1 appears to be the most clinically useful measurement of lung function and the best available predictor of survival in patients with CF [4,51,52,53]. We adopted the ESCF classification to assign CF subjects into disease severity groups based on FEV1 percent predicted estimates and age [39,40], and found that the majority of CF subjects assigned to the Mild or Normal/mild severity group tested negative for mucoid P. aeruginosa and were pancreatic sufficient. Both pulmonary function tests and P. aeruginosa cultures were performed and recorded simultaneously during the same clinic visit. Chronic colonization of P. aeruginosa is associated with an accelerated decline of FEV1% percent predicted [54,55,56]. Our results may reflect current pulmonary status, but they may not predict future outcome. Similar to the results of previous studies [50], our results suggest that the severity classification of CFTR mutations that classically apply to pancreatic phenotypes may also apply to pulmonary phenotypes.
We observed higher expression of plasma-induced ABCC1 mRNA in CF subjects with rs504348 (CC/CG) compared to those with the ancestral GG genotype (Figure 1D). Others [22] have reported reduced transcriptional activity associated with the G allele of this polymorphism in four cell lines. The cis-eQTL effect of rs504348 has been validated in whole blood in previous genome-wide eQTL projects [37,38]. We showed that the distribution of rs504348 genotypes (Table 3) and expression levels (Figure 1D) were not significantly different between CF disease severity groups. Since ABCC1 expression analysis was measured at the mRNA level, our study may not describe the potential impact of the SNP on ABCC1 protein activity. Further study is warranted to assess the correlation between CF disease severity groups and ABCC1 protein expression or activity.
Our in silico analysis using HaploReg v4.1 reported a high cluster of ABCC1 promoter activity across various tissues (Table 5). Histone marks associated with active promoters (H3K4me3 and H3K9ac) and active enhancers (H3K4me1 and H3K27ac) [57] were enriched in the region spanning rs504348 in PBMCs (Figure 4). ABCC1 is ubiquitously expressed in several tissues [58], and no histone marks associated with inactive or methylated ABCC1 promoter in any tissue were reported by HaploReg v4.1 (Table 5). It is well established that the methylation of promoters is inversely proportional to their transcriptional activity [27,59,60]. As we observed no difference in ABCC1 promoter methylation status among CF subjects (Figure 3), it appears that DNA methylation may not be responsible for regulating ABCC1 expression in peripheral blood of CF patients. Although in a previous cancer study [28], no significant difference in ABCC1 promoter methylation was observed between normal and cancerous pancreatic tissues, whether DNA methylation regulates ABCC1 expression in tissues other than peripheral blood in CF remains unclear and warrants further study.
Plasma-induced transcriptional signatures are capable of characterizing CF disease severity [31]. We observed plasma-induced ABCC1 expression in PBMCs to be significantly higher with the plasma of CF subjects compared to that of healthy controls (Figure 1A). In CF, long-term treatment with azithromycin results in improved lung function [61,62,63]. The macrolide is suggested to be efficacious in CF via upregulation of ABCC1 [23,64]. Similarly, previous studies [15] suggested that the improved lung function observed in CF patients following treatment with antitumor drugs was due to upregulation of ABCC1. Since our study cohorts comprised of a clinically diverse group of CF patients undergoing standard CF care, we were unable to ascertain whether any medication was solely responsible for the differences in ABCC1 expression. It is worth noting that ABCC1 plays a crucial role in the efflux of several drugs conjugated with glutathione and other anions [22,58,65]. Polymorphisms in the glutathione S-transferase (GST) family of genes have been associated with CF disease severity [66,67]. Early death and more severe disease are typical for GSTM1-deleted CF patients [68]. An enzyme belonging to the GST family of enzymes produces glutathione adducts and helps the human body by forming a detoxification system against electrophilic compounds and oxidative stress [69]. Both CFTR (ABCC7) and MRP1 (ABCC1) are suggested to share this function of detoxifying natural or xenobiotic conjugates by exporting glutathione conjugates [15]. Hence, these genes are likely capable of complementing each other [15,23,64]. Further, since many drugs are good substrates for ABCC1, its overexpression results in multidrug resistance, especially in cancer [22,58]. CF patients are known to have a high prevalence of P. aeruginosa [55,70,71], which readily evolves resistance after long-term treatment with antibiotics [72]. Multidrug resistance isolates of P. aeruginosa [73,74], and other bacteria [75,76,77] are common in CF, making CF therapy challenging. Future studies to assess increased expression of ABCC1 in CF patients, and its association with multidrug resistance to antibiotics, may shed light on the molecular underpinnings of recurrent P. aeruginosa infection in CF.
In summary, we demonstrated that plasma-induced transcriptional signatures are useful tools for investigating candidate genes associated with CF. The higher ABCC1 mRNA levels induced by the plasma of CF patients may be related to the development of multidrug resistance to CF therapy; the increased activity of ABCC1 may complement diminished CFTR. Although we confirmed the eQTL effect of rs504348 on plasma-induced ABCC1 expression in PBMCs, there was no correlation between CF phenotypes and rs504348 genotypes. Histone modifications rather than DNA methylation may play a role in regulating ABCC1 expression. Further study to identify regulatory networks and impact of therapies on ABCC1 expression and role in CFTR activity may enhance our understanding of the clinical heterogeneity in CF.
4. Materials and Methods
4.1. Study Sample Characteristics
All CF subjects and healthy controls were recruited at the Children’s Hospital of Wisconsin (Milwaukee, WI, USA) and the Ann & Robert H. Lurie Children’s Hospital of Chicago (Chicago, IL, USA) with approval from the relevant Institutional Review Boards (IRB# CHW 07/72, CTSI 847 and 2015-400 respectively). Peripheral blood was drawn into citrate dextrose solution A or K+ ethylenediaminetetraacetic acid (EDTA) anticoagulant for each CF subject and plasma was isolated using Ficoll Histopaque (Sigma-Aldrich Corporation, St. Louis, MO, USA). Genomic DNA was extracted using the Puregene DNA Isolation kit (Gentra Systems Inc., Minneapolis, MN, USA) according to the manufacturer’s recommendations.
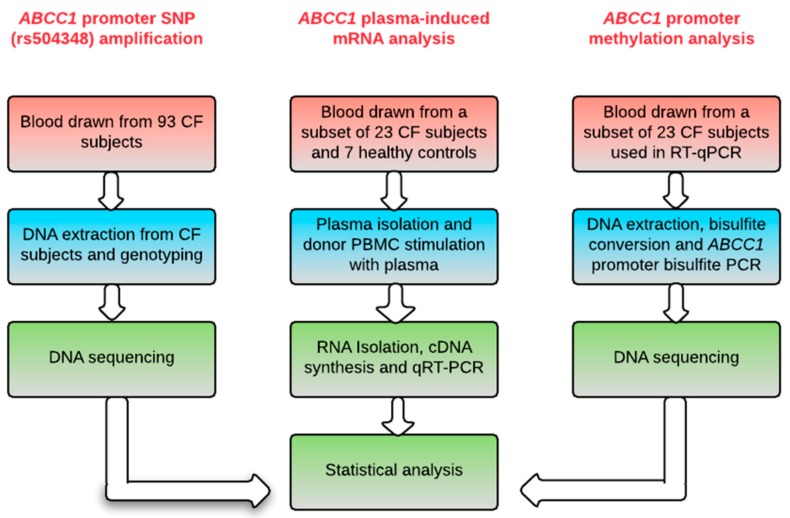
CF subjects were diagnosed based on the results of the pilocarpine iontophoresis sweat test (sweat chloride ≥ 60 mmol/L) and/or CFTR genotype, as previously described [31]. Demographic and clinical information, including pancreatic status, sweat chloride level, and P. aeruginosa infection status, was recorded for each CF subject. We adopted two definitions of CF disease severity categories in this study. First, the Epidemiologic Study of Cystic Fibrosis (ESCF) classification [39,40] was adopted to assign CF subjects into four severity groups based on measurements of FEV1 and age at recruitment. The four age groups/categories were: 6–12 years (severe, FEV1 ≤ 88.7% predicted; moderate, FEV1 > 88.7–94.5% predicted; mild, FEV1 > 94.5–99.0% predicted; very mild/normal, FEV1 > 99.0% predicted); 13–17 years (severe, FEV1 ≤ 76.5% predicted; moderate, FEV1 > 76.5–81.1% predicted; mild, FEV1 > 81.1–87.7% predicted; very mild/normal, FEV1 > 87.7% predicted); 18–29 years (severe, FEV1 ≤ 58.1% predicted; moderate, FEV1 > 58.1–63.9% predicted; mild, FEV1 > 63.9–70.7% predicted; very mild/normal, FEV1 > 70.7% predicted); and >30 years (severe, FEV1 ≤ 45.5% predicted; moderate, FEV1 > 45.5–50.9% predicted; mild, FEV1 > 50.9–59.8% predicted; very mild/normal, FEV1 > 59.8% predicted). The second definition of disease severity was based on the CF subject’s combination of CFTR class of mutations: the pancreatic insufficient group (severe disease) with CF subjects carrying functional mutations (class I, II, and III) [47] and a pancreatic sufficient (PS) group [47] (milder disease) with CF subjects who have a mild mutation (Class IV and V) [48,49]. We then assessed the association between disease severity groups and clinical status to determine whether our cohort data supported previous findings. A total of 93 CF subjects were genotyped at the ABCC1 SNP rs504348 locus. Plasma-induced mRNA expression levels were analyzed from a subset of the CF subjects and healthy controls, and bisulfite PCR was used to assess promoter methylation status (Figure 5).
Figure 5.
Schematic of experiment workflow. Peripheral blood collected from each CF subject was the primary source of genomic DNA and plasma. Genomic DNA extracted from 93 CF subjects was used for ABCC1 promoter SNP amplification. Commercial donor PBMCs from Cellular Technology Limited (Shaker Heights, OH, USA) (UPN 727) stimulated with plasma from a subset of 23 CF subjects and an additional 7 healthy control samples were used for RNA isolation and gene expression analysis via qRT-PCR. ABCC1 promoter methylation status of the subset of 23 CF samples used in qRT-PCR was determined using bisulfite PCR.
4.2. Genotyping
Genomic DNA extracted from 93 CF subjects were genotyped for ABCC1 promoter SNP rs504348. A previously described set of forward (ABCC1F: 5′-CAGGATGAAATGAGGGCACAG-3′) and reverse (ABCC1R: 5′-GAAGCGCCTGGGATCTTTGG-3′) primers [22] were used for PCR with the GeneAmp® PCR System 9700 (Applied Biosystems, Foster City, CA, USA). The 24 µL reaction mixture consisted of HotStarTaq Master Mix (Qiagen, Hilden, Germany), 5–15 ng/µL DNA, and 10 µM forward and reverse primers. Previously described [78] thermocycling conditions for the reaction were set: 1 min at 95 °C, followed by 35 cycles of 15 s at 95 °C, 15 s at 56 °C, 1.5 min at 72 °C, and a final cycle of 5 min at 72 °C.
4.3. PBMC Culture, Total RNA Isolation, and qRT-PCRRT-qPCR
Commercial cryopreserved PBMCs from a healthy Caucasian HLA-A2 male donor (UPN727) were acquired from the ePBMC donor library provided by Cellular Technology Limited (CTL; Shaker Heights, OH, USA). PBMCs were washed and thawed according to the manufacturer’s recommendations. PBMCs were cultured for 9 h at 37 °C in 5% CO2 with 20% autologous plasma, CF plasma, CF parent plasma, or healthy control plasma using protocols previously described [31]. Following culture, total RNA was isolated using TRIzol Reagent (Invitrogen Life Technologies, Waltham, MA, USA) in accordance with the manufacturer’s recommendations prior to quantification on a ND-1000 spectrophotometer (Nanodrop, Wilmington, DE, USA). First-strand cDNA was synthesized from 5 ng of the isolated total RNA using the iScript™ cDNA Synthesis Kit (BioRad, Hercules, CA, USA) according to the manufacturer’s recommendations. qRT-PCR was conducted using the ABI 7500 Fast Real-time PCR system (Applied Biosystems) with Fast SYBR Green Master Mix (Applied Biosystems) using previously published primers for ABCC1 mRNA [28]. The transcription of GAPDH was used as an internal control for normalization, and relative expression of the target gene for each sample was analyzed using the 2−ΔΔCt method [79].
4.4. Bisulfite PCR
EMBOSS Cpgplot (https://www.ebi.ac.uk/Tools/seqstats/emboss_cpgplot/) was used to predict CpG islands within the ABCC1 promoter, which became the primary targets for methylation analysis after bisulfite PCR amplification. Bisulfite primers targeting CpG islands in the promoter region were designed using MethPrimer [80]. Genomic DNA extracted from the subset of 23 CF subjects used for qRT-PCR were used for methylation analysis. Genomic DNA was first converted to bisulfite-modified DNA using the EZ DNA Methylation-Gold™ Kit (Zymo Research, CA, USA) according to the manufacturer’s recommendations. Double-round bisulfite PCR was performed using the BF2 forward primer (5′-GTGATTTTGGGTAGAGGGAATTATT-3′) and the BR2 reverse primer (5′-CCCAAATCCTCCAAAACTTAAA-3′) in a 24 µL reaction mixture consisting of HotStarTaq Master Mix (Qiagen), 2 µL bisulfite-modified DNA, and 10 µM BF2 and BR2 primers. The second-round of PCR was performed using 1 µL of PCR products from the first round. The following thermocycling conditions were used: 10 min at 94 °C, followed by 35 cycles of 30 s at 95 °C, 30 s at 60 °C, 30 s at 72 °C, and a final cycle of 7 min at 72 °C.
4.5. SNP Annotation Data Query
We performed in silico analysis to assess the interaction between rs504348 with regulatory features and expression to validate our findings of our study. Functional annotation was performed using the 15-state and 25-state chromHMM models in HaploReg v4.1 [36]. HaploReg v4.1 (http://archive.broadinstitute.org/mammals/haploreg/) uses SNP information from the 1000 Genomes Project to map known genetic variants to data derived from ENCODE and Roadmap Epigenomics, unveiling SNP effects on regulatory features. Histone marks reported to contribute to the chromatin-state assignment at rs504348 by HaploReg v4.1 were visualized using the WashU Epigenome Browser v42 [81]. The cis-eQTL effect of rs504348 was explored using the GTEx Portal (http://gtexportal.org/home/) and the Blood eQTL browser [37].
4.6. DNA Sequencing and Statistical Analysis
Purified PCR amplicons were sequenced using ABI 3730 (Applied Biosystems, CA, USA) by ACGT Inc. (Wheeling, IL, USA). Overall, statistical analysis was conducted using SAS 9.4 (SAS Institute, Cary, NC, USA). Deviation from Hardy–Weinberg equilibrium was tested with the χ2 test for rs504348. A Shapiro-Wilk test was used to examine the normality of the continuous variables. Median and IQR were used if the data were not normally distributed. As appropriate, a t-test with equal or unequal variance was used for comparison between two groups. A Chi-square test or Fisher’s exact test was used to investigate the association between the categorical variables. p < 0.05 was considered significant. Bisulfite DNA sequences were analyzed with MethTools 2.0 [35].
Acknowledgments
We gratefully acknowledge the patients who participated in this study. We thank the staff of Cystic Fibrosis Centers at The Ann & Robert H. Lurie Children’s Hospital of Chicago and Children’s Hospital of Wisconsin for their support and assistance during recruitment and sample collection. This work was supported by National Institutes of Health (DP2OD007031 to HL) and the Stanley Manne Children’s Research Fund (939001 to HL).
Author Contributions
Justin E. Ideozu, Xi Zhang, and Hara Levy designed and coordinated the study. Justin E. Ideozu and Xi Zhang performed experimental work. Justin E. Ideozu, Xi Zhang, Amy Pan, Pippa Simpson, and Hara Levy performed the statistical analysis. Zainub Ashrafi and Katherine J. Woods coordinated sample collection. Justin E. Ideozu, Xi Zhang, and Hara Levy drafted the manuscript. All authors contributed to writing and critically commented on the final version. All authors read and approved the final manuscript submitted.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Kerem B., Rommens J.M., Buchanan J.A., Markiewicz D., Cox T.K., Chakravarti A., Buchwald M., Tsui L.C. Identification of the cystic fibrosis gene: Genetic analysis. Science. 1989;245:1073–1080. doi: 10.1126/science.2570460. [DOI] [PubMed] [Google Scholar]
- 2.Xue R., Gu H., Qiu Y., Guo Y., Korteweg C., Huang J., Gu J. Expression of cystic fibrosis transmembrane conductance regulator in ganglia of human gastrointestinal tract. Sci. Rep. 2016;6:30926. doi: 10.1038/srep30926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Levy H., Farrell P.M. New challenges in the diagnosis and management of cystic fibrosis. J. Pediatr. 2015;166:1337–1341. doi: 10.1016/j.jpeds.2015.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Corvol H., Blackman S.M., Boelle P.Y., Gallins P.J., Pace R.G., Stonebraker J.R., Accurso F.J., Clement A., Collaco J.M., Dang H., et al. Genome-wide association meta-analysis identifies five modifier loci of lung disease severity in cystic fibrosis. Nat. Commun. 2015;6:8382. doi: 10.1038/ncomms9382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Drumm M.L., Konstan M.W., Schluchter M.D., Handler A., Pace R., Zou F., Zariwala M., Fargo D., Xu A., Dunn J.M., et al. Genetic modifiers of lung disease in cystic fibrosis. N. Engl. J. Med. 2005;353:1443–1453. doi: 10.1056/NEJMoa051469. [DOI] [PubMed] [Google Scholar]
- 6.Park J.E., Yung R., Stefanowicz D., Shumansky K., Akhabir L., Durie P.R., Corey M., Zielenski J., Dorfman R., Daley D., et al. Cystic fibrosis modifier genes related to pseudomonas aeruginosa infection. Genes Immun. 2011;12:370–377. doi: 10.1038/gene.2011.5. [DOI] [PubMed] [Google Scholar]
- 7.McKone E.F., Shao J., Frangolias D.D., Keener C.L., Shephard C.A., Farin F.M., Tonelli M.R., Pare P.D., Sandford A.J., Aitken M.L., et al. Variants in the glutamate-cysteine-ligase gene are associated with cystic fibrosis lung disease. Am. J. Respir. Crit. Care Med. 2006;174:415–419. doi: 10.1164/rccm.200508-1281OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hurbain I., Sermet-Gaudelus I., Vallee B., Feuillet M.N., Lenoir G., Bernaudin J.F., Edelman A., Fajac A. Evaluation of MRP 1–5 gene expression in cystic fibrosis patients homozygous for the delta f508 mutation. Pediatr. Res. 2003;54:627–634. doi: 10.1203/01.PDR.0000090926.16166.3F. [DOI] [PubMed] [Google Scholar]
- 9.Liu F., Zhang Z., Csanády L., Gadsby D.C., Chen J. Molecular structure of the human CFTR ion channel. Cell. 2017;169:85–95. doi: 10.1016/j.cell.2017.02.024. [DOI] [PubMed] [Google Scholar]
- 10.Vasiliou V., Vasiliou K., Nebert D.W. Human ATP-binding cassette (ABC) transporter family. Hum. Genom. 2009;3:281–290. doi: 10.1186/1479-7364-3-3-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dean M., Rzhetsky A., Allikmets R. The human ATP-binding cassette (ABC) transporter superfamily. Genome Res. 2001;11:1156–1166. doi: 10.1101/gr.GR-1649R. [DOI] [PubMed] [Google Scholar]
- 12.Schwiebert E.M., Benos D.J., Egan M.E., Stutts M.J., Guggino W.B. CFTR is a conductance regulator as well as a chloride channel. Physiol. Rev. 1999;79:S145–S166. doi: 10.1152/physrev.1999.79.1.S145. [DOI] [PubMed] [Google Scholar]
- 13.Akabas M.H. Cystic fibrosis transmembrane conductance regulator. Structure and function of an epithelial chloride channel. J. Biol. Chem. 2000;275:3729–3732. doi: 10.1074/jbc.275.6.3729. [DOI] [PubMed] [Google Scholar]
- 14.Hwang T.C., Kirk K.L. The CFTR ion channel: Gating, regulation, and anion permeation. Cold Spring Harb. Perspect. Med. 2013;3:a009498. doi: 10.1101/cshperspect.a009498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lallemand J.Y., Stoven V., Annereau J.P., Boucher J., Blanquet S., Barthe J., Lenoir G. Induction by antitumoral drugs of proteins that functionally complement CFTR: A novel therapy for cystic fibrosis? Lancet. 1997;350:711–712. doi: 10.1016/S0140-6736(05)63510-6. [DOI] [PubMed] [Google Scholar]
- 16.Linsdell P., Hanrahan J.W. Substrates of multidrug resistance-associated proteins block the cystic fibrosis transmembrane conductance regulator chloride channel. Br. J. Pharmacol. 1999;126:1471–1477. doi: 10.1038/sj.bjp.0702458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu Q., Center M.S. Cloning and sequence analysis of the promoter region of the MRP gene of HL60 cells isolated for resistance to Adriamycin. Cancer Res. 1994;54:4488–4492. [PubMed] [Google Scholar]
- 18.Cole S.P., Deeley R.G. Multidrug resistance-associated protein: Sequence correction. Science. 1993;260:879. doi: 10.1126/science.8098549. [DOI] [PubMed] [Google Scholar]
- 19.Kang X.-L., Zhang M., Wang K., Qiao X.-F., Chen M.-H. Molecular cloning, expression pattern of multidrug resistance associated protein 1 (MRP1, ABCC1) gene, and the synergistic effects of verapamil on toxicity of two insecticides in the bird cherry-oat aphid. Arch. Insect Biochem. Physiol. 2016;92:65–84. doi: 10.1002/arch.21334. [DOI] [PubMed] [Google Scholar]
- 20.Pradal U., Delmarco A., Morganti M., Cipolli M., Mini E., Cazzola G. Long-term azithromycin in cystic fibrosis: Another possible mechanism of action? J. Chemother. 2005;17:393–400. doi: 10.1179/joc.2005.17.4.393. [DOI] [PubMed] [Google Scholar]
- 21.Siedlinski M., Boezen H.M., Boer J.M., Smit H.A., Postma D.S. ABCC1 polymorphisms contribute to level and decline of lung function in two population-based cohorts. Pharmacogen. Genom. 2009;19:675–684. doi: 10.1097/FPC.0b013e32832f5eff. [DOI] [PubMed] [Google Scholar]
- 22.Wang Z., Wang B., Tang K., Lee E.J., Chong S.S., Lee C.G. A functional polymorphism within the MRP1 gene locus identified through its genomic signature of positive selection. Hum. Mol. Genet. 2005;14:2075–2087. doi: 10.1093/hmg/ddi212. [DOI] [PubMed] [Google Scholar]
- 23.Mafficini A., Ortombina M., Sermet-Gaudelius I., Lebecque P., Leal T., Iansa P., Reychler G., Dahan K., Pepermans X., Lenoir G., et al. Impact of polymorphism of multidrug resistance-associated protein 1 (ABCC1) gene on the severity of cystic fibrosis. J. Cyst. Fibros. 2011;10:228–233. doi: 10.1016/j.jcf.2010.10.007. [DOI] [PubMed] [Google Scholar]
- 24.Bell J.T., Pai A.A., Pickrell J.K., Gaffney D.J., Pique-Regi R., Degner J.F., Gilad Y., Pritchard J.K. DNA methylation patterns associate with genetic and gene expression variation in hapmap cell lines. Genome Biol. 2011;12:R10. doi: 10.1186/gb-2011-12-1-r10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 26.Illingworth R.S., Bird A.P. Cpg islands—‘A rough guide’. FEBS Lett. 2009;583:1713–1720. doi: 10.1016/j.febslet.2009.04.012. [DOI] [PubMed] [Google Scholar]
- 27.Yin Y., Morgunova E., Jolma A., Kaasinen E., Sahu B., Khund-Sayeed S., Das P.K., Kivioja T., Dave K., Zhong F., et al. Impact of cytosine methylation on DNA binding specificities of human transcription factors. Science. 2017;356:eaaj2239. doi: 10.1126/science.aaj2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen M., Xue X., Wang F., An Y., Tang D., Xu Y., Wang H., Yuan Z., Gao W., Wei J., et al. Expression and promoter methylation analysis of ATP-binding cassette genes in pancreatic cancer. Oncol. Rep. 2012;27:265–269. doi: 10.3892/or.2011.1475. [DOI] [PubMed] [Google Scholar]
- 29.Showe M.K., Vachani A., Kossenkov A.V., Yousef M., Nichols C., Nikonova E.V., Chang C., Kucharczuk J., Tran B., Wakeam E., et al. Gene expression profiles in peripheral blood mononuclear cells can distinguish patients with non-small cell lung cancer from patients with nonmalignant lung disease. Cancer Res. 2009;69:9202–9210. doi: 10.1158/0008-5472.CAN-09-1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bahr T.M., Hughes G.J., Armstrong M., Reisdorph R., Coldren C.D., Edwards M.G., Schnell C., Kedl R., LaFlamme D.J., Reisdorph N. Peripheral blood mononuclear cell gene expression in chronic obstructive pulmonary disease. Am. J. Respir. Cell Mol. Biol. 2013;49:316–323. doi: 10.1165/rcmb.2012-0230OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Levy H., Wang X., Kaldunski M., Jia S., Kramer J., Pavletich S.J., Reske M., Gessel T., Yassai M., Quasney M.W. Transcriptional signatures as a disease-specific and predictive inflammatory biomarker for type 1 diabetes. Genes Immun. 2012;13:593–604. doi: 10.1038/gene.2012.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pascual V., Allantaz F., Arce E., Punaro M., Banchereau J. Role of interleukin-1 (Il-1) in the pathogenesis of systemic onset juvenile idiopathic arthritis and clinical response to il-1 blockade. J. Exp. Med. 2005;201:1479–1486. doi: 10.1084/jem.20050473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaldunski M., Jia S., Geoffrey R., Basken J., Prosser S., Kansra S., Mordes J.P., Lernmark Å., Wang X., Hessner M.J. Identification of a serum-induced transcriptional signature associated with type 1 diabetes in the biobreeding rat. Diabetes. 2010;59:2375–2385. doi: 10.2337/db10-0372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gurram B., Salzman N.H., Kaldunski M.L., Jia S., Li B.U., Stephens M., Sood M.R., Hessner M.J. Plasma-induced signatures reveal an extracellular milieu possessing an immunoregulatory bias in treatment-naive paediatric inflammatory bowel disease. Clin. Exp. Immunol. 2016;184:36–49. doi: 10.1111/cei.12753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grunau C., Schattevoy R., Mache N., Rosenthal A. Methtools—A toolbox to visualize and analyze DNA methylation data. Nucleic Acids Res. 2000;28:1053–1058. doi: 10.1093/nar/28.5.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ward L.D., Kellis M. Haploreg: A resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40:D930–D934. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Westra H.J., Peters M.J., Esko T., Yaghootkar H., Schurmann C., Kettunen J., Christiansen M.W., Fairfax B.P., Schramm K., Powell J.E., et al. Systematic identification of trans eqtls as putative drivers of known disease associations. Nat. Genet. 2013;45:1238–1243. doi: 10.1038/ng.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lonsdale J., Thomas J., Salvatore M., Phillips R., Lo E., Shad S., Hasz R., Walters G., Garcia F., Young N., et al. The genotype-tissue expression (GTEx) project. Nat. Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morgan W.J., Butler S.M., Johnson C.A., Colin A.A., FitzSimmons S.C., Geller D.E., Konstan M.W., Light M.J., Rabin H.R., Regelmann W.E. Epidemiologic study of cystic fibrosis: Design and implementation of a prospective, multicenter, observational study of patients with cystic fibrosis in the us and canada. Pediatr. Pulmonol. 1999;28:231–241. doi: 10.1002/(SICI)1099-0496(199910)28:4<231::AID-PPUL1>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 40.Levy H., Murphy A., Zou F., Gerard C., Klanderman B., Schuemann B., Lazarus R., Garcia K.C., Celedon J.C., Drumm M., et al. Il1b polymorphisms modulate cystic fibrosis lung disease. Pediatr. Pulmonol. 2009;44:580–593. doi: 10.1002/ppul.21026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Consortium G.P. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cardon L.R., Palmer L.J. Population stratification and spurious allelic association. Lancet. 2003;361:598–604. doi: 10.1016/S0140-6736(03)12520-2. [DOI] [PubMed] [Google Scholar]
- 43.Gomes K.F.B., Santos A.S., Semzezem C., Correia M.R., Brito L.A., Ruiz M.O., Fukui R.T., Matioli S.R., Passos-Bueno M.R., Da Silva M.E.R. The influence of population stratification on genetic markers associated with type 1 diabetes. Sci. Rep. 2017;7:43513. doi: 10.1038/srep43513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hale M.L., Burg T.M., Steeves T.E. Sampling for microsatellite-based population genetic studies: 25 to 30 individuals per population is enough to accurately estimate allele frequencies. PLoS ONE. 2012;7:e45170. doi: 10.1371/journal.pone.0045170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kerem E., Corey M., Kerem B.S., Rommens J., Markiewicz D., Levison H., Tsui L.C., Durie P. The relation between genotype and phenotype in cystic fibrosis—Analysis of the most common mutation (delta F508) N. Engl. J. Med. 1990;323:1517–1522. doi: 10.1056/NEJM199011293232203. [DOI] [PubMed] [Google Scholar]
- 46.Gibson-Corley K.N., Meyerholz D.K., Engelhardt J.F. Pancreatic pathophysiology in cystic fibrosis. J. Pathol. 2016;238:311–320. doi: 10.1002/path.4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ooi C.Y., Dorfman R., Cipolli M., Gonska T., Castellani C., Keenan K., Freedman S.D., Zielenski J., Berthiaume Y., Corey M. Type of CFTR mutation determines risk of pancreatitis in patients with cystic fibrosis. Gastroenterology. 2011;140:153–161. doi: 10.1053/j.gastro.2010.09.046. [DOI] [PubMed] [Google Scholar]
- 48.Ahmed N., Corey M., Forstner G., Zielenski J., Tsui L., Ellis L., Tullis E., Durie P. Molecular consequences of cystic fibrosis transmembrane regulator (CFTR) gene mutations in the exocrine pancreas. Gut. 2003;52:1159–1164. doi: 10.1136/gut.52.8.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Walkowiak J., Herzig K.H., Witt M., Pogorzelski A., Piotrowski R., Barra E., Sobczynska-Tomaszewska A., Trawinska-Bartnicka M., Strzykala K., Cichy W. Analysis of exocrine pancreatic function in cystic fibrosis: One mild CFTR mutation does not exclude pancreatic insufficiency. Eur. J. Clin. Investig. 2001;31:796–801. doi: 10.1046/j.1365-2362.2001.00876.x. [DOI] [PubMed] [Google Scholar]
- 50.Cleveland R.H., Zurakowski D., Slattery D., Colin A.A. Cystic fibrosis genotype and assessing rates of decline in pulmonary status. Radiology. 2009;253:813–821. doi: 10.1148/radiol.2533090418. [DOI] [PubMed] [Google Scholar]
- 51.Rosenbluth D.B., Wilson K., Ferkol T., Schuster D.P. Lung function decline in cystic fibrosis patients and timing for lung transplantation referral. Chest. 2004;126:412–419. doi: 10.1378/chest.126.2.412. [DOI] [PubMed] [Google Scholar]
- 52.Schluchter M.D., Konstan M.W., Drumm M.L., Yankaskas J.R., Knowles M.R. Classifying severity of cystic fibrosis lung disease using longitudinal pulmonary function data. Am. J. Respir. Crit. Care Med. 2006;174:780–786. doi: 10.1164/rccm.200512-1919OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Taylor-Robinson D., Whitehead M., Diderichsen F., Olesen H.V., Pressler T., Smyth R.L., Diggle P. Understanding the natural progression in % FEV1 decline in patients with cystic fibrosis: A longitudinal study. Thorax. 2012;67:860–866. doi: 10.1136/thoraxjnl-2011-200953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Com G., Carroll J.L., Castro M.M., Tang X., Jambhekar S., Berlinski A. Predictors and outcome of low initial forced expiratory volume in 1 second measurement in children with cystic fibrosis. J. Pediatr. 2014;164:832–838. doi: 10.1016/j.jpeds.2013.11.064. [DOI] [PubMed] [Google Scholar]
- 55.Ahlgren H.G., Benedetti A., Landry J.S., Bernier J., Matouk E., Radzioch D., Lands L.C., Rousseau S., Nguyen D. Clinical outcomes associated with staphylococcus aureus and pseudomonas aeruginosa airway infections in adult cystic fibrosis patients. BMC Pulm. Med. 2015;15:67. doi: 10.1186/s12890-015-0062-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jarad N.A., Higgs S., Jeffcote T., Giles K. Factors associated with reduced FEV1 in adult patients with cystic fibrosis in a relatively affluent area. Chronic Respir. Dis. 2005;2:133–137. doi: 10.1191/1479972305cd065oa. [DOI] [PubMed] [Google Scholar]
- 57.Consortium E.P. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Słomka M., Sobalska-Kwapis M., Korycka-Machała M., Bartosz G., Dziadek J., Strapagiel D. Genetic variation of the ABC transporter gene ABCC1 (multidrug resistance protein 1–MRP1) in the Polish population. BMC Genet. 2015;16:114. doi: 10.1186/s12863-015-0271-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jones P.A., Takai D. The role of DNA methylation in mammalian epigenetics. Science. 2001;293:1068–1070. doi: 10.1126/science.1063852. [DOI] [PubMed] [Google Scholar]
- 60.Bird A.P. Cpg-rich islands and the function of DNA methylation. Nature. 1986;321:209–213. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- 61.Jaffe A., Francis J., Rosenthal M., Bush A. Long-term azithromycin may improve lung function in children with cystic fibrosis. Lancet. 1998;351:420. doi: 10.1016/S0140-6736(05)78360-4. [DOI] [PubMed] [Google Scholar]
- 62.Saiman L., Marshall B.C., Mayer-Hamblett N., Burns J.L., Quittner A.L., Cibene D.A., Coquillette S., Fieberg A.Y., Accurso F.J., Campbell P.W., III Azithromycin in patients with cystic fibrosis chronically infected with pseudomonas aeruginosa: A randomized controlled trial. JAMA. 2003;290:1749–1756. doi: 10.1001/jama.290.13.1749. [DOI] [PubMed] [Google Scholar]
- 63.Cai Y., Chai D., Wang R., Bai N., Liang B.B., Liu Y. Effectiveness and safety of macrolides in cystic fibrosis patients: A meta-analysis and systematic review. J. Antimicrob. Chemother. 2011;66:968–978. doi: 10.1093/jac/dkr040. [DOI] [PubMed] [Google Scholar]
- 64.Altschuler E.L. Azithromycin, the multidrug-resistant protein, and cystic fibrosis. Lancet. 1998;351:1286. doi: 10.1016/S0140-6736(05)79350-8. [DOI] [PubMed] [Google Scholar]
- 65.Borst P., Evers R., Kool M., Wijnholds J. The multidrug resistance protein family. Biochim. Biophys. Acta (BBA) Biomembr. 1999;1461:347–357. doi: 10.1016/S0005-2736(99)00167-4. [DOI] [PubMed] [Google Scholar]
- 66.Galli F., Battistoni A., Gambari R., Pompella A., Bragonzi A., Pilolli F., Iuliano L., Piroddi M., Dechecchi M.C., Cabrini G. Oxidative stress and antioxidant therapy in cystic fibrosis. Biochim. Biophys. Acta (BBA) Mol. Basis Dis. 2012;1822:690–713. doi: 10.1016/j.bbadis.2011.12.012. [DOI] [PubMed] [Google Scholar]
- 67.Strange R.C., Spiteri M.A., Ramachandran S., Fryer A.A. Glutathione-S-transferase family of enzymes. Mutat. Res. Fundam. Mol. Mech. Mutagen. 2001;482:21–26. doi: 10.1016/S0027-5107(01)00206-8. [DOI] [PubMed] [Google Scholar]
- 68.Baranov V., Ivaschenko T., Bakay B., Aseev M., Belotserkovskaya R., Baranova H., Malet P., Perriot J., Mouraire P., Baskakov V. Proportion of the GSTM 10/0 genotype in some slavic populations and its correlation with cystic fibrosis and some multifactorial diseases. Hum. Genet. 1996;97:516–520. doi: 10.1007/BF02267078. [DOI] [PubMed] [Google Scholar]
- 69.Bertuzzo C.S., Ribeiro A.F., Ribeiro J.D. Polymorphisms in the glutathione pathway modulate cystic fibrosis severity: A cross-sectional study. BMC Med. Genet. 2014;15:27. doi: 10.1186/1471-2350-15-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Davies J.C. Pseudomonas aeruginosa in cystic fibrosis: Pathogenesis and persistence. Paediatr. Respir. Rev. 2002;3:128–134. doi: 10.1016/S1526-0550(02)00003-3. [DOI] [PubMed] [Google Scholar]
- 71.Van Ewijk B., Wolfs T., Fleer A., Kimpen J., Van der Ent C. High pseudomonas aeruginosa acquisition rate in cf. Thorax. 2006;61:641–642. doi: 10.1136/thx.2006.062372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yum H.-K., Park I.-N., Shin B.-M., Choi S.-J. Recurrent pseudomonas aeruginosa infection in chronic lung diseases: Relapse or reinfection? Tuberc. Respir. Dis. 2014;77:172–177. doi: 10.4046/trd.2014.77.4.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Conway S.P., Brownlee K.G., Denton M., Peckham D.G. Antibiotic treatment of multidrug-resistant organisms in cystic fibrosis. Am. J. Respir. Med. 2003;2:321–332. doi: 10.1007/BF03256660. [DOI] [PubMed] [Google Scholar]
- 74.Ang J.Y., Abdel-Haq N., Zhu F., Thabit A.K., Nicolau D.P., Satlin M.J., van Duin D. Multidrug-resistant pseudomonas aeruginosa infection in a child with cystic fibrosis. Antimicrob. Agents Chemother. 2016;60:5627–5630. doi: 10.1128/AAC.00705-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cândido P.H.C., de Souza Nunes L., Marques E.A., Folescu T.W., Coelho F.S., de Moura V.C.N., da Silva M.G., Gomes K.M., da Silva Lourenço M.C., Aguiar F.S. Multidrug-resistant nontuberculous mycobacteria isolated from cystic fibrosis patients. J. Clin. Microbiol. 2014;52:2990–2997. doi: 10.1128/JCM.00549-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Van Dorn A. Multidrug-Resistant Mycobacterium Abscessus Threatens Patients with Cystic Fibrosis. Elsevier; Amsterdam, The Netherlands: 2016. [DOI] [PubMed] [Google Scholar]
- 77.Bryant J.M., Grogono D.M., Rodriguez-Rincon D., Everall I., Brown K.P., Moreno P., Verma D., Hill E., Drijkoningen J., Gilligan P. Emergence and spread of a human-transmissible multidrug-resistant nontuberculous mycobacterium. Science. 2016;354:751–757. doi: 10.1126/science.aaf8156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ideozu E.J., Whiteoak A.M., Tomlinson A.J., Robertson A., Delahay R.J., Hide G. High prevalence of trypanosomes in European badgers detected using its-PCR. Parasites Vectors. 2015;8:480. doi: 10.1186/s13071-015-1088-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(t)(-delta delta C) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 80.Li L.-C., Dahiya R. Methprimer: Designing primers for methylation PCRs. Bioinformatics. 2002;18:1427–1431. doi: 10.1093/bioinformatics/18.11.1427. [DOI] [PubMed] [Google Scholar]
- 81.Zhou X., Maricque B., Xie M.C., Li D.F., Sundaram V., Martin E.A., Koebbe B.C., Nielsen C., Hirst M., Farnham P., et al. The human epigenome browser at Washington university. Nat. Methods. 2011;8:989–990. doi: 10.1038/nmeth.1772. [DOI] [PMC free article] [PubMed] [Google Scholar]