Abstract
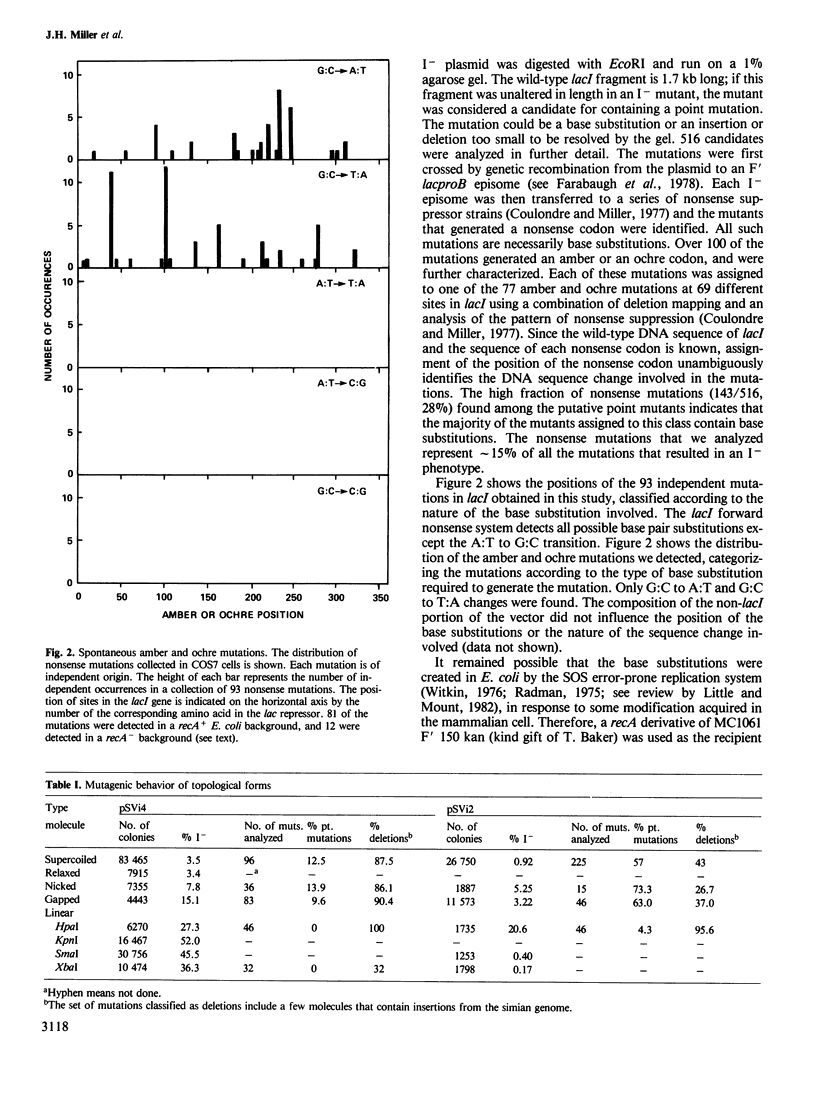
DNA transfected into mammalian cells is subject to the high mutation frequency of approximately 1% per gene. We present data bearing on the derivation of the two main classes of mutations detected, base substitutions and deletions. The DNA sequence change is reported for nearly 100 independent base substitution mutations that occurred in shuttle vectors as a result of passage in simian cells. All of the mutations occur at G:C base pairs and involve either transition to A:T or transversion to T:A. To identify possible mutational intermediates, various topological forms of the vector DNA were introduced separately. Supercoiled and relaxed DNA are mutated at equal frequencies. However, linearized DNA leads to a greatly elevated frequency of deletions. Nicked and gapped templates stimulate both deletions and base substitutions. We discuss a model involving intracellular degradation of the transfected DNA which explains these observations.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Calos M. P. DNA sequence for a low-level promoter of the lac repressor gene and an 'up' promoter mutation. Nature. 1978 Aug 24;274(5673):762–765. doi: 10.1038/274762a0. [DOI] [PubMed] [Google Scholar]
- Calos M. P., Lebkowski J. S., Botchan M. R. High mutation frequency in DNA transfected into mammalian cells. Proc Natl Acad Sci U S A. 1983 May;80(10):3015–3019. doi: 10.1073/pnas.80.10.3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clancy S., Mann C., Davis R. W., Calos M. P. Deletion of plasmid sequences during Saccharomyces cerevisiae transformation. J Bacteriol. 1984 Sep;159(3):1065–1067. doi: 10.1128/jb.159.3.1065-1067.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulondre C., Miller J. H. Genetic studies of the lac repressor. III. Additional correlation of mutational sites with specific amino acid residues. J Mol Biol. 1977 Dec 15;117(3):525–567. doi: 10.1016/0022-2836(77)90056-0. [DOI] [PubMed] [Google Scholar]
- Farabaugh P. J., Schmeissner U., Hofer M., Miller J. H. Genetic studies of the lac repressor. VII. On the molecular nature of spontaneous hotspots in the lacI gene of Escherichia coli. J Mol Biol. 1978 Dec 25;126(4):847–857. doi: 10.1016/0022-2836(78)90023-2. [DOI] [PubMed] [Google Scholar]
- Foster P. L., Eisenstadt E., Miller J. H. Base substitution mutations induced by metabolically activated aflatoxin B1. Proc Natl Acad Sci U S A. 1983 May;80(9):2695–2698. doi: 10.1073/pnas.80.9.2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gluzman Y. SV40-transformed simian cells support the replication of early SV40 mutants. Cell. 1981 Jan;23(1):175–182. doi: 10.1016/0092-8674(81)90282-8. [DOI] [PubMed] [Google Scholar]
- Greenfield L., Simpson L., Kaplan D. Conversion of closed circular DNA molecules to single-nicked molecules by digestion with DNAase I in the presence of ethidium bromide. Biochim Biophys Acta. 1975 Oct 15;407(3):365–375. doi: 10.1016/0005-2787(75)90104-5. [DOI] [PubMed] [Google Scholar]
- Helenius A., Kartenbeck J., Simons K., Fries E. On the entry of Semliki forest virus into BHK-21 cells. J Cell Biol. 1980 Feb;84(2):404–420. doi: 10.1083/jcb.84.2.404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967 Jun 14;26(2):365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A. Mutational specificity of depurination. Proc Natl Acad Sci U S A. 1984 Mar;81(5):1494–1498. doi: 10.1073/pnas.81.5.1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebkowski J. S., DuBridge R. B., Antell E. A., Greisen K. S., Calos M. P. Transfected DNA is mutated in monkey, mouse, and human cells. Mol Cell Biol. 1984 Oct;4(10):1951–1960. doi: 10.1128/mcb.4.10.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl T. DNA repair enzymes. Annu Rev Biochem. 1982;51:61–87. doi: 10.1146/annurev.bi.51.070182.000425. [DOI] [PubMed] [Google Scholar]
- Little J. W., Mount D. W. The SOS regulatory system of Escherichia coli. Cell. 1982 May;29(1):11–22. doi: 10.1016/0092-8674(82)90085-x. [DOI] [PubMed] [Google Scholar]
- Lusky M., Botchan M. Inhibition of SV40 replication in simian cells by specific pBR322 DNA sequences. Nature. 1981 Sep 3;293(5827):79–81. doi: 10.1038/293079a0. [DOI] [PubMed] [Google Scholar]
- McCutchan J. H., Pagano J. S. Enchancement of the infectivity of simian virus 40 deoxyribonucleic acid with diethylaminoethyl-dextran. J Natl Cancer Inst. 1968 Aug;41(2):351–357. [PubMed] [Google Scholar]
- Miller J. H. Carcinogens induce targeted mutations in Escherichia coli. Cell. 1982 Nov;31(1):5–7. doi: 10.1016/0092-8674(82)90398-1. [DOI] [PubMed] [Google Scholar]
- Miller J. H., Ganem D., Lu P., Schmitz A. Genetic studies of the lac repressor. I. Correlation of mutational sites with specific amino acid residues: construction of a colinear gene-protein map. J Mol Biol. 1977 Jan 15;109(2):275–298. doi: 10.1016/s0022-2836(77)80034-x. [DOI] [PubMed] [Google Scholar]
- Miller J. H., Low K. B. Specificity of mutagenesis resulting from the induction of the SOS system in the absence of mutagenic treatment. Cell. 1984 Jun;37(2):675–682. doi: 10.1016/0092-8674(84)90400-8. [DOI] [PubMed] [Google Scholar]
- Mulligan R. C., Berg P. Expression of a bacterial gene in mammalian cells. Science. 1980 Sep 19;209(4463):1422–1427. doi: 10.1126/science.6251549. [DOI] [PubMed] [Google Scholar]
- Razzaque A., Chakrabarti S., Joffee S., Seidman M. Mutagenesis of a shuttle vector plasmid in mammalian cells. Mol Cell Biol. 1984 Mar;4(3):435–441. doi: 10.1128/mcb.4.3.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razzaque A., Mizusawa H., Seidman M. M. Rearrangement and mutagenesis of a shuttle vector plasmid after passage in mammalian cells. Proc Natl Acad Sci U S A. 1983 May;80(10):3010–3014. doi: 10.1073/pnas.80.10.3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaaper R. M., Kunkel T. A., Loeb L. A. Infidelity of DNA synthesis associated with bypass of apurinic sites. Proc Natl Acad Sci U S A. 1983 Jan;80(2):487–491. doi: 10.1073/pnas.80.2.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmeissner U., Ganem D., Miller J. H. Genetic studies of the lac repressor. II. Fine structure deletion map of the lacI gene, and its correlation with the physical map. J Mol Biol. 1977 Jan 15;109(2):303–326. doi: 10.1016/s0022-2836(77)80036-3. [DOI] [PubMed] [Google Scholar]
- Shearman C. W., Loeb L. A. Effects of depurination on the fidelity of DNA synthesis. J Mol Biol. 1979 Feb 25;128(2):197–218. doi: 10.1016/0022-2836(79)90126-8. [DOI] [PubMed] [Google Scholar]
- Strauss B., Rabkin S., Sagher D., Moore P. The role of DNA polymerase in base substitution mutagenesis on non-instructional templates. Biochimie. 1982 Aug-Sep;64(8-9):829–838. doi: 10.1016/s0300-9084(82)80138-7. [DOI] [PubMed] [Google Scholar]
- Wake C. T., Gudewicz T., Porter T., White A., Wilson J. H. How damaged is the biologically active subpopulation of transfected DNA? Mol Cell Biol. 1984 Mar;4(3):387–398. doi: 10.1128/mcb.4.3.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witkin E. M. Ultraviolet mutagenesis and inducible DNA repair in Escherichia coli. Bacteriol Rev. 1976 Dec;40(4):869–907. doi: 10.1128/br.40.4.869-907.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Duve C., de Barsy T., Poole B., Trouet A., Tulkens P., Van Hoof F. Commentary. Lysosomotropic agents. Biochem Pharmacol. 1974 Sep 15;23(18):2495–2531. doi: 10.1016/0006-2952(74)90174-9. [DOI] [PubMed] [Google Scholar]


