Abstract
Self-assembling peptides programed by sequence design to form predefined nanostructures are useful for a variety of biomedical applications. However, assemblies of classic ionic self-complementary peptides are unstable in neutral pH, while charged peptide hydrogels have low mechanical strength. Here, we report on the rational design of a self-assembling peptide system with optimized charge distribution and density for bioscaffold development. Our designer peptides employs a sequence pattern that undergoes salt triggered self-assembly into β-sheet rich cationic nanofibers in the full pH range (pH 0 to 14). Our peptides form nanofibrils in physiological condition at a minimum concentration that is significantly lower than has been reported for self-assembly of comparable peptides. The robust fiber-forming ability of our peptides results in the rapid formation of hydrogels in physiological conditions with strong mechanical strength. Moreover, fiber structure is maintained even upon dense conjugation with a model bioactive cargo OVA257–264 peptide. Nanofibers carrying OVA257–264 significantly enhanced CD8+ T cell activation in vitro. Subcutaneous immunization of our peptide fiber vaccine also elicited robust CD8+ T cell activation and proliferation in vivo. Our self-assembling peptides are expected to provide a versatile platform to construct diverse biomaterials.
Keywords: self-assembly, peptide, nanofiber, hydrogel, scaffold
Graphical abstract
1. Introduction
Synthetic peptides and their derivatives can be programed to self-assemble into predefined supramolecular nanostructures of varying complexity such as fibers or tapes, vesicles, tubes, webs and sheets [1–5]. The naturally biodegradable self-assembling peptide systems are extensively investigated as biomaterials for tissue engineering, drug delivery, and vaccination [6, 7]. The versatility of these biomaterials relies on the precise and flexible design of unit peptide building blocks and often requires specific conditions for self-assembly and stability. A majority of reported self-assembled peptides adopt a β-sheet secondary structure and form into long-range ordered nanofibers, which at high concentrations can produce hydrogels [6]. Amongst these, RADA16-I is a classic self-assembling β-sheet peptide and is commercially available (PuraMatrix™) for biomedical research and pre-clinical applications [6, 8]. RADA16-I spontaneously forms fibers in aqueous solutions by using a well-known structural motif of ionic self-complementarity [6, 9, 10]. Previous reports showed that the net neutral RADA16-I self-assembles at low pH into nanofibers that become unstable at physiological conditions [9–11]. Similarly, peptide P11-II also employs ionic self-complementarity for self-assembly, but gelation occurs slowly over 7 days to form a hydrogel at pH < 5 yet loses structural integrity above this pH [12].
Self-assembling peptides that are not ionic self-complementary, such as β-hairpin and multidomain peptides. have typically produced hydrogels that show low mechanical strength in physiological condition [13, 14]. Furthermore, the capacity of self-assembling peptides to undergo in situ hydrogelation is important for many biomedical applications such as drug delivery [8]. For example, an amphiphilc peptide conjugated with an anti-inflammatory drug resulted in hydrogels that could sustain drug release for over a month [15]. However, the pure conjugate alone had relatively weak fiber forming ability and was incapable of forming long-range nanofibers or hydrogels, which resulted in an overall low drug loading. Accordingly, expanding the repertoire of self-assembling peptides to include sequences that form robust fibers in physiological conditions, but also exhibit broad pH adaptability and produce hydrogels at low concentrations with high mechanical strength, may open many potential applications.
While most reported biomedical applications of self-assembled peptides involve drug delivery or tissue engineering, there is an emerging interest for their use as vaccine carriers and scaffolds. Peptide Q11 nanofibers containing B and T cell antigens were observed to elicit strong antibody responses in mice without the need for co-administering adjuvants although Q11 alone was non-immunogenic [16]. This self-adjuvanting effect was also found for antigens presented on KFE8 nanofibers, indicating that the structure and not the sequence of the self-assembling peptide conferred adjuvant properties [17]. Q11 nanofibers that carry antigenic epitopes have been proposed to resemble the fibrillar morphology formed by bacterial curli and flagellin proteins, which may be a reason for the acquired adjuvant effect [18]. Cytotoxic T lymphocyte (CTL) responses have also been elicited without additional adjuvants by self-assembling peptides that forms cylindrical micelles or nanofibers [19–21]. In addition to the potential self-adjuvanting effect attributed to specific supramolecular structures, self-assembling peptides can serve as a scaffold to enhance the potency of subunit vaccines [22, 23]. Consequently, supramolecular nanostructures formed from self-assembled peptides are emerging as a useful platform to present antigens to the immune system.
Here we report on the rational design and synthesis of a self-assembling peptide system based on a novel sequence pattern with optimized charge distribution and density tailored to generate biomaterials with specific targeted properties. First, we designed our self-assembling peptide sequences to have a strong ability to form nanofibers in physiological condition so that the fiber structure would be stable even at a very low concentration after in vivo injection. Second, we prioritized sequences with a high capacity for conjugating bioactive cargo while maintaining the fiber structure so as to enable efficient bioactive molecule presentation or drug delivery. Third, we selected peptides that exhibit good solubility, which is preferred for self-assembly and delivery of cargo since many potent epitopes and drugs are hydrophobic. Finally, we wanted triggered self-assembly behavior for easier sample handling and purification and for in situ self-assembly. We expect the versatility and robust performance of our peptide sequence pattern will have broad biomedical applications.
2. Materials and Methods
2.1. Peptide Synthesis and Purification
All peptides were synthesized at a 0.1 mmol scale with a CEM Liberty Blue automated microwave peptide synthesizer using standard Fmoc chemistry. Preloaded Wang resins (Novabiochem) were used to generate unprotected C-terminal peptides and Rink Amide MBHA resins (Novabiochem) were used for peptides with a C-terminal amide. 5 equiv. of standard Fmoc amino acids (Chempep), 5 equiv. of N,N′-Diisopropylcarbodiimide (Sigma), and 5 equiv. of Ethyl (hydroxyimino)cyanoacetate (Sigma) were used for coupling and 20% (by volume) piperidine in DMF (Sigma) was used for deprotection. The cleavage of peptides from the resin was accomplished with an Accent peptide cleavage system (CEM) in the cleavage cocktail (trifluoroacetic acid (TFA)/triisopropylsilane/2,2′-(Ethylenedioxy)diethanethoil/water (9.25:0.25:0.25:0.25 by volume)) for 30 min. After filtration, peptides were collected by the addition of cold diethyl ether and centrifugation. The crude peptides with the purity lower than 90% and that were used for biological study were purified by semi-preparative high performance liquid chromatography (HPLC) using a Prominence LC20AD HPLC (Shimadzu) with a Phenomenex Gemini C18 column (250 × 10 mm), eluting with water-acetonitrile (0.1% TFA) gradients. Analytical HPLC was performed with a Phenomenex Kinetex C18 column (250 × 4.6 mm). Crude and purified peptides were analyzed by analytical HPLC and matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometer (MS) (Bruker AutoFlex II), and results were listed in Table S1, Figure S1 and Figure S2.
2.2. Transmission Electron microscopy (TEM)
Peptides were dissolved in Milli-Q water at a concentration of 2 mM as stock solutions. Various solutions were used to dilute peptide stock solutions to generate peptide solutions to the final concentration in different conditions for TEM and circular dichroism (CD) tests. To make sure the fiber structure was stable in these conditions, the diluted peptide solutions were incubated overnight before TEM and CD test at room temperature. Negative stain specimens were placed onto carbon-coated copper grids, followed by water wash and 1% uranyl acetate staining. Then images were collected with FEI Tecnai TF20 TEM operating at 200 kV.
2.3. CD
CD spectra were gathered at room temperature on a Jasco J-720 spectropolarimeter equipped with 1 mm path length quartz cells (Starna). The scan speed is 1 nm/s and the bandwidth is 0.2 nm. Five-times scans were applied for averaging without smoothing. The obtained ellipticity in millidegrees was used to calculate mean residue ellipticity [θ].
2.4. Fourier Transform Infrared (FTIR)
FTIR spectra were recorded on a Bruker Vertex 70 FTIR spectrometer equipped with an attenuated total reflectance (ATR) sampling accessory at room temperature. 3 μL of 2 mM Z1 in D2O or deuterated phosphate-buffered saline (PBS) was placed on the diamond stage. Spectra represent averages of 16 scans. Spectra were analyzed using ORIGINPRO software (OriginLab, Northampton, MA). Curve fittings of amide I band were performed to estimate the components of secondary structures and corresponding percentages based on peak positions determined by second derivatives as calculated with Savitsky-Golay method (2nd degree, 50 points) [24–26].
2.5. Oscillatory Rheology
Rheology measurements were conducted using a TA AR-G2 rheometer with a 2 cm parallel plate geometry and 750 μm gap distance. Frequency sweep was measured at 0.1% strain, within linear viscoelastic (LVE) region, at frequencies ranging from 100 to 0.1 rad/s at 25 °C with a humidified chamber [27, 28]. On-site gelation was performed with 225 μL peptide in Milli-Q water and 25 μL 10× PBS to give the final 1× PBS environment except Z3 that could not form hydrogels in this way. Z3 gelation was performed using a well-developed method for ionic self-complementary peptides [9, 29]. Briefly, 250 μL Z3 in Milli-Q water was placed on the plate and then the geometry was lowered to have 750 μm gap distance. 500 μL PBS was then added around Z3 solution as bath solution to trigger gelation. The gelation process was very fast, but to make sure every sample had enough time to form hydrogel, all rheology measurements were performed after 1 h of the on-site gelation. The hydrogel demonstration in the inverted glass vials was also pictured after 1 h incubation at room temperature.
2.6. In Vitro OVA Presentation Assay
DC2.4 cells, a murine dendritic cell (DC) line [30] and B3Z T cells, a CD8+ T cell hybridoma cell line [31] that expressed β-galactosidase, specifically related to the presentation of SIINFEKL were kindly provided by Prof. James Lai (University of Washington) and cultured in RPMI 1640 supplemented with 10% fetal bovine serum, 2 mM L-glutamine, 100 U/mL penicillin, 100 μg/mL streptomycin, 55 μM 2-mercaptoethanol, 1× nonessential amino acids, and 10 mM HEPES. DC2.4 cells were seeded at 5×104 cells/well in flat-bottom 96-well cell culture plates and cultured with different OVA-containing formulations and controls (OVA+LPS (1 μg/mL) as the positive control and PBS as the negative control) for 24 h at 37°C in a 5% CO2 incubator. In each OVA-containing formulation, the equivalent concentration of OVA257–264 was 50 μM. Cells were washed three times with pre-warmed PBS. 5×104 B3Z T cells were then added to each well and co-cultured for 24 h in complete RPMI 1640. Culture media was then carefully removed. 150 μL Lysis buffer (0.15 mM chlorophenol red β-D-galactopyranoside, 9 mM MgCl2, 0.125% Nonidet P-40, 100 μM 2-mercaptoethanol in PBS) was added into each well. Plates were incubated at 37°C for 2 h and read at 570 nm using a Tecan plate reader.
2.7. Cytotoxicity Assay
DC2.4 cells were cultured in 96 well plates in the absence or presence of various concentrations (5 μM, 50 μM, or 500 μM) of OVA, Z1, or OVA–Z1. After two days in culture, cell viability was assessed using CellTiter-Blue® Cell Viability Assay (Promega) following manufacturer’s recommended procedures. Briefly, cells were incubated for four hours with 20 μL/well of CellTiter-Blue® Reagent and fluorescence was recorded at 560/590 nm. Cell viability was normalized to untreated group considered as 100%.
2.8. OT-I Adoptive Transfer and In Vivo Immunization
All procedures were approved by the University of Washington Institutional Animal Care and Use Committee. Splenocytes were collected from OT-I transgenic mice (8–10 weeks old, C57BL/6-Tg(TcraTcrb)1100Mjb/J) and red blood cells were lysed. Then, the cells were labelled with carboxyfluorescein succinimidyl ester (CFSE) (Life Technologies, Carlsbad, CA) as per manufacturer’s protocol, followed by adoptive transfer via tail vein injection (1.2 × 107 cells in 400 μl PBS) [32] into recipient mice (female C57BL/6J, 8–12 weeks old). After 24 hours, the recipient mice were injected subcutaneously in the flank with 50 μl of each OVA-containing formulation (25 nmol equivalent OVA257–264 per mouse) whereas control mice were injected with only PBS. Specifically, there was 25 nmol OVA in ‘OVA’, 25 nmol OVA and 10 μg LPS in ‘OVA+LPS’, 25 nmol OVA–Z1 in ‘OVA–Z1’, 25 nmol OVA and 25 nmol Z1 in ‘OVA+Z1’, and 25 nmol Z1 in ‘Z1’. 1, 3, or 5 days after immunization, spleens and inguinal lymph nodes (IGLN) were collected and processed for single-cell suspensions (two mice per treatment group were combined) as previously described with minor modification [33–35]. To accumulate intracellular cytokine, the cells were incubated in vitro for 5 hours with 3 hour-brefeldin A (BD Biosciences) treatment (5 μg/ml) at 37°C. Then the cells were stained for 20 minutes at room temperature with a Live/Dead Fixable stain kit (Life Technologies) and then resuspended in FACS buffer (1× sterile PBS supplemented with 1% (v/v) heat-inactivated FBS) to 106 cells/ml. Cells were blocked with anti-mouse Fcγ III/II receptor monoclonal antibody (clone 2.4G2; IgG2bκ), and stained with fluorescently conjugated anti-mouse monoclonal antibodies against CD8a (clone 53-6.7; IgG2aκ), CD69 (clone H1.2F3; IgG), or CD44 (clone IM7; IgG2bκ) (all from BioLegend, San Diego, CA). Stained cells were fixed in 2% paraformaldehyde and stored at 4°C overnight.
Next morning, cells were permeabilized with Perm/Wash™ (BD Biosciences) containing 5% rat serum (Sigma). Cells were then stained with fluorescently conjugated anti-mouse monoclonal antibody against IFN-γ (clone XMG1.2; IgG1κ) (BioLegend) for 30 minutes at room temperature in the dark. Stained cells were extensively washed and resuspended in FACS buffer. Isotype staining was performed to use as negative controls in flow cytometry analysis. Cells were analyzed by flow cytometry using a FACS-Canto II (BD Biosciences). Data were analyzed with FlowJo Software (Tree Star, Ashland, OR). Proliferation was measured by quantifying the percent change of CFSE+ CD8+ T cells relative to control mice that were not immunized (PBS) but still recipients of adoptive T cell transfer by tail vein. To observe significant differences between control and treatment groups, various statistical analysis methods were used. One-way ANOVA followed by a Kruskal-Wallis test and Dunn’s multiple comparison test (nonparametric) was used depending on experimental data sets. For all statistical methods, GraphPad PRISM (Version 5.04, La Jolla, CA) was used. Unless otherwise indicated, a p-value ≤ 0.05 was considered to be significant.
3. Results
3.1. Rational Design of Peptide Sequences that Self-assemble into Charged Nanofibers
Peptide self-assembly mediated by intermolecular non-covalent interactions can be precisely controlled by sequence design. Here, we develop a new sequence pattern based on a four amino acid repeating unit of alternating charged (+ or –), polar uncharged (±) and nonpolar residues (o) that self-assembles into cationic or anionic peptide fibers (Table 1). While hydrophobic interactions and intermolecular hydrogen bonds are favored for self-assembly as a result of entropic effects, the role of charged interactions in the self-assembly process are more complex to predict and can hinder the formation of charged supramolecular structures [36]. For example, intramolecular electrostatic attractions may induce conformations that are unfavorable for certain secondary structures that promote self-assembly [9, 10]. Our pattern that is not ionic self-complementary uses polar uncharged residues to enable incorporation of either negatively or positively charged amino acids without compromising β-sheets structure. Peptides based on our sequence exhibit an optimized net charge at neutral pH, confer good solubility and self-assemble into long-range ordered fiber nanostructures. A number of design rules were implemented to promote robust self-assembly of our peptide nanofibers. First, we included either positively or negatively charged residues as the charged components in the classic pattern of alternating hydrophobic and hydrophilic residues. Second, polar uncharged residues were placed between charged residues to maintain charge separation. Finally, acylation of the N-terminal and an unprotected C-terminal were incorporated to reduce the overall net charge on the molecule.
Table 1.
Sequences and patterns of the self-assembling peptides used in this research.
| Short Name | Sequence | Pattern | Net Charge at pH 7 |
|---|---|---|---|
| Z1a | Ac-RVQVRVQVRVQV-COOH | + o ± o + o ± o + o ± o | +2 |
| Z5a | Ac-KVQVKVQVKVQV-COOH | + o ± o + o ± o + o ± o | +2 |
| Z7a | Ac-RVNVRVNVRVNV-COOH | + o ± o + o ± o + o ± o | +2 |
| Z8a | Ac-RIQIRIQIRIQI-COOH | + o ± o + o ± o + o ± o | +2 |
| Z4a | NH2-QVEVQVEVQVEV-Am | ± o − o ± o − o ± o − o | −2 |
| Z3 | Ac-RVEVRVEVRVEV-Am | + o – o + o − o + o – o | 0 |
| Z9 | Ac-RVRVRVRVRVRV-COOH | + o + o + o + o + o + o | +5 |
| Z2 | Ac-RVQVRVQVRVQV-Am | + o ± o + o ± o + o ± o | +3 |
Residue charge: +, positive; −, negative; ±, polar uncharged; and o, hydrophobic. Ac: acetylation; Am: amidation.
Peptides with designed sequence pattern based on a four amino acid repeating unit of alternating charged (+ or −), polar uncharged (±) and hydrophobic residues (o).
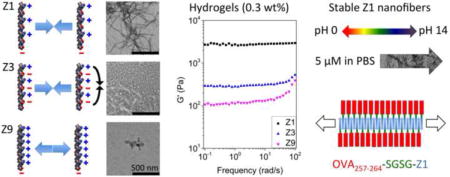
To test conditions in which these design rules were valid, we synthesized a series of peptides that could self-assemble into cationic or anionic nanofibers with different properties (Table 1). Using a core sequence pattern represented by the Z1 peptide that consists of the (RVQV) repeating unit, we observed that nanofibers were unable to form in water at a dilute concentration (100 μM, pH 4.5) (Figure 1a), and a representative TEM image of the sample detected fibers but at a relatively low density even at a high concentration (2 mM) (Figure S3a). However, in PBS (pH 7.4) Z1 assembled into long and unbranched nanofibers with a diameter of ~4 nm (Figure 1b). Salt-triggered self-assembly was also confirmed with CD and FTIR spectroscopy. The CD spectra indicated a mixed secondary structure for Z1 in water but the introduction of PBS induced a conformational change to a β-sheet with a negative peak at 216 nm (Figure 2e) [5, 9, 10]. FTIR analysis also demonstrated this conformational change from a mixture of α-helix (41.6%) and β-sheet (52.0%) in D2O (Figure 2g) to a predominately β-sheet (90.5%) structure in PBS (Figure 2h) [24–26].
Figure 1.
Charge distribution and density of peptide molecules significantly influenced their self-assembly behavior and the fiber forming ability. The net charges (z) of peptide molecules in water or PBS are indicated. (a) Dilute concentrations of peptides in water (pH 4.5) (Z1, Z2, and Z9) that have high charge density could not self-assemble into nanofibers due to the strong electrostatic repulsion, whereas the ionic self-complementary peptide Z3 with lower charge density formed well-defined nanofibers. (b) By increasing ionic strength with PBS (pH 7.4), the electrostatic repulsion of peptides with varied charge density was weakened due to charge shielding effect. Z1 with relatively lower charge density formed densely distributed nanofiber networks, whereas the fiber forming ability of Z2 and Z9 with higher charge density was partly or completely abrogated. A fraction of Z3 aggregated to form particles in PBS due to the intramolecular electrostatic attraction. The concentration of peptide dilute solutions is 100 μM. Scale bars represent 500 nm. (c) The stronger fiber forming ability of Z1 in dilute solutions formed much stronger hydrogels at higher concentration in PBS.
Figure 2.
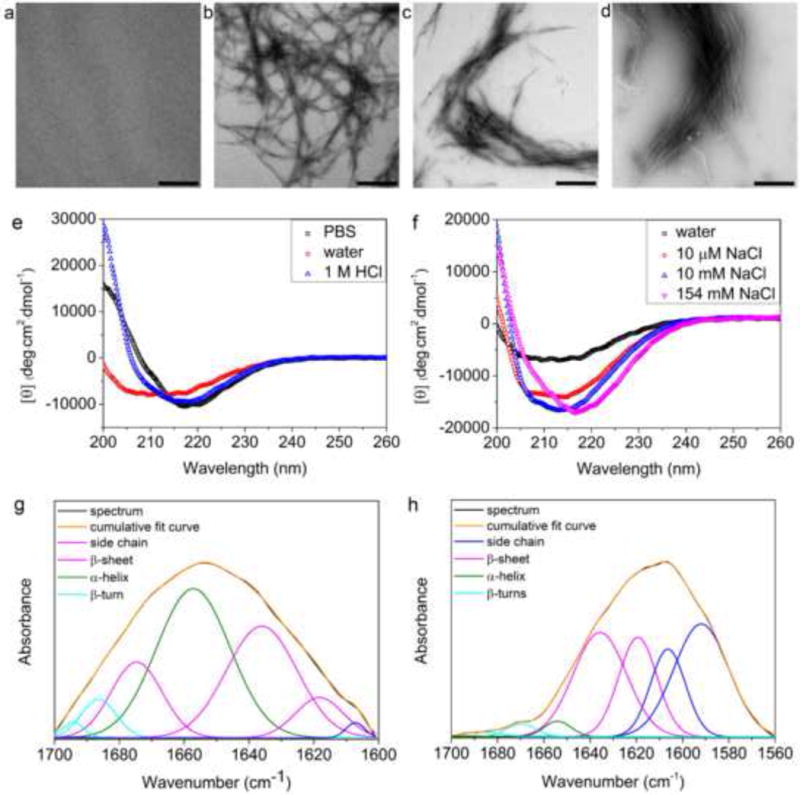
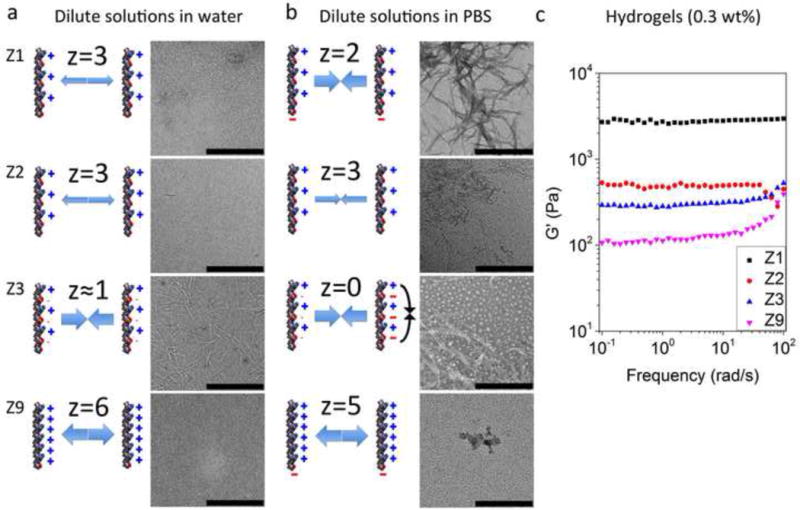
Salt-triggered self-assembly of Z1 characterized by TEM, CD and FTIR. TEM images of 100 μM Z1 in 10 μM NaCl pH 4.5 (a), 10 mM NaCl pH 4.5 (b), 1 M HCl pH 0 (c), and 1 M NaOH pH 14 (d) demonstrated the formation of nanofibers with salts. Scale bars represent 500 nm. CD spectra of 100 μM Z1 in water, PBS, or 1 M HCl (e) and in different concentrations of NaCl (f). FTIR analysis of the secondary structure of Z1 in D2O (g) and in PBS (prepared with D2O) (h). Curve fitting (Gaussian) was applied to decompose the experimental spectra (black) for quantitative analysis of secondary structures. (g) β-sheet (52.0%), α-helix (41.6%), β-turn (6.4%). (h) β-sheet (90.5%), α-helix (4.1%), β-turn (5.4%)[24–26].
We made amino acid substitutions to the Z1 core sequence pattern to test the robustness of fiber self-assembly. Replacing Arg of Z1 with Lys resulted in a peptide (Z5) that also exhibited salt-triggered self-assembly with PBS, suggesting this position is permissive to other positively charged residues (Figure S4a). Z1 with Arg is preferred over Z5 for the relatively higher β-sheet-forming propensity of Arg than Lys, which was supported with a wide shoulder at 208 nm (α-helix) in CD spectrum of Z5 (Figure S5a). Hydrophobic residues with bulky side chains also supported robust fiber formation. For instance, the Z8 peptide substitutes Val for Ile to generate the (RIQI)3 sequence that self-assembled into abundant nanofibers (Figure S4b). However, the Z1 peptide sequence is preferred over Z8 since Val favors β-sheet secondary structure and has greater solubility than Ile [37, 38]. When Gln in Z1 was replaced with Asn (Z7 peptide), we witnessed a significant morphological change to shorter fibers that remained rich in β-sheet secondary structure (Figure S4c and S5c). This substitution suggests that the size of the polar uncharged side chain could be used to influence fiber length. All of our cationic fiber sequences employed an unprotected C-terminal and a protected N-terminal to decrease the overall net charge on the peptide to +2. By applying the same design principles, we also generated anionic fibers. In this case, we designed Z4 by simply replacing the positively charged Arg in Z1 with the negatively charged Glu, and incorporating a protected C-terminus but an unprotected N-terminus. We also switched the position of the charged and polar uncharged residues. The resulting sequence Z4 has an overall net charge of −2 at neutral pH and readily self-assembled into anionic fibers rich in β-sheet (Figure S4d and Figure S5d). Accordingly, our sequence pattern exhibits flexibility to control properties of the resulting self-assembled materials such as charge, solubility, and fiber length.
We also synthesized a series of peptides that varied charge density from 0 to +5 (Z2, Z3, and Z9) to further probe the importance of our three design rules. First, we replaced the polar uncharged Gln in Z1 with an acidic Glu and amidated the C-terminus to generate the neutral Z3 peptide. This sequence exhibits the classic ionic self-complementary pattern widely used in other self-assembling peptide systems [2, 4, 16]. In water (pH 4.5), Z3 showed evidence of self-assembly into nanofibers but these fibers were unstable in PBS with the appearance of big shoulders at around 206 nm and 222 nm in CD spectra (α-helix). (Figure 1a, b), indicating the impact of electrostatic attractions on the fiber structure. Our observation is consistent with previous reports about the effect of pH on RADA16-I self-assembly [9–11], although considerable amount of nanofibers as well as β-sheet structure still existed for Z3 in PBS due to the high β-sheet forming propensity of Val compared with Ala. As predicted, Z3 self-assembly is favored under acidic conditions where Glu is predominately protonated and uncharged but the basic Arg is protonated and charged. The configuration of charges for Z3 under acidic conditions favors β-sheet structure (Figure S3c). Therefore, the pattern sequence of Z1 accomplishes both β-sheet structure at neutral pH while maintaining an overall +2 net charge. Next, we replaced all the hydrophilic residues of Z1 with a positively charged Arg, which resulted in a peptide with a net charge of +5 (Z9 peptide). In this case, the increased electrostatic repulsion created a high energetic barrier for self-assembly, and Z9 only formed fibers at a pH close to its isoelectric point (pH 13.2) or in high ionic strength (10× PBS) but not in normal PBS (Figure 1b and Figure S6). Finally, the Z2 peptide that differs from Z1 only by amidation of the peptide C-terminus, resulted in a net charge increase of +3 from +2, respectively. This increased net charge appeared to impede self-assembly of Z2 compared with Z1 and demonstrates the significance of the unprotected C-terminus in our design (Figure 1b). The superior fiber forming ability of Z1 with optimized charge density also manifested in hydrogels with significantly higher mechanical strength (Figure 1c).
We then investigated the affect of salt concentration on Z1 self-assembly. In 10 mM NaCl, Z1 exhibited densely distributed nanofibers whereas in 10 μM NaCl nanofibers in relatively low density were observed in the representative TEM image (Figure 2a, b). Correspondingly, a blue-shift in the β-sheet peak was observed for Z1 in low concentrations of NaCl (Figure 2f). When the NaCl concentration was increased to 154 mM (normal saline), more densely distributed fibers were observed (Figure S3b). The strong dependence of nanofiber formation on salt concentration is predicted from the charge shielding effect that arises in high ionic strength. We expect that self-assembly of the inherently charged Z1 peptide was abrogated in water due to electrostatic repulsion but addition of salt resulted in sufficient charge shielding to decrease the barrier for self-assembly and seed fiber formation. This hypothesis is further supported by the observation that fiber formation was impaired when using a Z1 peptide with a C-terminal amidation (Z2), which effectively increased the net charge of the peptide (Figure 1b).
3.2. Salt-Triggered Nanofibers are Ultrastable
Next, we looked at the effect of pH on self-assembly of our Z1 peptide sequence. A relatively low density of nanofibers was detected in 1 M HCl (pH 0) in the representative TEM image compared to neutral pH (Figure 2c). This result was expected due to the increased net charge of Z1 at pH 0 that arises from protonation of the C-terminal carboxyl group. The impaired fiber formation under this condition was also reflected in the CD spectra showing the appearance of a wide shoulder near 208 nm (α-helix) (Figure 2e). At pH 14, which is close to the isoelectric point of Z1 (pH 12.8), the decreased net charge was favorable for densely distributed fibers (Figure 2d). We think that the salt shielding effect in combination with the strong β-sheet forming tendency of our peptide was conducive for self-assembly in extreme pH. Accordingly, we demonstrate that Z1 self-assembles at a low concentration (100 μM) into nanofibers in the full pH range (pH 0 to 14) (Figure 2 and Figure S7).
Using PBS, we investigated the range of Z1 peptide concentrations that could support self-assembly. Z1 peptide concentrations as low as 5 μM resulted in β-sheet-rich nanofiber structures upon self-assembly at the final concentration rather than dilution from preformed fibers at high concentrations, but fibers appeared shorter and CD spectrum signal is weak as a result of the low concentration and indicates a decreased fraction of β-sheet content (Figure 3). This minimum concentration (5 μM) required for Z1 to form nanofibrils is much lower than the lowest reported concentration for self-assembly of other similar peptides (Table S2). PBS triggered assembly was also observed at 37 °C (Figure S8) and the fibers were stable after one month storage at room temperature or at 37 °C. In summary, our new peptide sequence pattern with optimized charge distribution and density represented by Z1 shows strong ability to form salt-triggered, β-sheet rich nanofibers that are stable within the full pH range and at extremely low concentrations.
Figure 3.
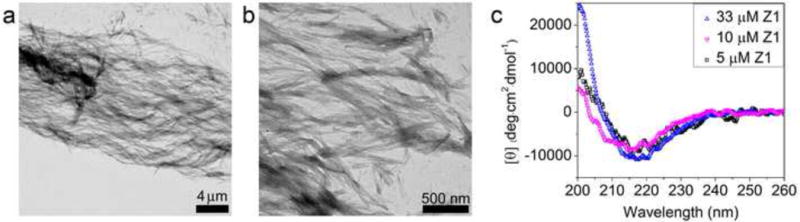
Z1 nanofibers and β-sheet conformation were detected in diluted solutions. (a) A representative large scale TEM image of Z1 in PBS at 5 μM. (b) Detailed nanofiber structure of Z1 in PBS at 5 μM at a high magnification. (c) CD spectra of Z1 in PBS at various concentrations.
3.3. Nanofibrils Form Hydrogels with High Mechanical Strength in Physiological Conditions
We hypothesized that the strong tendency of our Z1 peptide to self-assemble into nanofibers would also facilitate formation of hydrogels with strong mechanical properties. Z1 was prepared at 2 mM (~0.3 wt%) and the resulting gels were analyzed by oscillating frequency sweep rheometry. The measured storage modulus (G′) for Z1 hydrogels was at least one order of magnitude higher than the loss modulus (G″), both of which were independent of the frequency and indicate formation of rigid hydrogels (Figure 4a). The higher G′ of the hydrogels triggered by PBS or 0.1 M NaOH compared to normal saline (154 mM NaCl) may result from improved lateral association and entanglement of nanofibers caused by the pH increase. We also measured the effect of Z1 concentration on its storage modulus in PBS (Figure 4b). Z1 at a concentration of 1 wt% had a measured storage modulus of ~16 kPa, which makes it the strongest reported self-assembling peptide hydrogel in physiological conditions (Table S2).[9, 13, 14, 27] The high storage modulus of our Z1 hydrogels is also evidence of the strong tendency of Z1 to form fibers. We observed that G′ was constant within the sweep frequency from 0.1 to 100 rad/s when the concentration was equal to or higher than 0.05 wt%. However, hydrogelation of Z1 at 0.05wt% could not support its structure in an inverted glass vial, which required a minimum concentration of 0.1 wt% (Figure S9). Based on all current reports of self-assembling peptide hydrogels, 0.1 wt% appears to approach the lowest concentration for peptide fibers to form hydrogels [27, 29, 39]. The dependence of G′ on Z1 concentration scaled as G′~c1.42 (inset, Figure 4b), which is consistent with previously reported models for semiflexible macromolecular networks that are entangled rather than crosslinked [40]. For crosslinked polymer networks, a dependence stronger than G′~c2 is always observed [41, 42]. Therefore, the high storage modulus of our hydrogel is predominantly due to the strong fiber forming ability of Z1, which leads to increased fiber concentrations that can promote entanglement.
Figure 4.
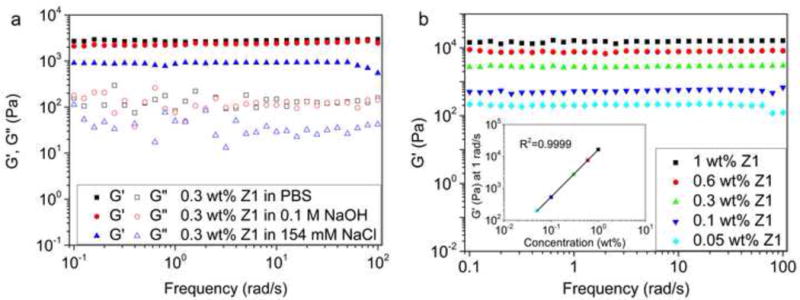
Z1 hydrogels formed upon salt-triggering had strong mechanical properties. (a) Frequency sweep of 0.3 wt% Z1 in PBS, 0.1 M NaOH, or 154 mM NaCl. (b) Frequency sweep of Z1 at different concentrations in PBS. The inset showed the dependence of the storage modulus at 1 rad/s on the concentration of Z1.
3.4. Nanofibers Displaying a CTL Epitope Enhances MHC-I Presentation In Vitro
Due to the strong fiber-forming ability of Z1, we tested its ability to serve as a scaffold for bioactive cargo conjugation. We conjugated a model OVA257–264 CTL epitope (SIINFEKL) to the N-terminus of Z1 via a hydrophilic spacer (SGSG), resulting in the self-assembling CTL epitope peptide OVA–Z1 (NH2-SIINFEKL-SGSG-RVQVRVQVRVQV-COOH). As with Z1 alone, OVA–Z1 in water produced rare fibers with an unordered conformation (Figure 5a, c). However, when triggered by PBS OVA–Z1 assembled into dense nanofiber networks with a conformational transition to β-sheets (Figure 5b, c). We detected a small peak around 206 nm, which is indicative of some unordered structure along the fiber showing the presence of the OVA257–264 peptide. The unordered cargo may sterically hinder the formation of β-sheet required for peptide self-assembly and fiber formation, which is evidenced by the mean residue ellipticity change at 216 nm between OVA–Z1 (Figure 5c) and Z1 (Figure 2e). OVA–Z1 could maintain fibrillar morphology and β-sheet conformation at 50 μM (Figure S10 and S11). The higher minimum fibrillization concentration of OVA–Z1 compared with Z1 is in consistent with the interrupted β-sheet conformation shown in CD spectra. In addition, OVA–Z1 could form a hydrogel in PBS but with a reduced storage modulus, indicating an excellent cargo loading capacity of Z1 scaffolds (Figure 5d). The weak hydrogel showed frequency-dependency of G′ at high frequency caused by network damage under high frequency shear stress. Fiber bundling may have an effect on the efficacy of displaying epitopes, which is not investigated in this work. In summary, fiber confirmation was detected for Z1 conjugated to OVA257–264 at equimole content at concentration as low as 50 μM.
Figure 5.
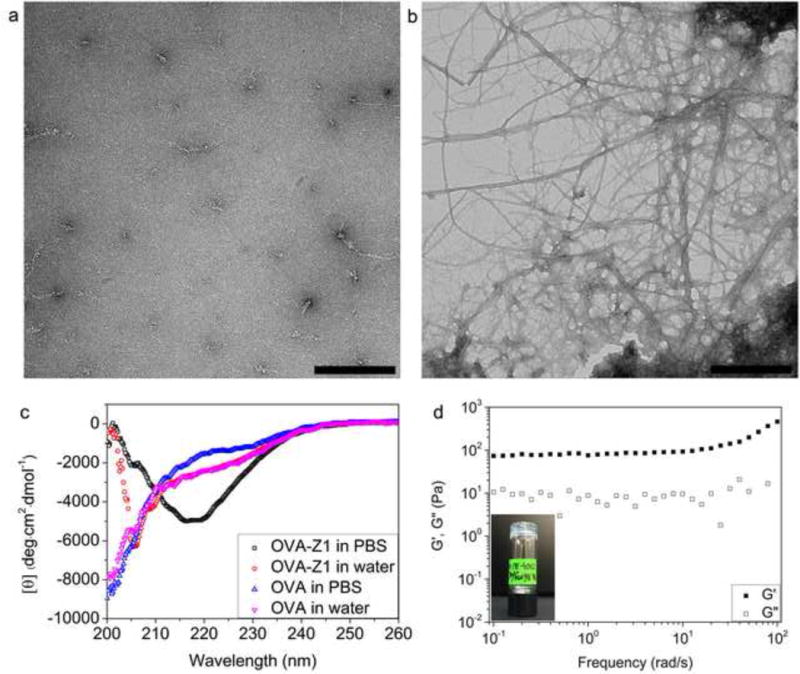
PBS triggered self-assembly of OVA–Z1. TEM images of 900 μM OVA–Z1 in water (a) and in PBS (b). Scale bars represent 500 nm. (c) CD spectra showing PBS triggered secondary structure transition of OVA–Z1 (500 μM) with OVA (500 μM) as the control. (d) PBS triggered hydrogel formation of 0.49 wt% OVA–Z1.
To determine the function of the CTL epitope displayed on nanofibers, we examined the capacity of antigen presenting cells loaded with OVA–Z1 fibers to activate B3Z CD8+ T cell as described previously [31]. A cytotoxicity assay confirmed that both OVA–Z1 and Z1 fibers had little to no toxicity on DC2.4 cells (Figure 6). The concentration of OVA–Z1 (50 μM) used to prime DC2.4 was high enough for fibrillar morphology (Figure S10b). We found that Z1 nanofibers without the CTL epitope did not activate B3Z CD8+ T cells (Figure 7a). Moreover, since no statistical improvement in antigen presentation was detected with a physical mixture of OVA257–264 and Z1 (OVA+Z1), we conclude that Z1 alone does not enhance antigen presentation. Notably, OVA–Z1 exhibited significant enhancement of OVA257–264 presentation on B3Z cells compared to soluble OVA257–264 peptide. This result supports that the OVA257–264 epitope on nanofibers was correctly processed and presented by DC2.4, and that the fibrillar epitope formulation activated DC2.4 comparable to the adjuvant lipopolysaccharides (LPS). Furthermore, the fact that the physical mixture of Z1 and OVA257–264 (OVA+Z1) failed to show this enhancement demonstrates the importance of conjugating the CTL epitopes onto the fibers. In summary, our Z1 pattern sequence accommodates high epitope loading without compromising self-assembly to form nanofibers that enhance the activation of B3Z CD8+ T cells in vitro.
Figure 6.
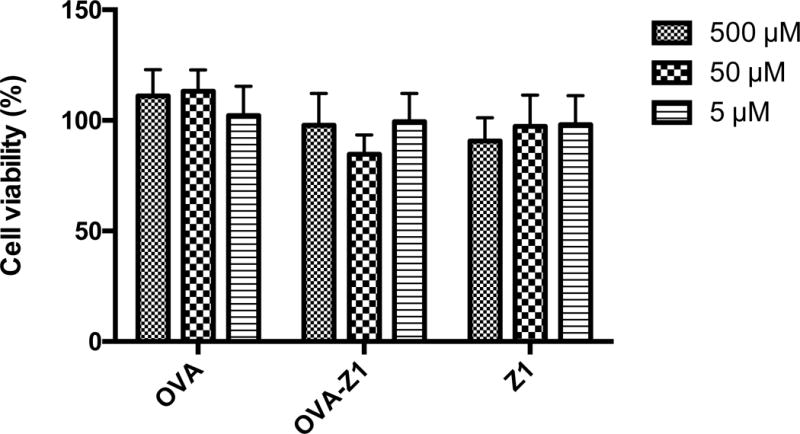
OVA-Z1 and Z1 fibers showed little to no cytotoxicity to DC2.4 at different concentrations. Data were presented as mean ± SD (n=6). Cell viability was normalized to untreated group considered as 100%.
Figure 7.
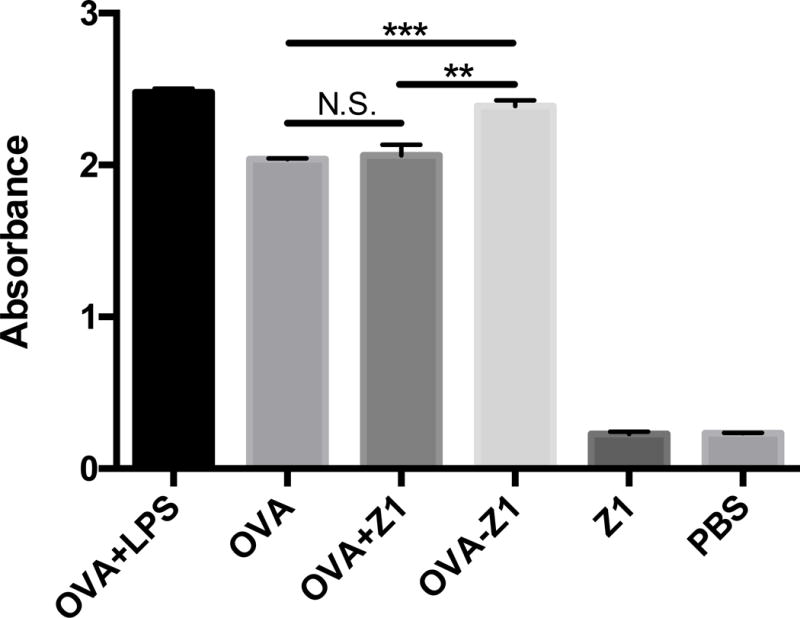
Conjugation of OVA257–264 to Z1 nanofibers enhanced MHC class I antigen presentation. (a) B3Z T cell activation detected based on β-galactosidase activity with the absorbance at 570 nm. The equivalent OVA concentrations of each group were the same (50 μM). The concentration of Z1 control was also 50 μM. The LPS concentration was 1 μg/mL. (b) The epitope density along nanofibers significantly affected B3Z activation. The three groups of OVA–Z1+Z1 had the same concentration of OVA–Z1 (50 μM), but varied concentrations of Z1. For both panels, B3Z activation data were presented as mean ± SD (n=3). Statistical analysis was conducted with student’s T test (*p<0.05, **p<0.01, ***p<0.001).
3.5. Subcutaneous Immunization of OVA–Z1 Elicits CD8+ T Cell Activation and Proliferation In Vivo
To further assess the ability of our OVA–Z1 nanofibers to elicit CTL responses, we adoptively transferred CFSE-labeled total splenocytes from OT-I transgenic mice into recipient naïve C57BL/6 mice one day prior to subcutaneous immunization with OVA–Z1 and then measured CD8+ T cell proliferation and activation. Due to rapid proliferation and dilution of donor CFSE+ cells, we compared proliferation of CFSE+ cells based on the frequencies of unproliferated CFSE+ cells as compared to the PBS control (Figure 7). Flow cytometry analysis of lymphocytes collected from IGLN showed significant dilution of CFSE-labeled CD8+ T cells relative to PBS for all treatment groups (Figure 8 and Figure S12). However, possibly due to high ratio of OVA dose to CFSE-labeled CD8+ T cells, the high dilution was unexpectedly observed for OVA257–264 treatment alone even after only 1 day. This high sensitivity of this model precluded our ability to observe an expected enhanced affect from adjuvanting OVA257–264 with LPS (OVA+LPS). Differently from the OVA peptide alone, OVA–Z1 delayed proliferations of CFSE+ CD8+ T cells over time for both IGLN and spleen. This delayed proliferation may reflect differences in diffusivity from the site of administration into systemic circulation of the antigenic peptide delivered from our conjugate compared to a soluble formulation.
Figure 8.
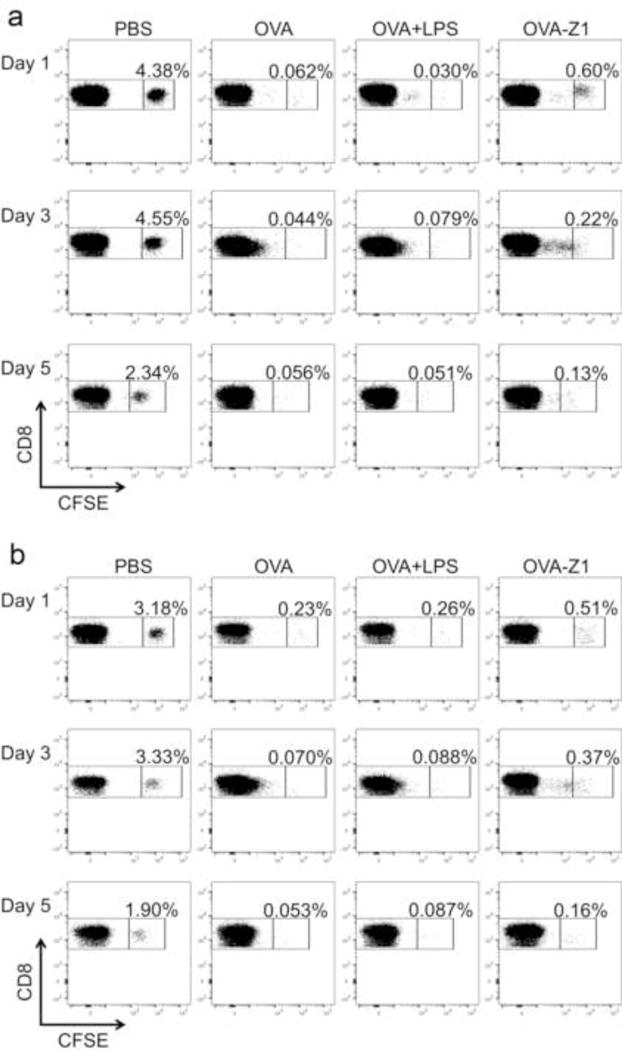
Immunization with peptide vaccines resulted in CD8+ T cell proliferation in vivo. The frequency of unproliferated CFSE labeled CD8+ cells was gated out in each plot as the evidence of proliferation of CFSE+ cells upon immunizations compared to PBS controls. PBS controls showed no CFSE dilution. OVA–Z1 treated groups showed delayed CD8+ T cell proliferation compared to OVA and OVA+LPS treated groups. Flow cytometry dot plots displaying the CFSE dilution of CD8+ cells from IGLN (a) and spleens (b). The frequency of unproliferated CFSE labeled CD8+ cells was gated out in each plot as the evidence of proliferation of CFSE+ cells upon immunizations compared to PBS controls. All PBS controls showed no CFSE dilution.
The quality of the in vivo CD8+ T cell response to OVA–Z1 immunization was also evaluated by measuring splenic CD8+ T cell expression of early stage activation (CD69+), effector-memory (CD44+) and pro-inflammatory and Th1 cytokine (IFN-γ) markers. The expression of activation markers on CD8+ splenocytes in OVA–Z1 treated mice were upregulated at levels comparable to OVA+LPS (Figure S13a and b). Moreover, IFN-γ production was largely augmented in CD8+ splenocytes and CD8+ CD44+ CD69+ splenocytes when treated with OVA–Z1 (Figure S13c and d). Therefore, delayed T cell proliferation does not necessarily indicate that OVA–Z1 is less immunogenic as compared to other controls and treatments. In summary, even though statistical significance was not reached with the limited time points and sample numbers in the study, subcutaneous immunization with OVA–Z1 elicits robust proliferation of IGLN and splenic CD8+ T lymphocytes that also show specific activations.
4. Discussion
Compared with other self-assembling peptides that have been widely studied, Z1 excels in both its fiber forming capacity in dilute solutions and the mechanical strength of its hydrogel (Table S2). Mechanical properties of tissue engineered scaffolds play a critical role in the regulation of mature cell behavior as well as stem cell differentiation pathways [43]. Therefore, self-assembling peptides that can form hydrogels with a wide range of mechanical strength tunable by concentration and gelation stimuli may serve as scaffolds for diverse cell types. The classic ionic self-complementary peptides, such as RADA16-I and P11-II, usually require acidic condition for self-assembly since electrostatic attraction at neutral pH impedes formation of β-sheets [9–12]. This impaired β-sheet formation at neutral pH might contribute to the relatively low G′ of the hydrogels formed by ionic self-complementary peptides [9, 28]. In contrast, the self-assembly of the classic cationic peptide (MAX8) requires basic pH due to strong electrostatic repulsion at acidic or neutral condition [13]. This strong electrostatic repulsion also significantly influenced the G′ of the cationic MAX8 peptide hydrogels (450 Pa at 0.5 wt% in 150 mM NaCl, pH 7.4) [13]. Similarly, the G′ of K2(QL)6K2 hydrogel (1 wt% in PBS) is reported to be 191 Pa, which is considerably weaker than hydrogels based on ionic self-complementary peptides [14]. Although, the Ac-LD6 peptide has been reported to form hydrogels with extremely high mechanical strength in water, at physiological ionic strength its storage modulus was significantly reduced to 1.5 kPa (1 wt% in 150 mM NaCl) [27]. We attribute the wide pH range for the self-assembly of Z1 to the distribution of charged residues. The use of only positively charged residues avoids interruption of β-sheet caused by opposite charges and maintains the low charge density compared with MAX8 and K2(QL)6K2. This configuration of charges in our Z1 peptide guarantees that electrostatic repulsion can be easily screened with modest ionic strength. In addition, fibers that form under low concentration have advantages in dilute environments, such as when used in vivo where there is a tendency for dilution by biological fluids, considering that fibrillization is reversible at low concentrations. Low fibrilization concentration may also be beneficial for delivering lower dose of toxic agents. The broad pH adaptability, self-assembly at low concentrations, and strong mechanical property of Z1 arose from our initial goal to make a peptide sequence that forms robust fibers in neutral pH. In summary, Z1 demonstrates excellent stability at low concentrations, broad pH adaptability, and produces hydrogels with high mechanical strength.
To evaluate the loading capacity of our Z1 peptide fibers for bioactive cargo, we conjugated a model CTL epitope (OVA257–264) to Z1 and obtained an equimole loading without disrupting fiber properties even at low concentrations. The B3Z T cell hybridoma has been engineered as a reporter cell line for detecting OVA257–264 antigen presentation by DC in the absence of costimulation or other activation signals [44]. The conjugated OVA–Z1 elicited significantly stronger CTL response in co-cultures of DC2.4 and B3Z T cells compared with either the soluble OVA257–264 peptide alone or the physical mixture of OVA+Z1 (Fig. 7a). Further study on DC activation profile would help identify the similarities and differences in B3Z activation state. Furthermore, OVA–Z1 immunized mice resulted in significant activation and dilution of CFSE-labeled CD8+ T cells, which supports the function of our nanofibers to present conjugated CTL epitopes for antigen processing in vivo (Fig. 8 and Fig. S12). The high sensitivity of this adoptive transfer model to OVA257–264 peptide was evidenced by a similar level of CD8+ T cell proliferation elicited by immunization with OVA257–264 peptide alone or with LPS. This rapid proliferation of CFSE labeled CD8+ T cells in response to OVA257–264 has been reported previously [45]. While we observe that OVA–Z1 shows a trend for increased expression of activation markers and cytokines compared to the other treatment groups, the differences are not statistically significant. In following studies, various OVA doses need to be examined with increased sample numbers to better understand immunological impacts of Z1. In addition, further investigation on long-term cellular and humoral immune responses in wild-type mice to our Z1 functionalized materials will be performed to better understand how our robust fibers as immune-functional scaffolds would benefit immunization, but these studies are beyond the scope of this report.
5. Conclusion
In conclusion, we rationally designed charged peptides employing a novel sequence pattern with optimized charge distribution and density that self-assembles into nanofiber biomaterials with targeted properties for in vivo applications. We show that peptide Z1 self-assembles into cationic nanofibers rich in β-sheet upon salt-triggering regardless of pH. The salt-triggering property of our self-assembled peptide system may facilitate precise and facile incorporation of multiple cargos with desired ratios for future development of more complex vaccines or for combination drug delivery. In addition, our cationic fibers have a potential to co-deliver negatively charged CpG adjuvants to further tailor the vaccine composition and resulting immunogenicity. As a result of the rationally designed sequence pattern, the strong fiber forming ability of our peptides guarantees retention of fiber structure and stability in physiological conditions at low concentrations, which is important for nanofiber vaccines and drug delivery systems. Also, the formation of dense fibers and the corresponding entanglements produced a strong hydrogel that can be useful for tissue engineering scaffolds. We expect our self-assembling peptide system will provide a platform to construct biomaterials for diverse applications.
Supplementary Material
Statement of Significance.
This work is an attempt of rational design of materials from molecular level for targeted properties and an exploration in molecular self-assembly. Current widely studied self-assembling peptides do not have stable nanofiber structures and form weak hydrogels under physiological conditions. To address this issue, we develop charged self-assembling peptides with a novel sequence pattern for strong fiber-forming ability under physiological conditions. Our designer peptides can undergo salt-triggered self-assembly into nanofibers that are ultrastable in extreme pH (0 to 14) and dilute solutions, and into hydrogels with strong mechanical strength. Upon conjugation with a model bioactive cargo, our self-assembled peptides exhibit great potential as bioscaffolds for multiple applications.
Highlights.
Self-assembling peptides are rationally designed for targeted properties.
Designer peptides undergo salt-triggered self-assembly into nanofibers.
Nanofibers are ultrastable in extreme pH (0 to 14) and dilute solutions (>5 μM).
Robust hydrogels are formed upon salt-triggering in physiological conditions.
Nanofibers carrying OVA257–264 enhance CD8+ T cell activation in vitro.
Acknowledgments
Supported by NIH grant HD075703. We thank J. Lai for the gifts of DC2.4 and B3Z cell lines and I. Suydam for the discussion on self-assembly.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare no conflicts of interest.
References
- 1.Santoso S, Hwang W, Hartman H, Zhang S. Self-assembly of Surfactant-like Peptides with Variable Glycine Tails to Form Nanotubes and Nanovesicles. nano letters. 2002;2:687–91. [Google Scholar]
- 2.Ruan L, Zhang H, Luo H, Liu J, Tang F, Shi Y-K, Zhao X. Designed amphiphilic peptide forms stable nanoweb, slowly releases encapsulated hydrophobic drug, and accelerates animal hemostasis. Proceedings of the National Academy of Sciences. 2009 doi: 10.1073/pnas.0900026106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jiang T, Xu C, Zuo X, Conticello VP. Structurally Homogeneous Nanosheets from Self-Assembly of a Collagen-Mimetic Peptide. Angewandte Chemie International Edition. 2014;53:8367–71. doi: 10.1002/anie.201403780. [DOI] [PubMed] [Google Scholar]
- 4.Aggeli A, Bell M, Boden N, Keen JN, Knowles PF, McLeish TC, Pitkeathly M, Radford SE. Responsive gels formed by the spontaneous self-assembly of peptides into polymeric beta-sheet tapes. Nature. 1997;386:259–62. doi: 10.1038/386259a0. [DOI] [PubMed] [Google Scholar]
- 5.Zhang S, Holmes T, Lockshin C, Rich A. Spontaneous Assembly of a Self-Complementary Oligopeptide to Form a Stable Macroscopic Membrane. Proceedings of the National Academy of Sciences. 1993;90:3334–8. doi: 10.1073/pnas.90.8.3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rad-Malekshahi M, Lempsink L, Amidi M, Hennink WE, Mastrobattista E. Biomedical Applications of Self-Assembling Peptides. Bioconjugate Chemistry. 2015 doi: 10.1021/acs.bioconjchem.5b00487. [DOI] [PubMed] [Google Scholar]
- 7.Parisa S, Soltani M, Reyhaneh N, Mousa J, Chen P. Self-Assembling Peptides: Potential Role in Tumor Targeting. Current Pharmaceutical Biotechnology. 2011;12:1089–100. doi: 10.2174/138920111796117409. [DOI] [PubMed] [Google Scholar]
- 8.Loo Y, Zhang S, Hauser CAE. From short peptides to nanofibers to macromolecular assemblies in biomedicine. Biotechnology advances. 2012;30:593–603. doi: 10.1016/j.biotechadv.2011.10.004. [DOI] [PubMed] [Google Scholar]
- 9.Zhang H, Luo H, Zhao X. Mechanistic Study of Self-Assembling Peptide RADA16-I in Formation of Nanofibers and Hydrogels. Journal of Nanotechnology in Engineering and Medicine. 2010;1:011007–6. [Google Scholar]
- 10.Ye Z, Zhang H, Luo H, Wang S, Zhou Q, Du X, Tang C, Chen L, Liu J, Shi Y-K, Zhang E-Y, Ellis-Behnke R, Zhao X. Temperature and pH effects on biophysical and morphological properties of self-assembling peptide RADA16-I. Journal of Peptide Science. 2008;14:152–62. doi: 10.1002/psc.988. [DOI] [PubMed] [Google Scholar]
- 11.Owczarz M, Casalini T, Motta AC, Morbidelli M, Arosio P. Contribution of Electrostatics in the Fibril Stability of a Model Ionic-Complementary Peptide. Biomacromolecules. 2015;16:3792–801. doi: 10.1021/acs.biomac.5b01092. [DOI] [PubMed] [Google Scholar]
- 12.Aggeli A, Bell M, Carrick LM, Fishwick CWG, Harding R, Mawer PJ, Radford SE, Strong AE, Boden N. pH as a Trigger of Peptide β-Sheet Self-Assembly and Reversible Switching between Nematic and Isotropic Phases. Journal of the American Chemical Society. 2003;125:9619–28. doi: 10.1021/ja021047i. [DOI] [PubMed] [Google Scholar]
- 13.Rajagopal K, Lamm MS, Haines-Butterick LA, Pochan DJ, Schneider JP. Tuning the pH Responsiveness of β-Hairpin Peptide Folding, Self-Assembly, and Hydrogel Material Formation. Biomacromolecules. 2009;10:2619–25. doi: 10.1021/bm900544e. [DOI] [PubMed] [Google Scholar]
- 14.Aulisa L, Dong H, Hartgerink JD. Self-Assembly of Multidomain Peptides: Sequence Variation Allows Control over Cross-Linking and Viscoelasticity. Biomacromolecules. 2009;10:2694–8. doi: 10.1021/bm900634x. [DOI] [PubMed] [Google Scholar]
- 15.Webber MJ, Matson JB, Tamboli VK, Stupp SI. Controlled release of dexamethasone from peptide nanofiber gels to modulate inflammatory response. Biomaterials. 2012;33:6823–32. doi: 10.1016/j.biomaterials.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rudra JS, Tian YF, Jung JP, Collier JH. A self-assembling peptide acting as an immune adjuvant. Proceedings of the National Academy of Sciences. 2010;107:622–7. doi: 10.1073/pnas.0912124107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rudra JS, Sun T, Bird KC, Daniels MD, Gasiorowski JZ, Chong AS, Collier JH. Modulating Adaptive Immune Responses to Peptide Self-Assemblies. ACS Nano. 2012;6:1557–64. doi: 10.1021/nn204530r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rudra JS, Mishra S, Chong AS, Mitchell RA, Nardin EH, Nussenzweig V, Collier JH. Self-assembled peptide nanofibers raising durable antibody responses against a malaria epitope. Biomaterials. 2012;33:6476–84. doi: 10.1016/j.biomaterials.2012.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Black M, Trent A, Kostenko Y, Lee JS, Olive C, Tirrell M. Self-Assembled Peptide Amphiphile Micelles Containing a Cytotoxic T-Cell Epitope Promote a Protective Immune Response In Vivo. Advanced Materials. 2012;24:3845–9. doi: 10.1002/adma.201200209. [DOI] [PubMed] [Google Scholar]
- 20.Ding Y, Liu J, Lu S, Igweze J, Xu W, Kuang D, Zealey C, Liu D, Gregor A, Bozorgzad A, Zhang L, Yue E, Mujib S, Ostrowski M, Chen P. Self-assembling peptide for co-delivery of HIV-1 CD8+ T cells epitope and Toll-like receptor 7/8 agonists R848 to induce maturation of monocyte derived dendritic cell and augment polyfunctional cytotoxic T lymphocyte (CTL) response. Journal of Controlled Release. 2016;236:22–30. doi: 10.1016/j.jconrel.2016.06.019. [DOI] [PubMed] [Google Scholar]
- 21.Chesson CB, Huelsmann EJ, Lacek AT, Kohlhapp FJ, Webb MF, Nabatiyan A, Zloza A, Rudra JS. Antigenic peptide nanofibers elicit adjuvant-free CD8+ T cell responses. Vaccine. 2014;32:1174–80. doi: 10.1016/j.vaccine.2013.11.047. [DOI] [PubMed] [Google Scholar]
- 22.Liu W, Chen Y-H. High epitope density in a single protein molecule significantly enhances antigenicity as well as immunogenicity: a novel strategy for modern vaccine development and a preliminary investigation about B cell discrimination of monomeric proteins. European Journal of Immunology. 2005;35:505–14. doi: 10.1002/eji.200425749. [DOI] [PubMed] [Google Scholar]
- 23.Jegerlehner A, Storni T, Lipowsky G, Schmid M, Pumpens P, Bachmann MF. Regulation of IgG antibody responses by epitope density and CD21-mediated costimulation. European Journal of Immunology. 2002;32:3305–14. doi: 10.1002/1521-4141(200211)32:11<3305::AID-IMMU3305>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 24.Kong J, Yu S. Fourier Transform Infrared Spectroscopic Analysis of Protein Secondary Structures. Acta Biochimica et Biophysica Sinica. 2007;39:549–59. doi: 10.1111/j.1745-7270.2007.00320.x. [DOI] [PubMed] [Google Scholar]
- 25.Zandomeneghi G, Krebs MRH, McCammon MG, Fändrich M. FTIR reveals structural differences between native β-sheet proteins and amyloid fibrils. Protein Science. 2004;13:3314–21. doi: 10.1110/ps.041024904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang H, Griggs A, Rochet J-C, Stanciu Lia A. In Vitro Study of α-Synuclein Protofibrils by Cryo-EM Suggests a Cu2+-Dependent Aggregation Pathway. Biophysical Journal. 2013;104:2706–13. doi: 10.1016/j.bpj.2013.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mishra A, Loo Y, Deng R, Chuah YJ, Hee HT, Ying JY, Hauser CAE. Ultrasmall natural peptides self-assemble to strong temperature-resistant helical fibers in scaffolds suitable for tissue engineering. Nano Today. 2011;6:232–9. [Google Scholar]
- 28.Jung JP, Jones JL, Cronier SA, Collier JH. Modulating the mechanical properties of self-assembled peptide hydrogels via native chemical ligation. Biomaterials. 2008;29:2143–51. doi: 10.1016/j.biomaterials.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yokoi H, Kinoshita T, Zhang S. Dynamic reassembly of peptide RADA16 nanofiber scaffold. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:8414–9. doi: 10.1073/pnas.0407843102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shen Z, Reznikoff G, Dranoff G, Rock KL. Cloned dendritic cells can present exogenous antigens on both MHC class I and class II molecules. The Journal of Immunology. 1997;158:2723–30. [PubMed] [Google Scholar]
- 31.Karttunen J, Sanderson S, Shastri N. Detection of rare antigen-presenting cells by the lacZ T-cell activation assay suggests an expression cloning strategy for T-cell antigens. Proceedings of the National Academy of Sciences. 1992;89:6020–4. doi: 10.1073/pnas.89.13.6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller MJ, Wei SH, Parker I, Cahalan MD. Two-Photon Imaging of Lymphocyte Motility and Antigen Response in Intact Lymph Node. Science. 2002;296:1869–73. doi: 10.1126/science.1070051. [DOI] [PubMed] [Google Scholar]
- 33.Zhao X, Deak E, Soderberg K, Linehan M, Spezzano D, Zhu J, Knipe DM, Iwasaki A. Vaginal Submucosal Dendritic Cells, but Not Langerhans Cells, Induce Protective Th1 Responses to Herpes Simplex Virus-2. The Journal of Experimental Medicine. 2003;197:153–62. doi: 10.1084/jem.20021109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Douillard P, Stoitzner P, Tripp CH, Clair-Moninot V, Aït-Yahia S, McLellan AD, Eggert A, Romani N, Saeland S. Mouse Lymphoid Tissue Contains Distinct Subsets of Langerin/CD207+ Dendritic Cells, Only One of Which Represents Epidermal-Derived Langerhans Cells. Journal of Investigative Dermatology. 2005;125:983–94. doi: 10.1111/j.0022-202X.2005.23951.x. [DOI] [PubMed] [Google Scholar]
- 35.Menning A, Walter A, Rudolph M, Gashaw I, Fritzemeier K-H, Roese L. Granulocytes and Vascularization Regulate Uterine Bleeding and Tissue Remodeling in a Mouse Menstruation Model. PLoS ONE. 2012;7:e41800. doi: 10.1371/journal.pone.0041800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Finkelstein AV, Ptitsyn O. Protein Physics: A Course of Lectures. San Diego: Academic Press; 2002. [Google Scholar]
- 37.Malkov S, Živković M, Beljanski M, Hall M, Zarić S. A reexamination of the propensities of amino acids towards a particular secondary structure: classification of amino acids based on their chemical structure. J Mol Model. 2008;14:769–75. doi: 10.1007/s00894-008-0313-0. [DOI] [PubMed] [Google Scholar]
- 38.Costantini S, Colonna G, Facchiano AM. Amino acid propensities for secondary structures are influenced by the protein structural class. Biochemical and Biophysical Research Communications. 2006;342:441–51. doi: 10.1016/j.bbrc.2006.01.159. [DOI] [PubMed] [Google Scholar]
- 39.Du X, Zhou J, Shi J, Xu B. Supramolecular Hydrogelators and Hydrogels: From Soft Matter to Molecular Biomaterials. Chemical Reviews. 2015;115:13165–307. doi: 10.1021/acs.chemrev.5b00299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hinner B, Tempel M, Sackmann E, Kroy K, Frey E. Entanglement, Elasticity, and Viscous Relaxation of Actin Solutions. Physical Review Letters. 1998;81:2614–7. [Google Scholar]
- 41.Rammensee S, Huemmerich D, Hermanson KD, Scheibel T, Bausch AR. Rheological characterization of hydrogels formed by recombinantly produced spider silk. Appl Phys A. 2006;82:261–4. [Google Scholar]
- 42.Gardel ML, Shin JH, MacKintosh FC, Mahadevan L, Matsudaira P, Weitz DA. Elastic Behavior of Cross-Linked and Bundled Actin Networks. Science. 2004;304:1301–5. doi: 10.1126/science.1095087. [DOI] [PubMed] [Google Scholar]
- 43.O’Brien FJ. Biomaterials & scaffolds for tissue engineering. Materials Today. 2011;14:88–95. [Google Scholar]
- 44.Hunzeker JT, Elftman MD, Mellinger JC, Princiotta MF, Bonneau RH, Truckenmiller ME, Norbury CC. A Marked Reduction in Priming of Cytotoxic CD8<sup>+</sup> T Cells Mediated by Stress-Induced Glucocorticoids Involves Multiple Deficiencies in Cross-Presentation by Dendritic Cells. The Journal of Immunology. 2011;186:183–94. doi: 10.4049/jimmunol.1001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hirosue S, Kourtis IC, van der Vlies AJ, Hubbell JA, Swartz MA. Antigen delivery to dendritic cells by poly(propylene sulfide) nanoparticles with disulfide conjugated peptides: Cross-presentation and T cell activation. Vaccine. 2010;28:7897–906. doi: 10.1016/j.vaccine.2010.09.077. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


