Summary
DNA methylation levels of certain CpG sites are thought to reflect the pace of human aging. Here, we developed a robust predictor of mouse biological age based on 90 CpG sites derived from partial blood DNA methylation profiles. The resulting clock correctly determines the age of mouse cohorts, detects the longevity effects of calorie restriction and gene knockouts, and reports rejuvenation of fibroblast-derived iPSCs. The data show that mammalian DNA methylomes are characterized by CpG sites that may represent the organism’s biological age. They are scattered across the genome, are distinct in human and mouse, and their methylation gradually changes with age. The clock derived from these sites represents a biomarker of aging and can be used to determine the biological age of organisms and evaluate interventions that alter the rate of aging.
Graphical abstract
Introduction
Organisms age at different rates, which are influenced by genotype, environment, and stochastic processes. Dietary, pharmacological and genetic interventions offer an opportunity to adjust these rates and examine, model, and ultimately regulate the process of aging in laboratory animals and, potentially, in humans. However, identifying such interventions is currently both time-consuming and cost-prohibitive. An accurate estimator of the biological age of organisms subject to an intervention, as compared to their chronological age, can resolve this problem and find many applications in biomedical science. Although no reliable molecular methods are currently available that could determine the biological age of model organisms, estimates of aging rates were recently developed for humans based on DNA methylation (DNAm) profiles (Hannum et al., 2013; Horvath, 2013; Weidner et al., 2014).
DNAm has pivotal roles in regulation of transcriptional programs and systematically varies as a function of age (Day et al., 2013; Jones, 2012; Jones et al., 2015; Maegawa et al., 2010; Schultz et al., 2015; Smith and Meissner, 2013; Zampieri et al., 2015). These changes start during embryogenesis and continue throughout the lifespan, affecting chromatin states, lineage specialization, gene expression, genome stability and self-renewal of stem cells (Beerman and Rossi, 2014; Benayoun et al., 2015; Consortium et al., 2015; Guo et al., 2014; Stelzer et al., 2015; Sun et al., 2014; Taiwo et al., 2013). The human DNAm clock model can predict certain health outcomes, such as increased future mortality (Chen et al., 2016; Christiansen et al., 2016; Horvath et al., 2015a; Marioni et al., 2015). In addition, accelerated DNAm aging was observed in patients with HIV infection and Down syndrome, and slower DNAm changes were reported for cerebellum aging (Horvath and Levine, 2015; Horvath et al., 2015b, 2015c). In this work, we sought to develop and employ a DNAm clock in the house mouse (Mus musculus), the main biological model of human aging and disease. The clock based on mouse blood DNA methylome was prepared and applied to test longevity interventions.
Results and Discussion
To assess global changes in the DNA methylome, we subjected whole blood DNA of 141 male C57BL/6 mice (16 age groups, ranging from 3- to 35-month-old animals) to a modified version of reduced representation bisulfite sequencing (RRBS) (Fig. 1A, Table 1). Among 3.9–13.0 million CpG sites detected in individual samples (Tables S1 and S2, related to Table 1), greater than 1.9 million sites were present in all samples (Fig. S1A, related to Fig. 1A and Table 1). These sites represented more than 8% of the mouse DNA methylome and mapped to 99% of UCSC RefGenes, 88% annotated CpG islands, 55% CpG island shores, 41% promoters, and 10% predicted enhancers (Fig. S1B). Overall, this RRBS procedure resulted in significant coverage of the mouse blood DNA methylome (Fig. S1A–C) (Boyle et al., 2012) and allowed us to assess its changes as a function of age.
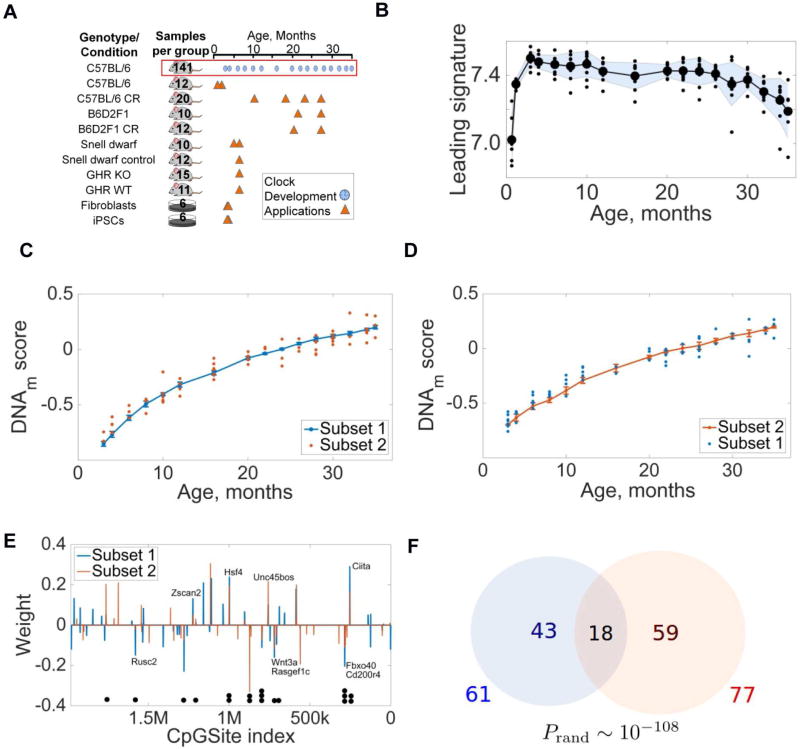
Figure 1. DNA methylation patterns predict the age of mice.
(A) Overview of mouse models used in the study. Blue circles show samples used to build the clock, and triangles models analyzed with the clock. Numbers indicate the number of animals or cell cultures for each genotype and cohort; see also Table S1.
(B) Behavior of the leading age-dependent DNAm signature for 141 C57BL/6 males with age.
(C) Behavior of the Subset 1 clock (blue). Orange dots correspond to samples from Subset 2. Goodness of fit R2 = 0.959, p = 1.16 · 10−49.
(D) Behavior of the Subset 2 clock (orange). Blue dots represent samples from Subset 1. Goodness of fit R2 = 0.959, p = 6.26 · 10−49.
(E) Weights and genome locations of CpG sites contributing to Subset 1 (blue) and Subset 2 (orange) clocks. Black dots below the graph point to 18 CpG sites common to both clocks.
(F) Number of CpG sites contributing to Subset 1 and 2 clocks. The probability to find common 18 sites in two random sets derived from 1.9 million sites is ~10−108.
Table 1. Characteristics of samples used in the study.
List of the strains, genotypes, sources, ages, and sexes of mice used in this study. NIH, National Institute on Aging; BWH, Brigham and Women’s Hospital; U of Michigan, University of Michigan.
| Strain/Treatment | Source | Age (months) | Sex | Number of samples |
|---|---|---|---|---|
| C57BL/6 | NIA/BWH | 0.67 | Male | 6 |
| C57BL/6 | NIA/BWH | 1.17 | Male | 6 |
| C57BL/6 | NIA | 3 | Male | 9 |
| C57BL/6 | NIA | 4 | Male | 9 |
| C57BL/6 | NIA | 6 | Male | 9 |
| C57BL/6 | NIA | 8 | Male | 9 |
| C57BL/6 | NIA | 10 | Male | 9 |
| C57BL/6 | NIA | 12 | Male | 10 |
| C57BL/6 | NIA | 16 | Male | 9 |
| C57BL/6 | NIA | 20 | Male | 9 |
| C57BL/6 | NIA | 22 | Male | 9 |
| C57BL/6 | NIA | 24 | Male | 9 |
| C57BL/6 | NIA | 26 | Male | 10 |
| C57BL/6 | NIA | 28 | Male | 9 |
| C57BL/6 | NIA | 30 | Male | 9 |
| C57BL/6 | NIA | 32 | Male | 7 |
| C57BL/6 | NIA | 34 | Male | 9 |
| C57BL/6 | NIA | 35 | Male | 6 |
| C57BL/6 (Calorie Restriction) | NIA | 10 | Male | 5 |
| C57BL/6 (Calorie Restriction) | NIA | 18 | Male | 5 |
| C57BL/6 (Calorie Restriction) | NIA | 23 | Male | 5 |
| C57BL/6 (Calorie Restriction) | NIA | 27 | Male | 5 |
| B6D2F1 | NIA | 20 | Male | 5 |
| B6D2F1 | NIA | 27 | Male | 5 |
| B6D2F1 (Calorie Restriction) | NIA | 21 | Male | 7 |
| B6D2F1 (Calorie Restriction) | NIA | 27 | Male | 5 |
| Snell Dwarf | U of Michigan | 5 | Male | 2 |
| Snell Dwarf | U of Michigan | 6 | Male | 4 |
| Snell Dwarf | U of Michigan | 6 | Female | 4 |
| Snell Dwarf Control | U of Michigan | 5 | Male | 2 |
| Snell Dwarf Control | U of Michigan | 6 | Male | 5 |
| Snell Dwarf Control | U of Michigan | 6 | Female | 5 |
| GHRKO | U of Michigan | 6 | Male | 9 |
| GHRKO | U of Michigan | 6 | Female | 6 |
| GHR WT | U of Michigan | 6 | Male | 8 |
| GHR WT | U of Michigan | 6 | Female | 3 |
To characterize the resulting DNA methylome, we extracted the leading age-dependent DNAm signature using non-negative matrix factorization and estimated the behavior of average DNAm with age. This revealed a global pattern of hypomethylation slowly developing with age, akin to that observed in humans (Hannum et al., 2013; Horvath, 2013). Following development, the leading DNAm signature gradually decreased (R2 = 0.545, bisquare robust linear fit) with age changing by ~5% (p = 1.1 · 10−4, two-sample t-test) between 4 and 35 months of age (Fig. 1B). The weights of different CpG sites in the leading signature suggested that the pattern of hypomethylation is determined by the dynamics of methylation levels on the whole DNA methylome.
To examine feasibility of the resulting partial DNA methylomes for describing the age of mice, we randomly divided 141 C57BL/6 mice into two subsets of 70 and 71 animals (Subsets 1 and 2). DNAm clocks were then constructed separately for Subsets 1 and 2 using elastic net regression with 10-fold cross-validation. The clock built using Subset 1 was then validated on the DNA methylation data from Subset 2 (Fig. 1C) and vice versa (Fig. 1D), revealing high correspondence between the methylation scores calculated using both Subsets. Precision of the clocks built using Subsets 1/2 and tested on Subsets 2/1 generally decreased with age (Fig. S2A related to Fig. 1C,D): in early life (<10 months) the chronological age corresponded to the DNA methylation age with precision 1–3 months, whereas the precision dropped to 5–7 months for the oldest mice. However, the relative precision of the clock, defined as the ratio of the clock error to the chronological age somewhat increased (Fig. S2B, related to Fig. 1C,D).
The Subset 1 clock was determined by the weighted average methylation on 61 different CpG sites, and the Subset 2 clock by methylation on 77 CpG sites (Fig. 1E). Interestingly, 18 sites were common to both clocks (Fig. 1F), and the values of their contributing weights were close. A relatively small overlap between the clocks built using Subsets 1 and 2 cannot be explained by a poor similarity of samples (Fig. S1D related to Fig. 1C,D). Instead, we suggest this is due to the fact that DNA methylation levels of many CpG sites are correlated with each other, and elastic net regularization picks relatively small subsets of these correlated sites (Fig. S1E related to Fig. 1C,D). We also examined the previously built human DNA methylation clocks (Hannum et al., 2013; Horvath, 2013) and observed several common CpG sites (3 sites for the 89 site clock; 2 for the 71 site clock), even though these clocks were built using different tissues. The data thus show that mouse and likely other mammalian genomes are characterized by certain CpG sites that may represent the age of organisms.
To develop a more precise mouse DNA methylation clock, we constructed a class of elastic net regression clocks based on the whole dataset representing 141 C57BL/6 mice. Among the clocks corresponding to different values of regularization parameter, we chose the model (further designated as mDNAm clock) that minimized both cross-validation error and the chance of overfitting (Fig. 2A). We further recalculated the DNAm score into the methylation age, AgeMet (Fig. 2B). On the training set, the estimated AgeMet of C57BL/6 mice coincided with the chronological age of the samples (R2 = 0.9971, p = 10−21) with the mean square difference between the mDNAm age and the chronological age of 16 days. To determine an out-of-sample lower bound estimate on the precision of the clock, we again used Subsets 1 and 2 (Fig. 1C,D). This estimation resulted in the lower out-of-sample bound on the precision of the clock R2 = 0.9008. High accuracy of the model may be attributed to the use of a single mouse strain and identical environmental conditions, although these factors may also introduce a source of bias. There is also a potential for confounding as a result of changes of blood cell composition as a function of age (Marioni et al., 2015).
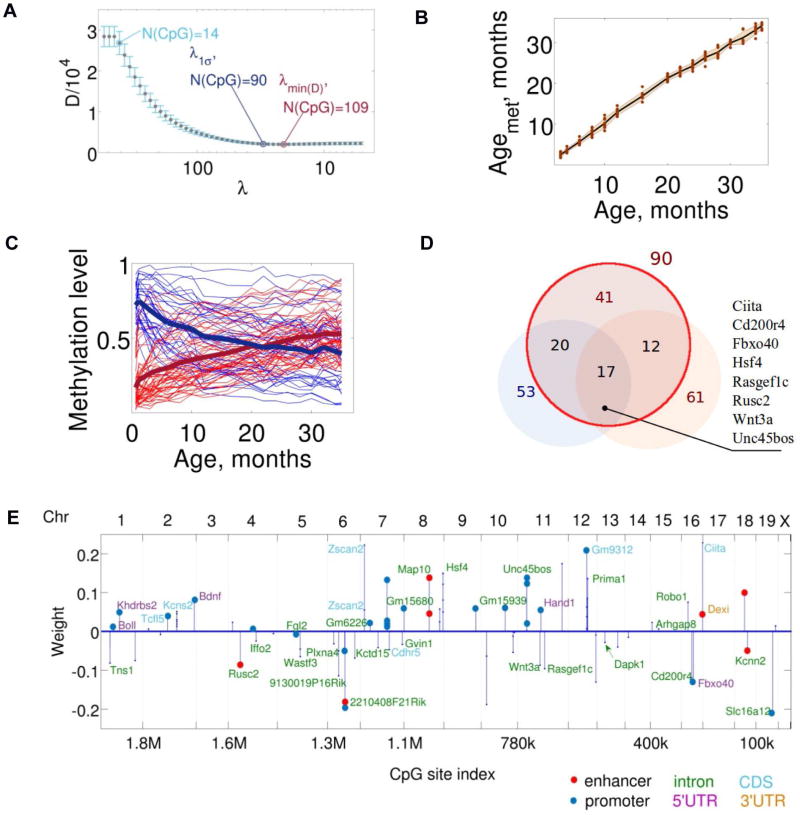
Figure 2. Development of the mDNAm clock.
(A) Selection of the optimally robust mDNAm clock. The clock corresponding to the minimum cross-validation deviation error is a weighted average of DNAm levels of 109 CpG sites. The optimally robust clock is a weighted average of DNAm levels of 90 CpG sites.
(B) Estimation of the mDNAm age of C57BL/6 control males.
(C) Trajectories of methylation levels of the 90 CpG sites that form the clock. Age-related increases in DNAm are shown in red, and decreases in blue. Solid dark blue and red lines correspond to the signal averaged over the CpG sites with methylation levels decreasing or increasing with age, respectively.
(D) The overlap between the CpG sites contributing to the 90-site mDNAm clock (red circle), Subset 1 (lower left) and Subset 2 (lower right) clocks. Gene list on the right shows genes that include 17 CpG sites common to all clocks.
(E) Distribution and weights of 90 CpG sites defining the clock along the genome. Positions of the contributing CpG sites within the genome are shown. CpGs were located within the bodies of particular genes, introns and untranslated regions, as well as in intergentic regions. Most mouse chromosomes (except 3, 17, X and Y) contain at least one CpG site contributing to the clock. The color scheme shows the indicated sequence/function elements within which the sites reside.
The mDNAm clock was defined by a weighted average DNA methylation over 90 CpG sites (Fig. 2C, Fig. S1F, related to Fig. 2C). The methylation state of these sites changed gradually, decelerating with age, with both hypomethylated and hypermethylated sites approaching more intermediate DNA methylation values (Fig. 2C, S1F). The sites contributed unequally to the mDNAm clock (Table S3 related to Fig. 2C) and formed several distinct clusters (~30) associated with genes Hsf4, Kcns2, Map10, Tns2, Wnt3a and Zscan2. 17 out of 18 CpG sites common to Subset 1 and 2 clocks were also present among 90 CpG sites of the mDNAm clock (Fig. 2D). Most of these 17 CpG sites were located within introns of Ciita, Cd200r4, Rasgef1c, Wnt3a, and Zscan2, and several were clustered. CpG sites in introns of particular genes often play a role of their secondary enhancers (Blattler et al., 2014), and we note that several identified genes are involved in development, differentiation and tissue morphogenesis, consistent with a program-like behavior (Gladyshev, 2013). The CpG sites contributing to the mDNAm clock were distributed across the chromosomes (Fig. 2E) and did not match the sites observed in human clocks (Hannum et al., 2013; Horvath, 2013). In addition, the pace of human and mouse clocks differed considerably, reflecting difference in the rates of aging in humans and mice.
We further tested if the mDNAm clock can detect the effects of interventions that are known to increase lifespan and delay age-dependent phenotypic changes, and thus plausibly serve as an index of the biological age of mice rather than their chronological age. Estimation of Agemet for calorie restricted C57BL/6 males (4 age groups, dietary intervention started at 14 weeks for all mice, Table 1) revealed on average 20% lower AgeMet than their chronological age (Fig. 3A), consistent with the effect of calorie restriction on lifespan in this strain (Turturro et al., 1999). The lower biological age of calorie restricted mice was highly significant in the tested age groups (Fig. 3A, one-sided two-sample t-test for ΔAgeMet in the combined age groups, p ≈ 7.17 · 10−12); however, the effect was less pronounced in younger animals (10 month-old mice) and was not observed in a separate experiment where 4 month-old mice were subjected to calorie restriction for 2 months (Fig. S3A related to Fig. 3A, p = 0.601 for the t-test of the control and calorie restricted group DNA methylation ages belonging to different distributions). This observation is congruent with the finding that severe weight loss in humans following bariatric surgery was not accompanied by a reduction of DNAm age (Horvath et al., 2014)). Thus, the pattern of DNA methylation indicative of the difference between biological and chronological age appears to develop gradually, suggesting a cumulative function.
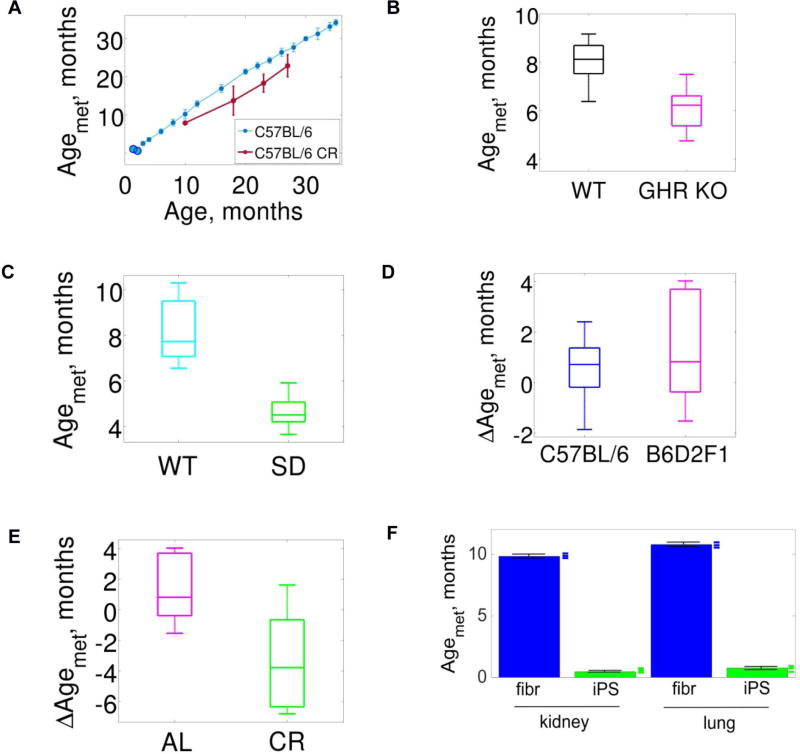
Figure 3. Applications of the mDNAm clock.
(A) Application of the mDNAm clock (light blue) to calorie restricted (CR) C57BL/6 males (red). Light blue blobs not connected to the clock correspond to 20- and 35-day old samples, each cohort including 6 mice (not used to construct the clock).
(B) Whole-body growth hormone receptor knockout (GHR KO) and WT ((C57BL/6J × BALB/cByJ)/F2) mice. The chronological age of GHR KO mice was 5.9 ± 0.3 months, and of WT mice 5.9 ± 0.4 months.
(C) Snell dwarf (SD) and control (WT; ((DW/J × C3H/HEJ)/F2) mice. The chronological age of both SD and WT mice was 5.9 ± 0.4 months.
(D) Comparing mDNAm ages of B6D2F1 and C57BL/6 mice. The differences ΔAgemet between chronological and mDNAm ages were calculated for cohorts of B6D2F1 and C57BL/6 mice.
(E) Comparing mDNAm ages of B6D2F1 control (AL) strain and the same calorie restricted (CR) strain. The differences ΔAgemet between chronological and mDNAm ages were calculated for AL and CR cohorts.
(F) Mouse lung and kidney fibroblasts and fibroblast-derived iPSCs. Fibroblasts were collected from C57BL/6 mice and grown in culture. iPSCs were then generated from them. Blue and green marks on the right of the bars represent individual samples.
We also examined genetic interventions of longevity. Full-body growth hormone receptor knockout (GHR KO) (Coschigano et al., 2003) showed a robust reduction in AgeMet as compared to control (one-sided two-sample t-test, p = 5.6 · 10−5, Fig. 3B). Application of the mDNAm clock to Snell dwarf mice and their respective wild-type control ((DW/J × C3H/HEJ)/F2) showed on average 50% lower age (one-sided two-sample t-test p = 0.0243) of Snell dwarf mice (Fig. 3C), consistent with 42% mean survival extension in this longevity model (Flurkey et al., 2001). Although the clock was developed using samples of male mice, we found that it also correctly predicted the age of female mice (Fig. S3B–E related to Fig. 3).
We further analyzed additional genetic backgrounds. The difference between AgeMet of B6D2F1 mice and their chronological age was not statistically significant (one-sided two-sample t-test, p = 0.0510, Fig. 3D). The AgeMet of calorie restricted B6D2F1 mice was 20% lower than that of the corresponding ad libitum mice (one-sided two-sample t-test, p = 1.0 · 10−5, Fig. 3E). We also analyzed the AgeMet of 20- and 35-day-old C57BL/6 mice, which were the ages not covered by the clock, and the model correctly estimated the age of these mice (Fig. 3A).
Finally, we collected kidney and lung fibroblasts from 10-week-old C57BL/6 mice, maintained/aged them in culture and prepared 6 independent iPSC lines from them (Fig. S4 related to STAR Methods, "Primary fibroblasts and generation of mouse iPSCs", "Characterization of mouse iPSCs"). The calculated AgeMet of kidney and lung fibroblasts was 295 ± 3 days and 323 ± 3 days, respectively, whereas all iPSCs were less than 1 month old (kidney: 14 ± 0.3 days; lung: 22 ± 1.2 days) (Fig. 3F). Although estimation of the exact age of fibroblasts and iPSCs may be influenced by the fact that the clock was developed based on mouse blood, the data shows that the iPSC procedure modifies mDNAm to resemble that seen in younger blood. A similar effect was previously observed in human studies (Horvath, 2013).
We think that identification of a relatively small set of CpG sites forming the clock is possible because of a very large number of observables (CpG sites) produced by the RRBS procedure and then subjected to elastic net regression, which extracts a combination of sites responsible for a given age-dependent pattern of behavior. Thus, presumably, accurate molecular clocks measuring biological age of a sample may also be developed based on other high-throughput approaches, such as metabolite profiling and RNAseq. The structure of the mDNAm clock may also change when the clock is developed across mouse tissues as was previously observed for the DNA methylation clocks in humans (Hannum et al., 2013; Horvath, 2013). We also suggest that researchers utilizing the clock use experimental and control groups for determining the difference between chronological and biological age as the RRBS procedure is not fully standardized. For example, a discrepancy of ~2 months between the chronological and estimated biological age of the GHR KO and Snell controls (Fig. 3B,C) can in principle be attributed to a lower precision on the methylation clock out-of-sample and/or a difference in genetic background. It should be expected that whole genome bisulfate sequencing data may further add to the power and resolution of the DNA methylation clocks due to a further increase in the number of age-dependent observables.
To conclude, we constructed a robust epigenetic clock based on the age-dependent behavior of blood DNA methylomes of C57BL/6 mice. The clock can estimate the age of cohorts of C57BL/6 mice, characterize the change in biological age in other mouse models, and evaluate the effects of genetic and pharmacological/dietary interventions on average lifespan. More broadly, we found that there are certain CpG sites in mammals, which are scattered across the genome, differ across species, and can form the biological clock. These sites can differentiate chronological and biological ages of organisms in a cumulative manner and represent a biomarker of aging. It may be expected that a related set of CpG sites may form a multi-tissue clock, similar to what was observed in humans (Horvath, 2013). We suggest that the mDNAm clock will find many applications in biomedical science, especially in areas where mouse models may be associated with changes in healthspan and lifespan.
STAR Methods
Contact for Reagent and Resource Sharing
Further information and requests for reagents should be directed to and will be fulfilled by the Lead Contact, Vadim Gladyshev (vgladyshev@rics.bwh.harvard.edu).
Method details
Mouse models
All mice used in this study, with the exception of Snell dwarf, GHR KO models and 3–5-week-old C57BL/6 mice, were obtained from the NIA Age Rodent Colony (Table 1). The oldest mice obtained from NIA were 32 months; to obtain mice 34 and 35 months of age, 32-month old mice were aged at Brigham & Women's Hospital for 2 and 3 months, respectively. Three of the six 35-month-old samples were from a different facility and denoted with an “R” (Table S1 related to Table 1). With the exception of mice at the 34 and 35 month time points, blood from all mice used for the clock was collected within 2 weeks of arrival from NIA. Snell dwarf and GHR KO models and their controls were housed at the University of Michigan. The 3–5-week-old C57BL/6 mice were bred at Brigham and Women's Hospital (BWH), Harvard Medical School, from parents obtained from NIA. Calorie restriction started at 14 weeks of age and continued until the time, when the animals were sacrificed. For all other animals, food was provided ad libitum. Blood was isolated from the inferior vena cavae, with the exception of 3–5-week-old mice and Snell dwarf and GHR KO mice (and their corresponding controls), in which blood was isolated directly from heart. The blood was immediately mixed with EDTA, and 100 µl was placed in 400 µl of lysis buffer (Zymo). In the case of Snell dwarf and GHR KO mice, including the corresponding controls, 100 µl EDTA mixed blood was placed in 100 µl of DNA/RNA Shield™ (2× concentrate) (Zymo).
Genomic DNA purification
DNA from samples in lysis buffer was purified by using Quick-gDNA™ Blood MiniPrep kit (Zymo) using manufacturer’s instructions. DNA from samples in DNA/RNA 2× concentrate was purified by using Quick-DNA™ Universal kit (Zymo). DNA was eluted from columns in 100 µl of 10 mM Tris-HCl buffer, pH 8.0. Then, 2 µl of RNase A (Life Technologies) was added to each sample. Samples were incubated at room temperature for 2 min, and DNA was prepared by using Genomic DNA Clean & Concentrator™-10 (Zymo). DNA was eluted in 25 µl of TE buffer (10 mM Tris-HCl, 0.1 mM EDTA, pH 8.0) and quantified using a Qubit 2.0 (Life Technologies).
RRBS
Reduced Representation Bisulfite Sequencing (RRBS) was performed as previously reported with few modifications (Boyle et al., 2012; Gu et al., 2011). MspI digestions were set up by adding 100 ng of DNA sample to 1 µl of MspI (20 U/µl) (New England Biolabs), 3 µl of 10× NEB buffer 2 (New England Biolabs), and nuclease-free water to obtain a reaction volume of 30 µl. Reactions were incubated at 37°C for 17 h. Then, 1 µl of Klenow fragment (3’–5’ exo-) (New England Biolabs) and 1 µl of dNTP mixture containing 10 mM dATP, 1 mM dCTP, and 1 mM dGTP were added to each sample for DNA end repair and A-tailing. The reactions were performed in a thermocycler without a heated lid. The program was set to 30°C for 20 min, 37°C for 20 min and 65°C for 20 min. Subsequently, ligation reactions were set up by adding 4.8 µl of T4 Ligation buffer, 1.6 µl of T4 DNA Ligase (New England Biolabs), and 3.2 of µl TruSeq Nano DNA LT adapter from Illumina that was diluted 1:20, and 6.4 µl of nuclease-free water to the DNA samples. We used 6–7 different indexed adaptors per library. The reactions were incubated at 16°C for 16 h.
After ligation, 1.2× SPRI bead clean-up was performed on the samples by adding 52 µl of water and 120 µl of Agencourt AMPure XP magnetic beads (Beckman Coulter). The mixture was pipetted 10 times and incubated at room temperature for 5 min. The samples were centrifuged and placed in 96-side magnet for 10 min. All liquid was removed without disturbing the bead pellets. Three washes were performed by adding 200 µl 80% ethanol to each sample still in the magnet, letting the samples sit at room temperature for 30 sec, and removing ethanol without disturbing the beads. After the final wash, beads were dried at room temperature for 15–20 min. The samples were removed from the magnet and 40 µl of 10 mM Tris-HCl buffer was added to elute the DNA. Beads were then re-suspended and samples were placed on the magnet for 10 min incubation at room temperature. Finally, the supernatant containing the eluted DNA was removed from the beads without disturbing them.
The purified DNA was quantified by qPCR using the Kapa Complete Universal Kit (KAPA Biosystems) following the manufacturer's instructions. Samples were pooled in libraries of 6 to 7. Subsequently, the libraries underwent bisulfite conversion using the EZ DNA Methylation-Gold™ Kit (Zymo) and eluted in 40 µl of buffer provided by the kit.
The bisulfite-converted samples were amplified by setting up 200 µl reactions consisting of 30 µl of library sample, 4 µl of Pfu Turbo Cx Hotstart DNA Polymerase, 2.5 U/µl (Agilent), 20 µl of 10× buffer (Agilent), 2 µl of dNTP mix (25 mM each, 100 mM total), 32 µl of Illumina TruSeq primer mix (2.5 µM each, 5.0 µM total), and 112 µl of nuclease-free water. The 200 µl reactions were then divided into aliquots of 50 µl and ran on a thermocycler as follows: Step 1: 95°C for 2 min; Step 2: for the least amount of cycles required for final amplification at 95°C for 30 sec, 65°C for 30 sec, and 72°C for 60 sec; Step 3: 72° for 7 min; Step 4: hold at 4°C. To determine the least amount of cycles needed, 10 µl test reactions (1 µl of library sample, 0.2 µl of Pfu Turbo Cx Hotstart DNA Polymerase, 2.5 U/µl (Agilent), 1 µl of 10× buffer (Agilent), 0.1 µl of dNTP mix (25 mM each, 100 mM total), 1.6 µl of Illumina TruSeq primer mix (2.5 µM each, 5.0 µM total), and 6.1 of µl nuclease-free water) were performed using 9, 11, and 13 cycles. After the final PCR amplification, each individual sample was pooled into a 1.5 ml tube. A 1.2× SPRI bead clean-up was performed on the samples by adding 240 µl of Agencourt AMPure XP magnetic beads (Beckman Coulter, Inc.). The mixture was pipetted 10 times and incubated at room temperature for 5 min. Then, the samples were centrifuged and placed on a magnetic rack for 10 min. All liquid was removed without disturbing the bead pellets. Two washes were performed by adding 1 ml of 80% ethanol still in the magnet, letting the sample sit at room temperature for 30 sec, and removing ethanol from the sample without disturbing the beads. After the final wash, beads were dried at room temperature for 15–20 min. The samples were removed from the magnet and 40 µl of 10mM Tris-HCl buffer was added to each sample to elute the DNA. Beads were re-suspended and samples were placed on the magnet for 10 min at room temperature. Finally, the supernatant containing the eluted DNA was removed from the beads without disturbing them. Libraries were sequenced on the Illumina HiSeq 2500 platform using 75 paired-end sequencing with v4 reagents. Since RRBS libraries generate low complexity libraries, the samples were spiked with ~10% phiX.
Equipment required for the construction of RRBS libraries included a qPCR machine, thermal cycler, NanoDrop spectrophotometer or Qubit fluorimeter with associated reagents, a DynaMag-2 magnet (Life Technologies), Bioanalyzer with associated reagents or TapeStation with High Sensitivity D1000 ScreenTapes (Agilent Technologies), and a DyanMag 96-side magnet (Life Technologies).
Primary fibroblasts and generation of mouse iPSCs
Primary fibroblasts were prepared from lungs and kidneys of 10-week-old C57BL/6 male mice. Cells were maintained in DMEM supplemented with 10% fetal bovine serum (FBS), 1% non-essential amino acids, 2 mM glutamine and antibiotics. Lentiviral vectors 4F2A and M2rtTA (Addgene plasmids #20321 and #20342) were used to reprogram fibroblasts (Carey et al., 2009). 48 h after transfection of HEK293T cells, supernatants containing viral particles were harvested, filtered, and directly used for transduction of fibroblasts. Infected fibroblasts were seeded onto a feeder layer in ESC medium in the presence of 2 mg/ml doxycycline (DMEM supplemented with 15% FBS, 2 × 106 units leukemia inhibitory factor, 0.1 mM β-mercaptoethanol, 1% nonessential amino acids, 2 mM glutamine and antibiotics (Buganim et al., 2014). On day 10, iPSC colonies were picked, dissociated by trypsin digestion, and plated into new culture dishes. These iPSCs were subcultured every 2–3 days. All cells were maintained in a humidified incubator at 37°C and 5% CO2. Testing for mycoplasma was routinely performed.
Characterization of mouse iPSCs
Alkaline phosphatase staining was performed using the Alkaline Phosphatase Staining Kit (Stemgent) according to manufacturer’s instructions. For fluorescence immunocytochemistry, iPSCs were cultured on mitotically inactivated MEFs for 2–3 days and fixed using PBS containing 4% PFA (Sigma) for 15 min at room temperature. After rinsing with PBS, fixed cells were blocked and permeabilised for 1 h in PBS containing 10% (v/v) goat serum (Sigma) in 0.1% Triton X-100-PBS. Primary antibodies were as follows: OCT4 (Santa Cruz Biotech), SOX2 (Millipore), NANOG (Millipore), and SSEA-1 (Santa Cruz Biotech), which were diluted 1:100 in blocking solution. Fixed cells were incubated with the primary antibody solution overnight at 4°C. After washing out the primary antibody solution, fixed cells were incubated with secondary antibodies (labeled with Alexa-488, 1:200, Jackson ImmunoResearch) for 1 h at room temperature. Nuclei were counterstained using 1 µg/ml Hoechst 33342 (Life Technologies). All fluorescence imaging was conducted using an LSM 700 confocal microscope (Zeiss).
Generation of embryoid bodies and analysis
Colonies growing on MEFs were detached using 0.05% trypsin and grown as a suspension culture on low adherent plates using ESC medium without LIF. After 1 week of suspension growth, cells were transferred to 12- or 24-well plates coated with 0.1% gelatin and grown in DMEM supplemented with 20% FBS. Embryoid bodies were grown for 1–2 weeks prior to fixation and immunofluorescence staining. Cultures were fixed and stained as described above using the following antibodies: AFP (1:200, Santa Cruz Biotech), GATA4 (1:200, Santa Cruz Biotech), α-actinin (1:200, Sigma), GFAP (1:500, Dako), and TUJ-1(1:500, Covance).
Quantification and Statistical Analysis
Analysis of sequence reads
Mouse genome assembly GRCm38.p2 and its annotation were downloaded from NCBI (ftp.ncbi.nih.gov). Locations of CpG islands for mm10 (Genome Reference Consortium Mouse Build 38) were obtained from UCSC (http://hgdownload.soe.ucsc.edu, released on 07-Mar-2012). CpG island shores were defined as regions 2 kp upstream and downstream of islands. Ensemble annotated features (release 82, ftp.ensemble.org) were utilized for the purpose of identification of promoters. Location of enhancers was based on a list of previously predicted enhancers (Yue et al., 2014). The quality of high throughput sequence libraries was verified using “FastQC v.0.10.1” package (www.bioinformatics.babraham.ac.uk/projects/fastqc/). Adapter removal and quality trimming were performed using Trim Galore! v.0.4.0. The TrimGalore tool (www.bioinformatics.babraham.ac.uk/projects/trim_galore/) was used with settings optimized for RRBS. Methylation sites were detected using Bismark v.0.14.5 according to the program’s manual. During post-processing, the identified methylation sites present in samples were extracted using R procedures and custom Perl scripts and used for further analyses.
Estimating the leading methylation signature and its age-dependent pattern
The leading methylation signature MSctotal was estimated by performing the dimensionality reduction (Antoulas and Sorensen, 2001) of the whole blood partial DNA methylomes of 141 C57BL/6 mice as follows:
all DNA methylomes/samples were combined into a single (N × M) dataset X, where N is the number of samples and M is the number of CpG sites, covered in each single observation (≈1.9M), keeping the rows of the matrix X chronologically ordered,
- the non-negative matrix factorization of the dataset X was performed:
(1)
(Applying non-negative matrix factorization to DNA methylomes as observables of interest (methylation levels) are always manifestly non-negative, and the methylation signatures should encode this.)
- the dimensionality reduction of the dataset X was performed, the result of the procedure approximating X by a rank 1 matrix
(2)
(The "low-rank" approximation above is valid in the sense of ratio of the Frobenius norms of the matrices X and X(red) being of the order 1. Namely, for the dataset of 141 DNA methylomes of C57BL/6 mice it was found that , where ‖X‖ denotes the Frobenius norm of a matrix X.)
- the age-dependent methylation signature MSctotal was defined as
(3) - the weights, which different CpG sites contribute to the leading age-dependent signature, were defined according to the prescription
(4)
We found a very large number of sites contributing to the leading age-dependent signature represented in Figure 1B. While no sites had weights larger than 1.4 · 10−3, ~194,000 CpG sites had weights larger than 1.3 · 10−3, ~450,000 CpG sites weights larger than 1.2 · 10−3, and ~570,000 CpG sites weights larger than 1.0 · 10−3, confirming that the age-related hypomethylation pattern is a characteristics of the mouse DNA methylome as a whole. The non-negative matrix factorization was performed in MATLAB using nnmf command.
To avoid possible confusion, it has to be noted that the procedure described here is different from the principal component analysis (PCA) in an important respect: the first leading NNMF component of the data does not encode the variance of the data and instead roughly corresponds to the mean of the data. The mean of the data was not subtracted before performing non-negative matrix factorization of the data; the latter was performed to check whether a good low-rank approximation of the data matrix is possible and to find such an approximation.
Controlling for batch effect
In order to minimize the possible batch effect on the behavior of age-dependent methylation signatures, all samples used to construct the mDNAm clock (144 C57Bl/6 males) were divided into 5 groups (sequenced by individual flow cells, Table S2 related to Table 1), with each group including samples from mice of different ages - from 3 to 36 months. Each flow cell had 8 sequencing lanes (columns in Table S6), and all sequencing lanes in any of the flow cells had samples representing different age groups. Other considered confounding variables were the adapter numbers and library numbers (Table S2). The values of other confounding variables (such as a researcher preparing samples) were the same for all samples used to build the clock and could not contribute to a possible batch effect. The adapter numbers were uncorrelated with the age group (correlation coefficient 0.0398, p = 0.6394) and accounting for the corresponding confounding variable did not contribute to the clock structure. By construction, library numbers had a high correlation with the flow cell number (correlation coefficient 0.9405, p = 10−67) and thus together were reduced to a single independent confounding variable. Although samples of different ages were randomized, there was ~30% redundant anti-correlation between the age-group and the flow cell number, so the possibility of the batch effect due to this correlation was studied further, and its significance was estimated as follows.
First, the PCA analysis of the data matrix Xij was performed. It was found that the leading PCA component (PCA1) explains about 6% of the total variance in the data, the second one (PCA2) contributes slightly more than 5% to the total variance, the 3rd - about 3%, etc. (Fig. S2C, related to Fig. 1B). The contribution of the first PCA component was then subtracted from the data, and the age-dependent behavior of the MSctotal score constructed using the remaining part of the data was found to be unchanged (Fig. S2D, related to Fig. 1B).
Second, the batch effect analysis analogous to the one described in (Johnson et al., 2007) was performed. Namely, the linear model establishing the correlation between DNA methylation data, the batch (flow cell) number and the age was constructed, assuming both additive batch contribution and the dependence of the error variance on the batch number. Again, it was found that the batch effect in the obtained data is negligible (Fig. S2E related to Fig. 1B).
Finally, a linear mixed-effects model (Age = β · M + Z · b + ε, where ε is the observation error vector) was constructed with methylation data as the fixed-effects variables M and flow cell numbers, adapter numbers and library numbers as random-effects (confounding) variables Z. The vector of weights β was then compared with a similar vector βbare constructed using a liner model Age = βbare · M + ε not taking into account the effect of confounding variables. It was found that the vectors β and βbare correlated perfectly with each other (correlation coefficient 0.999, ~ 10−97, Fig. S2F).
Constructing the elastic net regression clock and estimating the methylation age of samples
The elastic net regression clock describing a systematic age-dependent change in whole blood partial DNA methylomes of C57BL/6 males was defined as a vector w of weights of CpG sites contributing to the clock. The clock DNAm score at the chronological age of the sample t was then given by
| (5) |
where Mi(Age) is the DNAm level on a CpG site i contributing to the clock, estimated at the chronological age Age.
The vector w characterizing the clock was constructed as follows.
The m = 141 DNA methylome samples were combined into a single N × M matrix X, where the index M corresponds to the total number (≈ 1.9 M) of CpG sites of the overlap between different samples.
- Numerical minimization of the target function
(where N is the number of observations and n is the number of predictors) with different values of the Tikhonov regularization parameter λ was then performed with 20-fold cross-validation of the identified minimum. As usual, an k-fold cross-validation represented a random separation of the original dataset into k subsets, with k − 1 subsets used to train the clock and the remaining subset to validate it; the procedure was then repeated k times, the results of elastic net regression were averaged out over k repeats, and the deviation error D (cross-validation deviance) was estimated (Fig. 2A). The cross-validation deviance (as well as the mean square error in its estimation) was evaluated for every scanned value of λ, and the value of λ = λmin(D) corresponding to the minimum of cross-validation deviance was identified, Fig. 2A.
- To minimize the possibility of overfitting without much loss of predictability the value of λ = λ1σ = 33.1269 corresponding to a 1σ deviation from λ = λmin(D) was chosen. The age estimation by the clock was performed as follows. Behavior of the DNAm score on the chronological age was approximated (R2 = 0.9958) by the power law
where a = 0.1666 ± 0.1024, b = 0.4185 ± 0.0767 and c = −1.712 ± 0.3010. The constants a, b, c were found by performing the mean square regression of the DNAm scores of the actual samples calculated using the Eq. (5) to the power law (6) (a non-linear least squares fit was performed using trust-region-reflective algorithm employed in Matlab).(6)
The AgeMet of the samples was then calculated by inverting the functional dependence (6) as
| (7) |
where Δ(MSc) is the estimation error for the DNA methylation score.
It was found that the methylation clock corresponding to the minimum deviance is determined by methylation levels on 108 CpG sites (Fig. 2A), while the more robust (subject to lesser overfitting) clock corresponding to the choice λ = λ1σ is determined by methylation levels on 90 CpG sites homogeneously distributed across the genome (Fig. 2E). The elastic regression clock was constructed in MATLAB using the command lassoglm (MethylationLevels, ChronologicalAge, 'poisson', 'CV', 20, 'Alpha', 0.5), where the third argument implied a Poisson distribution of non-systematic variation of responses, the fourth argument - 20-fold cross-validation and the 6th argument enforced the form of the target function described above. The choice of α, which we used, corresponds to the equal weights of lasso (pure L1) and ridge (pure L2) optimizations. The lassoglm command uses the coordinate descent algorithm (Friedman et al., 2010); in our case, the default value 10−4 for the convergence threshold was used.
Statistical analysis of data represented on Figures 1–3
Application of standard t-tests for estimation of the effects of longevity interventions on the methylation age was validated by the distributions of methylation levels on individual CpG sites, which exhibited a distinct quasi-Gaussian peak. As the mDNAm clock is defined as a linear combination of methylation levels on n ≫ 1 sites, the stochastic variable defining the value of mDNAm age, subject to the central limit theorem, is normally distributed with non-zero mean.
Figure 1: (B) Dots represent individual samples, solid dots connected by a line - mean for a cohort of particular age, and blue shaded region - s.d. (C) Error bars are s.d. of the DNAm score from the mean over the cohort of particular age.
Figure 2: (B) Dots correspond to individual samples, the dark red solid line - to mean mDNAm age for cohort with a particular chronological age, and shared region - to the behavior of s.d.
Figure 3: For all box plots the center line is the median, the box boundaries are 25% and 75% percentile, and whiskers extend to the extreme data points, which are not considered as outliers. Outliers are plotted as individual points. (B) GHR KO: 9 males and 6 females combined into a single dataset, WT: 8 males and 3 females combined into a single dataset. (C) SD: 6 males and 4 females combined into a single dataset, WT: 7 males and 5 females combined into a single dataset. (D) For B6D2F1 strain, the latter included 20- and 27-month-old male mice (5 in each cohort), for C57BL/6 strain – 20-, 22-, 26- and 28- month-old mice (9 males in each cohort except the one aged 26 months, which included 10 mice). (E) The CR cohorts included 20- and 27-month-old male mice (5 in each cohort), while the AL cohorts – 21- and 27-month-old mice (7 and 5 males in the corresponding cohort). (F) Fibroblasts were collected from 3 C57BL/6 males.
Supplementary Material
Table S1. Related to Table 1. Detailed sample list. The table contains a list of all samples used in the study and the number of unique sequence reads per sample. Separate Excel file.
Table S2. Related to Table 1. Overview of sample pooling. The table contains information on sampling pooling and which samples and libraries were included in particular flow cells. Separate Excel file.
Table S3. Related to Fig. 2C. Analysis of CpG sites contributing to the mDNAm clock. The table contains a list of the 90 CpG sites contributing to clock, their locations, the associated genomic features, and elastic net regression weights. Separate Excel file.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| OCT-4 antibody (C-10) | Santa Cruz Biotech | Cat#sc-5279; RRID :AB_628051 |
| SOX2 antibody | Millipore | Cat#AB5603; RRID :AB_2286686 |
| NANOG antibody | Millipore | Cat#AB5731; RRID :AB_2267042 |
| SSEA-1 antibody | Santa Cruz Biotech | Cat#sc-21702; RRID :AB_626918 |
| AFP antibody (C-19) | Santa Cruz Biotech | Cat#sc-8108; RRID :AB_633815 |
| GATA4 antibody (C-20) | Santa Cruz Biotech | Cat#sc-1237; RRID :AB_2108747 |
| Anti-α-Actinin (Sarcomeric) antibody | Sigma | Cat#A7732; RRID :AB_2221571 |
| GFAPantibody | Dako | Cat#Z0334; RRID :AB_10013382 |
| TUJ-1 antibody | Covance | Cat#MMS-435P; RRID :AB_2313773 |
| Alexa Fluor® 488 AffiniPure Goat Anti-Rabbit IgG (H+L) | Jackson ImmunoResearch | Cat#111-545-144; RRID :AB_2338052 |
| Alexa Fluor® 488 AffiniPure Goat Anti-Mouse IgG (H+L) | Jackson ImmunoResearch | Cat#115-545-003; RRID :AB_2338840 |
| Alexa Fluor® 488 AffiniPure Mouse Anti-Goat IgG (H+L) | Jackson ImmunoResearch | Cat#205-545-108; RRID :AB_2339072 |
| Bacterial and Virus Strains | ||
| Biological Samples | ||
| Chemicals, Peptides, and Recombinant Proteins | ||
| MspI(20,000 units/ml) | New England Biolabs | Cat#R0106S |
| 10× NEB buffer 2 | New England Biolabs | Cat#B7002S |
| Klenow Fragment (3'→5' exo-) | New England Biolabs | Cat#M0212L |
| T4 DNA Ligase (2,000,000 units/ml) | New England Biolabs | Cat#M0202M |
| AgencourtAMPure XP - 60 ml | Beckman Coulter | Cat#A63881 |
| Pfu Turbo CxHotstart DNA Polymerase (500 units) | Agilent | Cat#600412 |
| DNA/RNA Shield (2× Concentrate) | Zymo Research | Cat#R1200-25 |
| Hoechst 33342 | Life Technologies | Cat#H3570 |
| Critical Commercial Assays | ||
| Quick-gDNA™ Blood MiniPrep kit | Zymo Research | Cat#D3072 |
| Quick-DNA™ Universal kit | Zymo Research | Cat#D4069 |
| Genomic DNA Clean & Concentrator™-10 | ZymoReserach | Cat#D4011 |
| Kapa Complete Universal Kit | KAPA Biosystems | Cat#KK4824 |
| EZ DNA Methylation-Gold™ Kit | Zymo Research | Cat#D5006 |
| Primers from TruSeq Nano LT Kits (Sets A and B) | Illumina | Cat#15041757 and 15041759 |
| Alkaline Phosphatase Staining Kit | Stemgent | Cat#00-0055 |
| Deposited Data | ||
| Raw and analyzed data | This paper | GEO: GSE80672 |
| Mouse reference genome NCBI build 38, GRCm38.p2 | Genome Reference Consortium | http://www.ncbi.nlm.nih.gov/projects/genome/assembly/grc/mouse/ |
| Location of CpG islands for mouse reference genome | UCSC | http://hgdownload.soe.ycsc.edu |
| Emsemble annotated mouse genome features, release 82 | Ensembl | http://useast.ensembl.org/Mus)musculus/Info/Index |
| Location of predicted enhancers in mouse genome | Yue et al., 2014 | N/A |
| Experimental Models: Cell Lines | ||
| Primary fibroblasts from kidney of C57BL/6 male mice | This paper | N/A |
| Primary fibroblasts from Lung of C57BL/6 male mice | This paper | N/A |
| Mouse iPSCs from kidney fibroblasts | This paper | N/A |
| Mouse iPSCs from Lung fibroblasts | This paper | N/A |
| HEK293T cells | ATCC | CRL-3216 |
| Experimental Models: Organisms/Strains | ||
| Mouse: C57BL/6 | Aging Mouse Colony from National Institute on Aging | N/A |
| Mouse: B6D2F1 | Aging Mouse Colony from National Institute on Aging | N/A |
| Mouse: GHR KO ((C57BL/6J × BALB/cByJ)/F2 | Laboratory of Richard Miller(Coschigano et al., 2003) | N/A |
| Mouse: Snell dwarf ((DW/J × C3H/HEJ)/F2) | Laboratory of Richard Miller(Flurkey et al., 2001) | N/A |
| Oligonucleotides | ||
| Primer: Illumina TruSeqprimer Forward: AATGATACGGCGACCACCGAGAT | This paper | N/A |
| Primer: Illumina TruSeqprimer Reverse: CAAGCAGAAGACGGCATACGA | This paper | N/A |
| Recombinant DNA | ||
| Plasmid: TetO-FUW-OSKM | Carey et al., 2009 | Addgene Plasmid #20321 |
| Plasmid: FUW-M2rtTA | Hockemeyer et al., 2008 | Addgene Plasmid #20342 |
| Software and Algorithms | ||
| Testing quality of high throughput sequence libraries: FastQC v.0.10.1 | N/A | http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ |
| Adapter removing and quality trimming: Trim Galore! v.0.4.0 | N/A | http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/ |
| Detection of methylation sites: Bismark v.0.14.5 | N/A | http://http://www.bioinformatics.babraham.ac.uk/projects/bismark/ |
| Dimensionality reduction by singular value decomposition: Matlab | Antoulas and Sorensen (2001) | https://www.mathworks.com/help/matlab/ref/svds.html |
| Elastic net regression: Matlab | Friedman et al. (2010) | https://www.mathworks.com/help/stats/lassoglm.html |
| Other | ||
Acknowledgments
We thank NIA (Age Rodent Colony) for providing the mice, and Adeline Augereau, Andrei Avanesov, Maxim Gerashchenko, Alaattin Kaya, Marco Mariotti, Zalan Peterfi, Alexei Mikhalchenko, Xuming Zhou and Sun Hee Yim for discussion and help with sample preparation. Supported by NIH AG047745, AG047200 and AG019899.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
V.N.G. supervised the study. D.A.P., D.I.P. and V.N.G designed experiments and interpreted results. D.A.P. collected all mouse samples, developed the adjusted RRBS protocol, sequenced the samples and contributed to bioinformatics analyses. D.I.P. developed the mDNAm clock and carried out other computational analyses. A.V.L. mapped sequencing reads, performed methylation calls and carried out additional bioinformatics analyses. S.G.L. prepared primary fibroblasts and iPSCs and contributed to mouse sample collection. R.A.M. provided growth hormone knockout and Snell dwarf models and the corresponding controls. D.I.P. and V.N.G. wrote the paper with significant contributions from D.A.P. and input from all authors.
Data and Software Availability
The data obtained in this study were deposited to GEO under accession number GSE80672.
References
- Antoulas A, Sorensen D. Approximation of large-scale dynamical systems: an overview. Int. J. Appl. Math. Comput. Sci. 2001;5:1093–1121. [Google Scholar]
- Beerman I, Rossi DJ. Epigenetic regulation of hematopoietic stem cell aging. Exp. Cell Res. 2014;329:192–199. doi: 10.1016/j.yexcr.2014.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benayoun BA, Pollina EA, Brunet A. Epigenetic regulation of ageing: linking environmental inputs to genomic stability. Nat. Rev. Mol. Cell Biol. 2015;16:593–610. doi: 10.1038/nrm4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blattler A, Yao L, Witt H, Guo Y, Nicolet CM, Berman BP, Farnham PJ. Global loss of DNA methylation uncovers intronic enhancers in genes showing expression changes. Genome Biol. 2014;15:469. doi: 10.1186/s13059-014-0469-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle P, Clement K, Gu H, Smith ZD, Ziller M, Fostel JL, Holmes L, Meldrim J, Kelley F, Gnirke A, et al. Gel-free multiplexed reduced representation bisulfite sequencing for large-scale DNA methylation profiling. Genome Biol. 2012;13:R92. doi: 10.1186/gb-2012-13-10-r92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buganim Y, Markoulaki S, van Wietmarschen N, Hoke H, Wu T, Ganz K, Akhtar-Zaidi B, He Y, Abraham BJ, Porubsky D, et al. The Developmental Potential of iPSCs Is Greatly Influenced by Reprogramming Factor Selection. Cell Stem Cell. 2014;15:295–309. doi: 10.1016/j.stem.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey BW, Markoulaki S, Hanna J, Saha K, Gao Q, Mitalipova M, Jaenisch R. Reprogramming of murine and human somatic cells using a single polycistronic vector. Proc. Natl. Acad. Sci. U. S. A. 2009;106:157–162. doi: 10.1073/pnas.0811426106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen BH, Marioni RE, Colicino E, Peters MJ, Ward-Caviness CK, Tsai P-C, Roetker NS, Just AC, Demerath EW, Guan W, et al. DNA methylation-based measures of biological age: meta-analysis predicting time to death. Aging (Albany. NY) 2016;8:1844–1865. doi: 10.18632/aging.101020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiansen L, Lenart A, Tan Q, Vaupel JW, Aviv A, McGue M, Christensen K. DNA methylation age is associated with mortality in a longitudinal Danish twin study. Aging Cell. 2016;15:149–154. doi: 10.1111/acel.12421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium RE, Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, Kheradpour P, Zhang Z, Wang J, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coschigano KT, Holland AN, Riders ME, List EO, Flyvbjerg A, Kopchick JJ. Deletion, but not antagonism, of the mouse growth hormone receptor results in severely decreased body weights, insulin, and insulin-like growth factor I levels and increased life span. Endocrinology. 2003;144:3799–3810. doi: 10.1210/en.2003-0374. [DOI] [PubMed] [Google Scholar]
- Day K, Waite LL, Thalacker-Mercer A, West A, Bamman MM, Brooks JD, Myers RM, Absher D. Differential DNA methylation with age displays both common and dynamic features across human tissues that are influenced by CpG landscape. Genome Biol. 2013;14:R102. doi: 10.1186/gb-2013-14-9-r102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flurkey K, Papaconstantinou J, Miller RA, Harrison DE. Lifespan extension and delayed immune and collagen aging in mutant mice with defects in growth hormone production. Proc. Natl. Acad. Sci. U. S. A. 2001;98:6736–6741. doi: 10.1073/pnas.111158898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman J, Hastie T, Tibshirani R. Regularization Paths for Generalized Linear Models via Coordinate Descent. J. Stat. Softw. 2010;33:1–22. [PMC free article] [PubMed] [Google Scholar]
- Gladyshev VN. The origin of aging: imperfectness-driven non-random damage defines the aging process and control of lifespan. Trends Genet. 2013;29:506–512. doi: 10.1016/j.tig.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu H, Smith ZD, Bock C, Boyle P, Gnirke A, Meissner A. Preparation of reduced representation bisulfite sequencing libraries for genome-scale DNA methylation profiling. Nat. Protoc. 2011;6:468–481. doi: 10.1038/nprot.2010.190. [DOI] [PubMed] [Google Scholar]
- Guo H, Zhu P, Yan L, Li R, Hu B, Lian Y, Yan J, Ren X, Lin S, Li J, et al. The DNA methylation landscape of human early embryos. Nature. 2014;511:606–610. doi: 10.1038/nature13544. [DOI] [PubMed] [Google Scholar]
- Hannum G, Guinney J, Zhao L, Zhang L, Hughes G, Sadda S, Klotzle B, Bibikova M, Fan J-B, Gao Y, et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol. Cell. 2013;49:359–367. doi: 10.1016/j.molcel.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013;14:R115. doi: 10.1186/gb-2013-14-10-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S, Levine AJ. HIV-1 Infection Accelerates Age According to the Epigenetic Clock. J. Infect. Dis. 2015;212:1563–1573. doi: 10.1093/infdis/jiv277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S, Erhart W, Brosch M, Ammerpohl O, von Schönfels W, Ahrens M, Heits N, Bell JT, Tsai P-C, Spector TD, et al. Obesity accelerates epigenetic aging of human liver. Proc. Natl. Acad. Sci. U. S. A. 2014;111:15538–15543. doi: 10.1073/pnas.1412759111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S, Pirazzini C, Bacalini MG, Gentilini D, Di Blasio AM, Delledonne M, Mari D, Arosio B, Monti D, Passarino G, et al. Decreased epigenetic age of PBMCs from Italian semi-supercentenarians and their offspring. Aging (Albany. NY) 2015a;7:1159–1170. doi: 10.18632/aging.100861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S, Garagnani P, Bacalini MG, Pirazzini C, Salvioli S, Gentilini D, Di Blasio AM, Giuliani C, Tung S, Vinters HV, et al. Accelerated epigenetic aging in Down syndrome. Aging Cell. 2015b;14:491–495. doi: 10.1111/acel.12325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S, Mah V, Lu AT, Woo JS, Choi O-W, Jasinska AJ, Riancho JA, Tung S, Coles NS, Braun J, et al. The cerebellum ages slowly according to the epigenetic clock. Aging (Albany. NY) 2015c;7:294–306. doi: 10.18632/aging.100742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- Jones MJ, Goodman SJ, Kobor MS. DNA methylation and healthy human aging. Aging Cell. 2015;14:924–932. doi: 10.1111/acel.12349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maegawa S, Hinkal G, Kim HS, Shen L, Zhang L, Zhang J, Zhang N, Liang S, Donehower LA, Issa J-PJ. Widespread and tissue specific age-related DNA methylation changes in mice. Genome Res. 2010;20:332–340. doi: 10.1101/gr.096826.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marioni RE, Shah S, McRae AF, Chen BH, Colicino E, Harris SE, Gibson J, Henders AK, Redmond P, Cox SR, et al. DNA methylation age of blood predicts all-cause mortality in later life. Genome Biol. 2015;16:25. doi: 10.1186/s13059-015-0584-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz MD, He Y, Whitaker JW, Hariharan M, Mukamel EA, Leung D, Rajagopal N, Nery JR, Urich MA, Chen H, et al. Human body epigenome maps reveal noncanonical DNA methylation variation. Nature. 2015;523:212–216. doi: 10.1038/nature14465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ZD, Meissner A. DNA methylation: roles in mammalian development. Nat. Rev. Genet. 2013;14:204–220. doi: 10.1038/nrg3354. [DOI] [PubMed] [Google Scholar]
- Stelzer Y, Shivalila CS, Soldner F, Markoulaki S, Jaenisch R. Tracing Dynamic Changes of DNA Methylation at Single-Cell Resolution. Cell. 2015;163:218–229. doi: 10.1016/j.cell.2015.08.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D, Luo M, Jeong M, Rodriguez B, Xia Z, Hannah R, Wang H, Le T, Faull KF, Chen R, et al. Epigenomic profiling of young and aged HSCs reveals concerted changes during aging that reinforce self-renewal. Cell Stem Cell. 2014;14:673–688. doi: 10.1016/j.stem.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taiwo O, Wilson GA, Emmett W, Morris T, Bonnet D, Schuster E, Adejumo T, Beck S, Pearce DJ. DNA methylation analysis of murine hematopoietic side population cells during aging. Epigenetics. 2013;8:1114–1122. doi: 10.4161/epi.26017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turturro A, Witt WW, Lewis S, Hass BS, Lipman RD, Hart RW. Growth curves and survival characteristics of the animals used in the Biomarkers of Aging Program. J. Gerontol. A. Biol. Sci. Med. Sci. 1999;54:B492–501. doi: 10.1093/gerona/54.11.b492. [DOI] [PubMed] [Google Scholar]
- Weidner CI, Lin Q, Koch CM, Eisele L, Beier F, Ziegler P, Bauerschlag DO, Jöckel K-H, Erbel R, Mühleisen TW, et al. Aging of blood can be tracked by DNA methylation changes at just three CpG sites. Genome Biol. 2014;15:R24. doi: 10.1186/gb-2014-15-2-r24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue F, Cheng Y, Breschi A, Vierstra J, Wu W, Ryba T, Sandstrom R, Ma Z, Davis C, Pope BD, et al. A comparative encyclopedia of DNA elements in the mouse genome. Nature. 2014;515:355–364. doi: 10.1038/nature13992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zampieri M, Ciccarone F, Calabrese R, Franceschi C, Bürkle A, Caiafa P. Reconfiguration of DNA methylation in aging. Mech. Ageing Dev. 2015;151:60–70. doi: 10.1016/j.mad.2015.02.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Related to Table 1. Detailed sample list. The table contains a list of all samples used in the study and the number of unique sequence reads per sample. Separate Excel file.
Table S2. Related to Table 1. Overview of sample pooling. The table contains information on sampling pooling and which samples and libraries were included in particular flow cells. Separate Excel file.
Table S3. Related to Fig. 2C. Analysis of CpG sites contributing to the mDNAm clock. The table contains a list of the 90 CpG sites contributing to clock, their locations, the associated genomic features, and elastic net regression weights. Separate Excel file.