Abstract
The purification, renewal and differentiation of native cardiac progenitors would form a mechanistic underpinning for unravelling steps for cardiac cell lineage formation, and their links to forms of congenital and adult cardiac diseases1–3. Until now there has been little evidence for native cardiac precursor cells in the postnatal heart4. Herein, we report the identification of isl1+ cardiac progenitors in postnatal rat, mouse and human myocardium. A cardiac mesenchymal feeder layer allows renewal of the isolated progenitor cells with maintenance of their capability to adopt a fully differentiated cardiomyocyte phenotype. Tamoxifen-inducible Cre/lox technology enables selective marking of this progenitor cell population including its progeny, at a defined time, and purification to relative homogeneity. Co-culture studies with neonatal myocytes indicate that isl1+ cells represent authentic, endogenous cardiac progenitors (cardioblasts) that display highly efficient conversion to a mature cardiac phenotype with stable expression of myocytic markers (25%) in the absence of cell fusion, intact Ca2+-cycling, and the generation of action potentials. The discovery of native cardioblasts represents a genetically based system to identify steps in cardiac cell lineage formation and maturation in development and disease.
In skeletal muscle, satellite cells are considered specialized endogenous muscle precursors (that is, myoblasts) that are preprogrammed to enter the skeletal muscle lineage5,6. Despite intensive inquiry, there has been no clear evidence for a native cardiac progenitor population with similar characteristics in the myocardium. Various cell surface markers have been used to purify precursor cells from heart muscle including c-kit and sca-1 (refs 7, 8), but the precise role of these cells in in vivo cardiogenesis is unclear and the efficiency of conversion to fully differentiated myocytes, in the absence of fusion, is relatively low9–11. Taking advantage of a developmental lineage marker for undifferentiated cardiogenic precursor cells as a requirement for a heart-specific origin, we have identified in the postnatal heart a novel cardiac cell type. The LIM-homeodomain transcription factor islet-1 (isl1) marks a cell population that makes a substantial contribution to the embryonic heart, comprising most cells in the right ventricle, both atria, the outflow tract and also specific regions of the left ventricle12.
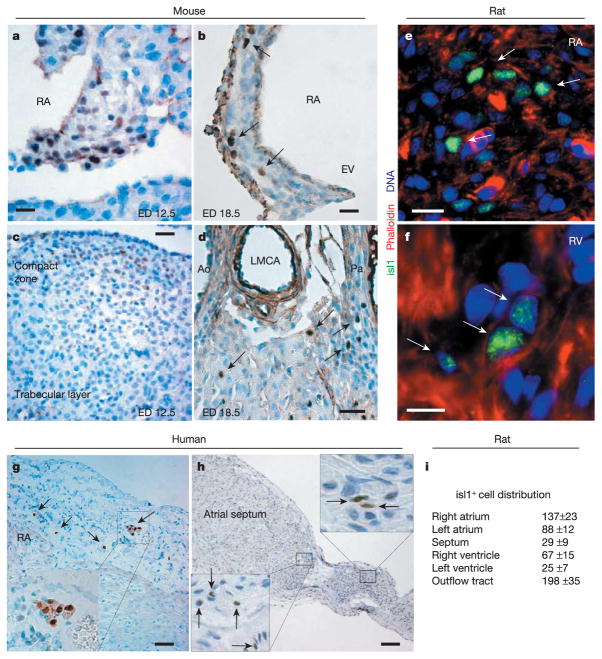
A subset of isl1+ undifferentiated precursors remains embedded in the embryonic mouse heart after its formation and their number decreases progressively from embryonic day 12.5 (ED12.5) to ED18.5 (Fig. 1a–d). After birth, relatively few isl1+ cardioblasts were still detectable, averaging 500 to 600 in the myocardium of a 1–5-day-old rat (Fig. 1e, f). Their organ distribution matched the defined contributions of isl1+ embryonic precursors (Fig. 1i), suggesting these cells are developmental remnants of the fetal progenitor population. Clusters of isl1+ cardioblasts were observed in both atria, whereas in the ventricles they occurred mostly as single cells (Fig. 1e, f). The localization of isl1+ progenitors in specific cardiac segments (atrial muscle wall, intra-atrial septum, conus muscle, right ventricle) was conserved among diverse species: mouse, rat and human (Fig. 1g, h; Supplementary Table 1).
Figure 1.
Isl1+ progenitors in the late embryonic and postnatal heart. a–d, Isl1+ progenitors in mouse sections at ED12.5 (a, atrial septum; c, ventricular tissue) and 18.5 (b, free atrial wall; d, ventricular arterial region). Scale bar, 20 μm (a, b, d), 50 μm (c). EV, eustachian valve; RA, right atrium; Ao, aorta; LMCA, left main coronary artery; Pa, pulmonary artery. e, f, Cluster of isl1+ cardiac precursors embedded in right atrial tissue and in the right ventricle of a postnatal day 1 rat heart. Scale bars, 10 μm (e), 15 μm ( f). g, h, Isl1+ progenitors in human right atrial tissue from an 8-day-old patient and intra-atrial septum from a 2-day-old patient. Scale bars, 75 μm (g), 150 μm (h). i, Quantification and localization of isl1+ cells in postnatal day 1 rat hearts. Mean values ±s.e.m. from three hearts. Arrows designate isl1+ cells in different species and insets represent a magnification of the areas of interest.
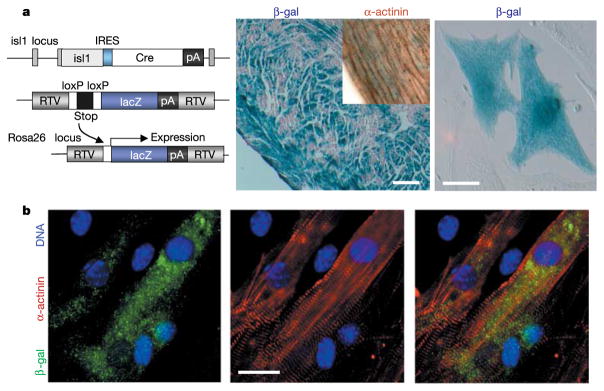
Using conditional genetic marking techniques in the mouse, we performed Cre-recombinase-triggered cell lineage tracing experiments to irreversibly mark isl1-expressing cells as well as their differentiated progeny during embryonic development (Fig. 2). Isl1-IRES-Cre mice13 were crossed into the conditional Cre reporter strain R26R14, in which Cre-mediated removal of a stop sequence results in the ubiquitous expression of the lacZ gene under the control of the endogenous Rosa26 promoter (Fig. 2a). In neonatal mice bearing both alleles a high proportion of right ventricular myocardium expressed β-galactosidase (β-gal) detected by 5-bromo-4-chloro-3-indolyl-β-D-galactoside (X-gal) staining (Fig. 2a). Around 30–40% of cardiac myocytes isolated from double heterozygous hearts stained positive for X-gal and displayed co-expression of β-gal and sarcomeric α-actinin (Fig. 2b), demonstrating that a significant proportion of myocytes originate from isl1+ cardiac progenitors.
Figure 2.
Genetic marking of isl1+ progenitors and myocytic cell fate. a, Mice carry one isl1-IRES-Cre allele and one R26R reporter gene. Cre expression catalyses excision of the stop cassette, resulting in selective lacZ expression and genetic marking of isl1-expressing cells and their differentiated progeny. RTV, retroviral integration sequence. Shown are β-gal+ cardiomyocytes in the right ventricle (middle panel; inset demonstrates α-actinin expression; scale bar, 180 μm) and after cell isolation (right panel; scale bar, 20 μm) from double heterozygous hearts of 4-month- or 1-day-old animals, respectively. b, Immunocytochemistry for β-gal and sarcomeric α-actinin in isolated cardiac myocytes from animals carrying both alleles. Scale bar, 15 μm.
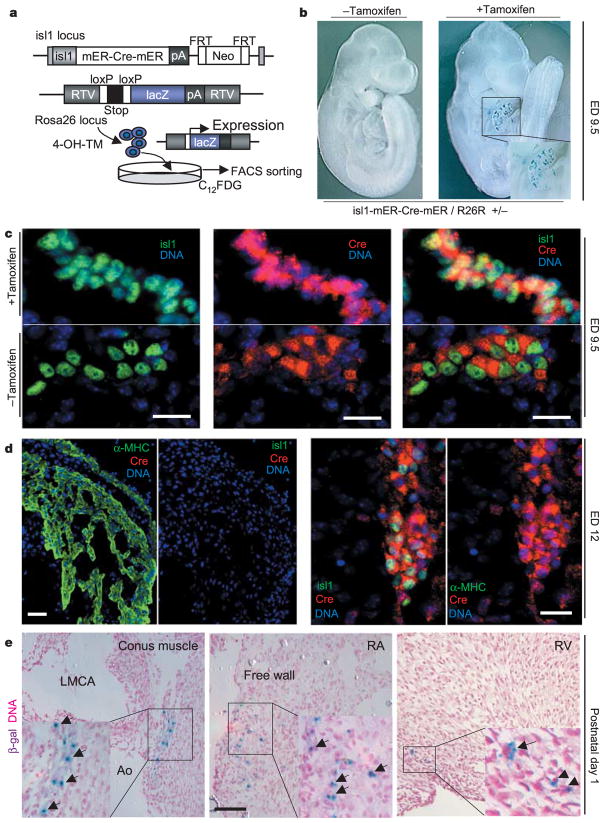
To achieve temporal and spatial control of Cre expression required for tracing the fate of isl1+ progenitors resident in the postnatal heart, we generated mice that harbour a knockin of the tamoxifen-dependent Cre recombinase (isl1-mER-Cre-mER) into the isl1 locus (Fig. 3). The presence of two mutated oestrogen-receptors (mERs) results in the sequestration of Cre in the cytoplasm15. Tamoxifen induces a rapid nuclear translocation of the mER-Cre-mER protein, which permits Cre-mediated recombination16,17. As a readout for Cre activity, we again used the reporter strain R26R14. Thus, application of tamoxifen to double heterozygous isl1-mER-Cre-mER/R26R mice leads to β-gal expression in cells expressing isl1 at the time of exposure (Fig. 3a). Experiments with isl1-mER-Cre-mER/R26R mice were performed to assess the specificity of cell labelling. β-gal+ cells are found only within the looping heart in double heterozygous animals after tamoxifen injection at ED7.5 and X-gal staining at ED9.5 (Fig. 3b). Double immunofluorescent analysis confirmed the co-expression of isl1 and Cre in undifferentiated cardiac progenitors and demonstrated that nuclear or cytosolic Cre localization was dependent on tamoxifen exposure (Fig. 3c). At ED12, sections of right ventricle and atrial tissue, heart regions that have been shown to originate from isl1+ precursors12, exhibited expression of α-myosin heavy chain but were negative for isl1 and Cre (Fig. 3d). Thus, tamoxifen injection leads to a highly specific marking of isl1-expressing progenitors and Cre protein is not detectable ~54 h after the promoter has been inactivated. After birth, β-gal+ cells can be detected with highest frequency in the conus muscle, the right atrium and the right ventricle by X-gal stain in isl1-mER-Cre-mER/R26R double heterozygous neonatal hearts after tamoxifen injection at ED17 (Fig. 3e), and represent a rare subset of cells in the adult right atrium and outflow tract (Supplementary Fig. 1).
Figure 3.
Specificity of recombination and distribution of β-gal+ cells in isl1-mER-Cre-mER/R26R hearts. a, Tamoxifen injection of isl1-mER-Cre-mER/R26R double heterozygous mice or administration of 4-OH-TM in culture results in heritable expression of lacZ. Labelling with C12FDG allows FACS purification of isl1+ cells. b, X-gal stain in whole-mount double heterozygous embryos with or without tamoxifen injection into pregnant mothers at ED7.5. c, Cre localization in isl1+ cells from isl1-mER-Cre-mER/R26R embryos is cytosolic without tamoxifen and nuclear with tamoxifen injection. Scale bar, 25 μm. d, Downregulation of isl1 and Cre in sequential sections of right ventricular and atrial tissue (left panels; scale bar, 80 μm) but not of dorsal mesocardium (right panels; scale bar, 30 μm). e, Sections from hearts of 1-day-old double heterozygous mice injected with tamoxifen at ED17, after X-gal stain and haematoxylin counterstain. LMCA, left main coronary artery; Ao, aorta; RA, right atrium; RV, right ventricle. Scale bar, 150 μm. Arrows designate β-gal+ cells and insets represent a magnification of the areas of interest.
Following cell isolation from postnatal isl1-mER-Cre-mER/R26R mouse hearts and reporter gene induction in vitro, most β-gal+ cells were found in the cardiac mesenchymal cell (CMC) fraction (Fig. 4a). Mesenchymal cells isolated from postnatal rat hearts also contain isl1+ progenitors (Supplementary Fig. 2). The CMC culture environment maintains isl1 expression in the progenitor population and promotes their expansion in culture without differentiation (Fig. 4b and Supplementary Fig. 2).
Figure 4.
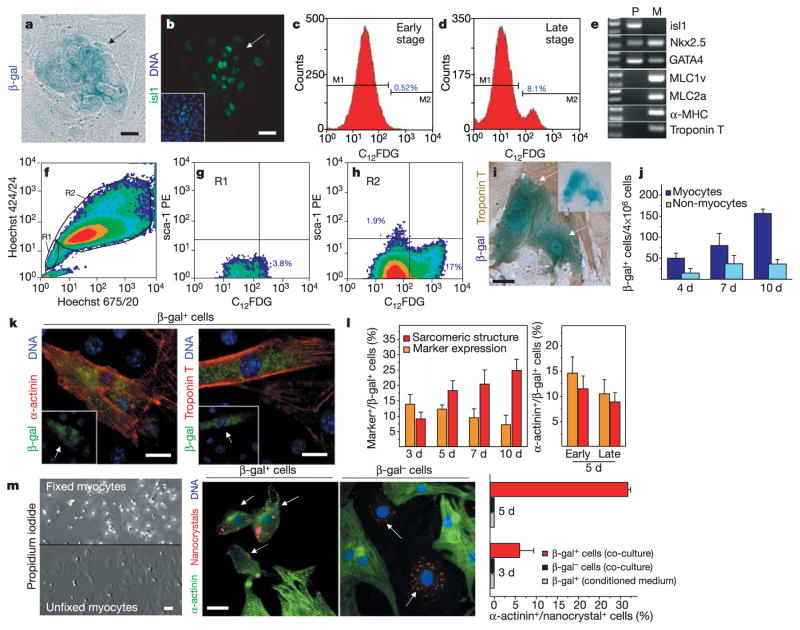
Amplification, characterization and myocytic differentiation of isl1+ cardioblasts in vitro. a, b, Mesenchymal cell fractions from isl1-mER-Cre-mER/R26R hearts at 10 days in culture after 4-OH-TM treatment. Arrows point to β-gal+ cardioblasts detected by X-gal stain (a) and isl1 expression (b). Scale bars, 15 μm. c, d, Histograms of FACS-sorted β-gal+-C12FDG-labelled cells in cardiac mesenchymal fractions after 5 days (c) and 14 days (d) in culture. e, RT–PCR analysis for myocytic and progenitor markers in FACS-sorted progenitors (P) and neonatal myocytes (M). f–h, Flow cytometry profile of Hoechst 33342 dye efflux in cardiac mesenchymal cell fractions from double heterozygous animals after C12FDG and sca-1 labelling. i, j, Neonatal myocytes isolated from isl1-mER-Cre-mER/R26R mice at 4 days in culture. Arrows designate β-gal+ cells coexpressing troponin T and inset shows the same cells before immunocytochemistry. Scale bar, 15 μm (i). Frequency of β-gal+ myocytes and non-myocytes over time ( j, mean value ±s.e.m., n = 3). Exposure to 1 μM 4-OH-TM was performed at the first day in culture. k, Images of differentiated β-gal+ progenitors in co-culture with wild-type neonatal myocytes. Arrows mark progenitor-derived β-gal+ myocytes. Scale bar, 15 μm. l, Quantification of differentiation events over time in co-culture of β-gal+ progenitors FACS-sorted at day 10–14 (left panel) and comparison at 5 days co-culture between β-gal+ progenitors expanded for 5 days (early) or 14 days (late) in culture (right panel). Mean values ±s.e.m. from six experiments (n = 1,200 cells per group). m, Cell fusion independent cardioblast-myocyte conversion. Propidium-iodide stain in pre-fixed myocytes (scale bar, 25 μm). Arrows point to QD655-labelled precursors expressing α-actinin in β-gal+, but not in β-gal− cells during co-culture with pre-fixed myocytes. Scale bar, 15 μm. The diagram represents a quantification of fusion-independent myocytic transition. Grey columns indicate the percentage of β-gal+ cells expressing α-actinin in absence of pre-fixed myocytes. Mean values ±s.e.m. (n = 3).
β-gal+ cells can be purified from the mesenchymal cells using fluorescence-activated cell sorting (FACS) after labelling with a fluorogenic, lipophilic β-gal substrate (C12FDG) (Fig. 4c, d). Isl1+ progenitors were isolated as a distinct population presenting seven to ten times higher C12FDG fluorescence than background at a purity of 90–95%. They corresponded to 0.5% of the mesenchymal cell fraction when FACS-sorted during an early stage in culture (4–6 days) and reached ~8% after 15–18 days culture (Fig. 4c, d). As phenotypic characteristics of an undifferentiated state, β-gal+ precursors expressed isl1 and early specification markers for cardiac mesoderm, Nkx2.5 and GATA-4, while lacking transcripts of mature myocytes (Fig. 4e).
FACS analysis of the FDG+ cell fraction, expanded in vitro over 10 days, revealed negative staining for sca-1 and no correlation between β-gal expression and the efflux of the DNA-binding dye Hoechst 33342, demonstrating that isl1+ cardioblasts are distinct from cardiac side population cells (Fig. 4f–h and Supplementary Table 2)8,18. In addition, isl1+ cells did not express c-kit.
After exposure to 4-hydroxytamoxifen (4-OH-TM) in vitro, a rare subset of β-gal+ cells coexpressing cardiac troponin T was observed at day 4 (n = 51 ± 12), day 7 (n = 81 ± 29) and day 10 (n = 157 ± 10) in the myocyte fraction (Fig. 4i, j), resulting in ~75% of the total β-gal+ cells. The fact that isl1 is repressed as soon as the cells acquire a differentiated phenotype12 and the lack of β-gal expression in the absence of 4-OH-TM (Fig. 3b) strongly suggest that isl1+ cardioblasts can spontaneously acquire myocytic characteristics when dispersed and cultured in the presence of differentiated myocytes.
To assess the potential of β-gal+ progenitors to adopt a myocytic phenotype, we performed co-culture experiments of FACS-sorted precursors with neonatal cardiac myocytes. After 3 days in co-culture, β-gal+ cells were found within the myocytic syncytium and expressed cardiac specific proteins, that is, α-sarcomeric actinin or troponin T in a partially or completely organized pattern (Fig. 4k, l). Moreover, a subset of β-gal+ cardioblasts showed contractile activity and intact electromechanical coupling to neighbouring cells, as documented by expression of the gap-junctional protein connexin 43 (data not shown). The differentiation capacity of β-gal+ precursors did not significantly differ between cells maintained in culture for only a few days (early stage) and cells expanded for 15–18 days (late stage) (Fig. 4l).
We addressed the role of cell fusion in the progenitor-to-cardiac myocyte transition, by performing co-culture experiments in which neonatal myocytes were pre-fixed with paraformaldehyde to stabilize cellular components and render the cells refractory to cell fusion19. Paraformaldehyde treatment disrupted the exclusion of propidium iodide by the myocytes, indicating that these cells were non-viable (Fig. 4m). FACS-sorted β-gal+ and β-gal− cells were labelled with semiconductor red-fluorescent nanocrystals (QD655)20 and co-cultured with pre-fixed myocytes in the presence of conditioned medium from viable myocytes. Under these conditions, within 5 days around 30% of the QD655-marked β-gal+ progenitors started to express α-actinin (Fig. 4m), indicating that cardioblasts enter the myocytic program independently from cell fusion. In contrast, all β-gal− cells stained negative for α-actinin, as well as β-gal+ precursors when cultured in the absence of pre-fixed myocytes, demonstrating that differentiation is cell-specific and requires secreted and membrane-bound factors (Fig. 4m).
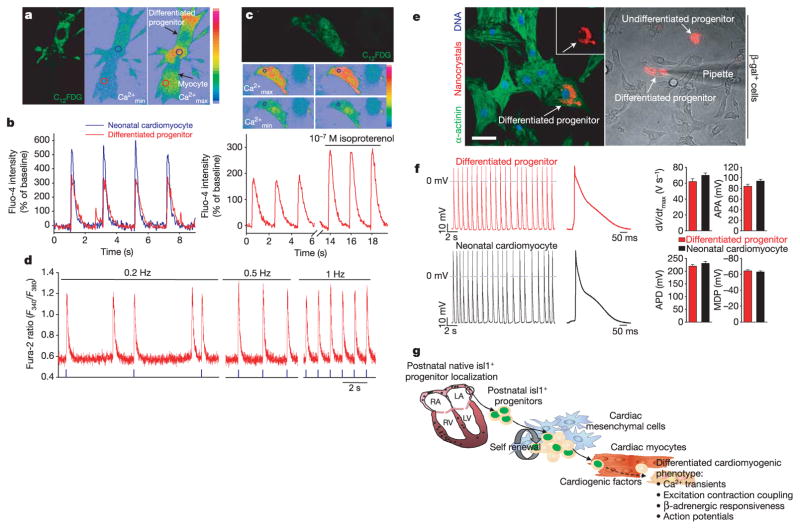
To characterize whether the β-gal+ cardioblasts could adopt a fully differentiated cardiac phenotype, C12FDG-labelled cells in co-culture were analysed by real-time intracellular calcium [Ca2+]i imaging. Differentiated C12FDG+ progenitors showed periodic [Ca2+]i oscillations similar to and synchronized with those in adjacent myocytes (Fig. 5a, b; Supplementary Video). Isoproterenol stimulation induced a 30% increase of the Ca2+ transient amplitude in differentiated β-gal+ cells, suggesting the existence of a functional β-adrenergic signalling pathway21 (Fig. 5c). The [Ca2+]i oscillations responded to increasing frequencies of external field stimulation (Fig. 5d). Of the C12FDG-labelled cardioblasts in the co-culture 2.3% showed active intracellular Ca2+ transients. Functional maturity was confirmed by analysing action potential characteristics in QD655-marked progenitors and neighbouring myocytes. Beating differentiated cardioblasts displayed continuous electrical activity or action potentials characterized by upstroke velocity (dV/dt max), amplitude (APA), duration (APD) and maximum diastolic potential (MDP) comparable to those of neonatal cardiac myocytes (Fig. 5e, f). Repeated impalements of individual QD655-marked differentiated progenitors revealed reproducible action potentials with distinct morphologies, nodal-like and atrial-like, supporting the concept that these cells are able to differentiate into a distinct subset of cardiac lineages, including conduction system myocytes22.
Figure 5.
Real time [Ca2+]i transients and action potentials in FACS-sorted β-gal+ precursors after myocytic differentiation in co-culture. a, b, Identification of β-gal+ cells by C12FDG labelling, before cell loading with the Ca2+ indicator fluo-4. Pseudo-colour images show minimal ( ) and maximal ( ) fluo-4 fluorescence intensity and circles indicate the Ca2+ measuring areas as outlined in the fluo-4 intensity traces in b. Eighteen measured cells: n = 11 differentiated progenitors, n = 7 neonatal myocytes. c, Increase in [Ca2+]i transient amplitude of a β-gal+ cell under isoproterenol (10−7 M) stimulation. d, Calcium transients of differentiated C12FDG+ precursor in response to electrical pacing at different frequencies after cell loading with the Ca2+ indicator fura-2. Six measured C12FDG+ cells. e, QD655-marked differentiated progenitors stained for α-actinin (left panel; scale bar, 20 μm) and impaled by an intracellular electrode to measure electrical activity (right panel). Inset represents the cell. f, Action potentials in a differentiated progenitor and a neonatal cardiomyocyte. Summarized data (means ± s.e.m.) showing dV/dt max (the maximum rate of rise of the action potential stroke), APA, APD90 (action potential duration at 90% of repolarization) and MDP (the maximum diastolic potential). We analysed 160 action potentials from four different cells for each cell type. g, Working model of isl1+ cardiac progenitors for self-renewal and cardiomyocytic differentiation.
We demonstrate that the mammalian heart harbours an organ-specific progenitor cell that can be localized, purified, expanded and differentiated into mature cardiac myocytes (Fig. 5g). It will be interesting to investigate whether isl1+ precursors can contribute to the cardiac lineage in the absence of fusion after cellular transplantation in vivo, and to analyse functionally on a single-cell level the differentiated progeny23. Cardioblasts may represent an in vitro system to study signalling pathways for cardiac cell lineage differentiation, which will ultimately give new insights into key steps that govern entry into specific contractile and conduction system lineages; recent work has now documented defects in later stages of ventricular lineage specification and forms of human congenital heart disease24. Furthermore, this study suggests the feasibility of isolating isl1+ cardiac progenitors from mammalian embryonic stem cell systems during cardiogenesis.
Methods
Mice
Isl1-IRES-Cre were provided by T. M. Jessel and have been previously described13. An IRES-Cre SV40 pA and a pgk-neomycin cassette were inserted into the exon encoding the second LIM homeodomain of isl1. The conditional Cre reporter mouse line R26R was generated by P. Soriano14. The isl1-mER-Cre-mER targeting construct was generated by an in-frame insertion of a mER-Cre-mER SV40 pA cassette15 along with a neo-selectable marker flanked by FRT sites into exon 1 of the genomic isl1 locus. The generation of isl1-mER-Cre-mER knockin mice will be described in detail elsewhere. Isl1-IRES-Cre/R26R and isl1-mER-Cre-mER/R26R double heterozygous mice were generated by crossing single heterozygous mice. Mice are in a mixed 129 × C57Bl/6 background. The isl1-mER-Cre-mER line showed exclusively a tamoxifen-dependent expression of Cre. Tamoxifen (Sigma) was dissolved in corn oil and injected twice intraperitoneally in the pregnant mothers at a dose of 75 μg g−1 body weight at ED17 and ED18, unless specified otherwise25. Animal care and all procedures were approved by the Institutional Animal Care Committee at UCSD.
Isolation of mouse cardiac progenitors and cell culture conditions
For cell isolation, we used 40–60 1–5-day-old pups that were double heterozygous for isl1-mER-Cre-mER and R26R alleles. Hearts were cut into four pieces, washed repetitively in ice-cold Hank’s balanced salt solution without Ca2+ (HBSS) and predigested overnight in 0.5 mg ml−1 trypsin-HBSS solution at 4 °C under constant shaking, to remove blood and dead cells. Cardiac cells were obtained by four rounds of 10 min digestions with 240 U ml−1 collagenase type II (Worthington) in HBSS at 37 °C. The mesenchymal cell fraction containing most β-gal+ progenitor cells was separated from myocytes and endothelial cells by two rounds of 1 h differential plating on plastic in DMEM/M199 (4:1 ratio) medium containing 10% horse serum and 5% fetal bovine serum at 37 °C in the presence of 5% CO2. Under these conditions preferentially mesenchymal and β-gal+ precursor cells attach on plastic. After rigorous washing to remove non-adherent cells, CMC and cardioblasts were cultured for 48 h in DMEM containing penicillin (100 U ml−1), streptomycin (100 μg ml−1), HEPES (25 mM), glutamine (2 mM), 10% newborn calf serum and 5% fetal bovine serum (FBS). Culture medium was exchanged to DMEM/F12 containing B27 supplement, 2% FBS and 10 ng ml−1 EGF on the second day in culture when the CMC reached confluency, to stimulate isl1 progenitor growth. 4-OH-TM (stock solution 1 mM in ethanol; Sigma) was applied in culture one day after cell plating at a concentration of 1 μM and maintained for 48 h.
Methods regarding flow cytometry analysis, co-culture experiments, immunocytochemistry and lacZ staining, polymerase chain reaction with reverse transcription (RT–PCR), detection of [Ca2+]i transients and electrophysiological action potential recordings are described in the Supplementary Methods section.
Supplementary Material
Acknowledgments
We thank W. Giles for discussions and comments on the real-time Ca2+ measurements and electrophysiology, the National Center for Microscopy and Imaging Research at UCSD for the use of the high-speed multi-photon laser-scanning microscope (Bio-Rad RTS2000) and custom plug-ins for the ImageJ software (H. Hakozaki, S. Chow and M. Ellisman), the UCSD Cancer Center Digital Imaging Shared Resource for deconvolution microscopy (S. McMullen and J. Feramisco). We would like especially to acknowledge D. Young for his expertise and help in the FACS sorting analysis (UCSD Cancer Center Flow Cytometry Laboratories). This work was supported by the NIH and the Jean Le Ducq Foundation. K.-L.L. is a Heisenberg Scholar of the German Research Foundation.
Footnotes
Competing interests statement
The authors declare that they have no competing financial interests.
Supplementary Information accompanies the paper on www.nature.com/nature.
References
- 1.Chien KR. Stem cells: lost in translation. Nature. 2004;428:607–608. doi: 10.1038/nature02500. [DOI] [PubMed] [Google Scholar]
- 2.Epstein JA. Developing models of DiGeorge syndrome. Trends Genet. 2001;17:S13–S17. doi: 10.1016/s0168-9525(01)02450-7. [DOI] [PubMed] [Google Scholar]
- 3.Chien KR, Olson EC. Converging pathways and principles in heart development and disease: CV@CSH. Cell. 2002;110:153–162. doi: 10.1016/s0092-8674(02)00834-6. [DOI] [PubMed] [Google Scholar]
- 4.Rosenthal N. Prometheus’s vulture and the stem cell promise. N Engl J Med. 2003;346:267–274. doi: 10.1056/NEJMra020849. [DOI] [PubMed] [Google Scholar]
- 5.Bischoff R. In: Myogenesis. Engel AG, Franszini-Armstrong C, editors. McGraw-Hill; New York: 1994. pp. 97–118. [Google Scholar]
- 6.Seale P, Rudnicki MA. A new look at the origin, function, and “stem-cell” status of muscle satellite cells. Dev Biol. 2000;218:115–124. doi: 10.1006/dbio.1999.9565. [DOI] [PubMed] [Google Scholar]
- 7.Beltrami AP, et al. Adult cardiac stem cells are multipotent and support myocardial regeneration. Cell. 2003;114:763–776. doi: 10.1016/s0092-8674(03)00687-1. [DOI] [PubMed] [Google Scholar]
- 8.Oh H, et al. Cardiac progenitor cells from adult myocardium: Homing, differentiation, and fusion after infarction. Proc Natl Acad Sci USA. 2003;100:12313–12318. doi: 10.1073/pnas.2132126100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Balsam LB, et al. Haematopoietic stem cells adopt mature haematopoietic fates in ischemic myocardium. Nature. 2004;428:668–675. doi: 10.1038/nature02460. [DOI] [PubMed] [Google Scholar]
- 10.Murry CE, et al. Hematopoietic stem cells do not transdifferentiate into cardiac myocytes in myocardial infarcts. Nature. 2004;428:664–668. doi: 10.1038/nature02446. [DOI] [PubMed] [Google Scholar]
- 11.Nygren JM, et al. Bone marrow-derived hematopoietic cells generate cardiomyocytes at a low-frequency through cell fusion, but not transdifferentiation. Nature Med. 2004;10:487–493. doi: 10.1038/nm1040. [DOI] [PubMed] [Google Scholar]
- 12.Cai CL, et al. Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Dev Cell. 2003;5:877–889. doi: 10.1016/s1534-5807(03)00363-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Srinivas S, et al. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nature Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 15.Verrou C, Zhang Y, Zurn C, Schamel WW, Reth M. Comparison of the tamoxifen regulated chimeric Cre recombinases MerCreMer and CreMer. Biol Chem. 1999;380:1435–1438. doi: 10.1515/BC.1999.184. [DOI] [PubMed] [Google Scholar]
- 16.Danielian PS, Muccino D, Rowitch DH, Michael SK, McMahon AP. Modification of gene activity in mouse embryos in utero by tamoxifen-inducible form of Cre recombinase. Curr Biol. 1998;8:1323–1326. doi: 10.1016/s0960-9822(07)00562-3. [DOI] [PubMed] [Google Scholar]
- 17.Schwenk F, Kuhn R, Angrand PO, Rajewsky K, Stewart AF. Temporally and spatially regulated somatic mutagenesis in mice. Nucleic Acids Res. 1998;26:1427–1432. doi: 10.1093/nar/26.6.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martin CM, et al. Persistent expression of the ATP-binding cassette transporter, Abcg2, identifies cardiac SP cells in the developing and adult heart. Dev Biol. 2004;265:262–275. doi: 10.1016/j.ydbio.2003.09.028. [DOI] [PubMed] [Google Scholar]
- 19.Wurmser AE, et al. Cell fusion-independent differentiation of neural stem cells to the endothelial lineage. Nature. 2004;430:350–356. doi: 10.1038/nature02604. [DOI] [PubMed] [Google Scholar]
- 20.Voura EB, Jaiswal JK, Mattoussi H, Simon SM. Tracking metastatic tumor cell extravasation with quantum dot nanocrystals and fluorescence emission-scanning microscopy. Nature Med. 2004;9:993–998. doi: 10.1038/nm1096. [DOI] [PubMed] [Google Scholar]
- 21.Maltsev VA, Ji GJ, Wobus AM, Fleischmann BK, Hescheler J. Establishment of β-adrenergic modulation of L-type Ca2+ current in the early stages of cardiomyocyte development. Circ Res. 1999;84:136–145. doi: 10.1161/01.res.84.2.136. [DOI] [PubMed] [Google Scholar]
- 22.He JQ, Ma Y, Lee Y, Thomson JA, Kamp TJ. Human embryonic stem cells develop into multiple types of cardiac myocytes. Circ Res. 2003;93:32–39. doi: 10.1161/01.RES.0000080317.92718.99. [DOI] [PubMed] [Google Scholar]
- 23.D’Amour KA, Gage FH. Are somatic stem cells pluripotent or lineage-restricted? Nature Med. 2002;8:213–214. doi: 10.1038/nm0302-213. [DOI] [PubMed] [Google Scholar]
- 24.Pashmforoush M, et al. Nkx2.5 pathways and congenital heart disease: loss of ventricular myocyte lineage specification leads to progressive cardiomyopathy and complete heart block. Cell. 2004;117:373–386. doi: 10.1016/s0092-8674(04)00405-2. [DOI] [PubMed] [Google Scholar]
- 25.Hayashi S, McMahon AP. Efficient recombination in diverse tissues by tamoxifen-inducible form of Cre: a tool for temporally regulated gene activation/inactivation in the mouse. Dev Biol. 2002;244:305–318. doi: 10.1006/dbio.2002.0597. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.