Abstract
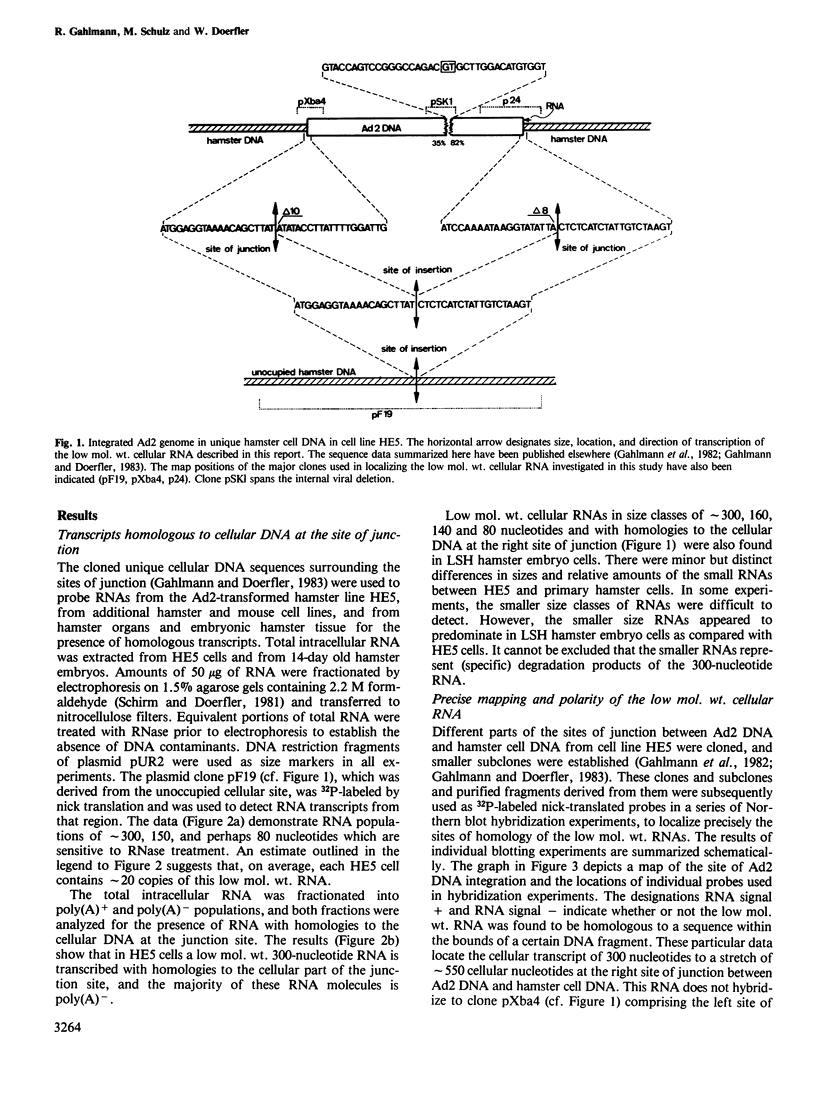
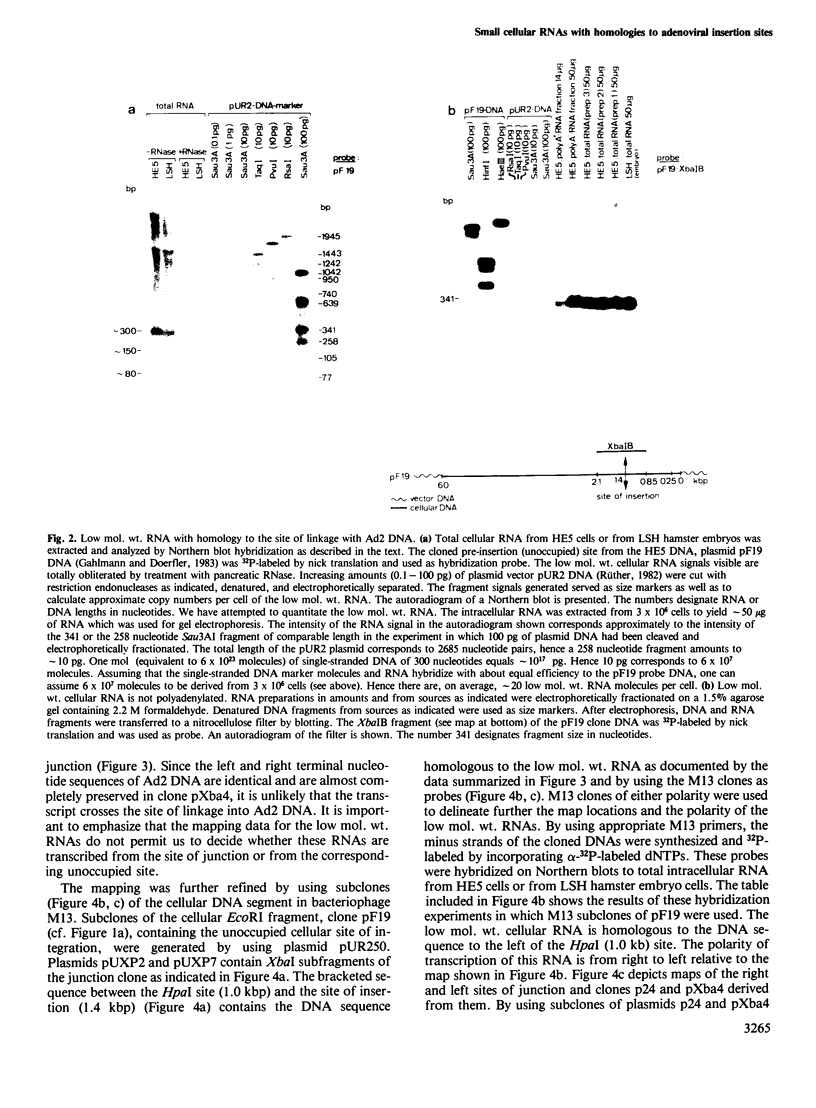
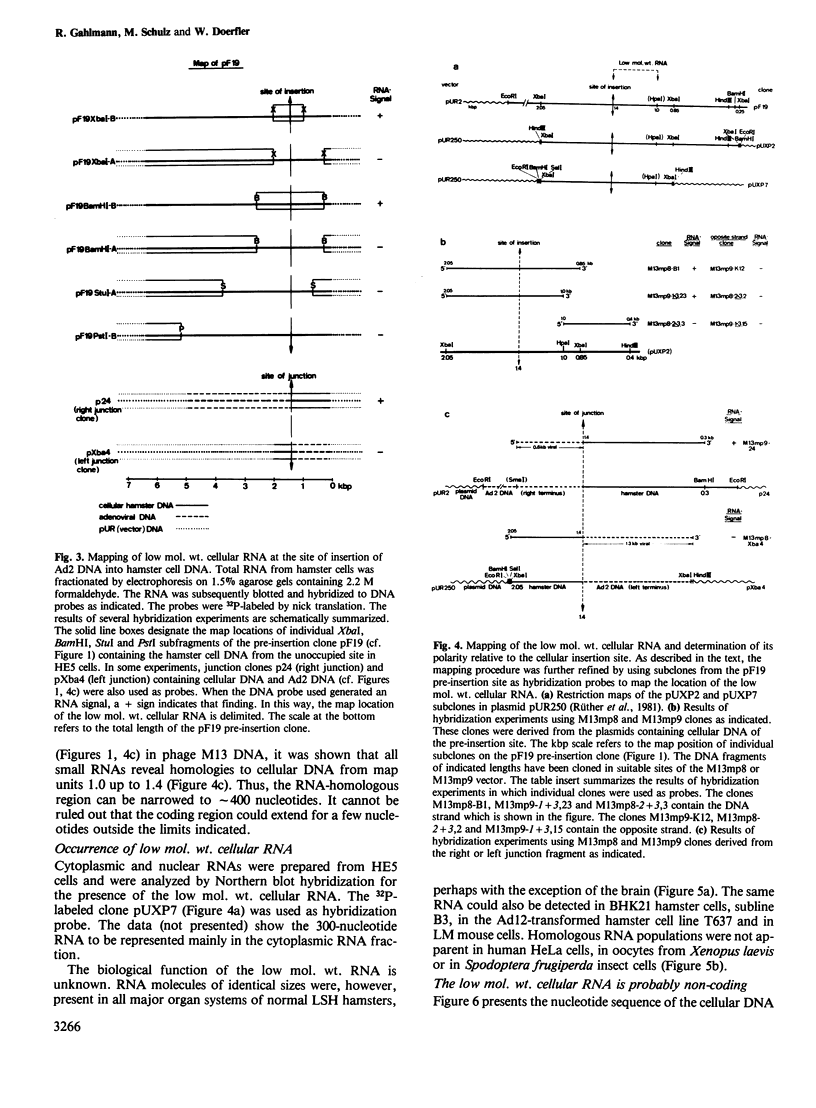
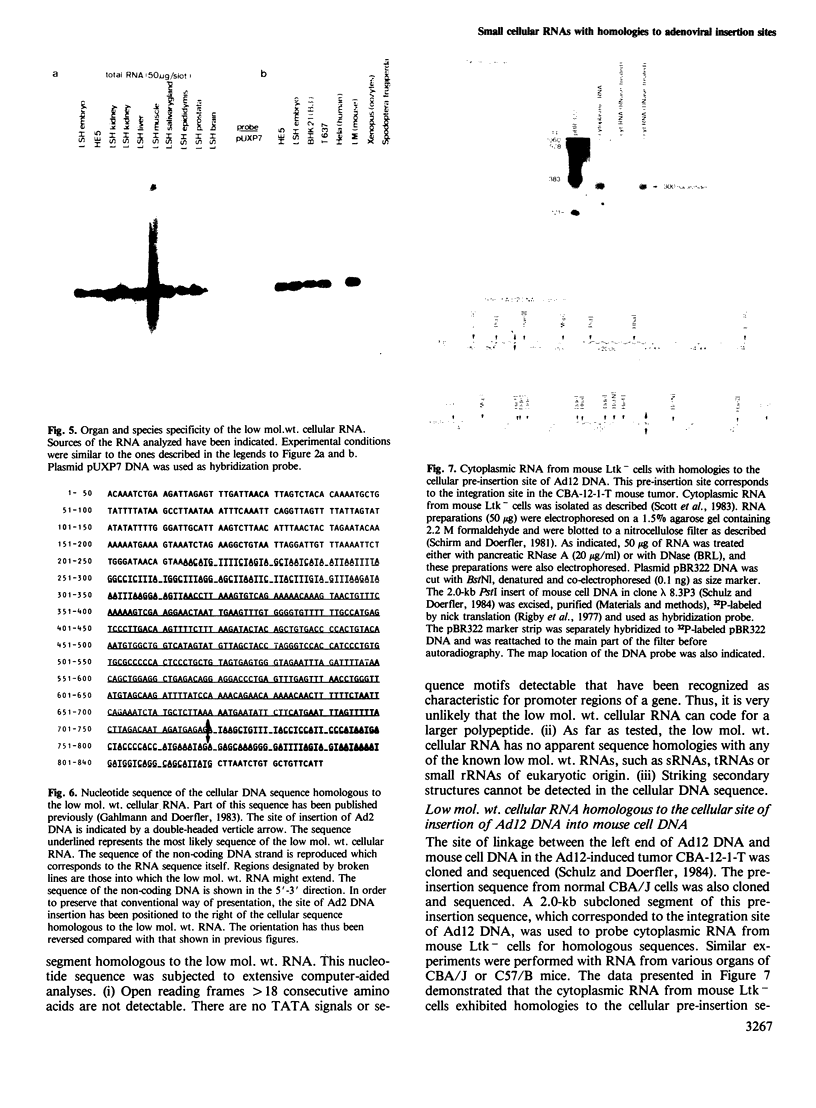
The adenovirus type 2 (Ad2)-transformed hamster cell line HE5 contains one or very few integrated copies of Ad2 DNA. At the site of insertion of Ad2 DNA, the cellular DNA sequence has been completely preserved and has homologies to small unpolyadenylated, cytoplasmic RNAs of 300 nucleotides in length and to minority populations of smaller RNAs present in HE5 cells and in normal hamster cells. The 300-nucleotide RNA is present on average in approximately 20 copies per cell. This RNA, and shorter RNAs, reveal homologies to the hamster DNA sequence of approximately 400 nucleotides to the right of the site of insertion of Ad2 DNA, which is present in one or very few copies per genome. The nucleotide sequence of the DNA segment homologous to this RNA does not contain open reading frames in excess of a sequence encoding 18 amino acids. Thus, it is unlikely that the small RNAs are actually translated and their function is unknown. The nucleotide sequence does not exhibit similarities to known low mol. wt. RNAs of eukaryotic origin. The low mol. wt. cellular RNA has been found in HE5 cells, in other hamster cell lines and organs, and also in mouse cells. There are differences with respect to size and abundance in the RNAs smaller than 300 nucleotides between HE5 cells and LSH hamster embryo cells. The adenovirus type 12 (Ad12)-induced mouse tumor CBA-12-1-T carries greater than 30 copies of integrated Ad12 DNA. The cellular DNA sequence at the site of Ad12 DNA insertion exhibits homologies to small RNAs (approximately 300 nucleotides long) from mouse cells.(ABSTRACT TRUNCATED AT 250 WORDS)
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Auffray C., Rougeon F. Purification of mouse immunoglobulin heavy-chain messenger RNAs from total myeloma tumor RNA. Eur J Biochem. 1980 Jun;107(2):303–314. doi: 10.1111/j.1432-1033.1980.tb06030.x. [DOI] [PubMed] [Google Scholar]
- Aviv H., Leder P. Purification of biologically active globin messenger RNA by chromatography on oligothymidylic acid-cellulose. Proc Natl Acad Sci U S A. 1972 Jun;69(6):1408–1412. doi: 10.1073/pnas.69.6.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BABLANIAN R., EGGERS H. J., TAMM I. STUDIES ON THE MECHANISM OF POLIOVIRUS-INDUCED CELL DAMAGE. I. THE RELATION BETWEEN POLIOVIRUS,-INDUCED METABOLIC AND MORPHOLOGICAL ALTERATIONS IN CULTURED CELLS. Virology. 1965 May;26:100–113. doi: 10.1016/0042-6822(65)90030-9. [DOI] [PubMed] [Google Scholar]
- Bernstein S. L., Gioio A. E., Kaplan B. B. Changes in gene expression during postnatal development of the rat cerebellum. J Neurogenet. 1983 Sep;1(1):71–86. doi: 10.3109/01677068309107073. [DOI] [PubMed] [Google Scholar]
- Chirgwin J. M., Przybyla A. E., MacDonald R. J., Rutter W. J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979 Nov 27;18(24):5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Cook J. L., Lewis A. M., Jr Host response to adenovirus 2-transformed hamster embryo cells. Cancer Res. 1979 May;39(5):1455–1461. [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerfler W., Gahlmann R., Stabel S., Deuring R., Lichtenberg U., Schulz M., Eick D., Leisten R. On the mechanism of recombination between adenoviral and cellular DNAs: the structure of junction sites. Curr Top Microbiol Immunol. 1984;109:193–228. doi: 10.1007/978-3-642-69460-8_9. [DOI] [PubMed] [Google Scholar]
- Dretzen G., Bellard M., Sassone-Corsi P., Chambon P. A reliable method for the recovery of DNA fragments from agarose and acrylamide gels. Anal Biochem. 1981 Apr;112(2):295–298. doi: 10.1016/0003-2697(81)90296-7. [DOI] [PubMed] [Google Scholar]
- Esche H. Viral gene products in adenovirus type-2 transformed hamster cells. J Virol. 1982 Mar;41(3):1076–1082. doi: 10.1128/jvi.41.3.1076-1082.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gahlmann R., Doerfler W. Integration of viral DNA into the genome of the adenovirus type 2-transformed hamster cell line HE5 without loss or alteration of cellular nucleotides. Nucleic Acids Res. 1983 Nov 11;11(21):7347–7361. doi: 10.1093/nar/11.21.7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gahlmann R., Leisten R., Vardimon L., Doerfler W. Patch homologies and the integration of adenovirus DNA in mammalian cells. EMBO J. 1982;1(9):1101–1104. doi: 10.1002/j.1460-2075.1982.tb01303.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu N., Messing J. The making of strand-specific M13 probes. Gene. 1982 Mar;17(3):271–277. doi: 10.1016/0378-1119(82)90143-3. [DOI] [PubMed] [Google Scholar]
- Johansson K., Persson H., Lewis A. M., Pettersson U., Tibbetts C., Philipson L. Viral DNA sequences and gene products in hamster cells transformed by adenovirus type 2. J Virol. 1978 Sep;27(3):628–639. doi: 10.1128/jvi.27.3.628-639.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langner K. D., Vardimon L., Renz D., Doerfler W. DNA methylation of three 5' C-C-G-G 3' sites in the promoter and 5' region inactivate the E2a gene of adenovirus type 2. Proc Natl Acad Sci U S A. 1984 May;81(10):2950–2954. doi: 10.1073/pnas.81.10.2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majors J. E., Varmus H. E. Nucleotide sequences at host-proviral junctions for mouse mammary tumour virus. Nature. 1981 Jan 22;289(5795):253–258. doi: 10.1038/289253a0. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Messing J., Vieira J. A new pair of M13 vectors for selecting either DNA strand of double-digest restriction fragments. Gene. 1982 Oct;19(3):269–276. doi: 10.1016/0378-1119(82)90016-6. [DOI] [PubMed] [Google Scholar]
- Nasmyth K. A. The regulation of yeast mating-type chromatin structure by SIR: an action at a distance affecting both transcription and transposition. Cell. 1982 Sep;30(2):567–578. doi: 10.1016/0092-8674(82)90253-7. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Rüther U., Koenen M., Otto K., Müller-Hill B. pUR222, a vector for cloning and rapid chemical sequencing of DNA. Nucleic Acids Res. 1981 Aug 25;9(16):4087–4098. doi: 10.1093/nar/9.16.4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rüther U. pUR 250 allows rapid chemical sequencing of both DNA strands of its inserts. Nucleic Acids Res. 1982 Oct 11;10(19):5765–5772. doi: 10.1093/nar/10.19.5765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirm S., Doerfler W. Expression of viral DNA in adenovirus type 12-transformed cells, in tumor cells, and in revertants. J Virol. 1981 Sep;39(3):694–702. doi: 10.1128/jvi.39.3.694-702.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubach W., Groudine M. Alteration of c-myc chromatin structure by avian leukosis virus integration. Nature. 1984 Feb 23;307(5953):702–708. doi: 10.1038/307702a0. [DOI] [PubMed] [Google Scholar]
- Schulz M., Doerfler W. Deletion of cellular DNA at site of viral DNA insertion in the adenovirus type 12-induced mouse tumor CBA-12-1-T. Nucleic Acids Res. 1984 Jun 25;12(12):4959–4976. doi: 10.1093/nar/12.12.4959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott M. R., Westphal K. H., Rigby P. W. Activation of mouse genes in transformed cells. Cell. 1983 Sep;34(2):557–567. doi: 10.1016/0092-8674(83)90388-4. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Stabel S., Doerfler W. Nucleotide sequence at the site of junction between adenovirus type 12 DNA and repetitive hamster cell DNA in transformed cell line CLAC1. Nucleic Acids Res. 1982 Dec 20;10(24):8007–8023. doi: 10.1093/nar/10.24.8007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storb U., Arp B., Wilson R. The switch region associated with immunoglobulin C mu genes is DNase I hypersensitive in T lymphocytes. Nature. 1981 Nov 5;294(5836):90–92. doi: 10.1038/294090a0. [DOI] [PubMed] [Google Scholar]
- Sutter D., Westphal M., Doerfler W. Patterns of integration of viral DNA sequences in the genomes of adenovirus type 12-transformed hamster cells. Cell. 1978 Jul;14(3):569–585. doi: 10.1016/0092-8674(78)90243-x. [DOI] [PubMed] [Google Scholar]
- Tjia S. T., Carstens E. B., Doerfler W. Infection of Spodoptera frugiperda cells with Autographa californica nuclear polyhedrosis virus II. The viral DNA and the kinetics of its replication. Virology. 1979 Dec;99(2):399–409. doi: 10.1016/0042-6822(79)90018-7. [DOI] [PubMed] [Google Scholar]
- Vardimon L., Doerfler W. Patterns of integration of viral DNA in adenovirus type 2-transformed hamster cells. J Mol Biol. 1981 Apr 5;147(2):227–246. doi: 10.1016/0022-2836(81)90439-3. [DOI] [PubMed] [Google Scholar]
- Wahl G. M., Stern M., Stark G. R. Efficient transfer of large DNA fragments from agarose gels to diazobenzyloxymethyl-paper and rapid hybridization by using dextran sulfate. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3683–3687. doi: 10.1073/pnas.76.8.3683. [DOI] [PMC free article] [PubMed] [Google Scholar]