Abstract
Background & Aims
Small intestinal neuroendocrine tumors (SI-NET) are serotonin-secreting well-differentiated neuroendocrine tumors believed to originate from enterochromaffin (EC) cells. Intestinal stem cell (ISC) are believed to contribute to formation of SI-NET, although little is known about tumor formation or development. We investigated the relationship between EC cells, ISCs, and SI-NETs.
Methods
We analyzed jejuno-ileal tissue specimens from 14 patients with familial SI-NETs enrolled in the Natural History of Familial Carcinoid Tumor study at the National institutes of Health from January 2009 to December 2014. Frozen and paraffin-embedded tumor tissues of different stages and isolated crypts were analyzed by in situ hybridization and immunohistochemistry. Tumor clonality was assessed by analyses of mitochondrial DNA.
Results
We identified multifocal aberrant crypt-containing endocrine cell clusters (ACEC) that contain crypt EC cell micro-tumors in patients with familial SI-NET. RNA in situ hybridization revealed expression of the EC cell and reserve stem cell genes TPH1, BMI1, HOPX, and LGR5low, in the ACECs and more advanced extra-epithelial tumor nests. This expression pattern resembled that of reserve EC cells that express ISC genes; most reside at the +4 position in normal crypts. The presence of multifocal ACECs from separate tumors and in the macroscopic tumor-free mucosa indicated widespread, independent, multifocal tumorigenesis. Analyses of mitochondrial DNA confirmed the independent origin of the ACECs.
Conclusion
Familial SI-NETs originate from a subset of EC cells (reserve EC cells that express ISC genes) via multifocal and polyclonal processes. Increasing our understanding of the role of these reserve EC cells in the genesis of multifocal SI-NETs could improve diagnostic and therapeutic strategies for this otherwise intractable disease.
Keywords: small intestine, cancer, malignancy, gene expression
Introduction
Small intestinal neuroendocrine tumors (SI-NET) (midgut carcinoid tumors) are usually well-differentiated, slow growing serotonin producing tumors of the distal small intestine. Although rare, SI-NETs are the most common malignant tumors of the small intestine 1. They display unique biological and genetic profiles that may account for their malignant characteristics and are distinct from other endocrine tumors 1, 2. Although SI-NETs are thought to originate from enterochromaffin (EC) cells 3, 4, their precise origin and development remain uncertain. Recent accumulating evidence suggests a critical role of the intestinal stem cell (ISC) in tumorigenesis of intestinal tumors as reported for colorectal cancers 5-8. However, the origin of intestinal neuroendocrine tumors has not been well investigated.
Currently two types of ISCs, rapidly cycling active and label-retaining quiescent reserve ISCs, have been proposed to play roles in normal growth and repair of the epithelial layer in the small intestine. Rapid and continuous generation of the intestinal epithelium is normally maintained by rapidly cycling active ISCs that reside at the crypt base 9, 10. Lgr5 is a well characterized, specifically expressed marker for rapidly cycling ISCs 9. However, upon loss or stress of Lgr5+ ISCs, quiescent reserve ISCs undergo asymmetric division to replenish rapidly cycling ISCs 11, 12. The label-retaining quiescent reserve ISCs are located just above the Paneth cells around +4 position 13. Current markers for the reserve ISCs include Bmi1 7, HopX14, DCLK115, Lrig1 16 and intermediate to low levels of Lgr5 (Lgr5low) 17. Studies using combined mouse models of genetic lineage tracing and transformation show that both Lgr5+ ISCs and Bmi1+ reserve ISCs can be transformed into tumor-initiating cells of origin for intestinal tumors. Thus, deletion of adenomatous polyposis coli (APC) in Lgr5+ cells in Lgr5-EGFP-IRES-creER; APCflox/flox mice induced adenoma formation in the small intestine and colon 5. In a similar fashion, conditional activation of the Wnt/β-catenin pathway in Bmi1-expressing cells by deletion of exon3 of β catenin in Bmi1-IRES-creER; Ctnnb1Ex3LoxP/+ mice induced multiple adenomas in the small intestine 7. However, unlike colon cancer, there is no animal model for SI-NETs.
We previously demonstrated that there was an ISC marker-expressing subset of enteroendocrine cells in the primary duodenal crypts from CCK-GFP transgenic mice 18. These cells resided at or below +4 position in the crypt and were identified immunohistochemically by their expression of ISC markers, Lgr5, CD133 or DCLK1. These ISC marker-expressing CCK-GFP cells were at the same time endocrine cells since they expressed endocrine markers Ngn3 and ChgA and functionally related peptides CCK, ghrelin, GIP and secretin. Recent multiple lines of evidence have since elucidated the stem cell biological nature of this ISC marker-expressing subset of endocrine cells. First, these +4 position enteroendocrine cells were shown to have stem cell properties in a mouse lineage tracing model using Cre-based recombination driven by the pro-endocrine Ngn3 promoter 19. Subsequent studies showed significant enrichment of multiple enteroendocrine markers along with reserve ISC markers Bmi1 and HopX in the reserve ISC-enriched population based on Sox9 gene expression 20. Further studies showed Dll1+ secretory precursors were able to revert back to the ISCs during radiation-induced injury 21. Consistent with these studies, label-retaining quiescent ISCs were identified to be the Lgr5-expressing precursors of secretory cell lineage that included enteroendocrine cells using Cyp1a1-H2B-YFP mice 22. Most recently, non-cycling Lgr5low/Ki67- +4 cells were found to express reserve ISC markers, Bmi1, HopX, Tert and Lrig1 as well as secretory cell markers including ChgA and suggest that they are secretory precursor cells with the capacity to revert to ISCs upon tissue injury 17.
Similar to other enteroendocrine cells, EC cells, the presumed cell of origin of the SI-NETs, differentiate from the ISCs and normally appear as well differentiated isolated single cells. A majority of EC cells are found at higher positions within the crypts and the villi and would eventually be sloughed as they reach the villus tips. However a small proportion of EC cells can be found below +4 position where an active stem cell niche maintains ISCs. Similar to our previous work in the duodenum of mice18 and the evidence for a secretory precursor cell capable of reversion to ISCs17,19,21,22, we wondered if the EC cells in the distal SI that reside below +4 position were derived from reserve ISCs and were susceptible to tumorigenesis in Familial SI-NET patients. Presently, it is unknown whether there is a subset of ISC marker-expressing enteroendocrine cells in the human small intestine and whether they are related to SI-NETs. Therefore, in the present study, we investigated the relationship between the expression of ISC marker genes in EC cells along the crypt-villus axis of the human ileum in normal subjects and patients with SI-NETs. For these studies, we focused on examining tumor tissue samples from patients with Familial SI-NETs which we recently identified as an autosomal dominant heritable disease 23. The familial form of SI-NET occurs on a germline basis with the majority of patients presenting with multiple synchronous primary tumors and thus provides an opportunity to investigate the varying stages of histogenesis of these otherwise rare sporadic unifocal tumors. This has allowed us to characterize not only later stage extra-epithelial tumor nests but also early stage cryptal micro-tumor formation from enterochromaffin cells 24, 25, namely aberrant crypt containing enteroendocrine cell clusters (ACEC), for the expression of the ISC marker genes by RNA in situ hydridization (ISH) and immunohistochemistry (IHC) as well as their multifocal origin using mitochondrial DNA (mtDNA)-based clonality analysis. Here we show that familial SI-NETs arise from polyclonal crypt based EC cells bearing a reserve stem signature and discuss likely mechanisms for SI-NET development from ACECs.
Materials and Methods
Human Tissue Specimens
Jejuno-ileal tissue specimens were obtained from patients with familial SI-NETs enrolled in the Natural History of Familial Carcinoid Tumor protocol registered with ClinicalTrials.gov (NCT00646022) and approved by the Institutional Review Board of the National Institute of Diabetes, Digestive and Kidney (NCT00646022). Patients were admitted to the NIH Clinical Research Center and underwent clinical evaluation and surgery as medically indicated. All of the patients in this study had a familial form of SI-NET unless specified otherwise. The clinical profiles of the patients are summarized in Supplementary Table 1. The patients F2, F4 and F5 are from the same family and carry an inositol polyphosphate multikinase (IPMK) mutation as described previously 23. Patients F7 and F14 are also related and presumed to carry an unidentified germline mutation. The remaining nine patients with familial SI-NET (F1, F3, F6, F8, F9, F10, F11, F12 and F13) are unrelated. A non-familial patient (S1) with a solitary sporadic tumor and a control patient without SI-NET (C1) were also included for comparison.
Details relating to RNA-ISH, immunohistochemistry and mtDNA-based analysis can be found in the Supplementary Appendix.
Results
The Majority of human EC cells at position 4 express Bmi1, HopX and NeuroD
Work from our lab 18 as well as others 17, 20, 22 in mice have suggested that a subset of position 4 cells having reserve stem cell markers may also express enteroendocrine lineage markers. Therefore, we examined the expression of ISC markers and differentiation genes in human EC cells as well as the relationship between their expression and position within the crypt. The genes tested were the rapidly cycling ISC gene Lgr5, reserve ISC genes Bmi1 and HopX, the pan ISC gene Sox9, and the early and late neuroendocrine transcription factors Ngn3 and NeuroD1, respectively. Consistent with many recent reports of Lgr5 expression in mice, Lgr5 mRNA expression was localized to the cells at the crypt base between the Paneth cells in human ileal mucosa (Supplementary Figure S1). EC cells identified by Tryptophan hydroxylase 1 (TPH1) mRNA expression (Supplementary Result section), co-expressed Lgr5 mRNA predominantly in EC cells located below position 4 (Figure 1). Bmi1 displayed a decreasing gradient pattern of expression from the crypt to the villus, but the expression was broad and uniform throughout the crypt at low levels (Supplementary Figure S1). Bmi1 showed no distinct, enriched pattern of expression at +4 position. However, when we examined its expression in the EC cells, the percent of EC cells expressing Bmi1 mRNA was the highest in the TPH1+ EC cells located at the +4 position (Figure 1). Over 70% of the +4 EC cells co-expressed Bmi1 gene. The expression of Bmi1 protein was consistent with this observation (Supplementary Figure S2). HopX exhibited a very similar pattern of expression to that of Bmi1. Thus, relatively higher HopX expression was seen around the crypt compared to the villus (Supplementary Figure S1). There was no distinct expression of HopX mRNA at the +4 position. Within the EC cells, again similar to Bmi1, the highest percent of EC cells expressing HopX was seen in the EC cells at the +4 position (Figures 1). Sox9 expression was high in the crypt area and gradually declined toward the villus (Supplementary Figure S1). Over 60% of the EC cells co-expressed Sox9 when the EC cells were located below position 10 (Figures 1). Ngn3 expression was seen at low levels in the crypt and also in the EC cells (Figures 1). Although the frequency was low, the percent of EC cells expressing Ngn3 seemed to be highest in the EC cells at the +4 position. We also noticed that TPH1 expression was consistently low in the Ngn3+ TPH1+ cells as seen in Figure 1. Interestingly, all the EC cells showed positive NeuroD1 expression regardless of their position (Figures 1). As expected, all the EC cells were positive for ChgA (Supplementary Figure S5A and S5B).
Figure 1.
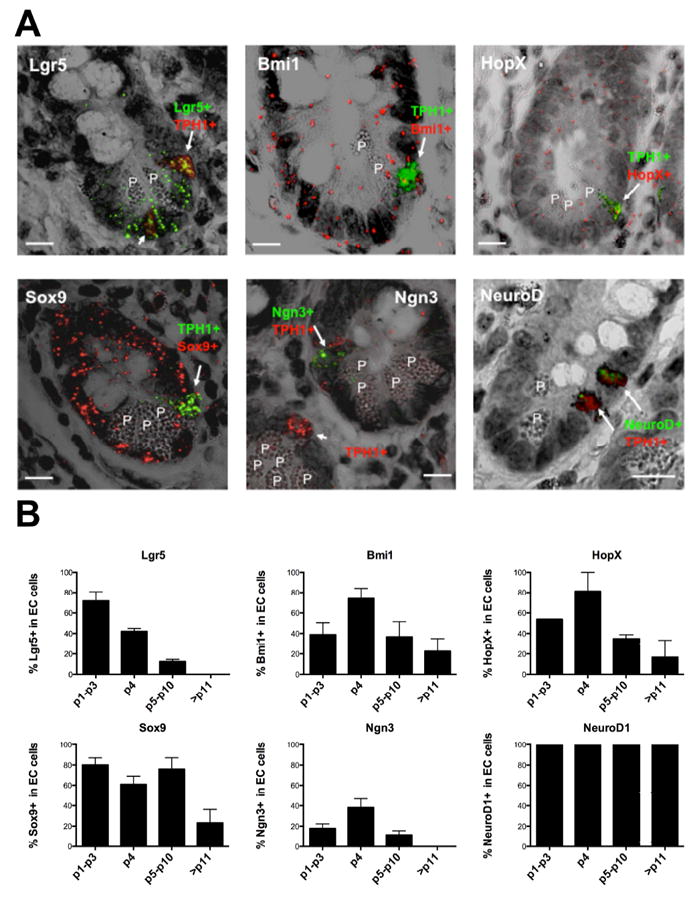
Expression of ISC and differentiation genes in human ileal EC cells. (A) Representative images of RNA-ISH of FFPE sections from normal ileal mucosa merging fluorescent and transmitted light images to visualize EC cell location and indicated gene expression. (Top left) Lgr5+ EC cell with high TPH1 (arrow) at position 4 and Lgr5+ EC cell with low TPH1 (short arrow) at crypt base. (Top center) Bmi1+/TPH1+ EC cell (arrow) at position 4. (Top right) HopX+/TPH1+ EC cell (arrow) at position 4. (Bottom left) Sox9+/TPH1+ EC cell (arrow) at position 4. (Bottom center) Ngn3+ cell with low TPH1+ expression (immature EC cell, arrow) at position 4 in the upper right crypt. Ngn3-/TPH1+ EC cell (mature EC, short arrow) in the left crypt. (Bottom right) Two NeuroD1+/TPH1+ EC cells at positions 4 and 6. EC cells were identified by the expression of TPH1 (fast red fluorescence, red) or (fast blue fluorescence, green). Expression of Bmi1, HopX and Sox9 were detected as fast red fluorescence (red). Lgr5 , Ngn3 and NeuroD1 were detected as fast blue fluorescence (green). Representative RNA-ISH images for Bmi1 and HopX were obtained from patient F1 and the other images from patient F2. P, Paneth cell. Scale bars, 10 μm (B) Graphical representation of the percent of EC cells (TPH1+) expressing the indicated genes in relation to their position within the crypt (mean+SE, n=2-4). The data were obtained from the normal ileal mucosa of the patients, C1(control without SI-NET), F1, F2 and F4. An average of 526 TPH1+ cells per patient and 105 TPH1+ cells per gene were examined.
Bmi1, HopX and NeuroD are constantly expressed in SI-NET cells
ISCs have been implicated in tumorigenesis of the intestinal epithelium 5, 7. Having identified a distinct subset of ISC marker-expressing EC cells, we sought to determine whether this population of EC cells could be the SI-NET cell of origin. Therefore, we examined the expression of the ISC markers Lgr5, Bmi1, HopX, Sox9 and differentiation genes Ngn3 and NeuroD1 and TPH1 in SI-NET cells. The tumor cells were identified by the expression of TPH1. Two-color RNA-ISH indicated that TPH1+ SI-NET cells highly express reserve ISC marker genes Bmi1 and HopX and negligible amount of the rapidly recycling ISC marker gene Lgr5 (Figures 2) and little to no expression of Sox9 or Ngn3 (Figures 2). NeuroD1 was highly expressed in all the tumor samples tested (Figure 2). To confirm the unusual co-expression of both reserve ISC genes and the differentiated EC marker gene TPH1, we examined the expression of other reserve ISC genes Lrig1 and DCLK1 (Figure 2) and differentiated EC cell genes Tac1 (Figure 2B) and ChgA (Supplementary Figures S5) and found that they were also highly expressed in tumors. The absence of expression of the non-EC cell GCG gene in both normal TPH1+ EC cells (Supplementary Figure S3B) and SI-NETs (Figure 2B) supports the specificity for the EC cell origin of the tumor. These results indicate that the pattern of expression of ISC marker genes and differentiation genes in SI-NET cells resembles reserve ISC marker gene expressing EC cells at the +4 position (Summarized in Supplementary Table 2).
Figure 2.
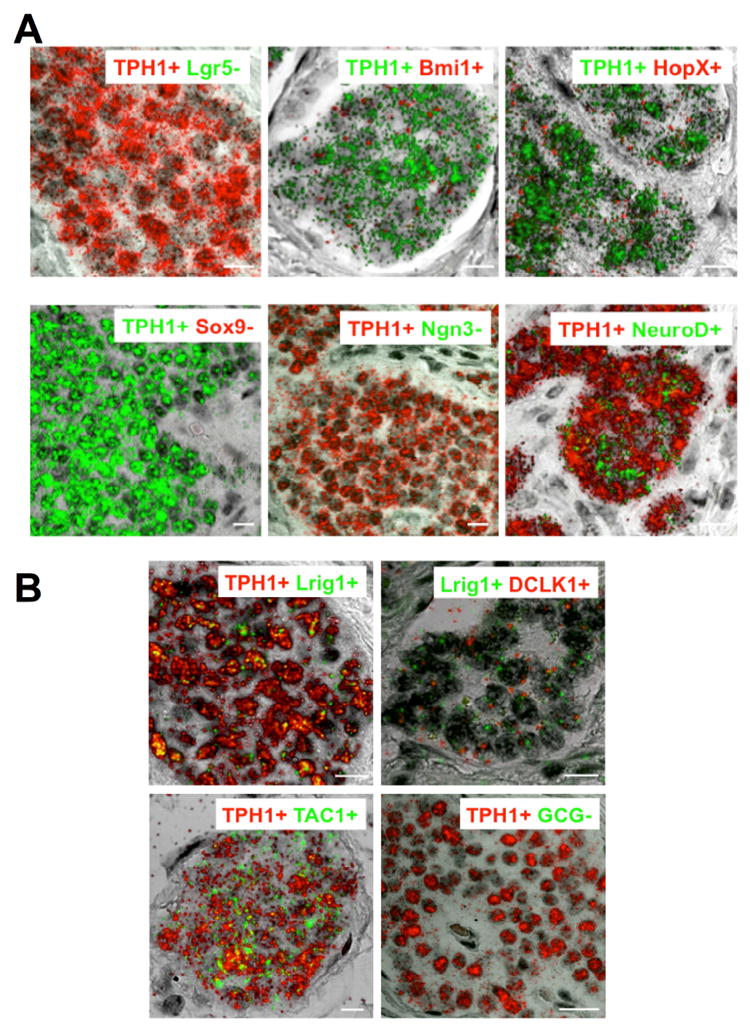
Expression of ISC and differentiation genes in SI-NETs in patients with Familial SI-NET. (A) Representative images of RNA-ISH of FFPE sections from an ileal SI-NET (patient F2) merging fluorescent and transmitted light images to visualize tumor cell location and indicated gene expression. Images show co-expression of Bmi1, HopX and NeuroD1 with TPH1 in the majority of the SI-NETs. There was no or negligible expression of Lgr5, Sox9 or Ngn3 mRNA in most of the TPH1+ tumors examined (See supplementary Tables). Expression of Bmi1, HopX and Sox9 was detected by fast red fluorescence (red) while expression of Lgr5, Ngn3 and NeuroD1 was detected by fast blue fluorescence (green). Expression of TPH1 was detected by either fast red (red) or fast blue (green) fluorescence. Scale bars, 10 μm. (B) Expression of ISC marker genes Lrig1and DCLK1 and differentiation marker genes GCG and Tac1 in the SI-NET cells. (Top left) Co-expression of Lrig1 and TPH1. (Top right) Co-expression of Lrig1 and DCLK1. (Bottom left) Co-expression of Tac1 and TPH1. (Bottom right) Absence of GCG expression in TPH1+ tumor cells. Expression of TPH1 and DCLK1 was detected by fast red fluorescence (red) and expression of Lrig1, Tac1 and GCG was detected by fast blue fluorescence (green). Scale bars in bottom right, 20 μm; for the rest, 10 μm. Representative RNA-ISH image for GCG was from patient F5. The images for Bmi1, Lrig1 and Tac1 were from patient F1 and the other images from patient F2.
SI-NET cells originate from the epithelium at the bottom of the crypt
Previous studies suggest that SI-NETs originate from intraepithelial enteroendocrine cells 24, 25. Consistent with this hypothesis, intraepithelial micro-tumors were found in six patients (F2, F10, F11, F12, F13 and F14, Supplementary Table 3 and Figure S4) in this study. This was most clearly demonstrated in patient F2 in whom TPH1 RNA-ISH in tumor sections showed several distinct micro-tumor formations within the epithelial layers of the adjacent crypt-like structures (Figure 3A and 3B). The epithelial layer where the tumor cells locate in Figure 3A was clearly continuous with the top epithelial layer of the villus. Villus structures were deformed due to the accumulation of the tumor nests in the lamina propria. Because it was difficult to determine the structural nature of the epithelial layers in these crypt-like structures in the absence of distinct Paneth cells, we examined the expression of Lgr5 as a marker of ISC cells residing at the crypt base. Lgr5 RNA-ISH in the next serial section revealed the location of Lgr5+ ISC cells at the bottom of the epithelial layer, indicating that the bottom was indeed the crypt base. Thus we could convincingly recognize the position of the TPH1+ cell cluster at the crypt bottom and identified this crypt as an early stage of ACEC. The presence of Lgr5+/TPH1+ doubly positive cells within the micro-tumor formation in this ACEC (Figure 3A) suggests that the micro-tumor originated from Lgr5+/TPH1+ EC cells. Furthermore, these doubly positive cells were specific to the micro-tumor in this early ACEC as no Lgr5 expression was seen in the tumor cells elsewhere within the tumor section. In the same section, a TPH1+ micro-tumor formation (Figure 3B, labeled as L in the square) consisted of a small cluster of tumor cells at the bottom and a larger cluster of tumor cells above, which are likely connected (intervening sections were not available) and more advanced than the early ACEC shown in Figure 3A. Interestingly, in contrast to the early ACEC (Figure 3A), this advanced ACEC (L, Figure 3B) showed no Lgr5 expression. The absence in Lgr5 expression in this advanced ACEC and positive expression in the ISCs of the same crypt base makes technical failure of Lgr5 ISH an unlikely explanation. Similarly, the tumor free crypt without a TPH1+ tumor cell cluster (Figure 3B, R) next to the tumor-filled crypt (Figure 3B, L) serves as a negative control for TPH1 ISH.
Figure 3.
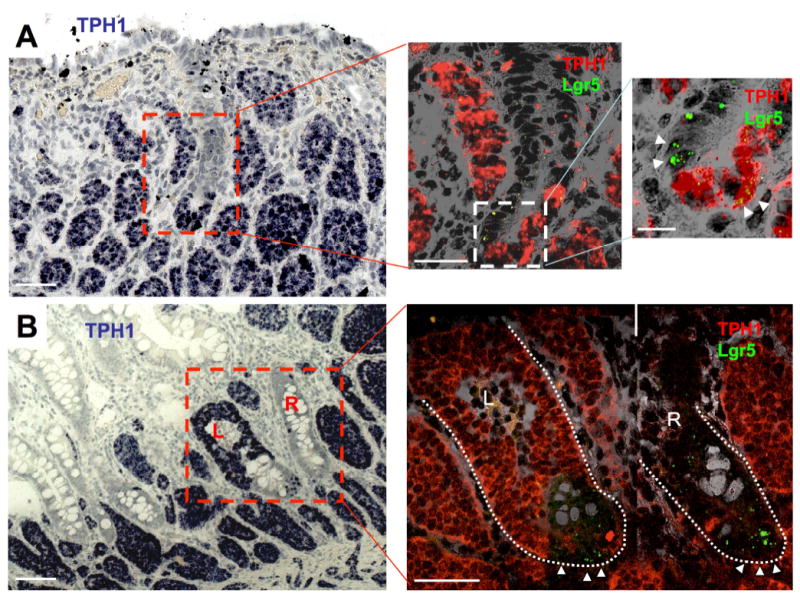
Early carcinoid tumorigenesis in a patient with Familial SI-NET. (A and B) Representative images of chromogenic (fast blue) RNA-ISH for expression of TPH1 in a FFPE section of an ileal SI-NET (patient F2) identifies multiple extra-epithelial tumor nests and a micro-tumor formation in the epithelium, ACEC, (center bottom in the red squared region, left panels). (A, center) TPH1 (fast red fluorescence, red) and Lgr5 (fast blue fluorescence, green) RNA-ISH of the next serial section in the area of the inset (outlined in red) from the left panel merged with the transmitted light image to illustrate the presence of expected TPH1-/Lgr5+ rapidly cycling ISCs and an unusual collection of TPH1+/Lgr5+ epithelial cells. (A, right) The enlarged image of the inset from the center panel more clearly shows the presence of TPH1-/Lgr5+ rapidly cycling ISCs at the crypt base (left two white triangles) and TPH1+/Lgr5+ putative tumor cells (right two white triangles) in the formation of TPH1+ tumor cluster at the crypt base. (B, right) TPH1 (fast red fluorescence, red) and Lgr5 (fast blue fluorescence, green) RNA-ISH of the next serial section in the area of the inset (outlined in red) from the left panel merged with the transmitted light image to illustrate an advanced ACEC (L) and a normal unaffected crypt (R). Triangles show TPH1(-)/Lgr5+ rapidly cycling ISCs at the crypt base. Scale bars in A-left and B-left, 100 μm; in A-center and B-right, 50 μm; in A-right, 10 μm.
Reserve stem cell markers are expressed in the early intraepithelial tumor formation within the ACEC
Double RNA-ISH for TPH1 and HopX from a tumor section from patient F2 contained an early ACEC (Figure 4A) that, unlike the intraepithelial formations in Figure 3A and B, contained Paneth cells marking the crypt bottom. The TPH1+ tumor cells lined up from position +4 to position 15 within the epithelium. The characteristics of these cells and the presence of Paneth cells suggested that this intraepithelial formation was at the initial stage of tumorigenesis. Most of the TPH1+ tumor cells including the cells at positions +4 and 6 expressed HopX (Figure 4A, white arrows). Bmi1 RNA-ISH of the next serial section also showed expression of Bmi1 in most of the TPH1+ tumor cells including the cells at positions +4 and 6 (Figure 4A, white arrows). Consistent with these findings, Bmi1 protein was expressed in most of the tumor cells in the early ACEC (Supplementary Figure S2). These results suggest that the reserve ISC marker genes were expressed in an early tumor formation within the ACEC. Unlike Lgr5 in Figure 3, Bmi1 and HopX were also expressed in extra-epithelial tumor nests in these sections.
Figure 4.
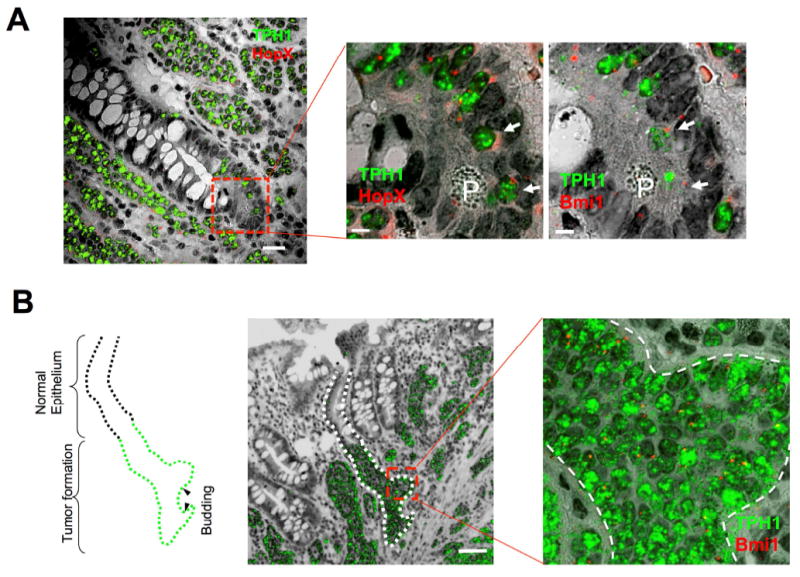
Expression of both reserve stem cell marker genes and differentiation genes in early tumorigenesis of SI-NETs. (A) Expression of HopX and Bmi1 in an early intraepithelial SI-NET formation, ACEC, in an FFPE section from an ileal tumor in a patient with familial SI-NET (F2). The merged image of HopX (fast red fluorescence, red) and TPH1 (fast blue fluorescence, green) with transmitted light image shows expression of TPH1 in multiple extraepithelial tumor nests and an early stage of ACEC (dashed square inset). (A, center) Enlarged image of the square inset region in the figure A-left illustrating a portion of abnormal cluster of doubly positive TPH1+/HopX+ cells including cells at positions 4 and 6 (white arrows). P indicates Paneth cell. (A, right) Enlarged image of a portion of abnormal cluster of Bmi1 (red) and TPH1 (green) expressing cells including cells at positions 4 and 6 (white arrows) obtained from a different RNA-ISH of the next serial section and same square inset shown in the A-center. P indicates Paneth cell. Scale bars in A-left, 20 μm; in A-center and -right, 5 μm. (B) Images of SI crypt budding of tumor cells from an intraepithelial SI-NET advanced ACEC, in the ileal epithelium. A merged image of a FFPE section from a SI-NET RNA-ISH of TPH1 expressing tumor cells (green) with the transmitted light image shows TPH1 positive SI-NET formation in the epithelium (outlined by dotted line). The outlined drawing (left) of this ACEC illustrates putative areas of nascent budding. Closer evaluation of budding areas designated by red square correspond to the enlarged image (right) of RNA-ISH of Bmi1 (red) and TPH1 (green) expression merged with transmitted light images. The dashed line in the illustration indicates the border of normal epithelium and tumor cell-filled epithelium. Arrowheads in the illustration mark putative areas of early budding. Scale bar, 50 μm.
The SI-NETs expand by budding
RNA-ISH in some sections from patient F2 showed early tumor spread suggestive of budding from the tumor–filled crypt (Figure 4B). This epithelial tumor formation in the ACEC was more advanced compared to the tumor formations in the ACECs present in Figures 3 and 4A. Bmi1 was expressed throughout this intraepithelial tumor formation including the regions of the buds (Figure 4B). In addition to patient F2, the budding formations were also observed in tumors from patients F11, F12 and F13 (Supplementary Table 3 and Figure S4). These findings suggest that SI-NETs develop at least in part by budding.
SI-NETs originate from the crypt bottom in a multifocal and polyclonal fashion
Multiple ACECs in a single SI-NET mass are shown in multiple sections (Figures 3 and 4) from the same paraffin-embedded block of a single tumor from patient F2. The presence of spatially separate multiple early ACECs, suggests a multifocal process of tumor formation from cells expressing EC and ISC marker genes at the crypt-bottom. To determine whether each ACEC arose independently or from a single cell-generated mutant patch, we analyzed two well-separated tumors that were unlikely to belong to the same patch formed from a single mutant cell. For this purpose, we examined multiple sections of two tumors from patient F10 that were separated by 7.5 cm along the ileum for the presence of ACECs (see mathematical explanation in the Supplementary Result and Discussion). ISH for ChgA and Bmi1 revealed multiple ACECs in both tumors (representative ACECs are shown in Figure 5) that were similar to ACECs seen in the patient F2 (Figures 3A and 4A) with well-confined clusters of cells in each ACEC. Therefore the presence of more than one ACEC in both tumor samples suggested that multifocal primary tumors originate independently in a polyclonal process.
Figure 5.
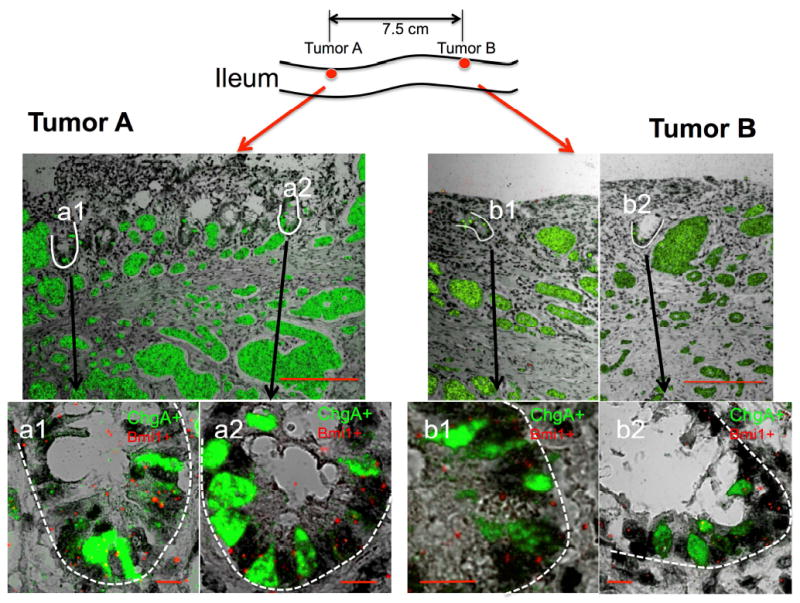
Presence of multifocal ACECs within two separate tumors. RNA-ISH shows expression of ChgA (green) in FFPE sections from ileal tumors A and B that were separated by 7.5 cm of grossly normal bowel in a patient (F10) with familial carcinoid. Scale bars in top panels, 200 μm. Bottom panels show enlarged images of the two outlined (white) ACECs (a1 and a2) in tumor A and two ACECs (b1 and b2) in tumor B. All images represent merged images of RNA-ISH for ChgA (green) and Bmi1 (red) doubly positive cells in abnormal clusters with transmitted light images. Scale bars in bottom panels, 10 μm.
To test this polyclonal tumor origin hypothesis, we used mitochondrial DNA (mtDNA)-based clonality analysis 26. The establishment of a homoplasmic mtDNA mutation within a single cell occurs through neutral drift and requires decades 26, 27. Once the homoplasmic mutation is established in an ISC and the ISC becomes dominant within a crypt (also through the process of neutral drift 28, 29), the mutation becomes traceable along the crypt-villus axis in the epithelial cells originating from this clone. Thus by tracing the mtDNA mutation we can distinguish the origin of the intestinal epithelial cells 26. The mtDNA-encoded cytochrome c oxidase (CCO) enzyme histochemistry mutations allow in vivo lineage tracing visualization 26. Using this technique, we examined the clonality of the ACECs and extraepithelial tumor nests in a 152-cm length of an isolated ileal segment containing more than 130 separate SI-NETs from patient F13. Frozen sections from 34 tumors and macroscopic tumor-free areas were assayed for CCO enzyme activity and the presence of ChgA in ACECs and tumors by immunohistochemistry. Some of the CCO normal (CCO+) and deficient (CCO-) tumor cells were laser-captured, micro-dissected and mtDNA sequenced to verify the presence or absence of mutations corresponding to the observed CCO enzymatic activity. In normal ileal epithelium, there were three groups of crypts; crypts containing 1) only CCO+ cells (brown), 2) only CCO- cells (blue) or 3) a mixture of CCO+ and CCO- cells (CCO+/-) (Figure 6A). The average frequency of CCO deficient (CCO- and CCO+/- combination) crypts was 2.2 % of 12,379 total crypts examined. There was a tendency toward increasing frequency of CCO- crypts in more distal areas of the ileum. Screening of these 12,379 CCO crypts for clusters of ChgA+ cells identified 0.27% that were ACECs. Approximately 6% of these ACECs that were randomly distributed over a 152 cm length of the tissue, consisted of a mixture of CCO+/-, ChgA+ cell clusters (Figure 6B). These results from CCO-based analysis suggest that the ACECs originated in a multifocal and polyclonal fashion.
Figure 6.
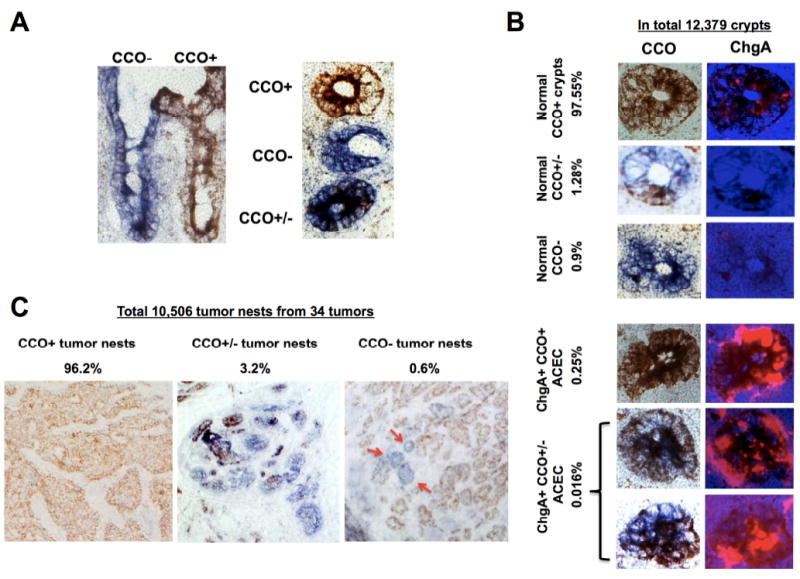
Frequency and distribution of cytochrome c oxidase (CCO) enzyme deficiency in human normal ileal mucosa and the SI-NETs. (A) Left: CCO enzyme histochemistry of representative frozen sagittal (left) and cross-sections (right) of normal ileal mucosa from a patient with familial SI-NET (F13) illustrating the presence of neighboring CCO- (blue) and CCO+ (brown) and CCO+/- (mixture) crypt-villus structures. (B) Representative images of five groups of crypts categorized on the basis of CCO histochemistry and ChgA immunofluorescence in an ileal segment from a patient with familial SI-NET (F13). The frequency of each category is given as a percent of the 12,379 crypts examined. (C) Representative images of three groups of tumor cell nests categorized on the basis of CCO histochemistry of ileal tumors from a patient with familial SI-NET (F13). The frequency of each category is given as a percent of the 10,506 tumor nests examined from 34 tumors. Tumor nests appear as either CCO+ (brown, left), CCO+/- (mixture of brown and blue, middle) or pure CCO- (blue, right). While the majority of tumor nests are CCO+ (brown), three notable CCO- (blue) tumor nests (red arrows) are present in this tumor.
We then examined extra-epithelial tumor nests in multiple tumors. Since extra-epithelial tumor nests were the consequence of ACECs, we expected that at least a small proportion of the tumor nests were CCO-deficient. In 34 examined tumors, 10,506 tumor nests larger than 50 μm2 were assessed for CCO activity. More than 96% of tumor nests were CCO+ (Figure 6C). As expected, however, 3.8% of 10,506 tumor nests were either CCO+/- (3.2%) or CCO- (0.6%). Therefore CCO enzyme-based lineage analysis of intraepithelial ACECs and more advanced extra-epithelial tumor nests indicate that both the ACECs and the tumors originated from both pure CCO+ crypts as well as from CCO deficient (CCO- or CCO+/-) crypts in this patient. These results from CCO-based lineage analysis of normal ileum tumor nests and especially early ACECs, indicate that familial SI-NETs originate from the crypt-bottom in a multifocal and polyclonal fashion.
Finally, we examined the macroscopic tumor-free areas (between tumors) for putative ACECs that might represent nascent tumor formation to characterize the three-dimensional organization of the cluster of ChgA+ cells within the ACEC. Anticipating rare ACECs, we prepared isolated crypts from large areas of macroscopic tumor-free ileal mucosa from patient F14 and screened for ACECs on the basis of IHC for ChgA (Figure 7). Two ACECs were identified among approximately 790 crypts examined. Three-dimensional confocal fluorescence image analysis revealed cell clusters consisting of at least 15 ChgA+ cells positioned at the base of the crypt. mtDNA sequencing of these isolated aberrant crypts identified a homoplasmic 7337 G>A mutation in MT-CO1 in one crypt while the other crypt was wild type G at this position. These findings indicate that these two ACECs originated from different stem cells and are consistent with earlier results suggesting that familial SI-NETs originate from the crypt-bottom in a multifocal and polyclonal fashion. These results also indicate that there were multifocal microscopic nascent tumor formations in a relatively large area of the small intestine that is consistent with the multifocal synchronous primary tumors characteristic of patients with Familial SI-NET.
Figure 7.
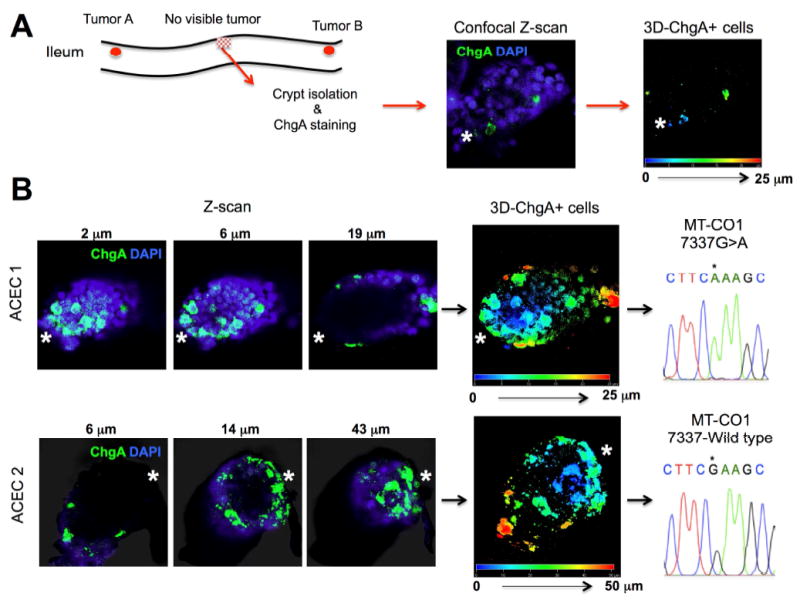
Confocal 3D image analysis and mtDNA sequencing of abnormal clusters of ChgA+ cells in the two isolated ACECs from the grossly normal appearing ileal mucosa from a patient with familial SI-NET (F14). (A) The crypts from grossly tumor-free mucosa were isolated from a patient with the familial SI-NET (F14), immunostained for ChgA, and screened for abnormal clusters of ChgA+ cells. Immuno-positive cyrpts were imaged and analyzed for the number and position of ChgA+ cells using 3D reconstruction from Z-stacked confocal images. (B) Two of approximately 790 crypts were found to contain abnormal clusters of ChgA + cells at the crypt bottom. Representative Z-scan images (ChgA in green and DAPI in blue) and 3D positions of ChgA+ cells appear in multiple colors corresponding to their depth in each 3D reconstructed ACEC. The approximate position of the crypt base is denoted by a white asterisk. Each of these crypts was manually isolated for sequencing of mtDNA. ACEC 1 harbored a homoplastic 7337 G>A (*) mutation in MT-CO1 while the ACEC 2 maintained a wild type G (*).
Discussion
The present RNA-ISH study found a subset of EC cells that express ISC marker genes in human ileal mucosa. As expected, these cells were found mostly at and below the +4 position. More than 70% of the EC cells located at the crypt base below +4 position expressed rapidly cycling ISC gene Lgr5 and at least 70% of the EC cells located at +4 position expressed reserve ISC genes Bmi1 and HopX. The presence of EC cells having overlapping expression of Bmi1, HopX and Lgr5 are mathematically apparent (Figure 1B). How these ISC gene-expressing EC cells differ from a majority of ISC marker gene-negative EC cells is currently unknown. However, it is suggested that these cells are the endocrine (secretory) lineage cells that possess stem cell as well as potentially tumorigenetic properties 8,17,22.
Since the SI-NETs examined in this study were positive for TPH1, Tac1, NeuroD1 and ChgA and negative for GCG or NTS, the results support the generally accepted proposal that the SI-NET originates from a group of serotonin and substance P producing EC cells 1-4. Our interest was to estimate if this cell of origin belongs to the ISC gene-expressing subset. Interestingly we have found that all of the familial SI-NETs examined in this study highly expressed reserve ISC genes Bmi1 and HopX. The tumors were also consistently positive for other reserve ISC genes Lrig1 and DCLK1. Therefore, these results suggest that SI-NETs may originate from EC cells expressing reserve ISC genes. Supporting this concept, we identified Bmi1+ and HopX+ tumor cells in nascent micro-tumor formations in the ACEC (Figure 4, Figure S2 and S4). While we identified Lgr5+ tumor cells in a micro-tumor formation in an ACEC (Figure 3A), there was little to no expression of Lgr5 in another micro-tumor formation in another ACEC (Figure 3B) as well as the majority of SI-NETs examined in this study. We hypothesize that initial Lgr5 expression in the micro-tumor was from the reserve ISCs as recently reported in the label retaining Lgr5+ cells 22 and Lgr5-low/Ki67- cells 17 and was subsequently lost during tumor progression as seen in the intestinal tumors from Lgr5-EGFP-creER × APC flox/flox mice 5. Whether this phenomenon is associated with the status of the Wnt active niche remains to be determined by further investigation. Overall RNA-ISH results from initial intraepithelial micro-tumor formations in the ACECs, progressive tumor nests and frank SI-NETs showed that the gene expression pattern of varying stages of the SI-NETs resemble the cells expressing EC cell marker genes TPH1, Tac1, NeuroD and ChgA and reserve ISC marker genes Bmi1, HopX and Lgr5low. These results therefore support the hypothesis that the cell of origin for SI-NET is the reserve ISC genes-expressing EC cell, the majority of which resides at +4 position. It should be noted that these findings were consistent across all the studied patients with familial SI-NET (Supplementary Table 2), including the three affected patients (F2, F4 and F5) from the recently identified family with a germline mutation in the IPMK gene 23. While further investigation will be necessary to determine whether this reserve ISC gene+ EC cell hypothesis applies to sporadic cases of SI-NETs, it is interesting to note that a similar ISC gene expression pattern was also observed in a sporadic case (S1) (Supplementary Table 2).
Consistent with the intraepithelial origin hypothesis by Lundqvist and Wilander 24, the early tumor formation at the crypt bottom in ACECs and distinct budding from the micro-tumor–filled crypt were evident as summarized in Supplementary Table 3 in this study. Thus the tumor originates from the crypt bottom, undergoes “bottom-up” morphogenesis and grows by budding and fission. Consistent with this observation, many of the submucosal tumor nests maintain crypt-like structures reminiscent of their origin (Figure S6) and thus show histologic architecture of so-called trabecular or insular patterns. In addition, within a single tumor, we identified a multifocal process of tumor formation, i.e., multifocal early and advanced ACECs (Figures 3 and 4). One possible explanation for this observation invokes a decades long process of establishment of a mutant patch 30 stemming from a single mutant cell through crypt fission cycles. In addition to this mechanism, a field cancerization effect 31, 25 by soluble transforming factors and exosomes 32 from the initial progenitor and/or its interaction with surrounding stromal and immune cells etc., may contribute to the multifocal genesis of ACECs.
The majority of familial SI-NET patients present with multiple synchronous primary tumors consistent with a germline mutant susceptibility gene 23. Following this logic, the multiple primary tumors are likely polyclonal in origin as observed in patients with familial adenomatous polyposis 33 or APC-mutant mouse models 34. The presence of multiple ACECs within two tumors that were separated by the distance that a mutant patch would not be able to span (Figure 5) in patient F10 suggests that these two tumors originated independently from different transformed cell clones rather than a single mutant cell-derived patch. More convincingly, the CCO histochemistry verified the polyclonal nature of the multifocal tumors in the patient F13 who had over 130 tumors. Finally, we also found nascent tumors in the form of ACECs, that were evidenced to be originated from different ISC clones by mtDNA-sequencing analysis, in the macroscopically normal appearing ileum between grossly detectable tumors in patient F14, thus suggesting wide-spread, polyclonal and multifocal genesis of the tumors.
In summary, we characterized different stages of tumor generation/progression in SI-NETS in patients with familial carcinoid and presented data suggesting that SI-NETs originate from EC cells expressing reserve ISC marker genes in a multifocal and polyclonal fashion (see proposed summary model in Supplementary Figure S7). Polyclonal ACECs in distal tumors and ACECs present in macroscopic tumor-free mucosa indicate a multifocal and polyclonal genesis of the tumors consistent with germline disease and the phenomenon of field cancerization 31. Investigating cell biological significance of reserve ISC gene-expressing EC cells, how it relates to the tumorigenesis and mechanisms of multifocal and polyclonal genesis of the tumors should help develop diagnostic and therapeutic inroads for this otherwise intractable disease.
Supplementary Material
Acknowledgments
Grant Support:
This work was supported by the Intramural Research Programs of the National Institute of Diabetes, Digestive and Kidney Diseases, Clinical Research Center, Frederick National Laboratory for Cancer Research and National Cancer Institute, National Institutes of Health.
Abbreviations in this paper
- ACEC
aberrant crypt containing endocrine cell clusters
- Bmi1
B lymphoma Mo-MLV insertion region 1 homolog
- ChgA
chromogranin A
- DCLK1
Doublecortin-like Kinase 1
- SI
small intestine
- NET
neuroendocrine tumor
- EC
enterochromaffin cell
- APC
adenomatous polyposis coli
- GFP
green fluorescent protein
- GIP
gastric inhibitory peptide
- HopX
homeodomain only protein x
- ISC
intestinal stem cell
- ISH
in situ hybridization
- Lgr5
Leucine-rich repeat-containing G-protein coupled receptor 5
- Lrig1
leucine-rich repeats and immunoglobulin-like domains protein 1
- TPH-1
tryptophan hydroxylase type 1
Footnotes
Authorship:
Study concept and design: Y.S., and S.A.W.
Acquisition of data: Y.S., J.F., X.Z., J.F., D.T., K.N., J.H., M.M.Q., M.S.H. and S.A.W.
Analysis and interpretation of data: Y.S., J.F., X.Z., J.F., D.T., K.N., J.H., M.M.Q., M.S.H. and S.A.W.
Manuscript drafting: Y.S., and S.A.W.
Disclosures:
The authors disclose no conflicts of interest.
Supplementary Material
Note: To access the supplementary material accompanying this article, visit the online version of Gastroenterology at www.gastrojournal.org.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Nilsson O. Profiling of ileal carcinoids. Neuroendocrinology. 2013;97:7–18. doi: 10.1159/000343232. [DOI] [PubMed] [Google Scholar]
- 2.Cunningham JL, Janson ET. The biological hallmarks of ileal carcinoids. Eur J Clin Invest. 2011;41:1353–60. doi: 10.1111/j.1365-2362.2011.02537.x. [DOI] [PubMed] [Google Scholar]
- 3.Lembeck F. The detection of 5-hydroxytryptamine in carcinoid metastases. Naunyn Schmiedebergs Arch Exp Pathol Pharmakol. 1954;222:235. [PubMed] [Google Scholar]
- 4.Lechago J. Neuroendocrine cells of the gut and their disorders. Monogr Pathol. 1990:181–219. [PubMed] [Google Scholar]
- 5.Barker N, Ridgway RA, van Es JH, et al. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature. 2009;457:608–11. doi: 10.1038/nature07602. [DOI] [PubMed] [Google Scholar]
- 6.Humphries A, Wright NA. Colonic crypt organization and tumorigenesis. Nat Rev Cancer. 2008;8:415–24. doi: 10.1038/nrc2392. [DOI] [PubMed] [Google Scholar]
- 7.Sangiorgi E, Capecchi MR. Bmi1 is expressed in vivo in intestinal stem cells. Nat Genet. 2008;40:915–20. doi: 10.1038/ng.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mills JC, Sansom OJ. Reserve stem cells: Differentiated cells reprogram to fuel repair, metaplasia, and neoplasia in the adult gastrointestinal tract. Sci Signal. 2015;8:re8. doi: 10.1126/scisignal.aaa7540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barker N, van Es JH, Kuipers J, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–7. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 10.Cheng H, Leblond CP. Origin, differentiation and renewal of the four main epithelial cell types in the mouse small intestine. V. Unitarian Theory of the origin of the four epithelial cell types. Am J Anat. 1974;141:537–61. doi: 10.1002/aja.1001410407. [DOI] [PubMed] [Google Scholar]
- 11.Tian H, Biehs B, Warming S, et al. A reserve stem cell population in small intestine renders Lgr5-positive cells dispensable. Nature. 2011;478:255–9. doi: 10.1038/nature10408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yan KS, Chia LA, Li X, et al. The intestinal stem cell markers Bmi1 and Lgr5 identify two functionally distinct populations. Proc Natl Acad Sci U S A. 2012;109:466–71. doi: 10.1073/pnas.1118857109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Potten CS, Kovacs L, Hamilton E. Continuous labelling studies on mouse skin and intestine. Cell Tissue Kinet. 1974;7:271–83. doi: 10.1111/j.1365-2184.1974.tb00907.x. [DOI] [PubMed] [Google Scholar]
- 14.Takeda N, Jain R, LeBoeuf MR, et al. Interconversion between intestinal stem cell populations in distinct niches. Science. 2011;334:1420–4. doi: 10.1126/science.1213214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.May R, Sureban SM, Hoang N, et al. Doublecortin and CaM kinase-like-1 and leucine-rich-repeat-containing G-protein-coupled receptor mark quiescent and cycling intestinal stem cells, respectively. Stem Cells. 2009;27:2571–9. doi: 10.1002/stem.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Powell AE, Wang Y, Li Y, et al. The pan-ErbB negative regulator Lrig1 is an intestinal stem cell marker that functions as a tumor suppressor. Cell. 2012;149:146–58. doi: 10.1016/j.cell.2012.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Basak O, van de Born M, Korving J, et al. Mapping early fate determination in Lgr5+ crypt stem cells using a novel Ki67-RFP allele. EMBO J. 2014;33:2057–68. doi: 10.15252/embj.201488017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sei Y, Lu X, Liou A, et al. A stem cell marker-expressing subset of enteroendocrine cells resides at the crypt base in the small intestine. Am J Physiol Gastrointest Liver Physiol. 2011;300:G345–56. doi: 10.1152/ajpgi.00278.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schonhoff SE, Giel-Moloney M, Leiter AB. Neurogenin 3-expressing progenitor cells in the gastrointestinal tract differentiate into both endocrine and non-endocrine cell types. Dev Biol. 2004;270:443–54. doi: 10.1016/j.ydbio.2004.03.013. [DOI] [PubMed] [Google Scholar]
- 20.Van Landeghem L, Santoro MA, Krebs AE, et al. Activation of two distinct Sox9-EGFP-expressing intestinal stem cell populations during crypt regeneration after irradiation. Am J Physiol Gastrointest Liver Physiol. 2012;302:G1111–32. doi: 10.1152/ajpgi.00519.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Es JH, Sato T, van de Wetering M, et al. Dll1+ secretory progenitor cells revert to stem cells upon crypt damage. Nat Cell Biol. 2012;14:1099–104. doi: 10.1038/ncb2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Buczacki SJ, Zecchini HI, Nicholson AM, et al. Intestinal label-retaining cells are secretory precursors expressing Lgr5. Nature. 2013;495:65–9. doi: 10.1038/nature11965. [DOI] [PubMed] [Google Scholar]
- 23.Sei Y, Zhao X, Forbes J, et al. A Hereditary Form of Small Intestinal Carcinoid Associated With a Germline Mutation in Inositol Polyphosphate Multikinase. Gastroenterology. 2015;149:67–78. doi: 10.1053/j.gastro.2015.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lundqvist M, Wilander E. A study of the histopathogenesis of carcinoid tumors of the small intestine and appendix. Cancer. 1987;60:201–6. doi: 10.1002/1097-0142(19870715)60:2<201::aid-cncr2820600214>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 25.Moyana TN, Satkunam N. A comparative immunohistochemical study of jejunoileal and appendiceal carcinoids. Implications for histogenesis and pathogenesis. Cancer. 1992;70:1081–8. doi: 10.1002/1097-0142(19920901)70:5<1081::aid-cncr2820700512>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 26.Wright NA. Stem cell identification--in vivo lineage analysis versus in vitro isolation and clonal expansion. J Pathol. 2012;227:255–66. doi: 10.1002/path.4018. [DOI] [PubMed] [Google Scholar]
- 27.Taylor RW, Barron MJ, Borthwick GM, et al. Mitochondrial DNA mutations in human colonic crypt stem cells. J Clin Invest. 2003;112:1351–60. doi: 10.1172/JCI19435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lopez-Garcia C, Klein AM, Simons BD, et al. Intestinal stem cell replacement follows a pattern of neutral drift. Science. 2010;330:822–5. doi: 10.1126/science.1196236. [DOI] [PubMed] [Google Scholar]
- 29.Snippert HJ, van der Flier LG, Sato T, et al. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell. 2010;143:134–44. doi: 10.1016/j.cell.2010.09.016. [DOI] [PubMed] [Google Scholar]
- 30.Greaves LC, Preston SL, Tadrous PJ, et al. Mitochondrial DNA mutations are established in human colonic stem cells, and mutated clones expand by crypt fission. Proc Natl Acad Sci U S A. 2006;103:714–9. doi: 10.1073/pnas.0505903103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dotto GP. Multifocal epithelial tumors and field cancerization: stroma as a primary determinant. J Clin Invest. 2014;124:1446–53. doi: 10.1172/JCI72589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Melo SA, Sugimoto H, O’Connell JT, et al. Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer Cell. 2014;26:707–21. doi: 10.1016/j.ccell.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thirlwell C, Will OC, Domingo E, et al. Clonality assessment and clonal ordering of individual neoplastic crypts shows polyclonality of colorectal adenomas. Gastroenterology. 2010;138:1441–54. 1454 e1–7. doi: 10.1053/j.gastro.2010.01.033. [DOI] [PubMed] [Google Scholar]
- 34.Thliveris AT, Schwefel B, Clipson L, et al. Transformation of epithelial cells through recruitment leads to polyclonal intestinal tumors. Proc Natl Acad Sci U S A. 2013;110:11523–8. doi: 10.1073/pnas.1303064110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


