Abstract
Inter-specific hybridization and backcrossing commonly occur in plants. The use of progeny generated from inter-specific hybridization and backcrossing has been developed as a novel model system to explore gene expression divergence. The present study investigated the analysis of gene expression and miRNA regulation in backcrossed introgression lines constructed from cultivated and wild rice. High-throughput sequencing was used to compare gene and miRNA expression profiles in three progeny lines (L1710, L1817 and L1730), with different plant heights resulting from the backcrossing of introgression lines (BC2F12) and their parents (O. sativa and O. longistaminata). A total of 25,387 to 26,139 mRNAs and 379 to 419 miRNAs were obtained in these rice lines. More differentially expressed genes and miRNAs were detected in progeny/O. longistaminata comparison groups than in progeny/O. sativa comparison groups. Approximately 80% of the genes and miRNAs showed expression level dominance to O. sativa, indicating that three progeny lines were closer to the recurrent parent, which might be influenced by their parental genome dosage. Approximately 16% to 64% of the differentially expressed miRNAs possessing coherent target genes were predicted, and many of these miRNAs regulated multiple target genes. Most genes were up-regulated in progeny lines compared with their parents, but down-regulated in the higher plant height line in the comparison groups among the three progeny lines. Moreover, certain genes related to cell walls and plant hormones might play crucial roles in the plant height variations of the three progeny lines. Taken together, these results provided valuable information on the molecular mechanisms of hybrid backcrossing and plant height variations based on the gene and miRNA expression levels in the three progeny lines.
Introduction
Rice (Oryza sativa), one of the most important food crops, feeds a large number of the world’s population. With the availability of reference genome sequences, rice has also become an eminent model system in plant research for monocotyledon species. The genus Oryza, including 2 cultivated species (O. sativa and O. glaberrima) and 22 wild species, was classified into 10 genome types, including the AA, BB, CC, BBCC, CCDD, DD(EE), FF, GG, HHJJ and HHKK genomes [1]. The AA genome contains 8 species in the genus Oryza, consisting of 2 cultivated species and 6 wild species (O. meridionalis, O. rufipogon, O. nivara, O. glumaepatula, O. barthii and O. longistaminata). The wild species in the genus Oryza are a valuable source of novel genetic variation, and the majority of genetic variation in the genus Oryza remains untapped [2]. O. sativa includes 2 primary groups, the indica and japonica subspecies. Rice cultivar 9311, an elite parental line of indica, was used to develop super hybrid rice in China, such as cultivar LYP9 [3]. O. longistaminata, a native to African rhizomatous perennial wild rice species, potentially diverged from O. glaberrima approximately 1.9 MYA [4]. Additionally, O. longistaminata contains many potentially useful characters, such as strong rhizomes, and strong resistance to stress [5, 6]. Several genes, such as Rhz2, Rhz3 and Xa21, have been identified and used for developing perennial rice and blight disease resistance rice [5, 7].
Hybrid rice with a combination of divergent genomes, leading to instant and profound genome modifications, has been effectively utilized to increase rice production based on their novel phenotypes, such as grain yield, stress tolerance, biomass production and plant height. As the majority of genetic variation primarily existed in wild species of the genus Oryza [2], the introduction of novel genes or gene-networks into cultivated rice from the wild species is an effective strategy to increase rice yield. To date, many backcross introgression lines (BILs), introgression lines (ILs) and near-isogenic lines (NILs) have been developed to introduce novel characters from the wild or resistant species into the elite breeding material in many crop species [8, 9]. Two sets of BC7F2 lines derived from RD23 (O. sativa) and O. longistaminata were generated to examine hybrid sterility and plant height, using genetic maps and molecular markers [9]. To detect the grain mineral concentration of of BILs in two environments, 2 elite indica varieties were crossed with a Zn-dense rice variety and 2 sets of BILs were raised [8]. A BC4F2 population of ILs was constructed to identify and map a novel blast resistance gene using SSR markers [6]. Studies on metabolic and gene expression levels in BILs and NILs have recently emerged. For example, to explore the impact of the introgression segment on a rice growth, the metabolic profiling and physiological trait analyses were performed between IL and its recurrent parent [10]. However, the roots and leaves of 2 pairs of NILs (drought-tolerant and susceptible rice) were used to unravel drought tolerant genes under different drought stress conditions through transcriptome analysis [11, 12]. Moreover, several genotype-specific drought-induced genes and higher expressed genes were identified as contributors to drought tolerate using comparative transcriptome sequencing [13]. Therefore, the BILs and ILs of rice are invaluable resources for rice genetic improvement, particularly those with wild rice as the donor parent. These studies on BILs and ILs of rice primarily focused on stress resistance but rarely examined plant height.
Micro-RNAs (miRNAs) of 20–30 nt are endogenously expressed short non-coding small RNAs (sRNAs) affecting gene expression through post-transcriptional mechanisms, similar to other classes of sRNAs. When miRNAs near-perfectly complement to their target mRNAs, target gene expression will be disturbed, or mRNA translation will be inhibited in plants [14]. To date, many miRNA studies have been conducted, and the essential roles of miRNAs in many biological processes, including various metabolic pathways, signal transduction and stress responses, have been characterized in plants [15–19]. In plant species, most miRNAs are highly conserved, but the expression levels of non-conserved miRNAs are lower, which is difficult to examine using small-scale sequencing projects. Recently, high throughput sequencing as an effective method have identified and estimated the expression profiles of miRNAs in different plant tissues [3, 18, 20, 21], and many low abundant and novel non-conserved miRNAs have been identified. In the rice miRBase, 713 miRNAs have been identified, including those associated with plant architecture [22], stem and grain development [20, 23].
Recently, studies on the integrated analysis of mRNA and miRNA expression profiling have been reported [18, 19, 21, 24–27]. A comprehensive interaction analysis of miRNA and mRNA revealed the regulatory mechanisms of plant height in Gossypium hirsutum [19], provided small RNA-mediated regulation mechanisms in hexaploid wheat [21], and identified a number of important regulators in tomato during Alternaria-stress responses [27]. The analysis of miRNAs and mRNAs expression levels between indica and japonica rice subspecies revealed that the target gene expression was more easily repressed by highly conserved miRNAs than by lowly conserved miRNAs [18]. However, little effort has been made toward exploring the mRNA and miRNA expression diversity of ILs rice and their parents to study the molecular mechanism associated with phenotypic changes, particularly plant height.
In the present study, next-generation high-throughput sequencing technology and bioinformatics were utilized to present an integrative analysis of the mRNA and its regulatory miRNA networks in three progeny lines of BILs and their parents. We focused on the following: (1) we examined mRNAs and miRNAs expression level changes in three progeny lines; (2) we determined whether the differentially expressed mRNAs and miRNAs in the three progeny lines were influenced by genome composition; and (3) we conducted an integrative analysis of the mRNA and miRNA regulation network in three progeny lines and their parents, which was essential to clarify the modulation of gene expression at the post-transcriptional level. These results will provide a comprehensive assessment of mRNA and miRNA expression levels, thereby increasing the current knowledge of the complex molecular mechanism of gene introgression that resulting in plant height variations in the three progeny lines.
Materials and methods
Plant materials and growth conditions
Three progeny lines of backcrossed introgression lines (BC2F12) and their parents, O. longistaminata and O. sativa ssp. indica cv. 9311, were used as plant materials in the present study (all plant materials were provided by State Key Laboratory of Hybrid Rice, Wuhan University, Wuhan, China). BC2F12 (containing 152 lines) were constructed from O. sativa as the female crossed with O. longistaminata to generate F1 hybrid; later, F1 as male was backcrossed with O. sativa to develop BC1F1, and 65 BC1F1 individuals were for backcrossing again with O. sativa to develop BC2F1 plants and subsequently BC2F1 using a single seed descent method by 11 generation self-fertilization to produce BC2F12. Three progeny lines, namely L1710, L1817 and L1730, with different plant height were used in subsequent studies. Most of the chromosome complements of the three progeny lines were inherited from O. sativa, and a fraction of the complements were inherited from O. longistaminata (15.72% in L1710, 10.32% in L1817 and 16.42% in L1730), which were calculated based on their SNP loci (unpublished, S1 Fig). Additionally, heterozygosity chromosome complements were most abundant in L1817 among the three progeny lines. Germinated seeds of the three progeny lines and O. sativa were sown in soil, and at approximately 30 days old, the seedlings were transplanted into plots in the greenhouse. The wild species O. longistaminata is a perennial plant, and rhizomes are essential organs for its rapid growth at the beginning of the next growing season. The rhizomes of O. longistaminata were planted in plots and clonally propagated in the same greenhouse.
Histological observation
For histological examination, the third internodes of natural growing plants, L1710, L1817, L1730, O. sativa and O. longistaminata, were collected and fixed with FAA (formalin: glacial acetic acid: 50% ethanol = 1: 1.2: 17.8) solution. After being dehydrated in a graded ethanol series and being rendered transparent in a grade transparent agent series, internodes were embedded in paraffin. Subsequently, 10 μm thick sections were sectioned on a rotary microtome (Leica Biosystems, Nussloch, Germany) and placed onto glass slides. The tissue sections were observed under a light microscope (BX51; Olympus, Tokyo, Japan), and the cell lengths were measured, after Safranine (Sinopharm Chemical Reagent Co., Ltd, Shanghai, China) and Fast Green staining (Sigma Aldrich, USA).
Construction of mRNA and small RNA libraries
Stem tissue from five individual plants of each line at jointing-booting stage was collected and immediately stored in liquid nitrogen for total RNA extraction. Total RNAs from each line were separately extracted from the five individual stem tissues using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) and purified using RNase-free DNase I (Fermentas, MD, USA), and the mRNAs were used to synthesize cDNAs, which were subsequently qualified and quantified, as previously described [28]. Equal amounts of cDNA from five individual plants of the same line were combined into one library for sequencing using the Illumina HiSeq 2000 platform.
Total RNA extraction for constructing a small RNA sequencing library was similar to that of mRNA library. The sRNA selection, library construction and sequencing were performed as previously described [15].
Analysis of mRNA and small RNA sequencing data
Illumina sequencing, data processing and RNA sequencing generated read mapping to the reference genome (the Rice Genome Annotation Project, http://rice.plantbiology.msu.edu) were performed as previously described [29]. The Illumina HiSeq2000 sequencing, data processing and small RNA sequencing generated read mapping to the same reference genome were followed as previously described [30].
Evaluations of genes in RNA-Seq data and miRNAs in small RNA-Seq data
Using ERANGE software (version 4.0) (http://woldlab.caltech.edu/gitweb/), we evaluated the expression of genes in RNA-Seq data by assigning reads to rice reference genomes. The gene expression levels were normalized in terms of RPKM [31].
The miRNA read count was normalized to TPM (transcripts per million) using the formula: normalized expression = actual miRNA count/total count of clean reads×1,000,000.
To group all annotated genes/miRNAs from the three progeny lines and their parents with similar expression patterns, hierarchical cluster analysis was performed. The cluster analysis of genes and miRNAs were conducted using the Heatmap and Cluster 3.0 software with Pearson correlation as the distance measure, and the Tree view tool was used to visualize results [32].
Analysis of differentially expressed genes and miRNAs
Differential expression analysis was performed using R packages of DESeq [33]. The false discover rate (FDR) was used to correct and determine the threshold of the P value for multiple comparisons. In the present study, genes that exhibited fold changes ≥5 (log2 Ratio ≥2.322) and FDR ≤0.001 were defined as differentially expressed genes (DEGs). The miRNAs with an absolute value of log2 Ratio ≥1 and P value ≤0.05 were determined to show significant differential expression.
Functional annotation analysis of genes and miRNA target genes
All miRNA sequences from five lines were mapped to the rice reference genome (http://rice.plantbiology.msu.edu). The criteria for target predicting used in the present study were previously described [34, 35].
Gene ontology (GO) annotation (http://www.blast2go.org/) of all expressed genes and all predicted target genes of miRNAs were performed using the Blast2GO program (version 2.3.5). A web gene ontology annotation plot (WEGO) server (http://wego.genomics.org.cn/cgi-bin/wego/index.pl) was utilized for GO functional classification.
In the KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway analysis, all DEGs were mapped to the KEGG database terms. Based on the RPKM of the genes, we identified up/down-regulated pathways in the three progeny lines compared with their parents. The results were analyzed and visualized using Cytoscape software (version 2.6.2) (www.cytoscape.org/).
Real-time quantitative PCR validation (qRT-PCR)
To validate the accuracy of the Illumina sequencing, 12 genes and 6 miRNAs were randomly selected for qRT-PCR reactions. In addition, the expressions of 6 differentially expressed miRNAs and their predicted target genes were validated by qRT-PCR reactions. Total RNAs were extracted from the plant stems, and oligo (dT) was used to reverse transcribe the RNAs of genes, and specific primers for RT-qPCR were designed using the Primer 5.0 software (Premier Biosoft International, Palo Alto, CA, USA, http://www.premierbiosoft.com/index.html). Since miRNAs are short and without polyA tails, the reverse-transcription of miRNAs was completed using a miRNA-specific stem-loop primer, according to stem-loop primer designing rules [36]. To normalize each gene threshold cycle reaction, actin1 was used as internal reference gene, and the U6 snRNA was selected as an internal control for miRNA expression. The qRT-PCR reaction was performed as previously described [15, 29]. The validation analysis of genes and miRNAs were performed in triplicate with three biological replicates. All primer sequences used in the present study were listed in the S1 and S2 Tables.
Results
Comparison of stem traits among the three progeny lines and their parents
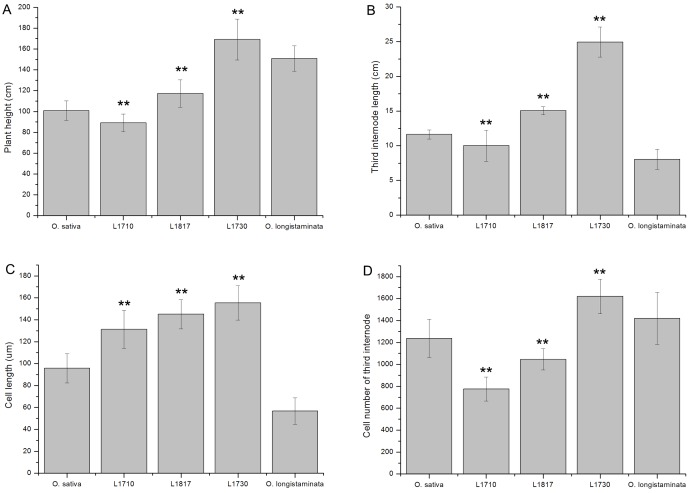
In the present study, the three progeny lines (L1710, L1817 and L1730) of a backcrossed introgression line (BC2F12), recurrent parent O. sativa and donor parent O. longistaminata were utilized. Compared with their parents, the plant height of L1730 was taller, and L1710 was shorter, while L1817 was taller than O. sativa, but shorter than O. longistaminata (Fig 1a). Rice plant height was primarily dependent on stem length, which was determined based on the number and length of internodes. In the present study, we observed that the total number of elongated internodes was the same in L1710, L1817, L1730 and O. sativa. Additionally, changes in the first, second, third, fourth, and fifth (uppermost) internode lengths of the three progeny lines and O. sativa showed similar trends. However, the number and length of internodes were unsteady in O. longistaminata. For the third internode length, L1817 and L1730 were longer than their parents, while L1710 was longer than O. longistaminata but shorter than O. sativa (Fig 1b). Internode elongation resulted from the cell division in the meristem and the cell elongation in the elongated region. To investigate whether variations of internode length were primarily attributed to the cell elongation and/or the cell division, the longitudinal sections of the third internodes of five lines were subjected to microscopic observation. Surprisingly, the cell length of three progeny lines was longer than that of their parents (Fig 1c). Among three progeny lines, the cell length of L1730 was longest, L1710 was shortest and L1817 was intermediate. The results showed that the cell length of the three progeny lines was consistent with their plant height variations. After calculating the cell number (internode length divided by the cell length of corresponding internode), we observed decreased cell number in L1710 compared with their parents, decreased cell number in L1817 compared with O. longistaminata, and increased cell number in L1730 compared with O. sativa (Fig 1d). These results showed that the increased cell length and cell number were important determinants of internode length in L1730, while L1817 internode length was primarily caused by increased cell length. However, the reduced cell number and increased cell length determined the internode length of L1710. To investigate the complex regulation of gene expression and characterize the various biological processes contributing to changes in plant architecture in the progeny lines, the gene and miRNA expression levels were characterized.
Fig 1. Morphological phenotypes characterizations of the three progeny lines and their parents.
(A) The rice height of five lines. (B) The third internode length of five lines. (C) The cell length of third internode in five lines. (D) The cell number of third internode in five lines. Asterisks indicate a significant difference (P< 0.01) between progeny line and their parents through t-test.
Analysis of gene expression in the progeny lines and their parents
To elucidate the gene expression patterns of stem in the three progeny lines and their parents, internodes at jointing-booting stage were used to construct RNA-Seq libraries sequenced using the Illumina HiSeq 2000 platform. After filtering low-quality reads and adaptor sequences, more than 11 million clean reads (11,959,710 in L1710, 12,636,297 in L1817, 11,832,864 in L1730, 11,792,127 in O. sativa and 11,800,115 in O. longistaminata) were obtained in each library constructed from these rice lines (S3 Table). Using SOAP aligner/soap2 software, 77.75% to 87.89% of all reads were matched to the reference genome, with no more than two base mismatches allowed in the alignment.
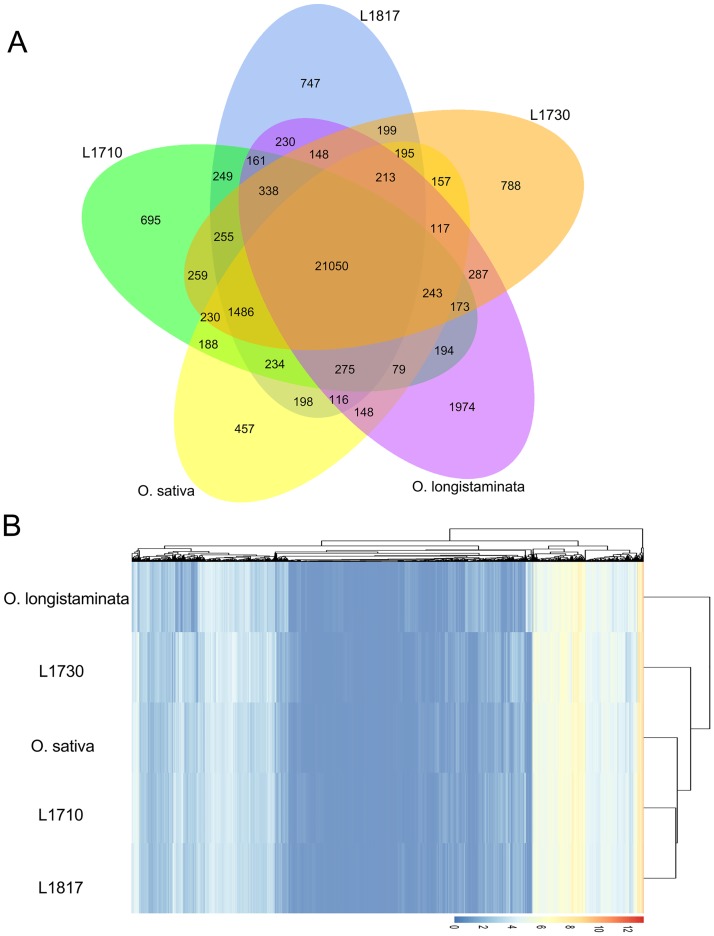
In the present study, a total of 26,110, 26,094, 26,138, 25,386 and 25,746 expressed genes were identified in the stems of L1710, L1817, L1730, O. sativa and O. longistaminata, respectively. Among the identified genes, 24,140 genes were longer assembled sequences, with gene lengths exceeding 1,000 bp (S4 Table). The Venn diagram showed the distribution of the expressed genes in the five lines. We observed that 21,050 (65.61%) genes were commonly expressed (co-expressed) in the five lines, and the number of co-expressed genes was obviously much higher than the number of specifically expressed genes (Fig 2a). In addition, the specifically expressed genes were most abundant in O. longistaminata (6.15%) and least abundant in O. sativa (1.42%) among the five lines. The results suggested that these co-expressed genes might act as housekeeping genes during rice growth and development stage.
Fig 2. Venn diagram analysis and hierarchical cluster analysis in the five lines.
(A) Venn diagram analysis of co-expressed and specially expressed genes in the five lines. (B) Hierarchical cluster analysis of all gene models in five lines. The branch length indicates the degree of variance, and the color represents the logarithmic intensity of expressed genes. Lines groups are shown as columns, individual expressed genes are arrayed in rows.
Heat maps for cluster analysis were used to investigate correlations among the 5 transcriptome profiles. In the present study, L1710 and L1817 were closely grouped together, and subsequently they were close to O. sativa, followed by L1730 (Fig 2b). Additionally, the gene expression profile of L1730 was closer to O. longistaminata than the other lines. The results of all gene expression profiles reflected that most of the gene expression patterns were closer to the recurrent parent in the three progeny lines, which might be influenced by their similar chromosome complements.
Differentially expressed genes (DEGs) among five lines
Many molecular mechanisms were involved in the reprogramming of interacting genomes in backcrossed progeny, and DEGs were key elements influencing the plant morphology changes. Interestingly, the three progeny lines, the nuclear genetic backgrounds of which were primarily inherited from O. sativa, showed obvious phenotypic variations in plant height. Thus, the present study was focused on stem gene expression patterns among five lines to explore genes, which may play crucial roles in progeny phenotypic variations.
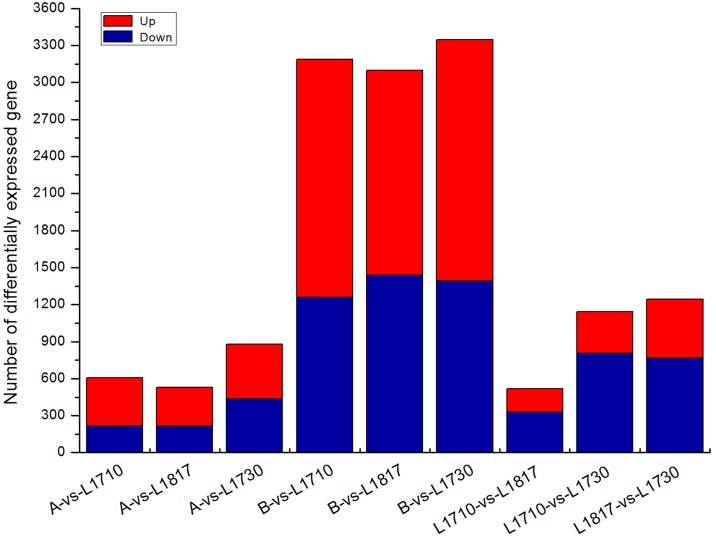
DEGs were analyzed to determine the phenotypic diversity at the molecular level among backcrossed progeny lines. Putative DEGs were identified using the following criteria: (1) the absolute value of the log2 ratio ≥2.322; and (2) P value ≤0.001 and FDR (false discovery rate) ≤0.001. In the present study, the DEGs in three progeny line/O. longistaminata comparison groups exceeded that in three progeny line/O. sativa comparison groups (Fig 3). For example, 3,188 DEGs were identified in the L1710/O. longistaminata comparison group, while only 605 DEGs were identified in the L1710/O. sativa comparison group. Among all comparison groups, the number of DEGs was the least in L1817 compared with their parents, which might reflect an additional heterozygous genome in L1817 than in the other lines. Additionally, more than 50% of the DEGs were up-regulated in the three progeny lines compared with their parents. Notably, most of the DEGs were down-regulated in the higher plant line in the comparison groups among the three progeny lines (Fig 3).
Fig 3. Differentially expressed genes (DEGs) in three progeny lines.
A and B stand for O. sativa and O. longistaminata, respectively.
GO analysis of DEGs in three backcrossed progeny lines
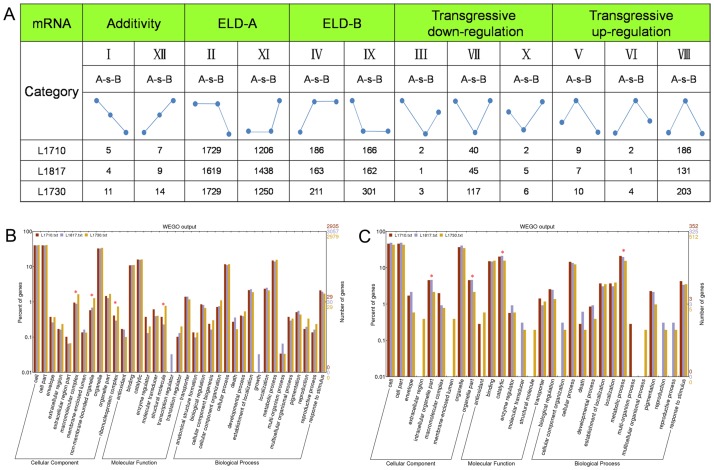
For a detailed analysis, DEGs between O. sativa and O. longistaminata were designated DEGPP, and the DEGs between progeny and their parents were designated DEGHP. For the DEGHP contained 2 classes, DEGO (shared by DEGHP and DEGPP) and DEGUHP (uniquely belonging to DEGHP), and DEGUHP might play roles in phenotype changes between progeny lines and their parents, hence we primarily focused on DEGUHP. In total, more DEGUHP was discovered in L1730 (1,461) than that in L1710 (1,083) and L1817 (1,001). GO analysis of DEGUHP on the WEGO website (http://wego.genomics.org.cn/cgi-bin/wego/index.pl) among the three progeny lines was performed to reveal important GO terms of three categories (cellular component, molecular function and biological process). We identified 23 GO terms in the three progeny lines that exhibited statistically significant differences (P <0.05), such as term ‘metabolic process’, ‘cellular process’, ‘organelle organization’, ‘regulation of cellular process’, ‘transport’, ‘biological regulation’ and ‘primary metabolic process’ (Fig 4a). Genes participating in these terms showed differential expression patterns. For example, OsXTH8 (LOC_Os08g13920) involved in ‘primary metabolic process’ was down-regulated in L1817 and L1730 compared with O. sativa, but up-regulated in L1710 and L1817 compared with O. longistaminata. Moreover, 2 terms in cellular component category, ‘ribonucleoprotein complex’ and ‘non-membrane-bounded organelle’, 2 terms in molecular function category, ‘structural molecule activity’ and ‘nucleic acid binding’, and 6 terms in biological process category, including terms ‘cellular component organization or biogenesis’, ‘macromolecular metabolic process’, ‘organelle organization’, ‘regulation of cellular process’, ‘biological regulation’ and ‘response to endogenous stimulus’, with the P <0.01, might contribute to plant height variation in the three progeny lines.
Fig 4. GO analysis of DEGs in the three progeny lines.
(A) GO analysis of DEGUHP in the three progeny lines. GO analysis of up/down-regulated DEGs in the L1710/L1817 comparison group (B), the L1710/L1730 comparison group (C) and the L1817/L1730 comparison group (D). The x-axis represents the name of the GO subcategories. The right y-axis indicates the number of genes expressed in a given sub-category. The left y-axis indicates log (10) scale, the percent of a specific category of genes in that main category. GO terms with P value<0.05 were denoted by one star, GO terms with P value<0.01 were denoted by two stars.
GO analysis of up/down-regulated DEGs between each progeny line was performed to identify the functional category that the DEGs were primarily involved in. As a result, 2 terms in the L1710/L1817 comparison, 8 terms in the L1817/L1730 comparison and 11 terms in the L1710/L1730 comparison showed statistically significant differences (Fig 4b–4d). Among these terms, 3 terms of molecular function category (‘catalytic activity’, ‘ion binding’ and ‘transferase activity’) and 4 terms of biological process category (‘cellular process’, ‘cellular metabolic process’, ‘metabolic process’ and ‘primary metabolic process’) showed statistically significant differences in the L1710/L1730 comparison and the L1817/L1730 comparison. In addition, ‘vesicle’ showed statistically significant differences in the L1710/L1730 comparison and the L1710/L1817 comparison. Furthermore, ‘cell’, ‘cell part’ and ‘hydrolase activity’ in the L1710/L1730 comparison, ‘membrane-bounded organelle’ in the L1710/L1817 comparison and ‘binding’ in the L1817/L1730 comparison showed statistically significant differences. In ‘metabolic process’, β-1,3-glucanase gene (Osg1, LOC_Os01g71930) was up-regulated in L1730 compared with L1817 and L1710. OsUGE1 (LOC_Os05g51670) involved in ‘vesicle’ and ‘primary metabolic process’ was down-regulated, while OsPIN10b (LOC_Os05g50140) involving in ‘vesicle’ was up-regulated in L1730 compared with L1817 and L1710. OsEXP1 (LOC_Os04g15840) and CYP714B1 (LOC_Os07g48330) involved in ‘vesicle’ were up-regulated in L1817. According to the above results, we speculated that the genes related to ‘vesicle’, ‘catalytic activity’, ‘ion binding’, ‘transferase activity’, ‘cellular process’, ‘cellular metabolic process’, ‘metabolic process’ and ‘primary metabolic process’ might contribute to the changes in plant height in the three progeny lines at the jointing stage.
KEGG pathway analysis of DEGs in the three progeny lines
Since genes typically interacted with other genes participating in certain biological functions, the DEGs were mapped to reference canonical pathways in the KEGG (http://www.genome.ad.jp/kegg/). KEGG pathway analysis was used to provide information to further understand the biological function of genes during rice growth process and identify the genes involved in significantly enriched pathways. All DEGs were assigned to more than 120 pathways in three progeny line/O. longistaminata comparison groups, while approximately 90 pathways were detected in three progeny line/O. sativa comparison groups (S5 Table). Most pathways were up-regulated in L1710 and L1817 compared with their parents, while more down-regulated pathways were observed in L1730. Furthermore, approximately 15% to 19% pathways were significantly enriched pathways (P ≤0.05) in the three progeny lines compared with O. sativa, which were fewer than that in three progeny line/O. longistaminata comparison groups (21–32%). Compared with progeny line/O. sativa comparison groups, more KEGG pathways and significantly enriched pathways existed in progeny line/O. longistaminata, which might be caused by more DEGs existed in three progeny line/O. longistaminata comparison groups.
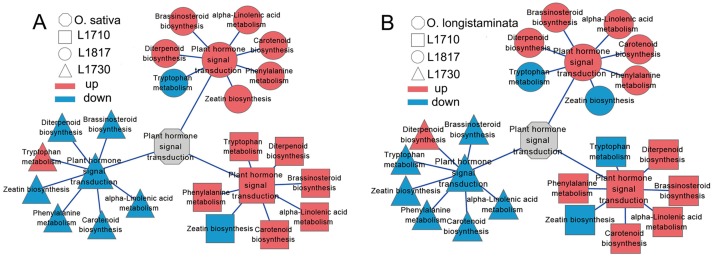
In addition, we pursued top 10 up/down-regulated pathways based on the RPKM of DEGs in three progeny lines. The results showed that ‘phenylalanine metabolism’ was up-regulated in L1710 and L1817, and down-regulated in L1730 (Table 1). The pathway ‘ribosome’ was up-regulated in most comparison groups, except in the L1710/O. longistaminata comparison group. Another pathway, ‘phenylpropanoid biosynthesis’ was down-regulated in L1730 compared with O. sativa, but was up-regulated in L1710 and L1817. In total, significantly enriched pathways (P ≤0.05) accounted for approximately 50% of the 20 pathways (top 10 up-regulated pathways and top 10 down-regulated pathways) in the L1710/O. sativa comparison group and L1730/O. longistaminata comparison group. In KEGG pathways, ‘plant hormone signal transduction pathway’ contained 7 secondary pathways, including ‘alpha-linolenic acid metabolism’, ‘brassinosteroid biosynthesis’, ‘carotenoid biosynthesis’, ‘diterpenoid biosynthesis’, ‘phenylalanine metabolism’, ‘tryptophan metabolism’ and ‘zeatin biosynthesis’ (Fig 5). We observed that this pathway was up-regulated in L1710 and L1817 compared with their parents, while its secondary pathways, ‘zeatin biosynthesis’ in L1710 and ‘tryptophan metabolism’ in L1817, were down-regulated. Additionally, ‘tryptophan metabolism’ in L1710 and ‘zeatin biosynthesis’ in L1817 were down-regulated compared with O. longistaminata. In contrast to L1710 and L1817, this pathway was down-regulated in L1730 compared with their parents, while ‘tryptophan metabolism’ was up-regulated in the L1730/O. sativa comparison group. In addition, we observed that ‘diterpenoid biosynthesis’ was up-regulated in the three progeny lines compared with O. longistaminata. The genes associated with plant hormone might play critical roles in the growth and development of the progeny lines. The up-regulation and down-regulation of these pathways were closely associated with the expression level changes of many genes. In the present study, most gene expression patterns were determined to be consistent with their pathway variations. For example, GH3 (LOC_Os07g40290) was up-regulated in L1710 and L1817 compared with their parents, but down-regulated in L1730 compared with L9311. Additionally, OsGSR1 (LOC_Os06g15620) and D62 (LOC_Os06g03710) were up-regulated in the three progeny lines.
Table 1. Top 10 up/down-regulated KEGG pathways in the three progeny lines.
| Up-regulated pathways | Down-regulated pathways | Up-regulated pathways | Down-regulated pathways | Up-regulated pathways | Down-regulated pathways | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A-vs-L1710 | B-vs-L1710 | A-vs-L1710 | B-vs-L1710 | A-vs-L1817 | B-vs-L1817 | A-vs-L1817 | B-vs-L1817 | A-vs-L1730 | B-vs-L1730 | A-vs-L1730 | B-vs-L1730 | |
| 1 | Biosynthesis of secondary metabolites (509.0) | Metabolic pathways (1,394.3) | Photosynthesis—antenna proteins (-2,221.5) | Photosynthesis—antenna proteins (-809.3) | Biosynthesis of secondary metabolites (1,052.9) | Plant hormone signal transduction (591.3) | Glyoxylate and dicarboxylate metabolism (-769.3) | Histidine metabolism (-1,290.5) | Ribosome (2,216.3) | Ribosome (2,824.4) | Metabolic pathways (-2,502.9) | Biosynthesis of secondary metabolites (-3,920.3) |
| 2 | Phenylpropanoid biosynthesis (464.6) | Biosynthesis of secondary metabolites (1,126.3) | Metabolic pathways (-1,233.0) | Protein processing in endoplasmic reticulum (-706.9) | Plant hormone signal transduction (517.1) | Carotenoid biosynthesis (521.2) | Photosynthesis (-459.4) | Biosynthesis of secondary metabolites (-925.9) | Base excision repair (214.9) | Base excision repair (271.9) | Biosynthesis of secondary metabolites (-1,455.7) | Metabolic pathways (-3,510.3) |
| 3 | Phenylalanine metabolism (416.6) | Glyoxylate and dicarboxylate metabolism (706.9) | Porphyrin and chlorophyll metabolism (-729.7) | Spliceosome (-497.2) | Plant-pathogen interaction (475.2) | Plant-pathogen interaction (398.0) | Carbon fixation in photosynthetic organisms (-136.2) | Protein processing in endoplasmic reticulum (-684.1) | Purine metabolism (174.8) | Oxidative phosphorylation (194.9) | Glyoxylate and dicarboxylate metabolism (-842.8) | Plant hormone signal transduction (-1,657.4) |
| 4 | Plant-pathogen interaction (356.8) | Phenylalanine metabolism (637.1) | Photosynthesis (-486.0) | Ubiquitin mediated proteolysis (-445.9) | Carotenoid biosynthesis (377.5) | Phenylalanine metabolism (300.2) | Propanoate metabolism (-112.4) | Metabolic pathways (-654.4) | Ribosome biogenesis in eukaryotes (163.2) | RNA transport (177.7) | Phenylpropanoid biosynthesis (-570.7) | Histidine metabolism (-1,373.8) |
| 5 | Plant hormone signal transduction (352.6) | Plant hormone signal transduction (503.6) | Carbon fixation in photosynthetic organisms (-330.7) | Tryptophan metabolism (-338.9) | ABC transporters (155.5) | Fatty acid eBation (213.0) | Pentose and glucuronate interconversions (-77.4) | Pyrimidine metabolism (-583.4) | Ubiquitin mediated proteolysis (131.8) | Mismatch repair (159.4) | Plant hormone signal transduction (-498.7) | Phenylpropanoid biosynthesis (-1,301.8) |
| 6 | Cysteine and methionine metabolism (346.2) | Cysteine and methionine metabolism (490.5) | Glyoxylate and dicarboxylate metabolism (-212.9) | Circadian rhythm–plant (-288.9) | Phenylalanine metabolism (150.3) | Diterpenoid biosynthesis (201.6) | Cyanoamino acid metabolism (-51.7) | Purine metabolism (-529.7) | Mismatch repair (124.4) | DNA replication (154.1) | Phenylalanine metabolism (-483.4) | Phenylalanine metabolism (-788.8) |
| 7 | Ribosome (140.0) | Phenylpropanoid biosynthesis (443.8) | Circadian rhythm–plant (-207.9) | Peroxisome (-274.2) | Fatty acid eBation (148.9) | ABC transporters (184.6) | Histidine metabolism (-36.4) | Cyanoamino acid metabolism (-525.8) | DNA replication (122.6) | Ribosome biogenesis in eukaryotes (150.9) | Photosynthesis—antenna proteins (-432.9) | Protein processing in endoplasmic reticulum (-691.1) |
| 8 | Phosphatidylinositol signaling system (109.0) | Plant-pathogen interaction (425.0) | Propanoate metabolism (-106.9) | Cyanoamino acid metabolism (-270.1) | Ribosome (141.8) | Cysteine and methionine metabolism (183.6) | Arginine and proline metabolism (-32.6) | Spliceosome (-453.5) | Nucleotide excision repair (113.4) | Homologous recombination (132.9) | Photosynthesis (-431.3) | Pyrimidine metabolism (-601.0) |
| 9 | alpha-Linolenic acid metabolism (103.3) | Phenylalanine, tyrosine and tryptophan biosynthesis (263.4) | Endocytosis (-34.6) | Endocytosis (-262.3) | Phenylpropanoid biosynthesis (141.5) | Glyoxylate and dicarboxylate metabolism (158.1) | Porphyrin and chlorophyll metabolism (-29.7) | Ubiquitin mediated proteolysis (-400.4) | RNA transport (111.7) | Nucleotide excision repair (125.7) | Endocytosis (-316.3) | Flavonoid biosynthesis (-539.2) |
| 10 | Stilbenoid, diarylheptanoid and gingerol biosynthesis (94.8) |
Diterpenoid biosynthesis (193.8) |
Zeatin biosynthesis (-34.0) | Glucosinolate biosynthesis (-222.8) | Metabolic pathways (141.4) | Ribosome (151.5) | Anthocyanin biosynthesis (-28.8) | Tryptophan metabolism (-314.1) | SNARE interactions in vesicular transport (91.1) | SNARE interactions in vesicular transport (125.0) | Protein processing in endoplasmic reticulum (-299.9) | Circadian rhythm–plant (-509.0) |
The numbers in parentheses represent the RPKM value (up or down) of all genes identified in one pathway of progeny and their parents. Pathways marked with bold font indicate significantly enriched pathways (P ≤0.05). A and B stand for O. sativa and O. longistaminata, respectively.
Fig 5. Pathway of plant hormone signal transduction analysis in the three progeny lines.
The up/down regulation of plant hormone signal transduction pathway in the progeny/O. sativa comparison groups (A) and the progeny/O. longistaminata comparison groups (B). Red standing for up-regulated and blue standing for down-regulated.
Parental expression level dominance analysis in three progeny lines
Recently, studies on hybrid progenies showed that a considerable part of genes expressed at a level approximate to that of one parent and different from that of the other parent, and have no correlation with the MPV (mid-parent value) and the additive expression. This phenomenon is defined as expression-level dominance (ELD) for the expressed genes in progenies. For a more detailed analysis, we categorized the genes into 5 major expression categories, according to previously defined criteria [37]. The 5 categories were further divided into 12 expression patterns based on the gene expression levels in the three progeny lines relative to their parents (Fig 6a). The 12 expression patterns were the expression-level dominance by O. sativa (II and XI, ELD-A), expression-level dominance by O. longistaminata (IV and IX, ELD-B), transgressive down-regulation (III, VII and X), equal to the average of O. sativa and O. longistaminata (I and XII), and transgressive up-regulation (V, VI and VIII) in the progeny line. On the whole, most genes displayed expression level dominance of one parent, and there were more ELD-A genes than ELD-B genes in the progeny lines (S6 Table). In L1710, we observed that the number of ELD-A genes (82.91%) was the highest, followed by ELD-B genes (9.94%) among the 5 expression categories. The proportional similarity of parental ELD genes were observed in L1817 and L1730. Thus, the genes displayed ELD toward the recurrent progenitor in the three progeny lines. The proportion of ELD-B genes (9.07% to 13.27%) in the three progeny lines may be affected by their different genome composition inherited from O. longistaminata which varied from 10.32% to 16.42%. In addition, the proportion of ELD-B genes in L1730 was higher than that in L1817 and L1710, which was consistent with the plant height changes of these progeny lines.
Fig 6. Parental expression level dominance (ELD) genes and GO analysis of ELD genes in progeny lines.
(A) Twelve patterns of DEGs in the three progeny lines. GO analysis of ELD-A genes (B) and ELD-B genes (C) in the three progeny lines. GO terms with P value<0.05 were denoted by one star. ELD-A gene indicate the gene expression level in progeny is similar to that in O. sativa, but different from that in O. longistaminata; ELD-B indicate the gene expression level in progeny is similar to that in O. longistaminata, but different from that in O. sativa.
To explore these functional differences, all parental ELD genes in the three progeny lines were assigned to the GO terms using GO functional classification analysis (WEGO). Thus, 4 GO terms of ELD genes were observed to have statistically significant differences among the three progeny lines (Fig 6b and 6c). Three terms belonging to cellular component category, i.e., ‘macromolecular complex’, ‘ribonucleoprotein complex’ and ‘non-membrane bounded organelle’, and one term of molecular function category, i.e., ‘structure molecule activity’, were statistically and significantly enriched in ELD-A genes among three progeny lines. However, ELD-B genes significantly enriched in two terms of cellular component category, ‘organelle part’ and ‘intracellular organelle part’, one term of molecular function category, ‘catalytic activity’ and one term of biological process category, ‘metabolic process’. In short, most genes in the GO analysis showed ELD-A patterns in three progeny lines, but some genes showed different patterns in different progeny lines. For example, OsEXP1 showed ELD-A in L1730 but showed ELD-B in L1710 and L1817. ELD genes participating in these GO terms might play different roles in the growth and development at the jointing stage in the three progeny lines.
To further pursue potential functional differences between ELD-A genes and ELD-B genes in each progeny line, GO analysis was performed (S2 Fig). In L1710, ELD-A genes and ELD-B genes enriched in 11 GO terms that showing statistically significant differences (P<0.05). In addition, we discovered ELD-A genes and ELD-B genes enriched in 15 GO terms in L1817 and 6 GO terms in L1730. Three GO terms, ‘non-membrane-bounded organelle’, ‘establishment of localization’ and ‘localization’ were only statistically significant differences in L1730. Moreover, we observed three GO terms (‘organelle’, ‘intracellular organelle part’ and ‘oxidoreductase activity’) in L1710 and one GO term (‘organelle’) in L1817 showed statistically significant differences, with P<0.01. Cellulose synthase subunit genes: OsCESA4 (LOC_Os01g54620), OsCESA7 (LOC_Os10g32980) and OsCESA9 (LOC_Os09g25490) in ‘glucan biosynthetic process’ showed ELD-A pattern. Some genes showed the same expression bias in all three progeny lines, for example, OsGI (LOC_Os01g08700) and OsCKX4 (LOC_Os01g71310) showed ELD-B pattern in three progeny lines. These genes showed different ELD patterns in these GO terms might differentially affect the cell growth and development.
Identification and expression analysis of miRNAs in five lines
Five small RNA libraries from the stem materials identical to the gene expression research were constructed, and the expression of sRNAs was detected using Illumina high-throughput sequencing technology. In total, nearly 58.74 million sRNA sequencing reads were generated in five libraries. After removing adaptor contaminations and low-quality reads, 58.25 million reads were identified as small RNA (S7 Table). The length distributions of sRNAs were similar in each of the five lines. Most fragment lengths were distributed between 21 and 24 nt (S3 Fig), and these results were consistent with previous studies that 24-nt sRNAs were most abundant [15, 19–21]. Subsequently, we mapped all clean reads to small RNA databases, and these reads were clustered into 10 RNA classes (including miRNA, rRNA, snRNA, snoRNA, siRNA, piRNA, tRNA, repeat associated sRNA and degraded tags of exon or intron) and unannotated group (S7 Table). For these small RNA reads, 1,285,091 (L1710), 762,856 (L1817), 757,171 (L1730), 1,323,683 (O. sativa) and 1,274,873 (O. longistaminata) were identified as miRNA reads.
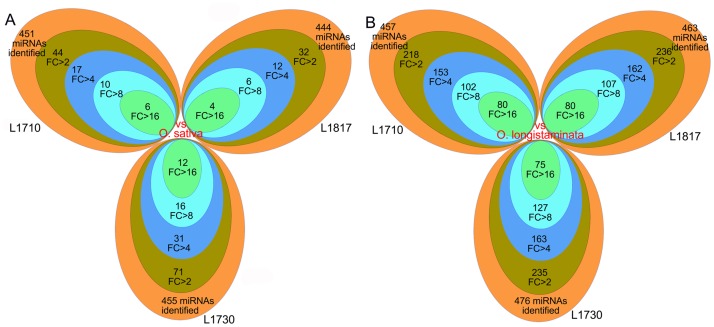
A total of 513 miRNAs (419, 411, 411, 404 and 379 in L1710, L1817, L1730, O. sativa and O. longistaminata, respectively) were expressed in the five lines. The relative expression level of miRNAs was detected from the frequency of their read counts using a deep-sequencing means. In the present study, the relative expression level of miRNAs varied from 1 to 856,496 reads among the five libraries (S8 Table). The expression levels of approximately 66% to 69% of the miRNAs were less than 100 reads in five lines, and the expression levels of 11–14% of the miRNA ranged from 100 to 500 reads, but the expression level of only 3% to 6% of the miRNAs surpassed 10,000 reads. Among these miRNAs, the expression levels of 11 miRNAs (osa-miR168a-5p, osa-miR528-5p, osa-miR166a-3p, osa-miR172d-3p, osa-miR166d-3p, osa-miR166f, osa-miR166b-3p, osa-miR166c-3p, osa-miR166j-3p, osa-miR166g-3p and osa-miR166h-3p) were more than 10,000 reads in five lines. The hierarchical clustering of all expressed miRNAs was performed to group five lines. We observed L1817 and O. sativa formed a single group, and clustered with L1710 and succeed to L1730 (S4a Fig). Based on the results of Venn diagrams, we analyzed the commonly and specifically expressed miRNAs in five lines (S4b Fig). Similar to mRNA analysis, 291 expressed miRNAs were shared by all five lines, accounting for 56.73% of the 513 miRNAs. These co-expressed miRNAs demonstrated conservation between progeny lines and their parents. The uniquely expressed miRNAs in each line only accounted for a small fraction of the miRNAs, showing the most abundance in O. longistaminata (21), subsequently followed by L1730 (19), L1710 (10), O. sativa (8) and L1817 (7).
Differentially expressed miRNAs between progeny lines and their parents
To explore the miRNAs expression patterns between progeny lines and their parents, each miRNA read count was normalized to transcripts per million (TPM). The number of differentially expressed miRNAs (log2Ratio≥1, P≤0.05) in progeny line/O. longistaminata comparison groups (more than 200) was much higher than that in progeny line/O. sativa comparison groups (less than 71) (Fig 7). The number of differentially expressed miRNAs in the L1710/L1730 comparison group was higher than that in the L1710/L1817 and the L1817/L1730 comparison groups (Table 2). This finding was consistent with the plant phenotypic changes, and more differentially expressed miRNAs were observed in the comparison group with greater phenotype differences. The proportion of down-regulated miRNAs were comparable to the up-regulated miRNAs in three progeny line/O. sativa comparison groups, while 67% of the differentially expressed miRNAs were down-regulated in three progeny lines compared with O. longistaminata (Table 2). Approximately 67% of the miRNAs were down-regulated in L1730 and L1817 compared with L1710, and approximately 60% of the miRNAs were up-regulated in L1730 compared with L1817. The differentially expressed miRNAs accounted for 47.70% to 50.97% of all expressed miRNAs in three progeny lines compared with O. longistaminata, but only accounted for 7.21% to 15.60% compared with O. sativa (Fig 7). Additionally, we calculated the differentially expressed miRNAs with a fold-change ≥16, accounting for 0.90% to 2.64% in progeny line/O. sativa comparison groups, but accounting for 15.76% to 17.51% in the progeny line/O. longistaminata comparison groups. These results suggested that both the number and fold-change of the differentially expressed miRNAs in progeny line/O. longistaminata comparison groups were higher than in the progeny line/O. sativa comparison groups. The proportion of differentially expressed miRNA among three progeny lines exhibited similarity, similar to the gene expression patterns, and this miRNA expression tendency may also be influenced by genome dosage.
Fig 7. The number of differentially expressed miRNAs in the three progeny lines.
FC, standing for fold change.
Table 2. Differentially expressed miRNAs and their target genes in three progeny lines.
| Comparison groups | Number of up-regulated miRNA | Number of down-regulated miRNA | Number of target genes of up-regulated miRNA | Number of target genes of down-regulated miRNA | Number of coherent target genes of up-regulated miRNA | Number of coherent target genes of down-regulated miRNAs |
|---|---|---|---|---|---|---|
| A-vs-L1710 | 27 | 17 | 772 | 454 | 1 | 4 |
| A-vs-L1817 | 14 | 18 | 321 | 702 | 1 | 7 |
| A-vs-L1730 | 35 | 36 | 549 | 1131 | 5 | 19 |
| B-vs-L1710 | 74 | 144 | 1655 | 2550 | 60 | 112 |
| B-vs-L1817 | 69 | 166 | 1584 | 2741 | 46 | 93 |
| B-vs-L1730 | 76 | 159 | 1693 | 2626 | 53 | 110 |
| L1710-vs-L1817 | 16 | 34 | 553 | 1069 | 6 | 4 |
| L1710-vs-L1730 | 31 | 61 | 789 | 1412 | 22 | 3 |
| L1817-vs-L1730 | 33 | 22 | 683 | 648 | 0 | 6 |
A and B stand for O. sativa and O. longistaminata, respectively.
For a more detailed analysis, we categorized differentially expressed miRNAs in three progeny lines into 5 major expression categories according to the gene parental expression level dominance analysis. Among the 12 expression patterns, more than 75% of the differentially expressed miRNAs showed parental ELD, similar to the observed gene expression patterns (S5 Fig). Differentially expressed miRNAs showing ELD-A (72% to 87%) significantly exceeded those showing ELD-B (5% to 9%) in each progeny line (S9 Table). In addition, the percentage of ELD-B miRNAs was clearly lower than their chromosome complements inherited from O. longistaminata in the three progeny lines. Furthermore, we observed that the number of down-regulated ELD-A miRNAs was higher than the up-regulated miRNAs, but an opposite trend was observed among the ELD-B miRNAs in the three progeny lines. In addition, similar to gene expression level dominance, the proportion of ELD-B miRNAs in L1730 was higher than that in L1817 and L1710.
Integrated miRNA/mRNA expression analysis in the three progeny lines and their parents
Gene expression level is affected by a variety of factors, primarily controlled by the relative rates of transcription and RNA degradation, and miRNA post-transcriptional regulated mechanism is one of the gene expression regulatory factors. To further characterize the potential functions of miRNAs in the five lines, the target genes of miRNAs were predicted using two softwares (psRobot and Target Finder) with default parameters. Altogether, 4,791, 5,027, 5,011, 4,855 and 4,068 genes were predicted as target genes in L1710, L1817, L1730, O. sativa and O. longistaminata, respectively. Among these genes, 1,226 (L1710), 1,023 (L1817) and 1,680 (L1730) genes were predicted as target genes of differentially expressed miRNAs in progeny lines compared with O. sativa, and more than 4,000 genes were predicted as target genes in each progeny line/O. longistaminata comparison group (Table 2). More than 3,000 genes were identified as target genes of ELD-A miRNAs in each progeny line, with an average number of 16.56 target genes per miRNA, and nearly 500 genes were identified as target genes of ELD-B miRNAs, with an average number of 28.83 target genes per miRNA (S10 Table).
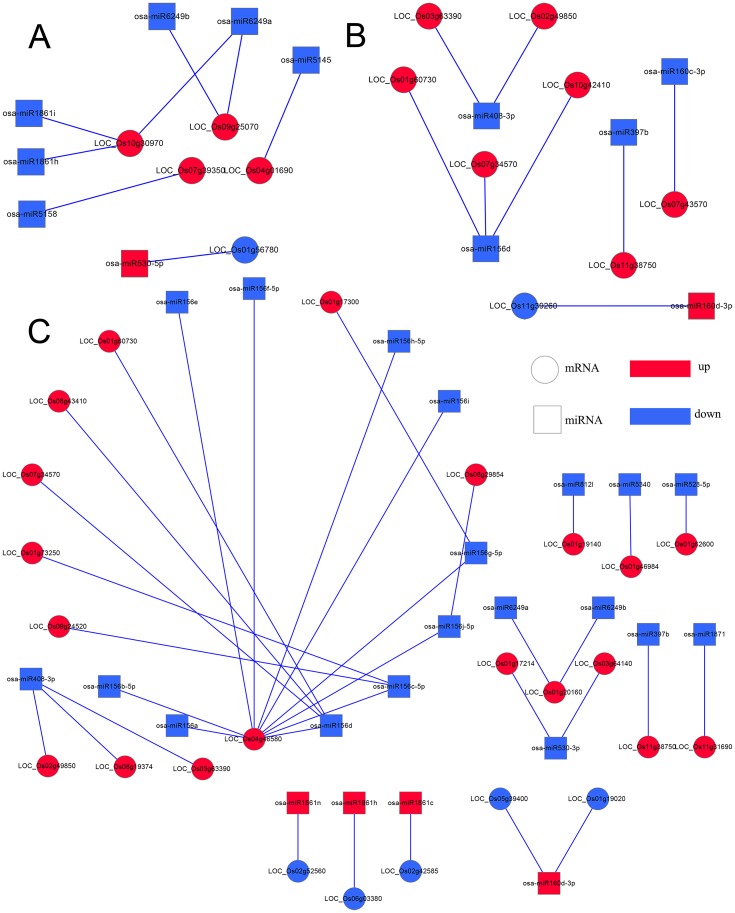
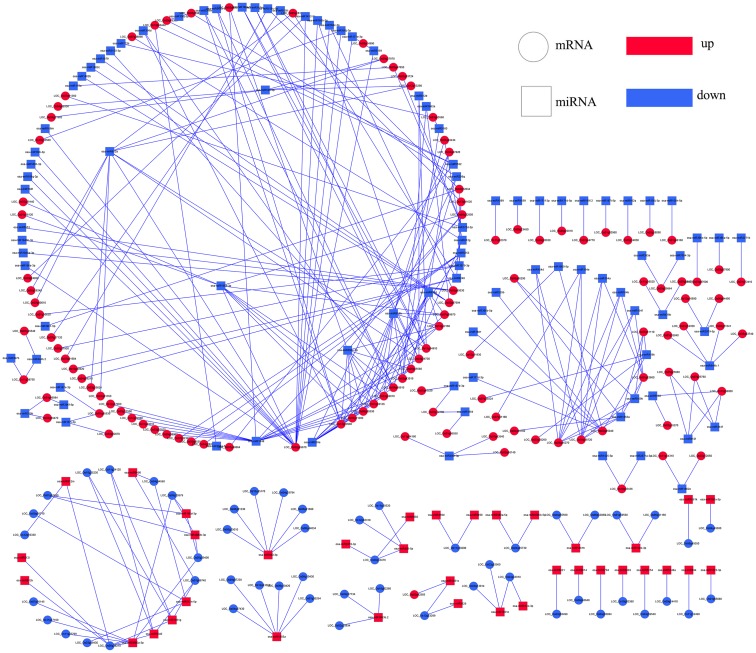
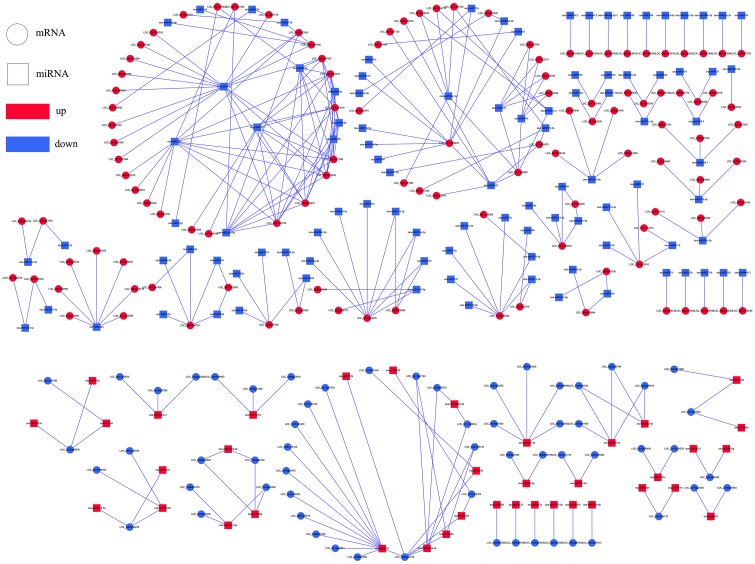
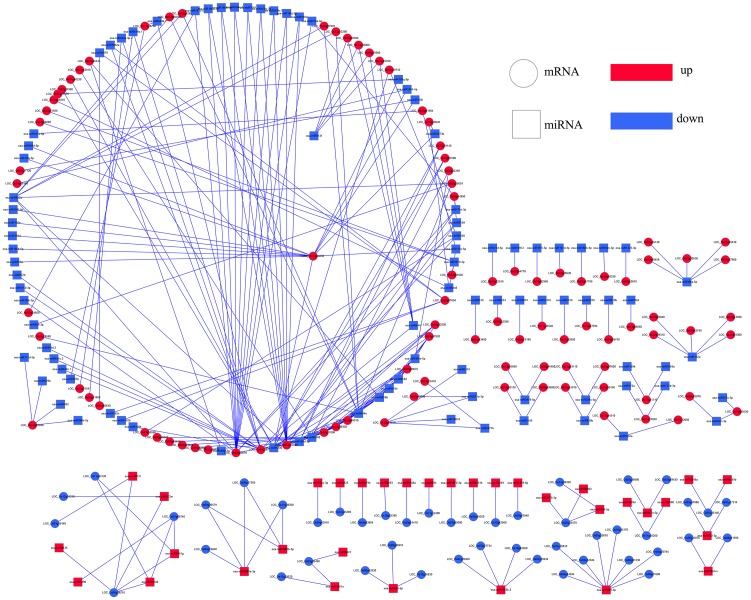
Among these target genes, we focused on coherent target genes inversely expressed with respect to miRNAs. One target gene of a miRNA was defined as a coherent target gene when its expression pattern was opposite that of the miRNA. For example, down-regulated genes were regarded as coherent target genes of up-regulated miRNAs, while other genes were defined as non-coherent target genes. Similarly, up-regulated genes were regarded as coherent genes related to the down-regulated miRNAs, and the remaining genes were considered non-coherent target genes. In the present study, 15.63%-63.76% of the differentially expressed miRNAs were predicted possessing coherent target genes in three progeny lines. In total, 5 (L1710), 8 (L1817) and 24 (L1730) target genes in three progeny line/O. sativa comparison groups, and 172 (L1710), 139 (L1817) and 163 (L1730) target genes in three progeny line/O. longistaminata comparison groups were regarded as coherent target genes (Table 2). Among those miRNAs, we observed the number of coherent target genes of down-regulated miRNAs was marked higher than up-regulated miRNAs in the three progeny lines (Figs 8–11). For example, the osa-miR1848, osa-miR396 and osa-miR818 families, corresponding to more than 10 coherent target genes in each progeny line, were down-regulated compared with O. longistaminata. The Osa-miR444 family possessing more than 20 coherent target genes, was down-regulated in L1710 and L1817 compared with O. longistaminata. The Osa-miR390 corresponding to more than 5 coherent target genes in each progeny line, was up-regulated compared with O. longistaminata. More than 50% of all the parental ELD miRNAs were observed with coherent target genes, and most of these miRNAs were ELD-A miRNAs and corresponded to less than 3 target genes (S6a–S6c Fig). Only 2 ELD-B miRNAs in L1730 had coherent target genes (S6d Fig). Some miRNAs showed similar expression patterns among three progeny lines compared with their parents. For example, ELD-A miRNAs osa-miR3980a-3p was up-regulated and osa-miR1848 was down-regulated in the three progeny lines compared with O. longistaminata. To validate the correlation between the differentially expressed miRNAs and their coherent target genes, 6 miRNAs and their target genes were examined using the qRT-PCR method. The expression patterns between the 6 miRNAs and their coherent targets were basically inversed among five lines, which was consistent with the RNA-seq data (S7 Fig). These results suggest that the target gene prediction of the differentially expressed miRNAs was accurate.
Fig 8. Integrated networks of differentially expressed miRNAs and their coherent target genes in three progeny lines compared with O. sativa.
(A), (B) and (C) represent in L1710, L1817 and L1730. Round rectangle represent miRNAs; Ellipse represent coherent target genes; red represent up-regulated; blue represent down-regulated.
Fig 11. Integrated network of differentially expressed miRNAs and their coherent target genes in L1730 compared with O. longistaminata.
Round rectangle represent miRNAs; Ellipse represent coherent target genes; red represent up-regulated; blue represent down-regulated.
Fig 9. Integrated network of differentially expressed miRNAs and their coherent target genes in L1710 compared with O. longistaminata.
Round rectangle represent miRNAs; Ellipse represent coherent target genes; red represent up-regulated; blue represent down-regulated.
Fig 10. Integrated network of differentially expressed miRNAs and their coherent target genes in L1817 compared with O. longistaminata.
Round rectangle represent miRNAs; Ellipse represent coherent target genes; red represent up-regulated; blue represent down-regulated.
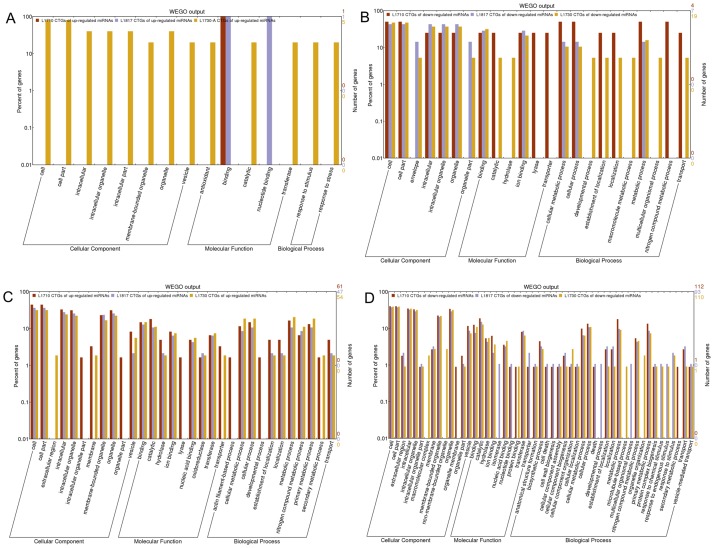
To determine the functional annotation of the differentially expressed miRNAs in each progeny line, GO analysis of miRNA coherent target genes was performed. In total, more GO terms involved in the target genes of differentially expressed miRNAs in three progeny line/O. longistaminata comparison groups were detected than in three progeny line/O. sativa comparison groups. In addition, the target genes of down-regulated miRNAs were assigned to more GO terms than that of up-regulated miRNAs in the three progeny lines. (Fig 12). The target genes of down-regulated miRNAs among three progeny lines compared with O. sativa assigned to 10 terms in three progeny lines, 6 terms in two progeny lines, and 7 terms in one progeny line. Three terms (‘lyase’, ‘transporter’ and ‘nitrogen compound metabolic process’) and 4 terms (‘hydrolase activity’, ‘developmental process’, ‘macromolecule metabolic process’ and ‘multicellular organismal process’) were only assigned by the target genes of down-regulated miRNAs in L1710 and L1730, respectively. Among the three progeny lines/O. longistaminata comparison groups, target genes of down-regulated miRNAs and up-regulated miRNAs were primarily enriched in 22 and 33 terms, respectively (Fig 12c and 12d). Some terms, such as ‘intracellular organelle’, ‘membrane-bounded organelle’, ‘oil binding’, ‘nucleic acid binding’, ‘oxidoreductase activity’, ‘transferase’ ‘cellular metabolic process’ and ‘primary metabolic process’, were enriched by not only target genes of up-regulated miRNAs, but also by that of down-regulated miRNAs in the three progeny lines. Most target genes participating in these terms corresponded to multiple miRNAs, indicating that these terms might be regulated by multiple miRNAs. For example, the NAC gene (OMTN4, LOC_Os06g46270) participated in ‘nucleic acid binding’ was up-regulated in L1730 compared with O. longistaminata, and was the target gene of osa-miR169k, osa-miR1863a, osa-miR164 family. NAC transcription factor gene (OsNAC2, LOC_Os04g38720) involved in ‘cell part’ term was up-regulated in L1710 and L1730 compared with O. longistaminata, and was the target gene of osa-miR164c, osa-miR164d, osa-miR164e, osa-miR169k, osa-miR1863a and osa-miR1861b. OsGH3-9 (LOC_Os07g38890) was the target gene of down-regulated osa-miR812g in the three progeny lines compared with O. longistaminata. In ‘hydrolase activity’, OsGRF12 (LOC_Os04g48510) and OsGRF7 (LOC_Os12g29980) up-regulated in L1730 were predicted to be target genes of osa-miR396 family, osa-miR5150-5p/3p and osa-miR818e. The LOG (LOC_Os01g40630) gene, involved in ‘cytokinin metabolic process’, was the coherent target gene of osa-miR1879 and was up-regulated in L1817 compared with O. longistaminata. The target genes of those down-regulated miRNAs were involved in more GO terms than that of up-regulated miRNAs, indicating that down-regulated miRNAs might play more important roles in gene expression regulation in three progeny lines.
Fig 12. GO analysis of coherent target genes of differentially expressed miRNAs in the three progeny lines.
Up-regulated (A) and down-regulated (B) miRNA in progeny lines compared with O. sativa. Up-regulated (C) and down-regulated (D) miRNA in progeny lines compared with O. longistaminata.
Owing to the small number of ELD-B miRNAs, no coherent target genes of ELD-B miRNAs were observed in L1710 and L1817, and 5 coherent target genes of ELD-B miRNAs were observed in L1730. Hence, we only performed GO analysis of the coherent target genes of ELD-A miRNAs to figure out the potential function in the three progeny lines (S8 Fig). The results revealed that only the coherent target genes in L1730 enriched in ‘macromolecular complex’ and ‘non-membrane-bounded organelle’, and the ELD-A genes number in these 2 terms in L1730 significantly exceeded that in L1710 and L1817 (Fig 4). Moreover, the coherent target genes specifically enriched in ‘developmental process’, ‘multicellular organismal development’ and ‘multicellular organismal process’ in L1817. Coherent target genes were enriched in ‘response to stimulus’, ‘response to chemical stimulus’ and ‘response to endogenous stimulus’ in L1817 and L1730. Three terms, ‘death’, ‘cell death’ and ‘cell wall biogenesis’, were enriched in coherent target genes in L1710 and L1817, without L1730. The number of target genes enriched in ‘cellular component organization’ in L1730 was much higher than that in L1710 and L1817, suggesting that miRNAs with coherent target genes might play key roles in cell growth and metabolism in L1730 during the rice jointing stage. These findings indicated that ELD-A miRNAs might regulate gene expression changes of cellular component category and biological process category.
Validation of the RNA-Seq data using qRT-PCR
To assess the validity and reliability of sequencing data, and confirm the expression profiles of genes and miRNAs among the five libraries, 12 expressed genes (S9 Fig) and 6 expressed miRNAs (S10 Fig) were randomly selected to examine the RNA-Seq analysis using qRT-PCR. The qRT-PCR results of genes and miRNAs were basically consistent with that of the RNA-Seq data, indicating that the sequencing data produced in the present study were reliable and could be subjected to further analysis.
Discussion
Using progeny lines of a backcross introgression line and their parental species as models to explore gene expression divergence is a novel means to address the mechanism of gene introgression affecting progeny phenotypic variations. The transcriptome and small RNA data generated in the present study were useful to understand the variations in gene expression between the progeny and their parents, and identify the complex molecular mechanism of plant height diversity in three progeny lines, which will provide additional insight into the mechanism underlying rice hybrid and backcross processes.
Expression of genes and miRNAs was primarily influenced by the genome dosage of recurrent parent in backcrossed progenies
Plant hybrid is a combination of divergent genomes, leading to instant and profound genome modifications in various ways, including both structural and epigenetic mechanisms [38], and subsequently leading to expression changes of gene [39, 40]. Many studies have been performed to decipher the potenial molecular basis for hybrids, allopolyploids and NILs through detailed annotation of the transcriptome and detection of the miRNA regulation mechanism in progenies and their parents [11, 12, 21, 39–41]. In hybrid rice LYP9, the transcriptome profiles of super-hybrid rice LYP9 were closer to the female parent at the early stages of development but similar to the male parent at later stages, consistent with their morphological characteristics [3]. In the present study, both the gene and miRNA expression patterns were similar to O. sativa in progeny lines, but their phenotypes were not all closer to O. sativa. The gene and miRNA expression profiles in L1710 and L1817 were closer to O. sativa than that in L1730. Additionally, gene and miRNA expression profiles in L1730 were closer to O. longistaminata, which was consistent with that the plant height of L1730 is closer to the O. longistaminata compared with the other lines. In previous studies, the number of co-expressed genes or miRNAs was higher than specifically expressed genes or miRNAs between progeny and its parents [15, 28]. However, the number of specifically expressed genes was higher than the co-expressed genes in super-hybrid rice LY2108 and its parents [42]. In the present study, the numbers of co-expressed genes and miRNAs were far more than that of specifically expressed genes and miRNAs in five lines. Additionally, more co-expressed genes and miRNAs existed between progeny line and O. sativa than that between progeny line and O. longistaminata. Furthermore, the number of specifically expressed genes and miRNAs in O. longistaminata were highest among the five lines, and followed by L1730. These co-expressed genes and miRNAs probably related to the basic growth that contained conservative function in rice jointing stage, while specifically expressed genes and miRNAs, probably contribute to the special growth and development processes in the five lines, particularly in L1730.
In previous studies, the maternal genome was demonstrated to have an advantage over the paternal genome, and the expression of DEGs and differentially expressed miRNAs were closely associated with the parental genome dosage in progeny [15, 21, 37]. In seedlings and spikes of synthetic hexaploid wheat and spikes of natural allohexaploids wheat, the genes and miRNAs showed expression-level dominance to the maternal parent (AB), but showing expression-level dominance to the maternal parent (D) in synthetic hexaploid wheat seeds [21]. However, the expression of triploid hybrid rice is closer to the paternal parent for the paternal subgenome (BC) comprised of two genomes and the maternal subgenome (A) was comprised of one genome [40]. In the present study, the obvious diversity of genes and miRNAs showed parental ELD patterns, indicating that genes and miRNAs of one parent (primarily O. sativa) were dominantly expressed in three progeny lines. These findings were consistent with previously reported about an inter-specific hybrid rice [40] and indicated that the O. sativa genome was globally dominant over the O. longistaminata genome due to dosage advantage after hybrid and backcross process. Together, this analysis further showed that, the expression of genes and miRNAs showed parental expression level dominance in progeny of BILs rice. More differentially expressed genes and miRNAs were observed in Brassica hexaploid (BBCCAA) compared with male parent B. rapa (AA) than that compared with female parent B. carinata (BBCC), which may be affected by its genome dosage [15, 28]. In the present study, more differentially expressed genes and miRNAs were observed between progeny and O. longistaminata, for most of the chromosome complements of progeny were inherited from O. sativa. These results suggested that the expression of genes and miRNAs in progeny was influenced by the parental genome dosage of the progeny, particular the differentially expressed genes and miRNAs.
The DEGs participated in complex pathways, such as ‘carbohydrate metabolism’, ‘plant hormone signal transduction’ and ‘photosynthesis-related pathways’, were associated with phenotypic variation in hybrid progeny [21, 40, 41]. In nascent hexaploid wheat, the expression of ‘development’ related genes are similar to the female parent and that of ‘adaptation’ related genes display similar to male parent in the progeny, which may be conducive to stress responses and photoperiod adaptability [21]. Furthermore, we noticed that DEGUHP were enriched in ‘cellular metabolic process’, ‘metabolic process’ and ‘primary metabolic process’ in all progeny lines, and these three terms were statistically significant differences among three progeny lines. The coherent target genes of differentially expressed miRNAs were primarily enriched in ‘cellular metabolic process and ‘primary metabolic process’ in three progeny lines compared with O. longistaminata. Thus, ‘cellular metabolic process’ and ‘primary metabolic process’ were differently expressed among three progeny lines. Nevertheless, genes displayed ELD toward O. sativa and the coherent target genes of miRNAs that displayed ELD toward O. sativa were significant enriched in ‘cellular metabolic process’ and ‘primary metabolic process’ in three progeny lines (Fig 6b, S8 Fig). Therefore, we speculated the primary metabolism and cellular metabolism related genes and miRNAs were primarily influenced by the genome dosage of O. sativa and might be contributed to the internode elongation and plant height variations among three progeny lines.
Regulation of genes and miRNAs related with plant hormones and cell wall might contribute to the height variations in backcrossed progeny
To achieve high-quality rice, both high yielding potential and optimal architecture, determined by plant height to a large extent, were required. Rice height is primarily determined according to rice stems, in which internode number and internode length play critical roles. Internode length is determined by cell division in the meristem and cell elongation in the elongated region, which is influenced by plant hormones and the cell wall components biosynthesis and metabolism [43–48]. The plant hormones, such as auxin, gibberellins (GA), cytokinin, jasmonic acid (JA) and brassinosteroid (BR), are important regulators that determine plant height by regulating internode elongation [43, 49–51]. The increased expression of auxin-responsive genes (GH3) inhibited the plant growth in cotton and result in dwarf plant [19]. Among these data, the expression of GH3 was up-regulated in L1710 and L1817 compared with O. longistaminata, but showed little change in L1730 compared with O. longistaminata. Additionally, GH3 was down-regulated in L1730 compared with O. sativa. PIN proteins as auxin output vector mediate auxin flow and the expression of PIN genes are influenced by exogenous auxin and other hormones [52]. OsPIN10b was monocot-specific PIN protein, and the expression of OsPIN10b was up-regulated in L1730 compared with L1817 and L1710 in the present study. BR biosynthesis-deficient mutants showed dwarf phenotypes with reduced cell elongation, shorter leaf sheaths and erect dark green leaves [44]. In the KEGG analysis, the ‘BR biosynthesis pathway’ was up-regulated in L1710 and L1817, but down-regulated in L1730, and ‘diterpenoid biosynthesis’ was up-regulated in progeny lines, except in the L1730/O. sativa comparison group. OsNAC2 directly participated in the GA pathway, delaying flowering time and reducing the length of the internodes in rice [53]. The expression of OsNAC2 was up-regulated and regulated by 6 miRNAs (osa-miR164c, osa-miR164d, osa-miR164e, osa-miR169k, osa-miR1863a and osa-miR1861b) in L1710, indicating that these miRNAs might participate in the regulation of internodes elongation. CYP714B1 encodes GA 13-oxidase and reduces the activity of GAs, thereby influencing the length of rice internodes [54]. In the present study, CYP714B1 was up-regulated in L1817 and L1730, indicating that its expression might contribute to the internode elongation of the three progeny lines. Moreover, some genes were coordinately regulated through GA and BR in rice, such as OsGSR1 and D62 [50, 55]. The OsGSR1, a positive regulate factor in GA signaling, acts key role in the interactions between BR and GA signaling pathways in rice [55]. In the BR-insensitive dwarf mutant, the BR-responsive gene (D62) affected the GA metabolism and mediated the interaction between GA and BR in rice [56]. Among these data, the expression of D62 and OsGSR1 were up-regulated in the three progeny lines, suggesting that the interaction between BR and GA signaling might be enhanced after hybridization and backcrossing process. These results indicated that the expression of genes and miRNAs related to BR biosynthesis and GA signaling pathways might contribute to hormone regulation and changes in plant height in the three progeny lines. Furthermore, LOG participates in bioactive cytokinin synthesis through encoding a cytokinin-activating enzyme [51]. Many studies have shown that miRNAs play important roles through regulating the target genes in the plants growth [16–18, 21, 27]. The study of Wen et al. provided important resources for the investigation of miRNA functions in rice domestication [18]. Among these data, LOG was involved in 2 GO terms, ‘intracellular membrane-bounded organelle’ and ‘cytokinin metabolic process’. In addition, LOG was the coherent target gene of osa-miR1879, and the expression level of osa-miR1879 in three progeny lines was dominance toward O. sativa but lower than that in O. longistaminata. Therefore, we speculated that osa-miR1879 might be a regulator in the regulation of ‘cytokinin metabolic process’ at the jointing stage of three progeny lines. In short, complex expression changes of these hormones related genes might contribute to the height variations in the three progeny lines after hybridization and backcrossing, and it will be interesting to explore more miRNAs who play a role as regulator in plant hormone signal transduction.
During plant growth and development, the cell wall plays an important role in plant architectural design, including influencing cell shape and mechanical strength. The plant cell wall is primarily comprised 3 polysaccharides, including cellulose, hemicellulose and pectin, and the biosynthesis of these polysaccharides is determined by the CESA (cellulose synthase active subunit) super-family and the CSL (cellulose synthase-like) super-family [48, 57]. Three genes (OsCESA4, OsCESA7 and OsCESA9) involving cellulose synthase subunit participated in the biosynthesis of secondary cell wall, and played considerable roles in plant growth [43, 58]. The expression of three OsCESA genes displayed ELD toward O. sativa and were highest in O. longistaminata, consistent with changes of the third internode cell length in the five lines, indicating that OsCESA genes might contribute to the various cell lengths of the three progeny lines. OsCSLD4 (LOC_Os12g36890) affects the cell-wall formation and plant growth, whose deficiency can lead to a significantly reduced plant height and the apparent structural defects in rice primary walls [59]. In the present study, the expression level of OsCSLD4 was highest in L1730, and lowest in L1710. Guevara et al. observed that OsUGE1 might play an important role in the distribution of carbohydrate by altering the contents of sucrose, cellulose, galactose and glucose in rice cell walls [60]. In the present study, OsUGE1 involved in ‘vesicle’ and ‘primary metabolic process’ likely participated in the biosynthesis of the internode cell walls in the three progeny lines. These results indicated that some cell wall related genes might play important roles in plant height by influencing cell division and formation. Through regulating the OsSPL14 (LOC_Os08g39890) gene osa-miR156 improved the grain yield of rice through changing its plant architecture [22]. In the present study, osa-miR156 was down-regulated in the three progeny lines compared with O. longistaminata, and the expression levels of OsSPL14 in the three progeny lines were higher than that in O. longistaminata. Os-GRF1 has been reported as potential regulator of stem elongation in rice [46]. In Arabidopsis, OsGRF genes participated in the cell division and differentiation during leaf development and were targeted by miR396 [61]. In the present study, up-regulated OsGRF12 and OsGRF7 were regulated through down-regulated osa-miR396 family, osa-miR5150-5p/3p and osa-miR818e in three progeny lines compared with O. longistaminata. Therefore, we predicted that miRNAs might participate in internode cell division and stem elongation through regulating the target genes that contributed to the variations of plant height in three progeny lines during rice jointing stage. Cell elongation is not only correlated with the biosynthesis of cellulose, hemicellulose and pectin, but is also influenced by cell-wall loosening, which is regulated by cell-wall loosening factors. Expansin was involved in cell expansion through regulating cell-wall loosening activity during cell-wall modification [62]. In the present study, expansin gene (OsEXP1) was up-regulated in L1817 compared with O. sativa, but down-regulated in L1730 compared with O. longistaminata. The construction of xyloglucan cross-links is influenced by xyloglucan endotransglucosylases/hydrolases (XTHs), XTH-related gene OsXTH8 is potentially related to cell elongation and affects the height of mutants [63]. In the present study, the expression of OsXTH8 was up-regulated in L1710 and L1817 compared with O. longistaminata, but down-regulated in L1817 and L1730 compared with O. sativa. Glucanases have been recognized as cell-wall loosening enzymes through hydrolyzing the major parts of the plant cell-wall (1,3;1,4)- β- D -glucans [64]. Plant β-1,3-glucanases are participated in the defense and development of plant, and the silence of Osg1 results in delayed development and reduced plant height in a japonica rice variety [65]. In this study, Osg1 involving in the ‘metabolic process’ was up-regulated in L1730 compared with L1817 and L1710. According to these results, we predicted that the genes and miRNAs related to plant hormones and cell wall might play important roles in cell division and elongation, contributing to the plant height of the three progeny lines at the jointing stage.
In summary, the transcription and sRNAs analysis utilizing HiSeq technology provided an integrative method to investigate molecular mechanism of hybridization and backcrossing. The present study produced sufficient and reliable data for the exploration of gene and miRNA expression variations in the three progeny lines of backcrossed introgression line (BC2F12). Most genes and miRNAs expression patterns were toward recurrent parent in the three progeny lines, which might be influenced by their genome components. In the present study, GO and KEGG pathway analysis revealed some genes involved in specific biological processes and pathways, which might contribute to the plant height changes of the three progeny lines. The genes participating in the synthesis of plant hormones and hormone signal transduction and the genes related to cell wall might act crucial roles in the plant height of the three progeny lines at the jointing stage. This information from the present study will enhance the current understanding of the molecular mechanisms of interspecific hybrid backcrossing and may be useful for further analysis of the molecular regulation in the variations of plant height among the three progeny lines.
Supporting information
The blue color standing for chromosome complements inherited from O. longistaminata; white color standing for chromosome complements inherited from O. sativa; green color standing for heterozygosity chromosome complements inherited from O. longistaminata and O. sativa.
(TIF)
The x-axis represents the name of the GO subcategories. The right y-axis indicates the number of genes expressed in a given sub-category. The left y-axis indicates log (10) scale, the percent of a specific category of genes in that main category. GO terms showed statistically significant differences (P value<0.05) were denoted by stars. A and B stand for O. sativa and O. longistaminata, respectively.
(TIF)
(TIF)
(A) Hierarchical cluster analysis of expressed miRNAs. (B) Co-expressed and specially expressed miRNAs. The branch length indicates the degree of variance, and the color represents the logarithmic intensity of expressed miRNAs. Species groups are shown as columns, and individual expressed miRNAs are arrayed in rows.
(TIF)
A and B stand for O. sativa and O. longistaminata, respectively. ELD-A miRNA indicated miRNA expression level is similar to O. sativa, but is differential compared with O. longistaminata; ELD-B miRNA indicated miRNA expression level is similar to O. longistaminata, but is differential compared with O. sativa.
(TIF)
(A), (B) and (C) Integrated ELD-A miRNAs and their coherent target genes network in L1710, L1817 and L1730; (D) Integrated ELD-B miRNAs and their coherent target genes network in L1730. A and B stand for O. sativa and O. longistaminata, respectively; Round rectangle represent miRNAs; Ellipse represent coherent target genes; red represent up-regulated; blue represent down-regulated.
(TIF)
Actin1 and U6 snRNA are used as internal reference gene and small RNA, respectively.
(TIF)
A stand for O. sativa.
(TIF)
Actin1 is used as internal reference gene. The data of quantitative RT-PCR validation are expressed as the mean SD after normalization. Error bars indicate the standard deviation (±SD) of three replicates. Blue polylines are derived from RNA-Sqe data.
(TIF)
U6 snRNA is used as a reference small RNA. The data of real-time qPCR validation are expressed as the mean SD after normalization. Error bars indicate the standard deviation (±SD) of three replicates. Blue polylines are derived from RNA-Seq data.
(TIF)
(XLSX)
(XLSX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(XLSX)
Acknowledgments
This work was supported by the State Key Basic Research and Development Plan of China (2013CB126900).
Data Availability
The mRNA and miRNA sequences used in this study have been submitted to the NCBI’s Sequence Read Archive Database. The accession numbers of ten runs are SRR5754959, SRR5754960, SRR5754961, SRR5754962, SRR5754963, SRR5759259, SRR5759260, SRR5759261, SRR5759262 and SRR5759263.
Funding Statement
This work was supported by the State Key Basic Research and Development Plan of China (2013CB126900). The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Ge S, Sang T, Lu BR, Hong DY. Phylogeny of rice genomes with emphasis on origins of allotetraploid species. Proc Natl Acad Sci USA. 1999;96(25):14400–5. doi: 10.1073/pnas.96.25.14400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tanksley SD, McCouch SR. Seed banks and molecular maps: unlocking genetic potential from the wild. Science. 1997;277(5329):1063–6. doi: 10.1126/science.277.5329.1063 [DOI] [PubMed] [Google Scholar]
- 3.Wei G, Tao Y, Liu GZ, Chen C, Luo RY, Xia HA, et al. A transcriptomic analysis of superhybrid rice LYP9 and its parents. Proc Natl Acad Sci USA. 2009;106(19):7695–701. doi: 10.1073/pnas.0902340106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang YS, Zhang SL, Liu H, Fu BY, Li LJ, Xie M, et al. Genome and comparative transcriptomics of African wild rice Oryza longistaminata provide insights into molecular mechanism of rhizomatousness and self-incompatibility. Mol Plant. 2015;8(11):1683–6. doi: 10.1016/j.molp.2015.08.006 [DOI] [PubMed] [Google Scholar]
- 5.Hu FY, Tao DY, Sacks E, Fu BY, Xu P, Li J, et al. Convergent evolution of perenniality in rice and sorghum. Proc Natl Acad Sci USA. 2003;100(7):4050–4. doi: 10.1073/pnas.0630531100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu P, Dong LY, Zhou JW, Li J, Zhang Y, Hu FY, et al. Identification and mapping of a novel blast resistance gene Pi57 (t) in Oryza longistaminata. Euphytica. 2015;205(1):95–102 [Google Scholar]
- 7.Song WY, Wang GL, Chen LL, Kim HS, Pi LY, Holsten T, et al. A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science. 1995;270(5243):1804–6. doi: 10.1126/science.270.5243.1804 [DOI] [PubMed] [Google Scholar]
- 8.Xu Q, Zheng TQ, Hu X, Cheng LR, Xu JL, Shi YM, et al. Examining two sets of introgression lines in rice (Oryza sativa L.) reveals favorable alleles that improve grain Zn and Fe concentrations. PLoS ONE. 2015;10(7):e0131846 doi: 10.1371/journal.pone.0131846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen ZW, Hu FY, Xu P, Li J, Deng XN, Zhou JW, et al. QTL analysis for hybrid sterility and plant height in interspecific populations derived from a wild rice relative, Oryza longistaminata. Breeding Sci. 2009;59(4):441–5. doi: 10.1270/jsbbs.59.441 [Google Scholar]
- 10.Zhao XQ, Zhang GL, Wang Y, Zhang F, Wang WS, Zhang WH, et al. Metabolic profiling and physiological analysis of a novel rice introgression line with broad leaf size. PLoS ONE. 2015;10(12):e0145646 doi: 10.1371/journal.pone.0145646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moumeni A, Satoh K, Kondoh H, Asano T, Hosaka A, Venuprasad R, et al. Comparative analysis of root transcriptome profiles of two pairs of drought-tolerant and susceptible rice near-isogenic lines under different drought stress. BMC Plant Biol. 2011;11:174 doi: 10.1186/1471-2229-11-174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moumeni A, Satoh K, Venuprasad R, Serraj R, Kumar A, Leung H, et al. Transcriptional profiling of the leaves of near-isogenic rice lines with contrasting drought tolerance at the reproductive stage in response to water deficit. BMC Genomics. 2015;16(1):1110 doi: 10.1186/s12864-015-2335-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang LY, Zhang F, Zhang F, Wang WS, Zhou YL, Fu BY. Comparative transcriptome sequencing of tolerant rice introgression line and its parents in response to drought stress. BMC Genomics. 2014;15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Voinnet O. Origin, biogenesis, and activity of plant microRNAs. Cell. 2009;136(4):669–87. doi: 10.1016/j.cell.2009.01.046 [DOI] [PubMed] [Google Scholar]
- 15.Shen YY, Zhao Q, Zou J, Wang WL, Gao Y, Meng JL, et al. Characterization and expression patterns of small RNAs in synthesized Brassica hexaploids. Plant Mol Biol. 2014;85(3):287–99. doi: 10.1007/s11103-014-0185-x [DOI] [PubMed] [Google Scholar]
- 16.Unver T, Parmaksiz I, Dundar E. Identification of conserved micro-RNAs and their target transcripts in opium poppy (Papaver somniferum L.). Plant Cell Rep. 2010;29(7):757–69. doi: 10.1007/s00299-010-0862-4 [DOI] [PubMed] [Google Scholar]
- 17.Biswas S, Hazra S, Chattopadhyay S. Identification of conserved miRNAs and their putative target genes in Podophyllum hexandrum (Himalayan Mayapple). Plant Gene. 2016;6:82–9. doi: 10.1016/j.plgene.2016.04.002 [Google Scholar]
- 18.Wen M, Xie M, He L, Wang Y, Shi S, Tang T. Expression variations of miRNAs and mRNAs in rice (Oryza sativa). Genome Biol Evol. 2016;8(11):3529–44. doi: 10.1093/gbe/evw252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.An W, Gong W, He S, Pan Z, Sun J, Du X. MicroRNA and mRNA expression profiling analysis revealed the regulation of plant height in Gossypium hirsutum. BMC Genomics. 2015;16:886 doi: 10.1186/s12864-015-2071-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zong Y, Huang L, Zhang T, Qin Q, Wang W, Zhao X, et al. Differential microRNA expression between shoots and rhizomes in Oryza longistaminata using high-throughput RNA sequencing. The Crop J. 2014;2(2–3):102–9. doi: 10.1016/j.cj.2014.03.005 [Google Scholar]
- 21.Li A, Liu D, Wu J, Zhao X, Hao M, Geng S, et al. MRNA and small RNA transcriptomes reveal insights into dynamic homoeolog regulation of allopolyploid heterosis in nascent hexaploid wheat. Plant Cell. 2014;26(5):1878–900. doi: 10.1105/tpc.114.124388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiao Y, Wang Y, Xue D, Wang J, Yan M, Liu G, et al. Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat Genet. 2010;42(6):541–4. doi: 10.1038/ng.591 [DOI] [PubMed] [Google Scholar]
- 23.Zhang JW, Long Y, Xue MD, Xiao XG, Pei XW. Identification of microRNAs in response to drought in common wild rice (Oryza rufipogon Griff.) shoots and roots. PLoS ONE. 2017;12(1):e0170330 doi: 10.1371/journal.pone.0170330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu C, Chen Y, Zhang H, Chen Y, Shen X, Shi C, et al. Integrated microRNA-mRNA analyses reveal OPLL specific microRNA regulatory network using high-throughput sequencing. Sci Rep. 2016;6:21580 doi: 10.1038/srep21580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ye BY, Wang RH, Wang JB. Correlation analysis of the mRNA and miRNA expression profiles in the nascent synthetic allotetraploid Raphanobrassica. Sci Rep. 2016;6:37416 doi: 10.1038/srep37416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Q, Li Y, Moose SP, Hudson ME. Transposable elements, mRNA expression level and strand-specificity of small RNAs are associated with non-additive inheritance of gene expression in hybrid plants. BMC Plant Biol. 2015;15:168 doi: 10.1186/s12870-015-0549-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sarkar D, Maji RK, Dey S, Sarkar A, Ghosh Z, Kundu P. Integrated miRNA and mRNA expression profiling reveals the response regulators of a susceptible tomato cultivar to early blight disease. DNA Res. 2017;24:235–250. doi: 10.1093/dnares/dsx003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao Q, Zou J, Meng JL, Mei SY, Wang JB. Tracing the transcriptomic changes in synthetic trigenomic allohexaploids of Brassica using an RNA-Seq approach. PLoS ONE. 2013;8(7):e68883 doi: 10.1371/journal.pone.0068883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gao Y, Xu H, Shen YY, Wang JB. Transcriptomic analysis of rice (Oryza sativa) endosperm using the RNA-Seq technique. Plant Mol Biol. 2013;81(4–5):363–78 doi: 10.1007/s11103-013-0009-4 [DOI] [PubMed] [Google Scholar]
- 30.Yang LY, Wu Y, Wang WL, Mao BG, Zhao BR, Wang JB. Genetic subtraction profiling identifies candidate miRNAs involved in rice female gametophyte abortion. G3-Genes Genom Genet. 2017;7(7):2281–93. doi: 10.1534/g3.117.040808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5(7):621–8. doi: 10.1038/nmeth.1226 [DOI] [PubMed] [Google Scholar]
- 32.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95(25):14863–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang LK, Feng ZX, Wang X, Wang XW, Zhang XG. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics. 2010;26(1):136–8. doi: 10.1093/bioinformatics/btp612 [DOI] [PubMed] [Google Scholar]
- 34.Allen E, Xie Z, Gustafson AM, Carrington JC. MicroRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005;121(2):207–21. doi: 10.1016/j.cell.2005.04.004 [DOI] [PubMed] [Google Scholar]
- 35.Schwab R, Palatnik JF, Riester M, Schommer C, Schmid M, Weigel D. Specific effects of microRNAs on the plant transcriptome. Dev Cell. 2005;8(4):517–27. doi: 10.1016/j.devcel.2005.01.018 [DOI] [PubMed] [Google Scholar]
- 36.Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33(20):e179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yoo MJ, Szadkowski E, Wendel JF. Homoeolog expression bias and expression level dominance in allopolyploid cotton. Heredity. 2012;110(2):171–80. doi: 10.1038/hdy.2012.94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiang B, Lou Q, Wu Z, Zhang W, Wang D, Mbira KG, et al. Retrotransposon-and microsatellite sequence-associated genomic changes in early generations of a newly synthesized allotetraploid Cucumis x hytivus Chen & Kirkbride. Plant Mol Biol. 2011;77(3):225–33. doi: 10.1007/s11103-011-9804-y [DOI] [PubMed] [Google Scholar]
- 39.Chen FF, He GM, He H, Chen W, Zhu XP, Liang MZ, et al. Expression analysis of miRNAs and highly-expressed small RNAs in two rice subspecies and their reciprocal hybrids. J Integr Plant Biol. 2010;52(11):971–80. doi: 10.1111/j.1744-7909.2010.00985.x [DOI] [PubMed] [Google Scholar]
- 40.Wu Y, Sun Y, Wang X, Lin X, Sun S, Shen K, et al. Transcriptome shock in an interspecific F1 triploid hybrid of Oryza revealed by RNA sequencing. J Integr Plant Biol. 2016;58(2):150–64. doi: 10.1111/jipb.12357 [DOI] [PubMed] [Google Scholar]
- 41.Zhai RR, Feng Y, Zhan XD, Shen XH, Wu WM, Yu P, et al. Identification of transcriptome SNPs for assessing allele-specific gene expression in a super-hybrid rice Xieyou9308. PLoS ONE. 2013;8(4):e60668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Song GS, Zhai HL, Peng YG, Zhang L, Wei G, Chen XY, et al. Comparative transcriptional profiling and preliminary study on heterosis mechanism of super-hybrid rice. Mol Plant. 2010;3(6):1012–25. doi: 10.1093/mp/ssq046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang DB, Wang SG, Zhang BC, Shang-Guan K, Shi YY, Zhang DM, et al. A gibberellin-mediated DELLA-NAC signaling cascade regulates cellulose synthesis in rice. Plant Cell. 2015;27(6):1681–96. doi: 10.1105/tpc.15.00015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu X, Feng ZM, Zhou CL, Ren YK, Mou CL, Wu T, et al. Brassinosteroid (BR) biosynthetic gene lhdd10 controls late heading and plant height in rice (Oryza sativa L.). Plant Cell Rep. 2016;35(2):357–68. doi: 10.1007/s00299-015-1889-3 [DOI] [PubMed] [Google Scholar]
- 45.Tong H, Xiao Y, Liu D, Gao S, Liu L, Yin Y, et al. Brassinosteroid regulates cell elongation by modulating gibberellin metabolism in rice. Plant Cell. 2014;26(11):4376–93. doi: 10.1105/tpc.114.132092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van der Knaap E, Kim JH, Kende H. A novel gibberellin-induced gene from rice and its potential regulatory role in stem growth. Plant Physiol. 2000;122(3):695–704. doi: 10.1104/pp.122.3.695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ayano M, Kani T, Kojima M, Sakakibara H, Kitaoka T, Kuroha T, et al. Gibberellin biosynthesis and signal transduction is essential for internode elongation in deepwater rice. Plant Cell Environ. 2014;37(10):2313–24. doi: 10.1111/pce.12377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fagard M, Desnos T, Desprez T, Goubet F, Refregier G, Mouille G, et al. PROCUSTE1 encodes a cellulose synthase required for normal cell elongation specifically in roots and dark-grown hypocotyls of Arabidopsis. Plant Cell. 2000;12(12):2409–23. doi: 10.1105/tpc.12.12.2409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yamamuro C IY, Wu X. Loss of function of a rice brassinosteroid insensitive1 homolog prevents internode elongation and bending of the lamina joint. Plant Cell. 2000. doi: 10.1105/tpc.12.9.1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang G, Komatsu S. Microarray and proteomic analysis of brassinosteroid-and gibberellin-regulated gene and protein expression in rice. Genomics, Proteomics & Bioinformatics. 2004;2(2):77–83. doi: 10.1016/s1672-0229(04)02013-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kurakawa T, Ueda N, Maekawa M, Kobayashi K, Kojima M, Nagato Y, et al. Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature. 2007;445(7128):652–5. doi: 10.1038/nature05504 [DOI] [PubMed] [Google Scholar]
- 52.Wang JR, Hu H, Wang GH, Li J, Chen JY, Wu P. Expression of PIN genes in rice (Oryza sativa L.): tissue specificity and regulation by hormones. Mol Plant 2009; 2(4):823–31. doi: 10.1093/mp/ssp023 [DOI] [PubMed] [Google Scholar]
- 53.Chen X, Lu S, Wang Y, Zhang X, Lv B, Luo L, et al. OsNAC2 encoding a NAC transcription factor that affects plant height through mediating the gibberellic acid pathway in rice. Plant J. 2015;82(2):302–14. doi: 10.1111/tpj.12819 [DOI] [PubMed] [Google Scholar]
- 54.Magome H, Nomura T, Hanada A, Takeda-Kamiya N, Ohnishi T, Shinma Y, et al. CYP714B1 and CYP714B2 encode gibberellin 13-oxidases that reduce gibberellin activity in rice. Proc Natl Acad Sci USA. 2013;110(5):1947–52. doi: 10.1073/pnas.1215788110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang L, Wang Z, Xu Y, Joo SH, Kim SK, Xue Z, et al. OsGSR1 is involved in crosstalk between gibberellins and brassinosteroids in rice. Plant J 2009;57(3):498–510 doi: 10.1111/j.1365-313X.2008.03707.x [DOI] [PubMed] [Google Scholar]
- 56.Li W, Wu J, Weng S, Zhang Y, Zhang D, Shi C. Identification and characterization of dwarf 62, a loss-of-function mutation in DLT/OsGRAS-32 affecting gibberellin metabolism in rice. Planta. 2010;232(6):1383–96 doi: 10.1007/s00425-010-1263-1 [DOI] [PubMed] [Google Scholar]
- 57.Arioli T. Molecular analysis of cellulose biosynthesis in Arabidopsis. Science. 1998;279(5351):717–20. [DOI] [PubMed] [Google Scholar]
- 58.Tanaka K. Three distinct rice cellulose synthase catalytic subunit genes required for cellulose synthesis in the secondary wall. Plant Physiol. 2003;133(1):73–83. doi: 10.1104/pp.103.022442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li M, Xiong G, Li R, Cui J, Tang D, Zhang B, et al. Rice cellulose synthase-like D4 is essential for normal cell-wall biosynthesis and plant growth. Plant J. 2009;60(6):1055–69. doi: 10.1111/j.1365-313X.2009.04022.x [DOI] [PubMed] [Google Scholar]
- 60.Guevara DR, El-Kereamy A, Yaish MW, Mei-Bi Y, Rothstein SJ. Functional characterization of the rice UDP-glucose 4-epimerase 1, OsUGE1: a potential role in cell wall carbohydrate partitioning during limiting nitrogen conditions. PLoS ONE. 2014;9(5):e96158 doi: 10.1371/journal.pone.0096158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang L, Gu X, Xu D, Wang W, Wang H, Zeng M, et al. miR396-targeted AtGRF transcription factors are required for coordination of cell division and differentiation during leaf development in Arabidopsis. J Exp Bot. 2011;62(2):761–73. doi: 10.1093/jxb/erq307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sampedro J, Cosgrove DJ. The expansin superfamily. Genome Biol. 2005;6(12):242 doi: 10.1186/gb-2005-6-12-242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jan A, Yang G, Nakamura H, Ichikawa H, Kitano H, Matsuoka M, et al. Characterization of a xyloglucan endotransglucosylase gene that is up-regulated by gibberellin in rice. Plant Physiol. 2004;136(3):3670–81. doi: 10.1104/pp.104.052274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fincher GB. Revolutionary times in our understanding of cell wall biosynthesis and remodeling in the grasses. Plant Physiol. 2009;149(1):27–37. doi: 10.1104/pp.108.130096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wan LL, Zha WJ, Cheng XY, Liu C, Lv L, Liu CX, et al. A rice β-1,3-glucanase gene Osg1 is required for callose degradation in pollen development. Planta. 2011;233(2):309–23. doi: 10.1007/s00425-010-1301-z [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The blue color standing for chromosome complements inherited from O. longistaminata; white color standing for chromosome complements inherited from O. sativa; green color standing for heterozygosity chromosome complements inherited from O. longistaminata and O. sativa.
(TIF)
The x-axis represents the name of the GO subcategories. The right y-axis indicates the number of genes expressed in a given sub-category. The left y-axis indicates log (10) scale, the percent of a specific category of genes in that main category. GO terms showed statistically significant differences (P value<0.05) were denoted by stars. A and B stand for O. sativa and O. longistaminata, respectively.
(TIF)
(TIF)
(A) Hierarchical cluster analysis of expressed miRNAs. (B) Co-expressed and specially expressed miRNAs. The branch length indicates the degree of variance, and the color represents the logarithmic intensity of expressed miRNAs. Species groups are shown as columns, and individual expressed miRNAs are arrayed in rows.
(TIF)
A and B stand for O. sativa and O. longistaminata, respectively. ELD-A miRNA indicated miRNA expression level is similar to O. sativa, but is differential compared with O. longistaminata; ELD-B miRNA indicated miRNA expression level is similar to O. longistaminata, but is differential compared with O. sativa.
(TIF)
(A), (B) and (C) Integrated ELD-A miRNAs and their coherent target genes network in L1710, L1817 and L1730; (D) Integrated ELD-B miRNAs and their coherent target genes network in L1730. A and B stand for O. sativa and O. longistaminata, respectively; Round rectangle represent miRNAs; Ellipse represent coherent target genes; red represent up-regulated; blue represent down-regulated.
(TIF)
Actin1 and U6 snRNA are used as internal reference gene and small RNA, respectively.
(TIF)
A stand for O. sativa.
(TIF)
Actin1 is used as internal reference gene. The data of quantitative RT-PCR validation are expressed as the mean SD after normalization. Error bars indicate the standard deviation (±SD) of three replicates. Blue polylines are derived from RNA-Sqe data.
(TIF)
U6 snRNA is used as a reference small RNA. The data of real-time qPCR validation are expressed as the mean SD after normalization. Error bars indicate the standard deviation (±SD) of three replicates. Blue polylines are derived from RNA-Seq data.
(TIF)
(XLSX)
(XLSX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(XLSX)
Data Availability Statement
The mRNA and miRNA sequences used in this study have been submitted to the NCBI’s Sequence Read Archive Database. The accession numbers of ten runs are SRR5754959, SRR5754960, SRR5754961, SRR5754962, SRR5754963, SRR5759259, SRR5759260, SRR5759261, SRR5759262 and SRR5759263.