Abstract
Colorectal cancer (CRC) is characterized by genome-wide alterations to DNA methylation that influence gene expression and genomic stability. Less is known about the extent to which methylation is disrupted in the earliest stages of CRC development. In this study we have combined laser-capture microdissection (LCM) with reduced representation bisulfite sequencing (RRBS) to identify cancer-associated DNA methylation changes in human aberrant crypt foci (ACF), the earliest putative precursor to CRC. Using this approach, methylation profiles have been generated for 10 KRAS-mutant ACF and 10 CRCs harboring a KRAS mutation, as well as matched samples of normal mucosa. Of 811 differentially methylated regions (DMRs) identified in ACF, 537 (66%) were hypermethylated and 274 (34%) were hypomethylated. DMRs located within intergenic regions were heavily enriched for AP-1 transcription factor binding sites and were frequently hypomethylated. Furthermore, gene ontology (GO) analysis demonstrated that DMRs associated with promoters were enriched for genes involved in intestinal development, including homeobox genes and targets of the Polycomb repressive complex 2 (PRC2). Consistent with their role in the earliest stages of colonic neoplasia, 75% of the loci harboring methylation changes in ACF were also altered in CRC samples, though the magnitude of change at these sites was lesser in ACF. While aberrant promoter methylation was associated with altered gene expression in CRC, this was not the case in ACF, suggesting the insufficiency of methylation changes to modulate gene expression in early colonic neoplasia. Together, these data demonstrate that DNA methylation changes, including significant hypermethylation, occur more frequently in early colonic neoplasia than previously believed, and identify epigenomic features of ACF that may provide new targets for cancer chemoprevention or lead to the development of new biomarkers for CRC risk.
Keywords: DNA methylation, colorectal cancer, aberrant crypt foci, AP-1
Introduction
Colorectal cancer (CRC) is the second-leading cause of cancer-related deaths in the U.S 1. In addition to the established series of mutational events that accompany the adenoma-carcinoma sequence, a number of epigenetic aberrations have been identified in CRC, including altered DNA methylation and covalent histone modifications 2. Global DNA hypomethylation, first identified in cancers more than three decades ago, is now recognized as a common genetic feature of CRC 3. DNA hypomethylation promotes genomic instability 4, in many cases leading to an increased mutational load and activation of proto-oncogenes 4. On the other hand, gene-specific promoter hypermethylation has been shown to promote CRC by silencing the expression of key tumor suppressor genes such as CDKN2A, hMLH1 and CDH1 5.
While extensive epigenetic modifications are a common feature of CRC, their role in early neoplastic progression is less well defined. As reviewed by Sakai et al. 6, changes to DNA methylation have been found at early stages of cancer development, particularly in colorectal adenomas. Specific DNA methylation changes have even been found within normal colonic mucosa of patients with CRC 7,8, suggesting the possibility that epigenetic defects may predict subsequent cancer risk. Aberrant crypt foci (ACF) are the earliest morphologically identifiable mucosal abnormality in the colon, a subset of which may be precancerous and contribute to a “field defect” within the mucosa 9. Studies from our laboratory 10 and others 11–13 have identified methylation defects associated with a limited set of genes within human ACF, including the silencing of tumor suppressor genes by promoter hypermethylation. An early study by Chan et al. 11 showed that MINT1, MINT2, MINT31, and the tumor suppressor gene CDKN2A, were frequently methylated in ACF from patients with synchronous CRC. We later demonstrated that the tumor suppressor gene RASSF1A is silenced by promoter hypermethylation in distal colon ACF, even in the absence of synchronous tumors 10. More recently, Inoue et al.13 demonstrated that 6 genes are frequently hypermethylated in BRAF-mutant ACF, sessile serrated polyps (SSPs) and cancers located in the proximal colon, providing additional evidence for the role of aberrant methylation in the serrated pathway to CRC. Together, these observations suggest that aberrant DNA methylation patterns may be established prior to adenoma formation and may be important for the promotion of early colonic neoplasia.
While BRAF-mutant tumors are associated with the CpG island methylator phenotype (CIMP)14, and are thus thought to be driven by epigenetic dysregulation, there is also evidence for the role of aberrant methylation in the progression of KRAS-mutant neoplasia. Several studies15–18 have demonstrated that KRAS-mutant CRCs are associated with a distinct set of DNA methylation changes. Notably, Yagi et al.18 have shown that many of the changes present in KRAS-mutant CRCs can also be detected in KRAS-mutant adenomas. Furthermore, Chan et al.11 have shown that the hypermethylation of tumor suppressor genes in ACF is strongly associated with KRAS codon 12 mutations. Together, these findings suggest a role for aberrant methylation in the establishment and progression of KRAS-mutant colonic neoplasia. However, compared to BRAF-mutant lesions, little is known about the specific loci effected by aberrant methylation in the earliest stages of KRAS-mutant colonic neoplasia.
Our limited understanding of the molecular alterations present in premalignant lesions represents an important barrier to the development of new cancer detection and prevention strategies 19. To address this issue, the present study was undertaken to develop a more in-depth understanding of the DNA methylation changes that are present throughout the genome in human ACF. To achieve the requisite sensitivity for analysis of isolated colonic epithelial cells, we combined reduced representation bisulfite sequencing (RRBS) with laser-capture microdissection (LCM) of ACF harboring mutations in exon 2 of the KRAS oncogene. RRBS is an efficient, high-throughput sequencing technique that combines Na-bisulfite conversion of unmethylated cytosines with MspI restriction digestion to enrich samples for CpG-rich regions of the genome 20. Using this approach, we have shown that KRAS-mutant ACF harbor extensive DNA methylation changes, many of which are also present in primary CRCs. In addition to promoter hypermethylation we identify a large number of hypomethylated intergenic regions, which are significantly enriched for AP-1 transcription factor binding sites. Furthermore, ACF-associated methylation changes were enriched in genes involved in cellular identity and differentiation, including a set of homeobox genes and targets of PRC2. These observations extend the scope of aberrant methylation in the earliest stages of colonic neoplasia and define features of an epigenomic landscape that may provide new targets for CRC detection and chemoprevention.
Results
Genome-wide DNA methylation changes in colon cancers and ACF
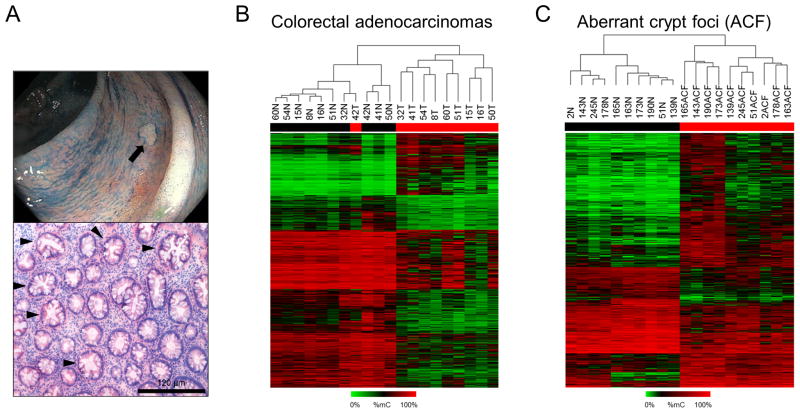
To investigate the epigenomic landscape of early colonic neoplasia, RRBS analysis was used to identify DNA methylation changes in stage III–IV CRCs and ACF biopsied from the distal colon; representative images of an ACF visualized during endoscopy and its histological appearance are shown in Figure 1A. As shown in Supplemental Figures S1A and S1B, data obtained using RRBS from two independent library preparations were highly reproducible. Differentially methylated regions (DMRs), defined as genomic regions exhibiting an average change in methylation ≥ 15% compared to matched normal that achieved a false-discovery-adjusted p-value < 0.05, were used for all subsequent analyses. The complete list of ACF- and cancer-associated DMRs is included in Supplemental Tables S1 and S2. The heat-map shown in Figure 1B, generated by unsupervised clustering, shows DMRs identified in CRC. A total of 23 745 DMRs were identified, consistent with previous reports that aberrant DNA methylation is a common feature of CRC 6. 5 995 (25%) of these DMRs were hypermethylated, while 17 750 (75%) were hypomethylated. Methylation changes were also found in ACF (Figure 1C); 811 regions were differentially methylated compared to matched normal mucosa. However, in ACF, 537 (66%) of DMRs were hypermethylated, indicating a greater tendency towards increased methylation at aberrantly methylated loci in ACF. The difference in net methylation change between ACF and CRC suggests that a shift in global methylation status may be important for progression from early neoplasia to invasive CRC.
Figure 1. DNA methylation patterns in Stage III–IV CRCs and ACF.
(A) Gross appearance of a human ACF during HD-chromoendoscopy (top) and H&E-stained section of an ACF biopsy with serrated morphology (bottom). Serrated crypts, characterized by their star-shaped lumen in cross section, are indicated with black arrows. (B) Heat-map depicting differentially methylated regions (DMRs), defined as genomic regions with a change in methylation relative to matched normal >15% and a FDR-adjusted P-value<0.05, in CRCs. Tumor samples are generally segregated from their matched normal mucosa by unsupervised clustering. Of the 23,745 DMRs detected in cancer samples, 5,995 (25%) were hypermethylated, while 17,750 (75%) were hypomethylated. (C) Heat-map depicting DMRs in ACF. ACF are segregated from normal mucosal samples by unsupervised clustering. 811 DMRs were identified, 537 (66%) of which were hypermethylated and 274 of which (34%) were hypomethylated.
To gain a better understanding of the functional significance of methylation changes found in ACF, GSEA was used to compare the set of all ACF DMRs to annotated functional gene sets stored in the Molecular Signatures Database (Broad Institute, Cambridge, MA) 21; GSEA results are summarized in Supplemental Table S3. GSEA identified enrichment for transcription factors involved in intestinal development, including APC2, FOXF1, NKX2-3, NOTCH1, PAX6, and PDGFA (Supplemental Figure S2) 22–27. In addition, this set of genes was enriched for targets of PRC2 identified in human embryonic stem cells (hESCs) 28. PRC2 is a chromatin-remodeling complex involved in the maintenance of stem cell plasticity 29. Furthermore, ACF-associated DMRs showed enrichment for genes known to be trimethylated at lysine 27 of histone subunit 3 (H3K27me3, the “polycomb mark”) in normal adult colonic mucosa 30. Finally, genes with frequent methylation changes in ACF contained a number of HOX genes (HOXA3, HOXC9, HOXC10, HOXC11, and HOXC13), a family of early developmental regulators whose aberrant expression has previously been implicated in human carcinogenesis 31. These findings demonstrate that genes involved in normal intestinal development and genes targeted by PRC2 in embryonic stem cells frequently exhibit methylation changes in human ACF and suggest a role for the epigenetic disruption of these genes in the establishment of early neoplasia.
Cancer-associated methylation changes are present in ACF
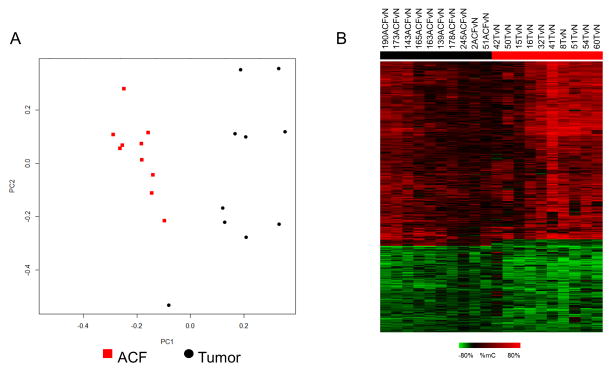
In the following analyses, methylation profiles of DMRs present in CRCs and ACF samples were normalized to matched normal-appearing mucosa. As shown in Figure 2A, data were clearly separated by PCA, indicating distinct methylation profiles, although ACF showed tighter overall clustering than tumors. Despite segregation by PCA, a subset of genomic regions was classified as DMRs in both cancer and ACF samples (Figure 2B). Of these shared DMRs, the majority exhibited methylation changes in the same direction (66% hypermethylated, 32% hypomethylated) and are referred to as “overlapping” DMRs. In general, these methylation changes were of greater magnitude in cancers than in ACF, regardless of the direction of change or the genomic location of the altered region. As shown in Supplemental Figure S3, aberrant methylation of a representative DMR associated with the gene RSPO2 was validated in an independent set of ACF and CRC samples using Combined Bisulfite Restriction Analysis (COBRA).
Figure 2. Direct comparison of DNA methylation changes in Stage III–IV CRCs and ACF.
(A) Principal component analysis (PCA) bi-plot of DNA methylation changes demonstrates a clear separation of ACF (red) and tumor samples (black). CRCs are less tightly clustered than ACF, indicating a greater variability of DNA methylation. (B) Heat-map depicting overlapping DMRs detected in both CRCs and ACF. Of 608 shared DMRs, 66% were hypermethylated and 32% were hypomethylated; a small subset of DMRs (2%) were hypermethylated in ACF, but hypomethylated in cancers. The magnitude of these overlapping methylation changes was typically greater in CRCs than in ACF.
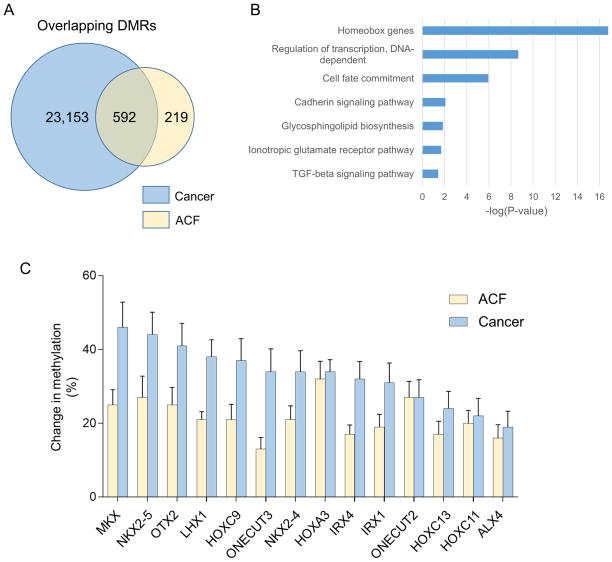
As shown in Figure 3A, 592 DMRs were classified as overlapping; a complete list of overlapping DMRs is provided in Supplemental Table S4. Bootstrapping analysis confirmed that this degree of overlap between ACF and CRC DMRs was highly significant (Supplemental Figure S1C). Approximately 75% of all DMRs identified in ACF overlapped with CRC DMRs. Hypermethylated overlapping DMRs were evenly distributed among promoters (139, 35%), gene bodies (147, 37%) and intergenic regions (113, 28%) (Supplemental Figure S4A). In contrast, hypomethylated overlapping DMRs occurred mainly within gene bodies (84; 44%) and intergenic regions (104; 53%); only 5 (3%) were located in promoter regions (Supplemental Figure S4A). As shown in Figure 3B, GSEA revealed that overlapping DMRs were frequently associated with homeobox genes. As shown in Figure 3C, overlapping DMRs associated with homeobox genes were frequently hypermethylated.
Figure 3. Functional enrichment analysis of overlapping DMRs.
(A) 592 DMRs exhibited a methylation change in the same direction in both stage III–IV cancers and ACF; these DMRs are referred to as “overlapping” DMRs. (B) Gene ontology (GO) analysis for overlapping DMRs. These DMRs were significantly enriched (FDR-adjusted P-value <0.05) for homeobox genes, as well as genes involved in the regulation of transcription and cell fate commitment. (C) Representative homeobox genes that show increased methylation in both CRCs and ACF. All of the genes identified in this panel exhibit significantly increased methylation in both CRCs and ACF compared to matched normal tissues. Error bars represent means +/− SEM.
Non-promoter DMRs undergo a switch in methylation status during cancer development
To better define the methylation landscape of early and late colonic neoplasia, all DMRs were separated according to their genomic location into gene body, intergenic, and promoter DMRs. 8 849 (37%) cancer-associated DMRs were located within gene bodies, 12 170 (51%) were located in intergenic regions, and 2 726 (11%) were located within known promoter regions, indicating more extensive methylation changes within non-promoter regions in CRC (Supplemental Figure S4B). Similarly, of the 811 DMRs found in ACF, 349 (43%) were located within gene bodies, 297 (37%) were located in intergenic regions, and 165 (20%) were located within promoters (Supplemental Figure S4B).
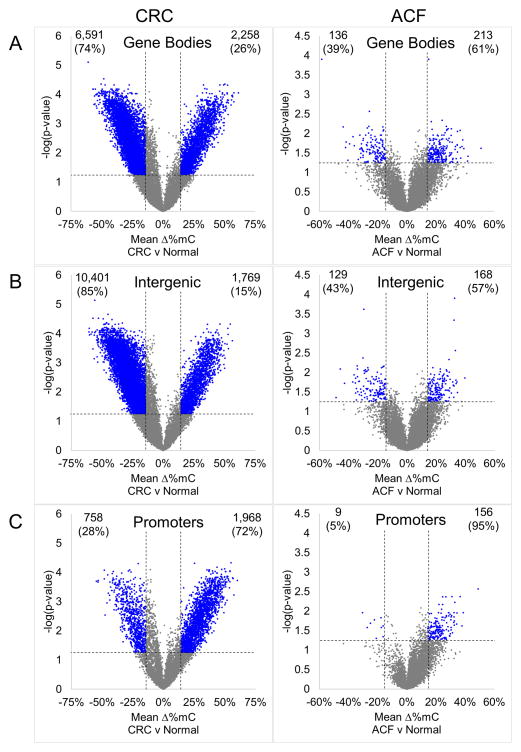
In cancers, DMRs in non-promoter regions were primarily hypomethylated compared to matched normal; 6 591 (74%) gene body-associated DMRs were hypomethylated (Figure 4A) and 10 401 (85%) intergenic DMRs were hypomethylated (Figure 4B). In contrast, 213 (61%) gene body-associated DMRs were hypermethylated (Figure 4A) and 168 (57%) intergenic regions were hypermethylated (Figure 4B) in ACF samples. Together, these results suggest that genome-wide DNA hypomethylation occurs during later stages of CRC progression. In contrast to gene bodies and intergenic regions, the majority of promoter DMRs in both cancers (1 968, 72%) and ACF (165, 95%) were hypermethylated (Figure 4C).
Figure 4. Volcano plots depicting DMRs in colon cancers and ACF grouped by genomic region.
DMRs identified in CRCs and ACF were segregated according to their genomic location, and the methylation changes in gene bodies, intergenic and promoter regions were examined; DMRs are depicted as blue dots. (A) DMRs located within gene bodies. In cancer tissue, 12,258 DMRs (74%) were hypomethylated and 6,591 (26%) were hypermethylated. In contrast, in ACF, 136 gene body DMRs (39%) were hypomethylated and 213 (61%) were hypermethylated. (B) Intergenic DMRs in colon cancers and ACF. In cancers, 10,401 (85%) intergenic DMRs were hypomethylated while only 1,769 (15%) were hypermethylated. Of the 297 intergenic DMRs identified in ACF, 129 (43%) were hypomethylated, while 168 (57%) were hypermethylated. (C) Promoter-associated DMRs in colon cancers and ACF. In cancers, 758 DMRs (28%) were hypomethylated while 1,968 (72%) were hypermethylated. The same pattern was observed in promoter DMRs identified in ACF; only 9 promoter DMRs (5%) were hypomethylated, while 156 (95%) were hypermethylated.
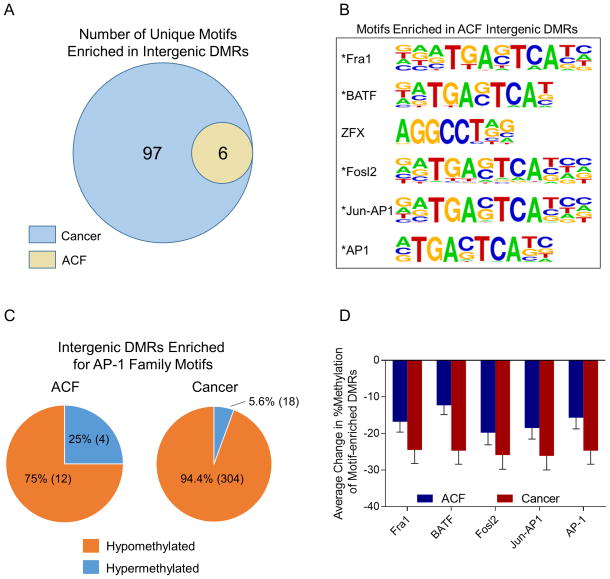
Intergenic DMRs are enriched for AP-1 family transcription factor binding motifs
As shown in Figure S3 and Figure 4, a significant fraction of ACF and cancer DMRs were located in noncoding intergenic regions of the genome. To determine whether these DMRs were enriched for regulatory features, we used Hypergeometric Optimization of Motif EnRichment (HOMER) analysis 32 to identify sequence elements within DMRs that correspond to known transcription factor binding motifs. As shown in Figure 5A, intergenic DMRs in CRCs were enriched for 103 motifs, including 6 that were also enriched in ACF intergenic DMRs. Furthermore, a number of motifs were also enriched in DMRs located within gene bodies in cancer samples; complete results of the HOMER analysis are provided in Supplemental Table S5. As shown in Supplemental Figure 1D, bootstrapping analysis indicated that these results were highly statistically significant. As shown in Figure 5B, 5 of the 6 motifs enriched in ACF intergenic DMRs corresponded to binding sequences for AP-1 family transcription factors (Figure 5B). The majority of DMRs enriched for AP-1 motifs were hypomethylated in both ACF and cancer samples (Figure 5C); however, a greater number of AP-1 sites were affected in cancer samples than in ACF. Furthermore, as shown in Figure 5D, the average degree of hypomethylation at AP-1 motif-associated DMRs was greater in cancers than in ACF.
Figure 5. Enrichment of AP-1 binding motifs in intergenic DMRs.
Intergenic DMRs from cancers and ACF were subjected to Hypergeometric Optimization of Motif EnRichment (HOMER) analysis to determine whether they were enriched for known regulatory elements. (A) 103 motifs were enriched in intergenic DMRs in cancers, including 6 motifs which were also enriched in intergenic DMRs in ACF. (B) Motifs enriched in ACF intergenic regions. Five of the 6 motifs enriched in ACF intergenic DMRs corresponded to the binding sequences of AP-1 transcription factor family members, indicated with an asterisk. (C) Change in methylation of DMRs enriched for AP-1 binding motifs in ACF and cancer. In both sample sets, the DMRs containing AP-1 binding motifs were significantly hypomethylated compared to their respective matched normal samples. This pattern was especially pronounced in CRCs, where 95% of AP-1 enriched DMRs were hypomethylated. Furthermore, a greater number of AP-1 sites were affected in CRCs than in ACF. (D) Average change in percent methylation of DMRs enriched for AP-1 family motifs. Intergenic DMRs containing AP-1 binding motifs exhibited a ~15% reduction in methylation in ACF and a ~25% reduction in cancer. Bars represent average change in methylation of all DMRs containing the indicated motif. Error bars represent means +/− SEM.
Aberrant DNA methylation is associated with altered gene expression in colon cancers, but not in ACF
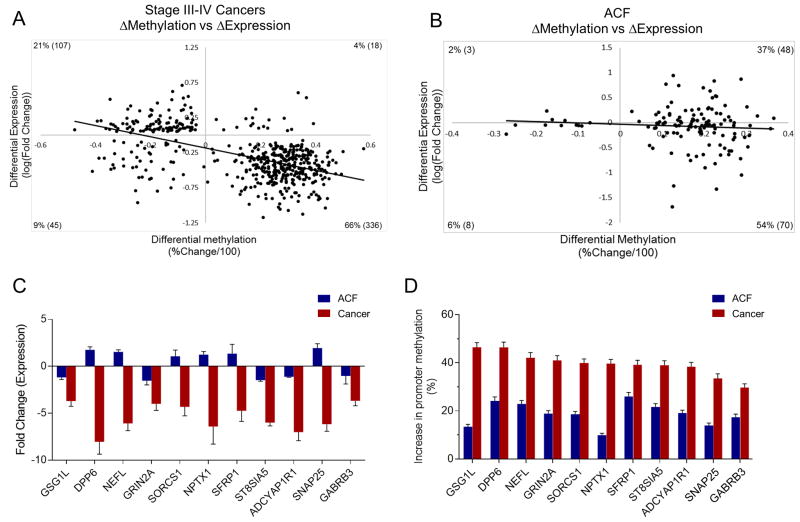
Promoter methylation is an important mechanism for regulating gene expression 5. To assess the functional consequences of aberrant promoter methylation, genome-wide RNA-Seq analysis was performed on CRC and ACF samples. Scatter-plots depicting the correlation between promoter methylation and gene expression are shown in Figure 6. In CRCs, promoter methylation was negatively correlated with gene expression (Pearson’s r = −0.55, P=0.005) (Figure 6A), consistent with the transcriptional silencing commonly found in advanced neoplasia 5. However, when ACF were subjected to a similar analysis, there was no correlation observed between promoter methylation status and changes in gene expression (Pearson’s r = −0.07, P=0.7) (Figure 6B).
Figure 6. Promoter methylation is correlated with gene expression in CRCs but not ACF.
(A) Scatter-plot depicting the correlation between changes in promoter methylation and gene expression in CRCs (Pearson’s correlation coefficient = −0.55), indicating increased promoter methylation associated with gene silencing. (B) Scatter-plot depicting the lack of correlation between changes in promoter methylation and gene expression in ACF (Pearson’s correlation coefficient = −0.07). (C) Selected genes with promoter DMRs identified in both CRCs and ACF. GSG1L, DPP6, NEFL, GRIN2A, SORCS1, NPTX1, SFRP1, ST8SIA5, ADCYAP1R1, SNAP25, and GABRB3 were significantly down-regulated in cancer samples (at least 3-fold reduction in expression), but not significantly altered in ACF. (D) These same genes exhibit promoter hypermethylation in both stages of neoplasia, but the extend of hypermethylation is greater in CRC. Bars represent the average percentage change in methylation of promoter DMRs for indicated genes. Error bars represent 95% confidence interval and SEM in (C) and (D), respectively.
This lack of correlation between promoter methylation and gene expression in ACF may be related to the magnitude of methylation changes. To address this possibility, 12 genes with promoter hypermethylation in both CRC and ACF, but reduced expression in CRC only, were selected for further analysis of their promoter methylation statuses. As shown in Figure 6C, the expression of GSG1L, DPP6, NEFL, GRIN2A, SORCS1, NPTX1, SFRP1, ST8SIA5, ADCYAP1R1, SNAP25 and GABRB3 were each significantly reduced by at least 3-fold in Stage III–IV cancers, but had no significant change in expression in ACF. These genes showed an average increase in promoter methylation of ~40% in CRCs, but an increase of only ~20% in ACF (Figure 6D), suggesting that the extent of promoter methylation in early neoplasia is insufficient to alter gene expression.
Overexpression of EZH2, the catalytic subunit of PRC2, has been identified in CRC and is frequently associated with a poor prognosis 33. To determine whether the enrichment of ACF DMRs for PRC2 targets may be due to the overexpression of components of the PRC2 complex, we interrogated our RNA-Seq data for changes in expression of the Polycomb genes EZH2, SUZ12, and EED. As shown in Supplemental Figure S5A, EZH2 was overexpressed in Stage III–IV CRCs (1.55-fold increase, p=0.03), but not in ACF (Supplemental Figure S5B). We also interrogated the RNA-Seq dataset for the expression levels of the DNA methyltransferases (DNMTs), a group of genes (DNMT1, DNMT3A and DNMT3B) that are often up-regulated in CRC 34. As shown in Supplemental Figure S5A&B, DNMT1 and DNMT3B are overexpressed in cancer samples, but not in ACF. Taken together, these results suggest that while overexpression EZH2, DNMT1 and DNMT3B may promote hypermethylation and cancer progression, their overexpression is not required for the establishment of early colonic neoplasia. Alternatively, higher expression of DNMT1, DNMT3B and EZH2 may accompany increased cell proliferation in CRC but may not be causative for the DNA methylation changes.
Discussion
The disruption of DNA methylation patterns has been shown to play an important role in the pathogenesis of CRC 6. However, the epigenomic landscape at the earliest stages of colonic neoplasia has not been clearly defined. Several studies have investigated methylation defects at pre-defined targets within ACF and identified changes associated with a limited set of genes 7,8,10,11,13. However, an investigation of DNA methylation changes occurring in early human colonic neoplasia using next-generation sequencing and an integrated genomics approach has not yet been reported. In the present study, we have applied a highly sensitive genome-wide approach by combining LCM with RRBS to compare DNA methylation changes present in early colonic neoplasia with the extensive modifications found in advanced CRC.
As expected, the majority of DMRs identified in Stage III–IV cancers were hypomethylated. This global loss of DNA methylation was particularly common within intergenic regions, in which ~85% of DMRs were hypomethylated. These findings are consistent with previous reports of genome-wide hypomethylation in a number of different human cancers 3,35. DNA hypomethylation also occurs in low-grade adenomas 35,36, indicating that this epigenetic alteration precedes the development of more advanced malignancies. However, our present study has uncovered an unexpected finding; the majority of DMRs (across all genomic locations) in ACF samples are hypermethylated, indicating a global gain of DNA methylation at the earliest stages of tumor initiation. Since the majority of ACF are self-limiting and unlikely to progress to malignancy 37, genome-wide hypermethylation may in fact provide a mechanism for restricting ACF progression, in part by reducing the likelihood of genomic instability associated with DNA hypomethylation 4. Because our investigation was limited to KRAS-mutant ACF, the present study is unable to rule out the possibility that this hypermethylation phenotype is restricted to early neoplastic lesions with KRAS mutations. Thus, these results warrant future study of the global methylation status of ACF harboring other common mutations, such as BRAF V600E.
Promoter DMRs in both CRC and ACF were enriched in genes known to be targeted by PRC2 in embryonic stem cells (ESCs). PRC2 is a histone-modifying complex expressed in embryonic stem cells that plays an important role in maintaining ‘stemness’ 29 by repressing genes required for differentiation via the methylation of histone H3K27 38. Since many genes targeted by PRC2 in human ESCs become hypermethylated in CRC and other cancers 39,40, it is thought that PRC2 occupancy and/or histone methylation will increase the susceptibility of genes to aberrant hypermethylation 41,42. Widschwendter et al. hypothesized that cancer-associated promoter hypermethylation of PRC2 targets originates in stem cells during the earliest stages of carcinogenesis, and predisposes these cells to neoplastic transformation by “locking in” a stem cell phenotype 40. Our data support the timing of this hypothesis by demonstrating for the first time that hypermethylation of PRC2 targets occurs as early as the ACF stage. While H3K27 methylation of PRC2 targets was originally thought to be restricted to ESCs, Rada-Iglesias et al. 43 and Hahn et al. 44 showed that this modification is also found in normal adult colonic epithelial cells. Together, these observations suggest that DNA hypermethylation in ACF is directed by the Polycomb complex, or by its associated histone marks that are present in normal cells prior to early neoplastic initiation.
In addition, the present study has uncovered extensive promoter and gene body hypermethylation of homeobox genes in both cancers and ACF, many of which belong to the HOX family. The expression of homeobox genes, which is regulated to a large extent by epigenetic modifications 45,46, contributes to the maintenance of cellular identity and adult tissue morphology 47. Aberrant hypermethylation of homeobox-containing genes has been described in breast and lung cancers, and several HOX genes (HOXA7, HOXA9, and HOXB13) are reportedly hypermethylated in CRC 39,48,49. However, the large number of differentially methylated homeobox genes identified in our panel of primary Stage III–IV cancers (Figure 3) is an unexpected result. A subset of homeobox genes was also hypermethylated in ACF, suggesting that this epigenetic change is an early event in colonic neoplasia. CpG islands associated with homeobox genes are commonly hypermethylated in early stage ductal carcinomas (DCIS) and early-stage lung carcinomas 39,49, suggesting that this change is an early event in other tissues as well. Together, these results suggest that dysregulation of homeobox genes via hypermethylation occurs more extensively in CRC than previously thought, and that this epigenetic aberration is established in early neoplasia.
Finally, our results indicate that aberrant DNA methylation commonly occurs within non-transcribed, intergenic regions in both ACF and CRCs. Upon further analysis, we have found that intergenic DMRs are enriched for transcription factor binding sites, especially those corresponding to members of the AP-1 transcription factor family. AP-1 is a nuclear transcription factor that controls many critical cellular functions, including proliferation, differentiation and apoptosis 50. AP-1 also plays an important role in oncogenesis; activating mutations in the KRAS gene are thought to promote tumor development by increasing AP-1 activity, with concurrent up-regulation of proliferative and anti-apoptotic genes 51. Notably, all of the ACF and CRC samples examined in this study contained mutant KRAS. Recent evidence indicates that transcriptional activation by AP-1 is controlled, in part, by DNA methylation 52. As shown by Park et al., the DNA binding activity of AP-1 is significantly reduced when CpGs in close proximity to its DNA binding motif are methylated 52. A genome-wide methylation study of a single KRAS-mutant stage III colon adenocarcinoma by Berman et al. 53 suggested that epigenetic regulation of AP-1 is disrupted in cancer by demonstrating that genomic regions with cancer-specific hypomethylation are significantly enriched for AP-1 binding motifs. Our study has confirmed this observation in a larger panel consisting of 10 KRAS-mutant colon cancers, substantiating a role for hypomethylation of AP-1 sequences in CRC. Furthermore, our data extend this hypothesis by demonstrating that a small number of AP-1 sites are hypomethylated in ACF, suggesting that this epigenetic phenomenon begins early in the neoplastic process. However, the number of affected AP-1 sites, and the average degree of hypomethylation at these sites, was significantly greater in cancer samples than in ACF. These observations suggest that the expansion of AP-1 site hypomethylation may promote neoplastic progression.
In summary, the present study demonstrates that cancer-associated DNA methylation changes are more abundant in ACF than previously thought. These changes frequently affect genes involved in cellular identity and differentiation, suggesting that disrupted regulation of cell fate determination may contribute to the establishment of early colonic neoplasia. Methylation of these genes, including those targeted by PRC2 or containing homeobox sequences, may be useful targets for novel chemopreventive interventions. In addition, we identified significant hypomethylation associated with AP-1 transcription factor binding sites, suggesting a role for epigenetic dysregulation of AP-1 activity in the development of CRC. Furthermore, the present study identifies epigenetic changes specifically associated with advanced neoplasia, including a switch in the global methylation status of non-promoter regions, which may serve as useful biomarkers for cancer risk. Finally, while aberrant promoter methylation was associated with altered gene expression in CRC, no such association was detected in ACF. Combined with the observation that the magnitude of change in methylation was greater, on average, in CRC than ACF, these findings suggest that DNA methylation changes are insufficient to alter gene expression at the ACF stage. Our results provide new insights into the role of DNA methylation in the development of early colonic neoplasia, and provide target candidates for the use of epigenetic profiling in CRC detection and prevention.
Materials and Methods
Subject selection and human tissue collection
All ACF subjects included in the present study were selected from a larger population of patients enrolled in an ongoing clinical study at John Dempsey Hospital (JDH, UConn Health). All patients who met the Amsterdam criteria for familial adenomatous polyposis (FAP) or hereditary non-polyposis CRC (HNPCC) were excluded from this overarching study. In order to control for the effects of age and smoking on DNA methylation, all subjects selected for the present study were non-smokers between the ages of 50 and 65. 10 ACF were biopsied from the distal colons of individual subjects as previously described 54. Briefly, ACF were identified and biopsied during high-definition, magnifying chromoendoscopy using indigo carmine dye-spray 54. For each subject, a sample of normal-appearing mucosa was also obtained from the distal colon. Biopsies were immediately embedded in OCT freezing medium (Neg 50, ThermoFisher Scientific, Waltham, Massachusetts, USA), flash-frozen, and stored at −80°C. Frozen tissues were sectioned onto polyethylene naphthalate (PEN, ThermoFisher Scientific) membrane slides using a Leica CM1900 Cryostat. Frozen sections were stained with hematoxylin and eosin (H&E) and routine histologic analyses were performed. All biopsies collected from ACF subjects were histologically confirmed by a board-certified human gastrointestinal pathologist blinded according to previously established criteria 54.
Twenty Stage III–IV CRCs resected from the distal colons of individual subjects were obtained from the City of Hope (COH) frozen tumor bank. In addition, 10 histologically-confirmed matched normal samples, collected from the margins on either side of the resected tumor, were obtained. Tissue sections were stained with H&E and reviewed by a board-certified pathologist to confirm the presence and histopathology of the lesions. This study was conducted with the written, informed consent of each subject, as well as Institutional Review Board approval from both the University of Connecticut Health and COH (IRB Protocols IE-10-068OSJ-3 and 97134, respectively).
Laser-capture microdissection
LCM of ACF and matched normal biopsies was performed as previously described 54. Briefly, an ArcturusXT Laser Capture Microdissection system (ThermoFisher Scientific) was used to isolate a minimum of 1 mm2 of tissue (~5000 cells) from 12 μm-thick frozen serial sections. LCM caps were stored at −80°C until nucleic acid extraction.
DNA methylation profiling and mutation screening
Genomic DNA was isolated from laser-captured colonic crypt epithelium using phenol/chloroform extraction. All ACF samples collected as part of the overarching ACF study currently ongoing at JDH undergo routine screening for KRAS and BRAF mutations using Sanger sequencing. In addition, the 20 Stage III–IV CRC samples obtained from the COH Tumor Bank were screened for KRAS and BRAF mutations at the COH Integrative Genomics Core. KRAS/BRAF mutation screening was done using PCR to amplify target regions (KRAS Exon 2: 5′GGTCCTGCACCAGTAATATG and 5′AACCTTATGTGTGACATGTTCTAA, BRAF V600E Region: 5′AACTCTTCATAATGCTTGCTCTGA and 5′CAGACAACTGTTCAAACTGATGGGACC) followed by Sanger sequencing. Ten ACF and 10 cancer samples, each with a mutation in KRAS Exon 2 (KRASG12D or KRASG12V) and no detectable BRAF mutation, were selected for further analysis by RRBS. RRBS was done as previously described 55. Briefly, purified DNA was digested by overnight incubation with MspI at 37°C. Following restriction digest, DNA fragments were subjected to end repair and A-tailing, followed by linker ligation. Bisulfite conversion was done using the EZ DNA Methylation-Gold Kit (Zymo Research, Irvine, California, USA) according to the manufacturer’s instructions. RRBS libraries were sequenced on the HiSeq 2500 (Illumina, San Diego, California, USA) using 100 bp paired-end sequencing according to the manufacturer’s protocol. Alignment of bisulfite sequencing reads and determination of the methylation state at each cytosine was done using Bismark (version 0.13.0) with default parameters. A minimum of 14 million reads/sample were aligned to human reference genome assembly hg19 and cytosines with fewer than 5 reads in any sample were removed from subsequent analyses. Per-sample sequencing depth metrics are listed in Supplemental Table S6. For RRBS analysis, genomic regions were defined as sequences containing a minimum of 2 CpGs separated by no more than 100 bp. Differentially methylated regions (DMRs) were defined as genomic regions exhibiting an average change in methylation ≥15% compared to matched normal, with a false discovery rate (FDR)-adjusted p-value < 0.05. Bootstrapping analysis was performed as a control for the identification of overlapping DMRs by generating 1 000 sets of 811 randomly selected DMRs, calculating the average number of overlapping DMRs in these sets and comparing the expected value to the observed number of overlapping DMRs. Changes in methylation status were validated in an independent set of samples using Combined Bisulfite Restriction Analysis (COBRA) according to standard protocol56.
Transcriptome profiling
RNA was isolated from cancer samples using the mirVana miRNA Isolation Kit (ThermoFisher) and from laser-captured ACF samples using the Arcturus PicoPure Frozen RNA Isolation Kit (ThermoFisher) according to the manufacturers’ instructions. Sequencing libraries were prepared from RNA isolated from cancer samples using the TruSeq Stranded Total Library Preparation Kit and the RiboZero Gold rRNA Removal Kit (Illumina). For RNA extracted from ACF, depletion of rRNA was done using the RiboZero Magnetic Kit according to the “Protocol for Removal of rRNA from Small Amounts of Total RNA” (Clontech, Mountain View, California, USA). Subsequently, dscDNA library preparation and amplification were done using the SMARTer Stranded RNA-Seq Kit (Clontech) according to the manufacturer’s protocol. RNA sequencing for both sample types was done on the HiSeq 2500 (Illumina) using 40-bp paired-end sequencing according to the manufacturer’s protocol. A minimum of 40 million paired-end reads per sample were aligned to the human reference genome assembly hg19.
Bioinformatics Analysis
Gene ontology (GO) analysis was conducted using the Database for Annotation, Visualization, and Integrated Discovery (DAVID) according to standard protocol 57. Briefly, GO analysis was done using the statistical overrepresentation test with Bonferroni correction for multiple testing. Gene set enrichment analysis (GSEA) was performed by calculating the overlap of gene sets of interest with annotated gene sets stored in the Molecular Signatures Database (MSigDB) version 5.1 (Broad Institute, Cambridge, Massachusetts, USA)21. Comparison of DMRs to H3K4me-marked enhancer regions was done using GEO dataset GSM621670. Hypergeometric Optimization of Motif EnRichment (HOMER) analysis 32 was used to evaluate the enrichment of known transcription factor binding motifs located within differentially methylated regions of the genome. Bootstrapping analysis was performed as a control for HOMER by generating 1 000 sets of 297 randomly selected intergenic regions, calculating the expected percent enrichment for motifs of interest and comparing this value to the observed percent enrichment. RRBS and RNA-seq datasets have been deposited in the NCBI Gene Expresson Omnibus, and may be retrieved using accession number GSE95656.
Statistical Analysis
The set of genomic regions with frequently altered DNA methylation was subjected to complete linkage clustering using a Euclidean distance measure. Correlation between changes in promoter methylation and changes in gene expression were estimated by Pearson’s correlation analysis. Statistical analyses of differentially methylated regions (DMRs) and differentially expressed genes were performed using Student’s t-test followed by false discovery rate (FDR) correction using the Benjamini-Hochberg procedure. Principal component analysis (PCA) and biplot generation were done using the R programming language and software environment.
Supplementary Material
(A) Parallel sequencing runs of two independent library preparations from the same ACF sample show a high correlation between the number of successfully mapped reads per genomic region. (B) Correlation between multiple sequencing runs from the same library for 2 ACF and 2 samples of normal colonic mucosa. Correlation coefficients were calculated for the total number of successfully mapped reads for individual CpGs with a minimum of 10 mapped reads. Excluding Msp1 restriction sites (CCGG) increased between-run correlations. (C) Histogram depicting the results of bootstrapping control analysis for overlapping DMR analysis. On average, randomly generated genomic region sets contained 9.6 ± 3.1 overlapping DMRs, significantly fewer than the 608 observed real overlapping DMRs (P<0.001), indicating that the probability of identifying these overlapping DMRs by random chance is very low. (D) Density plot of motif scores for AP-1 family binding motifs generated via bootstrapping analysis. Asterisks indicate the actual enrichment score for each motif observed in intergenic ACF DMRs.
These DMRs tended to overlap with H3K4me peaks identified in normal colonic mucosa, suggesting that they occur within active enhancer regions. H3K4me data was obtained from the ENCODE project (Accession: GSM621670).
(A) Several DMRs exhibiting increased methylation relative to patient-matched normal were identified by RRBS within the body of RSPO2. (B) Hypermethylation of the DMR associated with Exon 2 of RSPO2 was validated in an independent set of human colorectal tumors using the COBRA assay. The appearance of a second band in the BstUI-digested lanes (+) indicates the presence of methylation at this site. (C) Hypermethylation of this same DMR was also validated in an independent set of ACF samples using COBRA.
(A) Distribution of the genomic location of DMRs with altered methylation identified in both ACF and CRC. Overlapping hypermethylated DMRs were evenly distributed amongst promoters (35%), gene bodies (37%) and intergenic regions (28%). Overlapping DMRs exhibiting hypomethylation were rarely located within promoters (3%), but were common in gene bodies (44%) and intergenic regions (53%). (B) Distribution of the genomic location of all DMRs (increased and decreased methylation) in ACF and CRC. While the genomic distribution of DMRs identified in ACF was very similar to the distribution of all sequenced regions, DMRs identified in cancer samples exhibited a bias towards non-promoter localization.
Expression of PRC2 components and DNA methyltransferases in cancer and ACF samples. (A) Significant overexpression of the PRC2 components EZH2 (1.55-fold, P<0.05), SUZ12 (1.12-fold, P<0.05) and EED (1.1-fold, P<0.05) were detected in cancer samples. Furthermore, overexpression of the DNA methyltransferase enzymes DNMT1 (1.2-fold, P<0,05) and DNMT3B (1.45-fold, P<0.05) were detected in cancers. (B) Despite similarities in the patterns of aberrant methylation in cancer and ACF, including the enrichment of promoter DMRs for PRC2-target genes in ACF, no change in the expression of any PRC2 component or DNMT enzyme was detected in ACF samples.
Acknowledgments
This work was supported by NIH grant CA159976 to DWR, and NIH grants CA084469 and CA160965 to GPP.
Footnotes
Conflicts of Interest:
The authors state that there are no conflicts of interest to disclose.
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc).
References
- 1.American Cancer Society. Cancer Facts & Figures 2016. Am Cancer Soc. 2016 [Google Scholar]
- 2.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ehrlich M. DNA hypomethylation in cancer cells. Epigenomics. 2009;1:239–259. doi: 10.2217/epi.09.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eden A, Gaudet F, Waghmare A, Jaenisch R. Chromosomal instability and tumors promoted by DNA hypomethylation. Science. 2003;300:455. doi: 10.1126/science.1083557. [DOI] [PubMed] [Google Scholar]
- 5.Esteller M. CpG island hypermethylation and tumor suppressor genes: a booming present, a brighter future. Oncogene. 2002;21:5427–5440. doi: 10.1038/sj.onc.1205600. [DOI] [PubMed] [Google Scholar]
- 6.Sakai E, Nakajima A, Kaneda A. Accumulation of aberrant DNA methylation during colorectal cancer development. World J Gastroenterol. 2014;20:978–987. doi: 10.3748/wjg.v20.i4.978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kawakami K, Ruszkiewicz A, Bennett G, Moore J, Grieu F, Watanabe G, et al. DNA hypermethylation in the normal colonic mucosa of patients with colorectal cancer. Br J Cancer. 2006;94:593–598. doi: 10.1038/sj.bjc.6602940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Silviera ML, Smith BP, Powell J, Sapienza C. Epigenetic differences in normal colon mucosa of cancer patients suggest altered dietary metabolic pathways. Cancer Prev Res Phila Pa. 2012;5:374–384. doi: 10.1158/1940-6207.CAPR-11-0336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chai H, Brown RE. Field effect in cancer-an update. Ann Clin Lab Sci. 2009;39:331–337. [PubMed] [Google Scholar]
- 10.Greenspan EJ, Jablonski MA, Rajan TV, Levine J, Belinsky GS, Rosenberg DW. Epigenetic alterations in RASSF1A in human aberrant crypt foci. Carcinogenesis. 2006;27:1316–1322. doi: 10.1093/carcin/bgi373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chan AO-O, Broaddus RR, Houlihan PS, Issa J-PJ, Hamilton SR, Rashid A. CpG island methylation in aberrant crypt foci of the colorectum. Am J Pathol. 2002;160:1823–1830. doi: 10.1016/S0002-9440(10)61128-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suzuki H, Watkins DN, Jair K-W, Schuebel KE, Markowitz SD, Dong Chen W, et al. Epigenetic inactivation of SFRP genes allows constitutive WNT signaling in colorectal cancer. Nat Genet. 2004;36:417–422. doi: 10.1038/ng1330. [DOI] [PubMed] [Google Scholar]
- 13.Inoue A, Okamoto K, Fujino Y, Nakagawa T, Muguruma N, Sannomiya K, et al. B-RAF mutation and accumulated gene methylation in aberrant crypt foci (ACF), sessile serrated adenoma/polyp (SSA/P) and cancer in SSA/P. Br J Cancer. 2015;112:403–412. doi: 10.1038/bjc.2014.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weisenberger DJ, Siegmund KD, Campan M, Young J, Long TI, Faasse MA, et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet. 2006;38:787–793. doi: 10.1038/ng1834. [DOI] [PubMed] [Google Scholar]
- 15.Shen L, Toyota M, Kondo Y, Lin E, Zhang L, Guo Y, et al. Integrated genetic and epigenetic analysis identifies three different subclasses of colon cancer. Proc Natl Acad Sci. 2007;104:18654–18659. doi: 10.1073/pnas.0704652104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yagi K, Akagi K, Hayashi H, Nagae G, Tsuji S, Isagawa T, et al. Three DNA Methylation Epigenotypes in Human Colorectal Cancer. Clin Cancer Res. 2010;16:21–33. doi: 10.1158/1078-0432.CCR-09-2006. [DOI] [PubMed] [Google Scholar]
- 17.Hinoue T, Weisenberger DJ, Lange CPE, Shen H, Byun H-M, Van Den Berg D, et al. Genome-scale analysis of aberrant DNA methylation in colorectal cancer. Genome Res. 2012;22:271–282. doi: 10.1101/gr.117523.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yagi K, Takahashi H, Akagi K, Matsusaka K, Seto Y, Aburatani H, et al. Intermediate methylation epigenotype and its correlation to KRAS mutation in conventional colorectal adenoma. Am J Pathol. 2012;180:616–625. doi: 10.1016/j.ajpath.2011.10.010. [DOI] [PubMed] [Google Scholar]
- 19.Campbell JD, Mazzilli SA, Reid ME, Dhillon SS, Platero S, Beane J, et al. The Case for a Pre-Cancer Genome Atlas (PCGA) Cancer Prev Res Phila Pa. 2016;9:119–124. doi: 10.1158/1940-6207.CAPR-16-0024. [DOI] [PubMed] [Google Scholar]
- 20.Gu H, Smith ZD, Bock C, Boyle P, Gnirke A, Meissner A. Preparation of reduced representation bisulfite sequencing libraries for genome-scale DNA methylation profiling. Nat Protoc. 2011;6:468–481. doi: 10.1038/nprot.2010.190. [DOI] [PubMed] [Google Scholar]
- 21.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zimmerman SG, Thorpe LM, Medrano VR, Mallozzi CA, McCartney BM. Apical constriction and invagination downstream of the canonical Wnt signaling pathway requires Rho1 and Myosin II. Dev Biol. 2010;340:54–66. doi: 10.1016/j.ydbio.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pabst O, Zweigerdt R, Arnold HH. Targeted disruption of the homeobox transcription factor Nkx2-3 in mice results in postnatal lethality and abnormal development of small intestine and spleen. Dev Camb Engl. 1999;126:2215–2225. doi: 10.1242/dev.126.10.2215. [DOI] [PubMed] [Google Scholar]
- 24.VanDussen KL, Carulli AJ, Keeley TM, Patel SR, Puthoff BJ, Magness ST, et al. Notch signaling modulates proliferation and differentiation of intestinal crypt base columnar stem cells. Dev Camb Engl. 2012;139:488–497. doi: 10.1242/dev.070763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hill ME, Asa SL, Drucker DJ. Essential requirement for Pax6 in control of enteroendocrine proglucagon gene transcription. Mol Endocrinol Baltim Md. 1999;13:1474–1486. doi: 10.1210/mend.13.9.0340. [DOI] [PubMed] [Google Scholar]
- 26.Ormestad M, Astorga J, Landgren H, Wang T, Johansson BR, Miura N, et al. Foxf1 and Foxf2 control murine gut development by limiting mesenchymal Wnt signaling and promoting extracellular matrix production. Dev Camb Engl. 2006;133:833–843. doi: 10.1242/dev.02252. [DOI] [PubMed] [Google Scholar]
- 27.Karlsson L, Lindahl P, Heath JK, Betsholtz C. Abnormal gastrointestinal development in PDGF-A and PDGFR-(alpha) deficient mice implicates a novel mesenchymal structure with putative instructive properties in villus morphogenesis. Dev Camb Engl. 2000;127:3457–3466. doi: 10.1242/dev.127.16.3457. [DOI] [PubMed] [Google Scholar]
- 28.Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, et al. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Enroth S, Rada-Iglesisas A, Andersson R, Wallerman O, Wanders A, Påhlman L, et al. Cancer associated epigenetic transitions identified by genome-wide histone methylation binding profiles in human colorectal cancer samples and paired normal mucosa. BMC Cancer. 2011;11:450. doi: 10.1186/1471-2407-11-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bhatlekar S, Fields JZ, Boman BM. HOX genes and their role in the development of human cancers. J Mol Med Berl Ger. 2014;92:811–823. doi: 10.1007/s00109-014-1181-y. [DOI] [PubMed] [Google Scholar]
- 32.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fluge Ø, Gravdal K, Carlsen E, Vonen B, Kjellevold K, Refsum S, et al. Expression of EZH2 and Ki-67 in colorectal cancer and associations with treatment response and prognosis. Br J Cancer. 2009;101:1282–1289. doi: 10.1038/sj.bjc.6605333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sarabi MM, Naghibalhossaini F. Association of DNA methyltransferases expression with global and gene-specific DNA methylation in colorectal cancer cells. Cell Biochem Funct. 2015;33:427–433. doi: 10.1002/cbf.3126. [DOI] [PubMed] [Google Scholar]
- 35.Beggs AD, Jones A, El-Bahrawy M, El-Bahwary M, Abulafi M, Hodgson SV, et al. Whole-genome methylation analysis of benign and malignant colorectal tumours. J Pathol. 2013;229:697–704. doi: 10.1002/path.4132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bariol C, Suter C, Cheong K, Ku S-L, Meagher A, Hawkins N, et al. The relationship between hypomethylation and CpG island methylation in colorectal neoplasia. Am J Pathol. 2003;162:1361–1371. doi: 10.1016/S0002-9440(10)63932-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rosenberg DW, Yang S, Pleau DC, Greenspan EJ, Stevens RG, Rajan TV, et al. Mutations in BRAF and KRAS differentially distinguish serrated versus non-serrated hyperplastic aberrant crypt foci in humans. Cancer Res. 2007;67:3551–3554. doi: 10.1158/0008-5472.CAN-07-0343. [DOI] [PubMed] [Google Scholar]
- 38.Benoit YD, Lepage MB, Khalfaoui T, Tremblay E, Basora N, Carrier JC, et al. Polycomb repressive complex 2 impedes intestinal cell terminal differentiation. J Cell Sci. 2012;125:3454–3463. doi: 10.1242/jcs.102061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rauch T, Wang Z, Zhang X, Zhong X, Wu X, Lau SK, et al. Homeobox gene methylation in lung cancer studied by genome-wide analysis with a microarray-based methylated CpG island recovery assay. Proc Natl Acad Sci U S A. 2007;104:5527–5532. doi: 10.1073/pnas.0701059104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Widschwendter M, Fiegl H, Egle D, Mueller-Holzner E, Spizzo G, Marth C, et al. Epigenetic stem cell signature in cancer. Nat Genet. 2007;39:157–158. doi: 10.1038/ng1941. [DOI] [PubMed] [Google Scholar]
- 41.Ohm JE, McGarvey KM, Yu X, Cheng L, Schuebel KE, Cope L, et al. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat Genet. 2007;39:237–242. doi: 10.1038/ng1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schlesinger Y, Straussman R, Keshet I, Farkash S, Hecht M, Zimmerman J, et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat Genet. 2007;39:232–236. doi: 10.1038/ng1950. [DOI] [PubMed] [Google Scholar]
- 43.Rada-Iglesias A, Enroth S, Andersson R, Wanders A, Påhlman L, Komorowski J, et al. Histone H3 lysine 27 trimethylation in adult differentiated colon associated to cancer DNA hypermethylation. Epigenetics. 2009;4:107–113. doi: 10.4161/epi.4.2.8038. [DOI] [PubMed] [Google Scholar]
- 44.Hahn MA, Li AX, Wu X, Yang R, Drew DA, Rosenberg DW, et al. Loss of the polycomb mark from bivalent promoters leads to activation of cancer-promoting genes in colorectal tumors. Cancer Res. 2014;74:3617–3629. doi: 10.1158/0008-5472.CAN-13-3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Noordermeer D, Leleu M, Splinter E, Rougemont J, De Laat W, Duboule D. The dynamic architecture of Hox gene clusters. Science. 2011;334:222–225. doi: 10.1126/science.1207194. [DOI] [PubMed] [Google Scholar]
- 46.Haberland M, Mokalled MH, Montgomery RL, Olson EN. Epigenetic control of skull morphogenesis by histone deacetylase 8. Genes Dev. 2009;23:1625–1630. doi: 10.1101/gad.1809209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Freschi G, Taddei A, Bechi P, Faiella A, Gulisano M, Cillo C, et al. Expression of HOX homeobox genes in the adult human colonic mucosa (and colorectal cancer?) Int J Mol Med. 2005;16:581–587. [PubMed] [Google Scholar]
- 48.Ahlquist T, Lind GE, Costa VL, Meling GI, Vatn M, Hoff GS, et al. Gene methylation profiles of normal mucosa, and benign and malignant colorectal tumors identify early onset markers. Mol Cancer. 2008;7:94. doi: 10.1186/1476-4598-7-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tommasi S, Karm DL, Wu X, Yen Y, Pfeifer GP. Methylation of homeobox genes is a frequent and early epigenetic event in breast cancer. Breast Cancer Res BCR. 2009;11:R14. doi: 10.1186/bcr2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shaulian E, Karin M. AP-1 as a regulator of cell life and death. Nat Cell Biol. 2002;4:E131–136. doi: 10.1038/ncb0502-e131. [DOI] [PubMed] [Google Scholar]
- 51.Ashida R, Tominaga K, Sasaki E, Watanabe T, Fujiwara Y, Oshitani N, et al. AP-1 and colorectal cancer. Inflammopharmacology. 2005;13:113–125. doi: 10.1163/156856005774423935. [DOI] [PubMed] [Google Scholar]
- 52.Kong HK, Yoon S, Park JH. The regulatory mechanism of the LY6K gene expression in human breast cancer cells. J Biol Chem. 2012;287:38889–38900. doi: 10.1074/jbc.M112.394270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Berman BP, Weisenberger DJ, Aman JF, Hinoue T, Ramjan Z, Liu Y, et al. Regions of focal DNA hypermethylation and long-range hypomethylation in colorectal cancer coincide with nuclear lamina-associated domains. Nat Genet. 2012;44:40–46. doi: 10.1038/ng.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Drew DA, Devers TJ, O’Brien MJ, Horelik NA, Levine J, Rosenberg DW. HD chromoendoscopy coupled with DNA mass spectrometry profiling identifies somatic mutations in microdissected human proximal aberrant crypt foci. Mol Cancer Res MCR. 2014;12:823–829. doi: 10.1158/1541-7786.MCR-13-0624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hahn MA, Li AX, Wu X, Pfeifer GP. Single base resolution analysis of 5-methylcytosine and 5-hydroxymethylcytosine by RRBS and TAB-RRBS. Methods Mol Biol Clifton NJ. 2015;1238:273–287. doi: 10.1007/978-1-4939-1804-1_14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xiong Z, Laird PW. COBRA: a sensitive and quantitative DNA methylation assay. Nucleic Acids Res. 1997;25:2532–2534. doi: 10.1093/nar/25.12.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Parallel sequencing runs of two independent library preparations from the same ACF sample show a high correlation between the number of successfully mapped reads per genomic region. (B) Correlation between multiple sequencing runs from the same library for 2 ACF and 2 samples of normal colonic mucosa. Correlation coefficients were calculated for the total number of successfully mapped reads for individual CpGs with a minimum of 10 mapped reads. Excluding Msp1 restriction sites (CCGG) increased between-run correlations. (C) Histogram depicting the results of bootstrapping control analysis for overlapping DMR analysis. On average, randomly generated genomic region sets contained 9.6 ± 3.1 overlapping DMRs, significantly fewer than the 608 observed real overlapping DMRs (P<0.001), indicating that the probability of identifying these overlapping DMRs by random chance is very low. (D) Density plot of motif scores for AP-1 family binding motifs generated via bootstrapping analysis. Asterisks indicate the actual enrichment score for each motif observed in intergenic ACF DMRs.
These DMRs tended to overlap with H3K4me peaks identified in normal colonic mucosa, suggesting that they occur within active enhancer regions. H3K4me data was obtained from the ENCODE project (Accession: GSM621670).
(A) Several DMRs exhibiting increased methylation relative to patient-matched normal were identified by RRBS within the body of RSPO2. (B) Hypermethylation of the DMR associated with Exon 2 of RSPO2 was validated in an independent set of human colorectal tumors using the COBRA assay. The appearance of a second band in the BstUI-digested lanes (+) indicates the presence of methylation at this site. (C) Hypermethylation of this same DMR was also validated in an independent set of ACF samples using COBRA.
(A) Distribution of the genomic location of DMRs with altered methylation identified in both ACF and CRC. Overlapping hypermethylated DMRs were evenly distributed amongst promoters (35%), gene bodies (37%) and intergenic regions (28%). Overlapping DMRs exhibiting hypomethylation were rarely located within promoters (3%), but were common in gene bodies (44%) and intergenic regions (53%). (B) Distribution of the genomic location of all DMRs (increased and decreased methylation) in ACF and CRC. While the genomic distribution of DMRs identified in ACF was very similar to the distribution of all sequenced regions, DMRs identified in cancer samples exhibited a bias towards non-promoter localization.
Expression of PRC2 components and DNA methyltransferases in cancer and ACF samples. (A) Significant overexpression of the PRC2 components EZH2 (1.55-fold, P<0.05), SUZ12 (1.12-fold, P<0.05) and EED (1.1-fold, P<0.05) were detected in cancer samples. Furthermore, overexpression of the DNA methyltransferase enzymes DNMT1 (1.2-fold, P<0,05) and DNMT3B (1.45-fold, P<0.05) were detected in cancers. (B) Despite similarities in the patterns of aberrant methylation in cancer and ACF, including the enrichment of promoter DMRs for PRC2-target genes in ACF, no change in the expression of any PRC2 component or DNMT enzyme was detected in ACF samples.