Abstract
Microfluidic partitioning of long genomic DNA fragments, and barcoding of shorter fragments derived from them, retains long-range information in short sequencing reads. Such read cloud approaches represent a powerful and cost-effective alternative to single-molecule long-read sequencing. We developed GROC-SVs, which uses read clouds for structural variant detection and assembly, and apply it to Illumina-sequenced 10× Genomics sarcoma and breast cancer data sets. Validation demonstrates substantial improvement in specificity of breakpoint detection compared to short-fragment sequencing, at comparable sensitivity, and vice versa. The long-range information also facilitates sequence assembly of breakpoints; importantly, consecutive breakpoints closer than the average length of the input DNA molecules can be assembled, with some events exhibiting remarkable complexity. We show that chromothriptic rearrangements occurred before copy number amplifications and that single-nucleotide and structural variants are not correlated. We predict significant advances in structural variant science using 10×/GROC-SVs and other read cloud-specific methods.
Introduction
Structural variants (SVs) represent the highly heterogeneous class of large-scale changes in the genome, including DNA deletions, duplications, inversions and translocations. Because each event affects a large genomic region, SVs are responsible for the majority of nucleotides varying between individuals1 and in many cancer genomes2,3.
Despite its importance in evolution and disease, structural variation remains difficult to comprehensively characterize. The number of SVs possible is huge because DNA breakage and subsequent fusions can connect any genomic locus to any other. Repetitive loci, uneven or biased sequencing coverage, and the typically short length of sequenced fragments complicate accurate detection. In addition, previous work has illuminated the potential complexity of SVs4–8. One example of large-scale complexity is chromothripsis3, in which a chromosome shatters into many pieces that are then apparently randomly reassembled, leading to massive rearrangements.
These and other types of complex events can be difficult to interpret using existing sequencing technologies. For example, analyses of short-fragment sequence data can only confidently relate breakpoints that are within the fragment size distribution, typically <500 bp. Longer-distance reconstruction (e.g. ref 2) requires the assumptions that downstream events occur in the same haplotype and that all breakpoints have been accurately identified. Single-molecule long-read approaches are better suited for detection of SVs, but throughput and cost are typically limiting, and the high per-base error rate is a drawback.
Read clouds marry the advantages of standard Illumina sequencing (high throughput and accuracy) with long-fragment information added through a barcode tag incorporated during a molecular partitioning step9–12. The recently released 10× Genomics platform produces read cloud libraries with dramatically higher numbers of partitions compared to previous methods, enabling new applications13. To prepare 10× Genomics libraries, long DNA fragments are diluted into 105 to 106 microfluidic droplets, each of which contains a unique barcode. Within each droplet, randomly primed amplification produces many identically barcoded short fragments templated off the handful of long fragments. When these barcoded short fragments are Illumina-sequenced, their alignments to the reference genome form clusters. We refer to the clusters of identically barcoded, linked reads as clouds. Each cloud allows us to infer the extent of an originating large DNA molecule even though the sparse sampling by short reads means we only directly observe a subset of positions within each long fragment.
The long-range information in read clouds can in principle be leveraged to identify, sequence-assemble, and reconstruct complex SVs. Using a novel method that we developed for this purpose, Genome-wide Reconstruction of Complex Structural Variants (GROC-SVs), we show that 10× data substantially improves detection of SVs compared to standard short-fragment sequencing and that it enables the reconstruction of large-scale complex SVs. In addition, we use the read cloud information to produce high-quality assemblies of the sequences spanning the breakpoints, enabling us to better interpret local complexity. We applied GROC-SVs to characterize chromothripsis and subsequent evolution of structural variation in a liposarcoma and to analyze SVs in a breast cancer cell line.
Results
Sequence Data Generation and Characteristics
We Illumina-sequenced 10× GemCode libraries from each of 7 spatially distinct sites within a well-differentiated liposarcoma, as well as a control sample from the same patient. For purposes of comparison and validation, we also sequenced PCR-free Illumina and long-insert (~7kb) mate-pair libraries.
Size-selection of sarcoma DNA resulted in a tight 10× fragment size distribution (mean > 30kb; 95th percentile=~80kb; Supplementary Figure 1 and Supplementary Table 1). Each genomic position was covered on average by ~250 long fragments, but this was accomplished at an overall sequence coverage of ~25× through sparse sampling of each long fragment by short reads. We also analyzed 2nd generation 10× Chromium data from the HCC1143 breast cancer and matched-normal cell lines. Prepared without size-selection, HCC1143 fragment sizes covered a wide distribution (mean=41kb, 95th percentile=148kb).
Overview of GROC-SVs
GROC-SVs first looks for statistical evidence for long fragments that span breakpoints by quantification of barcode similarity between all pairs of genomic locations (Supplementary Figure 2a). Levels of barcode similarity are highest between any two nearby loci since input long fragments tend to overlap both loci (Supplementary Figure 2a, diagonal). Loci separated by distances larger than the input fragment size share zero or only a small number of barcodes. (This is because each barcoded partition contains only a small number of fragments randomly drawn from the genome, and thus the chance that multiple partitions contain long fragments from the same two distant loci is small; Supplementary Figure 2a, background). Thus, the presence of multiple barcodes that are shared between two distant locations at a level higher than that background is indicative of a breakpoint where the two locations are joined (Supplementary Figure 2a, translocation). Subsequent to breakpoint identification and refinement we perform sequence assembly of the linked reads from the relevant breakpoints. This includes the reconstruction of complex events on the basis of breakpoints that are connected by long fragments (Supplementary Figure 2b).
Structural variant discovery with GROC-SVs: breakpoint detection
Barcode similarity is highest near a breakpoint, and drops off at distances proportional to the fragment size distribution (Figure 1; see Supplementary Figure 3 and Supplementary Note 1 for more detailed explanations). In the matched normal samples, the same region exhibits only low background similarity, indicative of a somatic SV (Figure 1b). Background levels are even lower when using larger numbers of molecular partitions (barcodes), and fragment lengths can be seen to extend further from each breakpoint when using higher molecular weight DNA (Supplementary Figure 4).
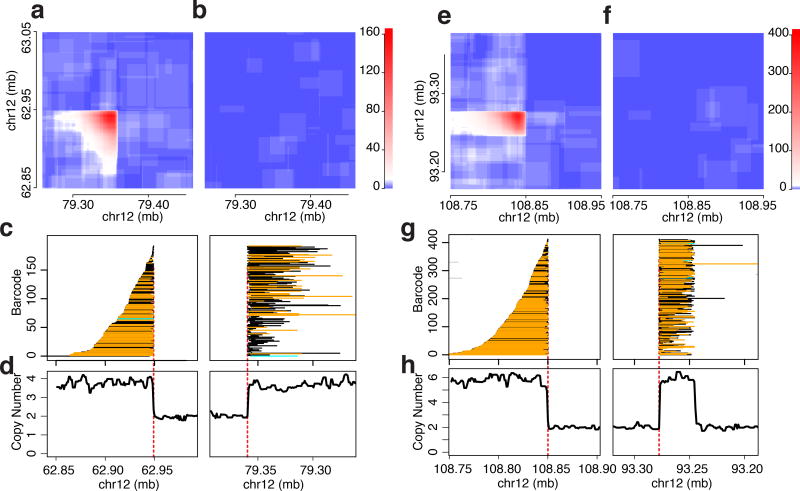
Figure 1.
Examples of breakpoint signals in 10× data. (a – d), a simple breakpoint in sarcoma sample 0, GemCode data. (e – h), two breakpoints in close proximity in sarcoma sample 0. (a, e), barcode similarity histograms in tumor. For each pair of genomic locations, the number of shared barcodes is color-coded according to the scales to the right, with the greatest signal forming a corner shape whose point is at the breakpoint coordinates. (b, f), same locations in the control samples. (c, g), inferred extent of breakpoint-supporting read clouds (corresponding to input fragments). Each row is one cloud, colored according to its assignment to a haplotype: supporting haplotype, orange; unassigned, black; cyan, non-supporting haplotype; and non-supporting cloud in the same barcode as a supporting cloud, grey (derived from independent fragments in the same molecular partition). As barcodes are ordered identically in the left and right panel for each event, the long fragments can be seen to tile across the breakpoint when ordered by their left-most position in the left panel. (d, h), copy number profiles based on the short fragment data in the sarcoma. Decreasing coordinates indicate depiction of minus strand.
All supporting read clouds end near the putative breakpoint location (Figure 1c), a signal that is used during breakpoint refinement. In size-selected samples (as in the sarcoma), the clouds, ordered by their position relative to the first side of the breakpoint, tile across the breakpoint such that those starting furthest from the breakpoint tend to extend the least into the second region, while those starting closest to the first side extend the furthest into the second. Short-fragment sequencing coverage profiles support changes in copy number at many structural variant breakpoints (Figure 1d).
A second example from the sarcoma illustrates the nature of the barcode similarity when two breakpoints are in close proximity. It shows the expected sudden dropoff in signal at the 108.85 mb breakpoint, but along the Y axis the signal ends abruptly not only at 93.27 Mb but also, in the other direction, at 93.25 Mb (Figure 1e; the control only exhibits background, Figure 1f). When tiling the read clouds, it becomes apparent that there are two breakpoints present at the 93 mb locus (Figure 1g), with copy number profiles exhibiting consistent levels that change abruptly at the breakpoint locations (Figure 1h). A substantial number of fragments appear to span from the first to the second breakpoint, suggesting that it is possible to use the 10× long-fragment information to directly link breakpoints that are in proximity to one another (see below).
Structural variant discovery with GROC-SVs: Sequence assembly of breakpoints and reconstruction of complex events
To better characterize breakpoints, GROC-SVs attempts to perform sequence assembly using the long-fragment information present in the barcoded reads. First, we identify barcodes that are shared among multiple breakpoints, suggesting some long fragments spanned across them; breakpoints that do not share barcodes are retained as singletons. For each such event or collection of events, we identify barcodes supporting each breakpoint and gather all reads marked by those barcodes (Supplementary Figure 2), including those that were unmappable or had low mapping quality in the initial genome-wide mapping. We then perform sequence assembly on these reads and align the resulting contigs to the reference genome to identify the precise breakpoint locations.
In many cases, we are able to directly use the sequence assemblies to reconstruct the order of multiple genomic segments within complex SVs. However, even when the sequence assemblies are incomplete, we can reconstruct complex events using the fact that adjacent genomic segments within a complex event will share more barcodes than distant segments within the same event.
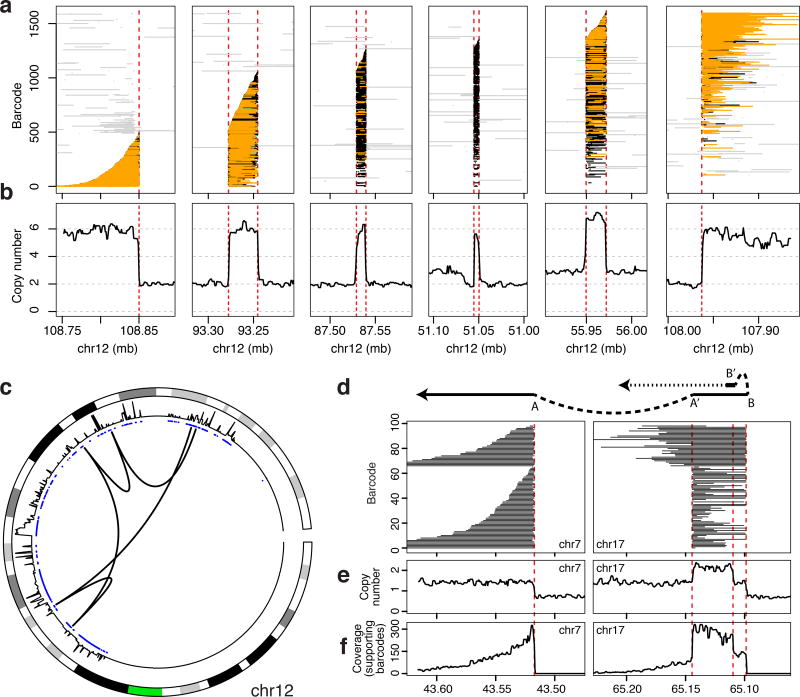
Using this approach, we identified 12 events with 4 or more breakpoints, and 60 events with 2 or 3 breakpoints in the sarcoma. As a fraction of all somatic breakpoints, 204/503 (41%) were assigned to complex events made up of at least 2 breakpoints. The ordering and assembly of 5 breakpoints comprising a sample complex event that spans 75 kb (Figure 2a–c and Supplementary Note 1) illustrates how the clouds tile and thereby connect neighboring breakpoints. Copy number profiles are consistent with the reconstruction (Figure 2b). Strikingly, the variant connects sequence from all over the long arm of chromosome 12 (Figure 2c).
Figure 2.
Reconstruction of complex events. (a – c) Read clouds that support a complex event in the sarcoma. Clouds, colored as in Figure 1, tile across 5 consecutive breakpoints (a) with consistent copy number profiles (b). Circos plot with arcs depicting breakpoint connections illustrates that the event connects distant segments from the long arm of chromosome 12 (c). From outside to inside, chromosome ideogram (green indicates the location of the centromere), then copy number profiles, then copy number aberration calls (blue for amplifications, red for deletions) are shown. (d – f) A complex event in cell line HCC1143 (d) and its corresponding sequence read coverage (e, f).
In the non-chromothriptic breast cancer cell line, we reconstructed 11 complex somatic events with a total of 24 breakpoints, including a large inverted repeat that illustrates both the potential complexity of structural variation and the power of read clouds to resolve it (Figure 2d–f).
Genome-wide SV discovery, comparison and validation
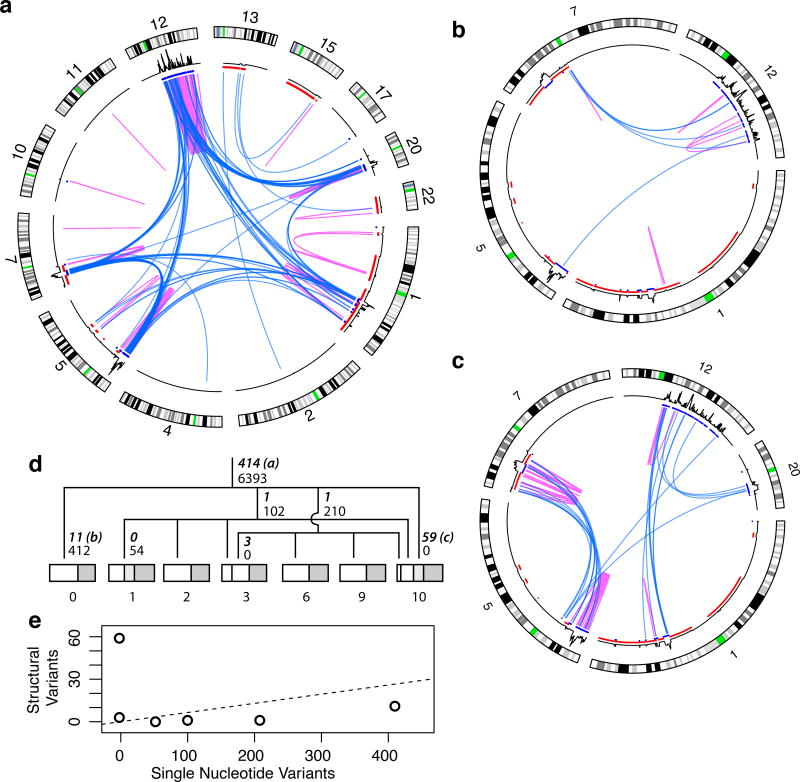
The sarcoma genome harbored substantial structural variation, represented by a total of 503 called somatic breakpoints (Figure 3a and Supplementary Figure 3a). The highest density of events occurred on the long arm of chromosome 12, involving 174 breakpoints (Figure 3a).
Figure 3.
Somatic genome evolution of the sarcoma. (a), Circos plots of the 414 ancestral (trunkal) events (b), events private to sample 0 and (c), events private to sample 10. Blue, interchromosomal events; magenta, intrachromosomal. Otherwise, as in 2c. (d) Lineage tree of the samples reconstructed from high-confidence somatic SNVs. Number of SNVs supporting each branch are in small font, number of breakpoints are in bold italic with circos plot panel letters indicated for plots a–c. Samples are subdivided proportionally to somatic allele frequencies to indicate subclone size. Portion corresponding to normal contribution (e.g., infiltrating lymphocytes) is in dark grey. (e) Number of SVs vs SNVs for each branch in lineage tree. The ancestral branch is shown as a dashed line indicating the rate at which SVs would accumulate relative to SNVs under a constant rate model.
One expectation regarding the detection of SVs using 10× data is that its high physical coverage improves the signal-to-noise ratio compared to standard short-read SV detection approaches. The number of SV-supporting 10× fragments correlated highly with the number of supporting mate-pairs (rho=0.89; Supplementary Fig. 6) and fairly well with short fragments (rho=0.71; Supplementary Fig. 6b). Strikingly, there was a median 3.2 times as many 10× barcodes as short fragments supporting an event. The overall rate of validation of our breakpoints by mate-pairs was 94.6% (424/448), and this increased to 98.6% (351/356) when examining only successfully assembled SVs. Sensitivity and specificity were lower for events substantially smaller than the average fragment lengths (Supplementary Note 2). To compare the read cloud approach to previous methods, we applied commonly used tools to our standard Illumina libraries to identify large-scale SVs14. We found that only 65.1% (375/576) of the short fragment-called somatic events were validated by mate-pair data (Supplementary Fig. 7).
Genome evolution within the sarcoma
The 414 breakpoints present in all sarcoma samples but not in the control arose before the last common ancestor of the samples' cells. These shared, ancestral events include the chromothripsis on chromosome 12, with the vast majority of the other events involving chromosomes 1, 5, 7 and 20. In addition, we found an ancestral rearrangement followed by high-level amplification harboring the characteristic liposarcoma driver gene, MDM2 (ref15).
We also identified 89 SVs that were present in certain subsets of the samples (but not in the control). The majority of these involved chromosomes 5, 7, and 12, and were private to one of the samples, marking subclone expansions that did not extend to the other samples: 59 in sample 10, 11 in sample 0 and 3 in sample 3. The remaining 19 SVs did not robustly determine subclones on their own.
The non-ancestral SVs and the inferred presence of subclones suggests that there was some evolutionary differentiation within the sarcoma that was captured by our sampling. We therefore set out to determine the evolutionary relationships amongst the samples and then analyze the dynamics of SV accumulation, based on the inferred phylogenetic tree. Because they are more common than SVs, we used somatic SNVs to build the samples' evolutionary tree16. In agreement with the SVs and copy number profiles, the majority (6393/7171) of high-confidence somatic SNVs were ancestral, originating before the last common ancestor of the samples' cells. We identified an additional four subclones based on the presence of SNVs shared between subsets of samples.
Analysis of the SVs on the basis of the tree suggests that SVs do not accumulate proportionally to the number of cell divisions and that they instead tend to occur in bursts, clustering in evolutionary time. If SVs accumulated gradually through evolutionary time (ie, with each cell division), we would expect a constant ratio between the number of SVs and SNVs present in a subclone. Instead, we see very low numbers of SVs compared to SNVs for most of the lower branches on the evolutionary tree, with the subclone that is private to sample 10 being a major outlier (Figure 3e). This utter lack of agreement between SNV and SV rates suggests that SV accumulation is episodic, similar to what has been observed for copy number variation in breast cancer17.
Further evidence for the episodic nature of SV accumulation is found in the differential localization of the breakpoints depending on exactly when they occurred during the evolution of the sarcoma. The 414 trunkal events are highly enriched for involvement of chromosome 12, mostly intrachromosomally, with some involvement of chromosomes 1, 5, 7 and 20 (Figure 3a). The private events in sample 1 mostly fell near regions of chromosomes 7 and 12 that harbor trunkal structural variation (Figure 3b). Strikingly, a large majority (43/59, 73%) of breakpoints present in the subclone private to sample 10 occurred within or between chromosomes 5 and 7 (Figure 3c). In contrast, only 30% of ancestral mutations occurred within or between those chromosomes. This enrichment was highly significant (p < 10−9, Fisher exact test), supporting the occurrence of a sudden series of events affecting a small portion of the tumor genome. These structural events thus likely occurred in a short enough time span that SNVs could not accumulate to substantial enough levels to directly observe the subclone.
Discussion
Based on the high rate of validation of breakpoints with mate-pair data, it is apparent that 10×/GROC-SVs provides a substantial improvement in the detection of large-scale structural variation when compared to standard analysis methods applied to short-fragment sequencing data. We note that because mate-pair libraries span a limited range of fragment sizes, they are not well-suited to reconstructing complex structural variants. We expect that other methods leveraging read cloud data for breakpoint detection will also show improved accuracy due to the substantially improved physical coverage and long distance information.
Breakpoint detection is only the first step in the characterization of SVs. We designed GROC-SVs to take full advantage of 10× data to perform simultaneous analysis of multiple breakpoints when it detects a complex SV, and it performs sequence assembly of SVs. Sequence assembly serves as both validation (as incorrect breakpoint calls would not lead to a consistent assembly), and base-pair level reconstruction. GROC-SVs thus differs substantially from the current version of 10× Genomics’ LongRanger package, which performs only the SV detection step. In addition, we note that GROC-SVs also supports multi-sample analysis, enabling accurate calling of somatic SVs when paired tumor and normal samples are available.
Because of the importance of large-scale rearrangements in tumor genomes, and the poor performance of short-fragment detection methods on these types of variants, we focused our efforts here (Supplementary Note 2). To-date, genome-scale reconstruction of complex SVs has been limited to cases where the breakpoints are spaced no longer than the fragment insert size (typically ~500bp), or has involved indirect inference that events are related, based on their proximity and orientation in the reference genome2,18. Using the 10× data, we were able to directly reconstruct the order of large scale genomic rearrangements involving many breakpoints without the need for any assumptions about pairs of breakpoints. In the sarcoma genome, where chromothripsis produced dramatic genomic change, we found that 40% of our breakpoints fell within complex SVs, with adjacent breakpoints frequently separated by tens of kb.
Most SVs in the sarcoma were shared across all 7 spatially distinct locations, and therefore must have occurred early in the evolution of the tumor. These ancestral events include the 174 chromosome-12 chromothripsis breakpoints and subsequent copy number amplifications as well an additional 240 breakpoints. In contrast, while 778 subclonal SNVs were detected, corresponding to 4 distinct subclone lineages, very few SVs other than the ancestral ones were shared across subclones. Thus, the sarcoma must have undergone an initial period of substantial structural instability, accumulating hundreds of rearrangements and copy number changes, before converging to a stable genomic configuration. Our results are concordant with a model recently proposed for liposarcomas, in which chromothripsis is followed by breakage-fusion-bridge cycles and subsequent chromosome linearization19. In addition to the ancestral SVs, we found a subclone private to sample 10 with 59 breakpoints that likely occurred in an additional, recent burst of genome instability.
In summary, using GROC-SVs, which we specifically developed for leveraging read cloud information, we show that 10× data allows for direct, data-driven reconstruction of complex structural variation. This is accomplished at high sensitivity and excellent specificity compared to short-fragment data, and at much lower laboratory effort and sample requirements than specialized libraries or mate pair approaches. Two distinct substrates, a chromothriptic sarcoma and a less highly rearranged breast cancer cell line, demonstrate wide applicability of the approach. Our evolutionary analysis of the sarcoma foreshadows substantial future advances in the related pursuits of reconstructing the full cancer genome and understanding each tumor's structural evolution.
Online Methods
Sample Preparation and Library Construction
Sections (0.5cm thick, 14cm diameter) of a well-differentiated liposarcoma tumor, obtained under informed consent from the Stanford Tissue Bank, were cut into multiple pieces, snap frozen with liquid nitrogen, and stored at −80°C. Genomic DNA was extracted from 7 spatially distinct sites of this sarcoma as well as from matched control kidney tissue of the same patient. We extracted genomic DNA from about 20 mg tissue using Gentra Puregene Tissue Kit (Qiagen, Cat 158667). Tissue was ground in liquid nitrogen, lysed in Cell Lysis Solution and Proteinase K, and digested with RNase A. Protein was pelleted and removed by adding Protein Precipitation Solution followed by centrifugation. Genomic DNA was precipitated with isopropanol and resuspended in buffer EB. Purified genomic DNA was aliquoted and stored at −20°C.
Genomic DNA was separated by running about 1µg DNA on a 1% low-melting-point agarose gel using Pulsed Field Gel Electrophoresis (PFGE). DNA of size 50–100 kb was then recovered by β-agarase I digestion and filter concentration (NEB, Cat M0392S). 1.2 ng of size-selected DNA was partitioned and barcoded using the 10× Genomics GemCode platform13. Libraries were then sequenced with a HiSeq2500 to ~25-fold sequence coverage.
Standard short-fragment Illumina libraries were prepared for all 7 sarcoma samples plus the matched normal control. Mate-pair libraries were prepared for sarcoma samples 0, 9 and 10 as well as the matched control.
For short-fragment DNA libraries, 1 µg of total genomic DNA was sheared to 350 bp. PCR-free libraries were then constructed using Illumina’s TruSeq DNA PCR-Free library preparation kit and sequenced with the Illumina HiSeqX system to ~35-fold sequence coverage.
For large-insert mate-pair libraries, 4 µg of total genomic DNA was fragmented with Tagment Enzyme and gel size-selected to build 7kb-insert mate-pair libraries using Illumina’s Nextera Mate Pair Sample Preparation Kit (FC-132-1001) (Tagmentation, Strand Displacement, Gel Size Selection, Circularization, Linear DNA Digestion, Circulated DNA Shearing, Streptavidin Bead Binding, End Repairing, A-Tailing, Adaptor Ligation, and PCR Amplification). Libraries were sequenced with HiSeq2500 to ~20-fold sequence coverage.
Breakpoint Detection
GROC-SVs is implemented as a multi-sample analysis pipeline, allowing the simultaneous analysis of multiple tumor and matched normal samples, or multiple related individuals.
10× Genomics sequencing libraries are first demultiplexed and droplet barcodes are called using the provided scripts, then reads are aligned to the reference genome using bwa mem20 or RFA12 (which has been implemented in the Long Ranger pipeline as the “Lariat” aligner). Barcodes are then ranked in decreasing order by the number of sequenced reads, and barcodes comprising 90% of all reads are retained while the remainder, which are enriched for experimental artifacts, are filtered. Next read clouds are identified as previously13. Briefly, reads with the same barcode are combined into a single barcode if the largest distance between any adjacent reads is less than a certain distance threshold and the reads on either end are of high map quality. This distance threshold was fixed as per ref13 at 60 kb for the data produced in this paper, but an appropriate threshold, typically ~20 kb for Chromium data, can be learned directly from the data in order to increase sensitivity for smaller events.
GROC-SVs begins SV detection by identifying all barcodes overlapping each 10 kb genomic window and then performing an all-by-all comparison. Some independent fragments with the same barcode can cause a low level of background similarity, typically <1 (Chromium) or 0–5 (GemCode) barcodes at any given pair of positions. A pair of loci (x,y) is considered a structural variant candidate if the number of shared barcodes exceeds that expected based on the total number of barcodes (proportional to copy number) at each locus. For computational efficiency, this initial test is performed as a binomial test (a more rigorous test is applied later for each structural variant).
Next, candidate SV loci are clustered, and candidate breakpoints are extracted based on peaks in the distribution of read cloud ends. This takes advantage of the fact that read clouds are expected to end suddenly near each of the breakpoints; performing this operation only on those barcodes that are shared between the two loci dramatically improves both the signal and reduces the background. Candidate breakpoints are identified in each sample separately.
At this point, the breakpoints have been identified typically to within several kb of the correct location. The next step is to perform refinement on the breakpoint coordinates to obtain approximately nucleotide-level accuracy. This step takes all read clouds within 20 kb of the candidate site and selects only those clouds with barcodes shared on both sides of the breakpoint. Then, for each breakend (the two half-open intervals that make up each breakpoint) separately, the maximum point of read cloud density is found, and then walked toward the putative breakpoint location until the read cloud density drops off suddenly to background levels, indicating the presence of the breakpoint location. We found that this procedure typically identifies the correct breakpoint location to within several nucleotides if the breakpoint is uniquely mappable with short reads. In the case that the breakpoint region is not uniquely mappable, the inferred breakpoint location will be the last well-mappable (mapq ≥ 30) position before the breakpoint. Breakpoint refinement occurs across samples together so all fragments spanning a breakpoint are used for refinement, even if the event is only present in a small subclone within a sample.
Copy numbers were not used in the detection of SVs and were only calculated to gain a better understanding of the context for SVs. Because the coverage profiles for the 1st generation 10× GemCode libraries showed substantial GC bias, we used standard PCR-free Illumina libraries to calculate copy number, normalized to the matched normal and normalized for DNA content within a sample. Coverage was typically higher for the tumor samples because of the many, large single-copy genomic regions.
Sequence assembly of breakpoints
Next, a permissive clustering step groups breakpoints together if they share a substantial proportion of their barcodes. This is formulated as a simple threshold using the Jaccard Index, defined as the number of barcodes shared between the loci divided by the total number of barcodes. This Jaccard Index can be viewed as a sort of “allele frequency,” where the numerator counts the number of fragments supporting the event, and the denominator counts the number of fragments in the reference and alternate alleles. This is however an approximation because it is difficult to confidently assign any individual fragment to one allele since both reference- and alternate-allele-supporting fragments can end near either breakpoint location. Theoretically, another confounder is the non-zero rate of “barcode collisions”, where one fragment occurs near breakpoint x and an independent fragment occurs near breakpoint y, both in the same barcode. However, barcode collisions typically contribute a negligible amount to the numerator since the average number of barcode collisions is very small for most genomic regions (< 1 for GemCode and <<1 for Chromium in normal copy number regions, and only appreciably higher for extreme copy number outliers).
Within each cluster, the barcodes supporting each event are pooled together, and all reads originating from these supporting barcodes are collected. Sequence assembly is then performed on the collected reads using idba_ud21. As each barcode marks multiple fragments, many of the reads do not derive from a breakpoint-supporting genomic region. However, because fragments are randomly assigned to a barcode, these non-supporting fragments should be distributed randomly throughout the genome. Thus, combined sequence coverage is highest near the breakpoints, which should be covered by every barcode, and low elsewhere. Therefore, most assembled contigs actually derive from the SV haplotype.
As with breakpoint refinement, sequence assembly is performed multi-sample, so spanning fragments can be used for assembly even if they occur in samples with very low allele frequency. idba_ud was selected because its good performance across a wide range of sequence coverage, which is highest near the breakpoints and then low farther away. Contigs are then aligned against the reference genome and breakpoint locations are called where appropriate. Note that this assembly process may discover additional breakpoints that were not significant in the genome-wide breakpoint detection step for various reasons.
Phasing germline haplotypes across SVs
In addition to providing high physical coverage of structural variant breakpoints, the long-fragment information in the 10× data allows for phasing of small variants with respect to the germline haplotypes13. Read clouds overlapping a heterozygous short variant can be assigned to one of the haplotypes. The low sequence coverage CR of each fragment means that some read clouds, especially shorter ones, will not cover a short variant informative for haplotype assignment. However, the high physical coverage CF results in a high total number of phased fragments for most genomic regions.
Because the structural variant breakpoints are distant from one another in the genome, the haplotypes are called independently for each side of the breakpoint, and so the standard phasing process does not uncover the phase arrangement for the tumor genome. However, nearly all informative fragments near each breakpoint support a single haplotype indicating that each side of the breakpoint only contributes a single haplotype to the event (Figure 1c,g). Thus we can use the predominant haplotype on either side of a breakpoint to locally phase the genomic regions that participate in the SV.
We identified 239 somatic breakpoints in the sarcoma with at least 20 phased read clouds supporting each side of the breakpoint. Of these events, the vast majority (229 or 96%) were supported by only a single haplotype combination, which is expected because the probability of the same exact SV occurring at the same position on both haplotypes is vanishingly small. In contrast, systematic errors resulting from, for example, genome repetitiveness, should affect all haplotypes equally. Therefore, the high percentage of events supported by only a single haplotype combination not only supports the validity of our phasing across breakpoints but also provides evidence that the breakpoint calls themselves do not result from substantial systematic biases.
Genome-wide reconstruction of complex events
Following sequence assembly, a more rigorous complex event reconstruction is performed. First, breakends sharing a substantial proportion of barcodes are again clustered together. The resulting clusters are represented as graphs with breakends represented as nodes, and connections between nearby (contiguous genomic segments) and distant (non-contiguous structural variants) breakends represented as edges. Because fragments may span many breakpoints at once, there may be barcode similarity between breakends that are separated by one or more breakpoints. Thus, for each breakend, we first select the assembly-supported breakpoint if one exists. The remaining breakpoints are selected based on the highest barcode support (nearby breakends should share more barcodes than distant ones). This process uses the high-quality information present in the sequence assemblies but can still perform complex event reconstruction even for breakpoints that cannot be sequence-assembled.
Post-processing
During post-processing, a more rigorous p-value is assigned to each breakpoint. This p-value is calculated by randomly sampling the correct number of barcodes for each breakend from the background distribution of fragments per barcode, then calculating the number of shared barcodes. Resampling is performed 100 times, then the significance of the observed vs resampled number of shared barcodes is calculated using a ranksum test. This resampling procedure takes into account the effect of differences in genome coverage as well as the non-uniform partitioning of fragments across barcodes.
Additional filters are applied, primarily for use when analyzing germline events to identify candidate confounding segmental duplications (segmental duplications should be present in both tumor and matched normal samples and are thus removed when analyzing somatic events). One filter of note compares the observed fragment lengths across breakpoints to those expected based on the background distribution. Structural variants should show long fragment support at 10s of kb away from each breakend. In contrast, segmental duplications and other repetitive genomic sequences often result in short supporting read clouds.
A final post-processing step assigns a present/absent call to each event for each sample. This genotype combines the resampling p-value calculated above as well as requiring a minimum allele frequency (again calculated using the Jaccard Index). Note that heterozygous and homozygous calls are not calculated because these are difficult to accurately define for the different types of structural variant and especially when copy numbers are variable. SV calls were considered to be somatic if there was no more than 1 supporting barcode in the control sample; results were nearly identical when using cutoffs of 0 or 2 barcodes instead.
Validation and comparison to short-fragment methods
Mate-pair validation was performed by counting the number of mate-pairs in the expected orientation and distance relative to the two breakends. We used only reads with a very conservative mapping quality filter of mapq ≥ 55. The rationale for this high mapq filter was that true events should typically have mates mapping several kb away from the breakend, escaping any local repetitiveness around a breakend. We analyzed the background distribution of random genomic regions, and found that the vast majority of regions shared zero mate-pairs, and thus we used a conservative cutoff of 50 mate-pairs to consider an event to be validated. We also tried a more lenient cutoff of 10 mate-pairs with similar results.
Identification of large-scale SVs from the standard short-fragment Illumina sequencing libraries was performed using LUMPY14. We also performed these analyses using delly22 but only the LUMPY SV calls are shown as these validate more consistently using mate-pair data. A threshold of <1 reads in the control sample supporting the event was used to filter out germline events and artifacts, with the remainder of the events therefore inferred to be somatic. This conservative threshold produced the most specific results possible, and a slightly less conservative threshold of <2 control reads did not substantially affect sensitivity while substantially decreasing specificity.
Evolutionary analysis
Evolutionary trees relating samples within the sarcoma were built as in16 and23. The alternate allele frequencies of the SNVs of the two phylogenetically informative classes are highly consistent with the allele frequencies of the ancestral SNVs. The frequencies of SNVs present in the mixed lineage samples (3 and 10) are consistent with one another, with their sums matching the ancestral frequencies. The mutation spectrum of the somatic SNVs (data not shown) closely matches that of germline events, suggesting that they were caused by replication errors without special mutational mechanisms, and that they accumulated at a rate proportional to the number of cell divisions. Finally, as expected, the most phylogenetically similar samples were in close spatial proximity to one another within the tumor (Supplementary Figure 5c). These lines of evidence support the idea that we were able to construct a robust evolutionary tree of our samples that could form the basis for interpreting the accumulation of SVs in this tumor (Figure 3d).
Data Availability and Accession Codes
GROC-SVs is open source and available at https://github.com/grocsvs/grocsvs. Raw sequencing data are available from dbGaP with accession code phs001255.v1.p1.
Supplementary Material
Acknowledgments
We thank K. Giorda, S. Kyriazopoulou-Panagiotopoulou, and M. Schnall-Levin for their assistance in preparing and analyzing the 10× data, and D. Ramazzotti for analyzing mutation spectra. This work was supported by the Stanford Center for Computational, Evolutionary and Human Genomics (NS), R01CA183904 (NIH/NCI; RBW, SB, AS), and the BRCA Foundation (AS). Certain commercial equipment, instruments, or materials are identified in this document. Such identification does not imply recommendation or endorsement by the National Institute of Standards and Technology, nor does it imply that the products identified are necessarily the best available for the purpose.
Footnotes
Author Contributions
NS, ZW, AB, RBW, JMZ, MS, SB and AS designed the experiments and/or analyses.
NS, ZW, JM and DC conducted the experiments. NS wrote analysis software. NS and AS analyzed the data. NS and AS wrote the manuscript with input from all authors.
Competing Financial Interests Statement
The authors declare no competing financial interests.
References
- 1.Weischenfeldt J, Symmons O, Spitz F, Korbel JO. Phenotypic impact of genomic structural variation: insights from and for human disease. Nat. Rev. Genet. 2013;14:125–138. doi: 10.1038/nrg3373. [DOI] [PubMed] [Google Scholar]
- 2.Yang L, et al. Diverse Mechanisms of Somatic Structural Variations in Human Cancer Genomes. Cell. 2013;153:919–929. doi: 10.1016/j.cell.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stephens PJ, et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144:27–40. doi: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baca SC, et al. Punctuated evolution of prostate cancer genomes. Cell. 2013;153:666–677. doi: 10.1016/j.cell.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chiang C, et al. Complex reorganization and predominant non-homologous repair following chromosomal breakage in karyotypically balanced germline rearrangements and transgenic integration. Nat. Genet. 2012;44:390–397. doi: 10.1038/ng.2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tupler R, et al. A complex chromosome rearrangement with 10 breakpoints: tentative assignment of the locus for Williams syndrome to 4q33----q35.1. J. Med. Genet. 1992;29:253–255. doi: 10.1136/jmg.29.4.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sudmant PH, et al. An integrated map of structural variation in 2,504 human genomes. Nature. 2015;526:75–81. doi: 10.1038/nature15394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Quinlan AR, Hall IM. Characterizing complex structural variation in germline and somatic genomes. Trends Genet. 2012;28:43–53. doi: 10.1016/j.tig.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Amini S, et al. Haplotype-resolved whole-genome sequencing by contiguity-preserving transposition and combinatorial indexing. Nat. Publ. Gr. 2014;46:1343–1349. doi: 10.1038/ng.3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peters BA, et al. Accurate whole-genome sequencing and haplotyping from 10 to 20 human cells. Nature. 2012;487:190–5. doi: 10.1038/nature11236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Voskoboynik A, et al. The genome sequence of the colonial chordate, Botryllus schlosseri. Elife. 2013;2013:2104–2105. doi: 10.7554/eLife.00569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bishara A, et al. Read clouds uncover variation in complex regions of the human genome. Genome Res. 2015;25:1570–1580. doi: 10.1101/gr.191189.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zheng GXY, et al. Haplotyping germline and cancer genomes with high-throughput linked-read sequencing. Nat. Biotechnol. 2016;34:303–11. doi: 10.1038/nbt.3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Layer RM, Chiang C, Quinlan AR, Hall IM. LUMPY: a probabilistic framework for structural variant discovery. Genome Biol. 2014;15:R84. doi: 10.1186/gb-2014-15-6-r84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oliner JD, Kinzler KW, Meltzer PS, George DL, Vogelstein B. Amplification of a gene encoding a p53-associated protein in human sarcomas. Nature. 1992;358:80–83. doi: 10.1038/358080a0. [DOI] [PubMed] [Google Scholar]
- 16.Newburger DE, et al. Genome evolution during progression to breast cancer. Genome Res. 2013;23:1097–1108. doi: 10.1101/gr.151670.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao R, et al. Punctuated copy number evolution and clonal stasis in triple-negative breast cancer. Nat. Genet. 2016;48:1119–1130. doi: 10.1038/ng.3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greenman CD, et al. Estimation of rearrangement phylogeny for cancer genomes. Genome Res. 2012;22:346–361. doi: 10.1101/gr.118414.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garsed DW, et al. The Architecture and Evolution of Cancer Neochromosomes. Cancer Cell. 2014;26:653–667. doi: 10.1016/j.ccell.2014.09.010. [DOI] [PubMed] [Google Scholar]
- 20.Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv Prepr. 2013 doi:arXiv:1303.3997 [q-bio.GN] [Google Scholar]
- 21.Peng Y, Leung HCM, Yiu SM, Chin FYL. IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics. 2012;28:1420–1428. doi: 10.1093/bioinformatics/bts174. [DOI] [PubMed] [Google Scholar]
- 22.Rausch T, et al. DELLY: Structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics. 2012;28 doi: 10.1093/bioinformatics/bts378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weng Z, et al. Cell-lineage heterogeneity and driver mutation recurrence in pre-invasive breast neoplasia. Genome Med. 2015;7:28. doi: 10.1186/s13073-015-0146-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
GROC-SVs is open source and available at https://github.com/grocsvs/grocsvs. Raw sequencing data are available from dbGaP with accession code phs001255.v1.p1.