Abstract
Room-temperature ionic liquids (RTIL) are a new class of organic salts whose melting temperature falls below the conventional limit of 100 °C. Their low vapor pressure, moreover, has made these ionic compounds the solvents of choice of the so-called green chemistry. For these and other peculiar characteristics, they are increasingly used in industrial applications. However, studies of their interaction with living organisms have highlighted mild to severe health hazards. Since their cytotoxicity shows a positive correlation with their lipophilicity, several chemical–physical studies of their interactions with biomembranes have been carried out in the last few years, aiming to identify the molecular mechanisms behind their toxicity. Cation chain length and anion nature of RTILs have seemed to affect lipophilicity and, in turn, their toxicity. However, the emerging picture raises new questions, points to the need to assess toxicity on a case-by-case basis, but also suggests a potential positive role of RTILs in pharmacology, bio-medicine and bio-nanotechnology. Here, we review this new subject of research, and comment on the future and the potential importance of this emerging field of study.
Keywords: Ionic liquids, Biomembranes, Phospholipid bilayers, Toxicity, Biomedicine, Nanotechnology
Introduction
Room-temperature ionic liquids (RTIL) are a vast class of ionic systems, usually consisting of an organic cation and either an organic or inorganic anion (Fig. 1), whose melting temperature falls below the conventional limit of 100 °C (Welton 1999). They have been intensively investigated for their potential applications as solvents, non-aqueous electrolytes, high-performance lubricants, and advanced engineering materials (Plechkova et al. 2009; Ranke et al. 2007b; Ghandi 2014). The widespread appeal of RTILs to some extent relies on their perceived low environmental impact, making these compounds one of the bases of the so-called green chemistry (Earle and Seddon 2000). Their introduction into industrial processes, together with their organic character, motivated the first studies of their interaction with biomolecules and bio-organisms (Petkovic et al. 2011). As a result, several studies have highlighted their toxicity on living organisms (Pretti et al. 2006; Bernot et al. 2005; Ranke et al. 2006, 2007a; Stolte et al. 2007; Kulacki and Lamberti 2008). Toxicity is also a measure of the high affinity between RTILs and biosystems. This affinity, together with the extremely tunable chemistry of RTILs, is the basis for the future of potential applications to pharmacology, bio-medicine, and bio-nanotechnology (Stoimenovski et al. 2012; Hough et al. 2007). It has been already shown, for example, that RTILs are able to:
-
(i)
Kill bacteria (O’Toole et al. 2012);
-
(ii)
Extract, purify, and even store DNA at ambient temperature (Clark et al. 2015, 2016);
-
(iii)
Stabilize proteins and enzymes (Kumar and Venkatesu 2014, Kumar et al. 2017);
-
(iv)
Assist or prevent protein amyloidogenesis, and in some cases even return amyloid fibers to functional proteins (Byrne et al. 2007, Byrne and Angell 2008, 2009);
-
(v)
Penetrate, create pores, and destroy biomembranes (Benedetto et al. 2014b, 2015, Bhattacharya et al. 2017, Evans 2008, Yoo et al. 2014, 2016a, b, Wang et al. 2015a, b, 2016; Drücker et al. 2017, Rühling et al. 2017); and
-
(vi)
Dissolve cellulose and other complex polysaccharides (Youngs et al. 2011, Zhang et al. 2017, Li et al. 2015, Cheng et al. 2011, Liu et al. 2010).
Fig. 1.
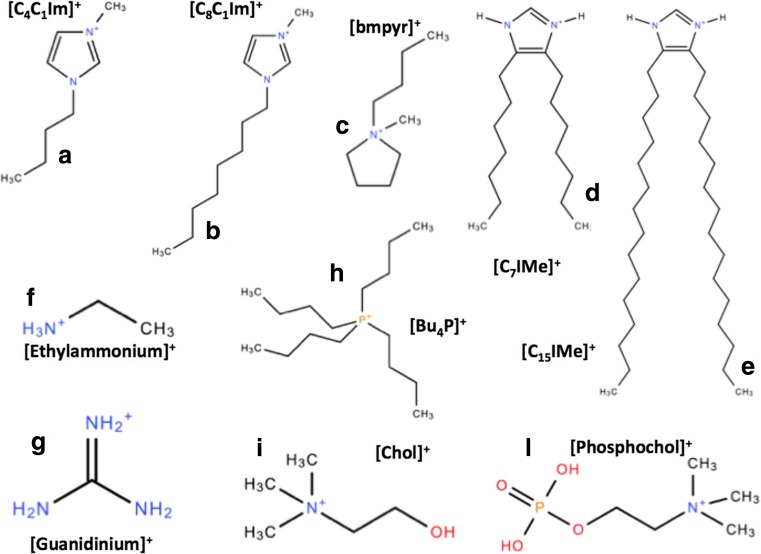
Chemical sketches of some selected RTILs cations: (a),(b),(c) the most common imidazolium and pyrridinium RTIL cations; (d),(e) the double-tail lipid-mimic imidazolium-based RTILs (Wang et al. 2015a); (f),(g) the ethylammonium and guanidinium RTIL cations that help and contrast protein amyloidogenesis, respectively (Byrne et al. 2007, Byrne and Angell 2008, 2009); (h) a phosphonium-based RTIL cation; and (i),(l) the choline and phosphocholine cations also used in RTILs made of amino acids (Benedetto et al. 2014a)
A general overview of the interaction between RTILs and several classes of biomolecules (e.g., proteins and peptides, mono- and polysaccharides, nucleic acids, and biomembranes) has been presented in two mini-reviews published in 2016 (Benedetto and Ballone 2016a, b). The main aim of these studies has been to link the biological effects of RTILs — usually detected by biochemical approaches — to the microscopic mechanisms of interaction between RTILs and biomolecules. Only a complete understanding of their microscopic mechanisms can provide the basis for the synthesizing of greener RTILs for industrial applications, and develop breakthrough applications in bio-nanotechnology. With this big-picture in mind, the interactions of RTILs with biomembranes is a relevant topic. Since the first encounter of any foreign chemical species with a living cell is likely to occur at its protective plasma membrane, this subject merges the two original aims of these investigations, i.e., assessing and reducing RTIL cytotoxicity, and developing bio-nanotechnologies. It has been shown or suggested that:
Controlling the poration of biomembranes by RTILs could result in the development of new drug delivery methods in pharmacology;
Their chemistry can be finely tuned in such a way that RTILs become lethal to bacteria at the same doses that are not harmful to eukaryotic cells, thus opening new antibacterial strategies (O’Toole et al. 2012); and finally,
Tuning their chemistry already enables RTILs to kill cancer cells and leave healthy cells almost unaffected (Wang et al. 2015a, b).
Moreover, the aqueous environment in which biomembranes reside is rich in a variety of inorganic ions that play a major role in promoting and regulating the biomembrane functions. The effects of simple ions such as Na+, K+, Cl−, etc. has been studied extensively using experimental and computational methods (Berkowitz and Vaćha 2012; Pabst et al. 2007; Böckmann et al. 2003) as well as empirical modeling (Aroti et al. 2007). It is natural, therefore, to turn the attention to RTILs, i.e., organic salts, whose complex structure and larger size provide many more ways to tune their interaction.
This review summarizes recent results from studies of the interactions between RTILs and biomembranes, with a special focus on their physico-chemical properties.
From complex biomembranes to phospholipids: their similarities with room-temperature ionic liquids
Biomembranes are complex and diverse biological supramolecular structures that separate the intracellular proteins, bio-complexes and bio-machineries, and nuclei from the extracellular environment. They play a role in the biochemistry of cells that is far more extensive than just representing a physical barrier. Biomembranes regulate the diffusion of chemical species into cells, either through specific (protein) channels or simply by absorption into their phospholipids. They also play a major role in cell replication processes. Biomembranes are the target of several antibiotics, since even small modifications of their structure, kinetics, and elastic properties can drastically affect the stability and viability of cells. Therefore, it is not surprising that biomembranes are one of the major and quite broad subjects of studies. Here, we review a small proportion of those studies that focused on chemical–physical investigations of phospholipid bilayers. Phospholipid bilayers are a well-accepted first-order model of biomembranes. They can be seen as the skeleton of any biomembrane into which proteins, extra lipids, saccharides, and, in general, biocomplexes can be absorbed to create more detailed and specific models of real biomembranes. As a result, assessing the effect of RTILs on phospholipid bilayers is the required first step along the path of the molecular-level comprehension of the biological effects of RTILs on biomembranes, and in cells. This modus operandi is corroborated by the fact that the cytotoxicity of RTILs measured by a variety of bioassays shows a clear positive correlation with their lipophilicity (Ranke et al. 2006, 2007a; Stolte et al. 2007). Moreover, phospholipid bilayers can be easily prepared in a controlled, and, thus reproducible, way in laboratories. In addition, their cost is affordable for even small research groups, making experimental investigations both possible and accurate.
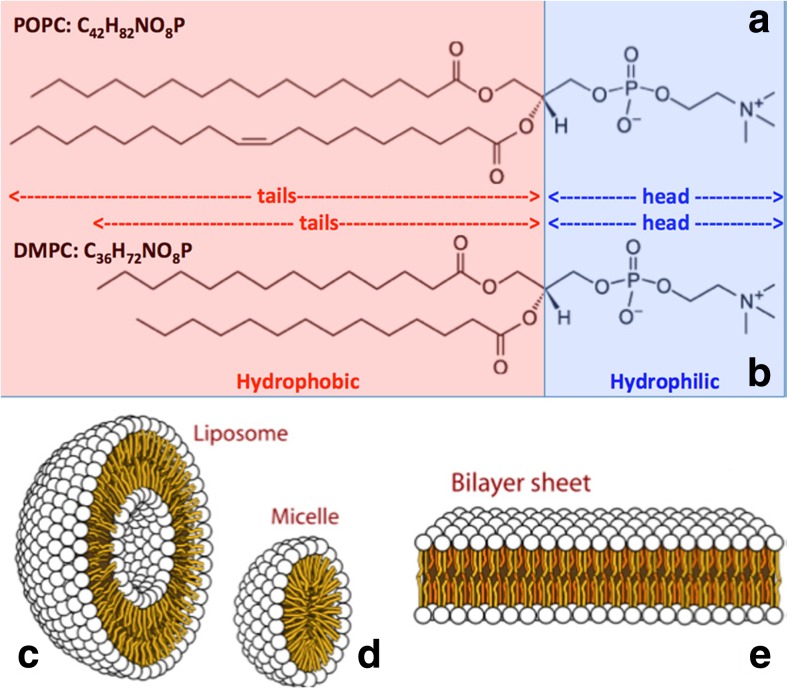
The basic units of any phospholipid bilayer are phospholipid molecules (Fig. 2a). They can be zwitterionic or ionic. They consist of hydrocarbon tails that are hydrophobic, and a hydrophilic head. As a result, in an aqueous environment, they arrange themselves into superstructures, minimizing the contact between their hydrophobic tails and water, and maximizing contact between their hydrophilic heads and water. Due to factors such as concentration of phospholipids in water, physico-chemical conditions, and geometry constraints, they can form uni- and multi-lamellar vesicles and micelles, and supported single-, bi- and multi-layers (Fig. 2b). Almost all of these geometries have been used when investigating the effect of RTILs on biomembranes. However, among all those systems, (i) supported bilayers, and (ii) vesicles are those that reproduce the double layer of biomembranes and, in turn, can be considered the best models to mimic them.
Fig. 2.
Two of the most common phospholipids: a 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) and b 1,2-Dimyristoyl-sn-glycero-3-phosphorylcholine (DMPC). They differentiate for the length of their (hydrophobic) hydrocarbon tails, whereas they share exactly the same (hydrophilic) head. When dispersed in aqueous environments, the hydrophobic-hydrophilic competition generates supramolecular structures such as uni-lamellar c liposomes, d micelles, and e bilayer sheets. Multi-lamellar structures can also be formed
A comparison of the phospholipids in Fig. 2a with the RTIL cations in Fig. 1 highlights their similarities. Both have an ionic/polar character, and both display hydrophilic and hydrophobic regions. The closest similarities are between phospholipids and the new RTILs synthetized by Galla and co-workers in Fig. 1d (Wang et al. 2015a). These have two hydrocarbon chains as phospholipids. A second intriguing overlap is with the ionic liquids of amino-acids (AAIL), in which anions are deprotonated amino-acids, and cations are either a protonated choline or phosphocholine that are present in phospholipid head groups (Benedetto et al. 2014a). In these two cases, but also in general, the similarities between phospholipids and RTILs are certainly responsible for their mutual affinity. Moreover, they suggest that a combination of: (i) electrostatic, (ii) dispersion interactions, (iii) hydrophobic and hydrophilic effects, and (iv) hydrogen bond structures and dynamics at the interface have to be taken into account. These describe the mechanisms of interaction that should result in a fine balance between all competing forces. It should then also be clear that even small changes in the chemistry of the molecules could affect the total balance of the forces, and, in turn, dramatically change the system properties. This circumstance highlights how a full comprehension of the microscopic mechanisms of RTIL–biomembrane interactions, together with the tunable character of the RTIL chemistry, is a key step for any major progress in this field. This observation contrasts with one of the main motivations underlying these investigations, namely to discover general rules for assessing the effects of RTILs on biomembranes. However, this “unfortunate” circumstance can be balanced by the almost immense playground we have in front of us to develop new breakthrough applications in bio-nanotechnology.
A snapshot of RTIL–biomembrane interactions: a joint neutron scattering and computational study
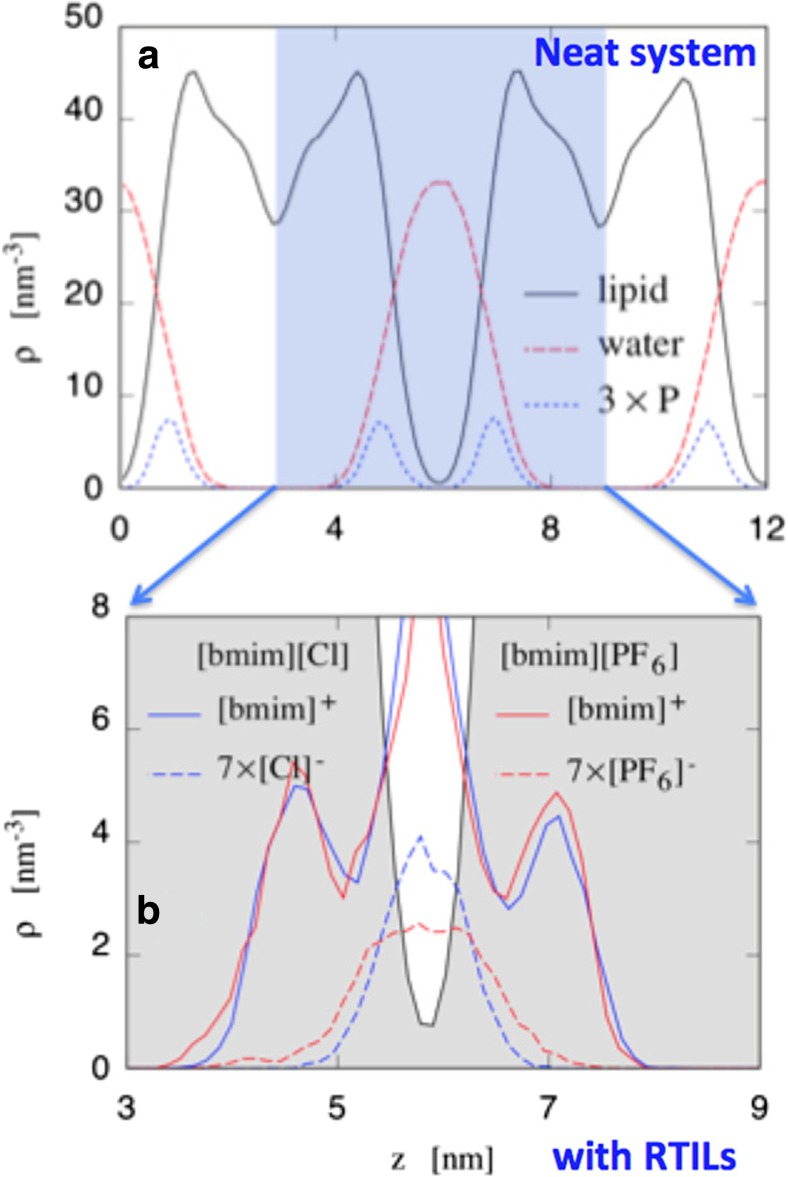
Different techniques, both experimental and computational, have been used over the last decade to study the interactions between RTILs and model biomembranes. Their aim is to determine the microscopic mechanisms behind their observed biological effects. Initial experimental studies carried out by Evans (2008) pointed to the marginal stability of phospholipid bilayers in contact with a solution of imidazolium and pyrrolidinium RTILs in water. The geometries of floating lipid vesicles and supported phospholipid bilayers have been studied by photoluminescence, atomic force microscopy (AFM), and quartz microbalance, respectively. All measurements revealed moderate to substantial damage to the bilayer, which increased with increasing length of the cation hydrocarbon tail. To the best of our knowledge, these are the first chemical–physical investigations of the interactions between RTILs and model biomembranes. However, the first molecular level experimental study to characterize the penetration of RTILs into model biomembranes has been done by means of neutron scattering (Benedetto et al. 2014b). More specifically, the density distribution of the chemical species (Fig. 3) has been assessed by neutron reflectometry and used to investigate the changes in the microscopic structure and stability of two model phospholipid bilayers made by POPC and DMPC, respectively, in contact with water solutions of two RTILs — [bmim][Cl] and [Cho][Cl]. It was observed that: (i) phospholipid bilayers maintain their characteristic 2D structure at the RTIL concentrations used in the experiments (up to 0.5 M), (ii) RTIL cations penetrate into the phospholipid region, staying in the first leaflet at the junction between the phosphonium polar head and neutral hydrocarbon tail of phospholipid molecules (red curve in Fig. 3), (iii) the phospholipid bilayer thickness shrinks by about 1 Å, and the area per lipid increases, (iv) the amount of cation absorbed in the lipid region is more for DMPC than for POPC, (v) the position of cations in the lipid region is independent of either the RTILs or the choice of phospholipids, (vi) for DMPC, the [bmim]+ cations have diffused into the inner layer as well, and (vii) the penetration of the RTIL cations is not fully reversible, since after rinsing with pure water, a non-negligible amount of cation remains in the lipid region (about 8% and 2.5% for DMPC and POPC respectively).
Fig. 3.
Density distribution profiles as a function of height z from the surface of the substrate obtained by fitting the neutron reflectivity data taken from (Benedetto et al. 2014b). Neutron reflectometry has allowed us to model each single supported phospholipid bilayers with four different density distributions accounting for: (i) the inner lipid heads layer (cyan), (ii) the inner lipid tail layer (blue), (iii) the outer lipid tail layer (blue), (iv) the outer lipid heads layer (cyan); and also (v) the density distribution of the cations (red), whereas the anion (Cl−) is almost invisible to neutrons. Three cases are here reported where two different phospholipid bilayers interact with aqueous solutions of two different RTILs at 0.5 M: a POPC and [Chol][Cl], b POPC and [bmim][Cl], and c DMPC and [bmim][Cl]. RTIL cations absorption accounts for 8%, 6.5%, and 11% of the lipid bilayer volume respectively. In c, the diffusion of the cations into the inner leaflet is apparent. In all the cases, phospholipid bilayers are in the liquid phase
Starting from the above experimental results, Benedetto and coworkers carried out classical atomistic molecular dynamics (MD) simulations on the same systems (Benedetto et al. 2015). The MD simulations reproduced the density distributions obtained by neutron reflectivity (Fig. 4), thus certifying the validity and quality of the model. This is something that was not obvious, and has to be checked. The empirical force fields used in these MD studies have never been tested properly for these ternary systems (phospholipids, water, and RTILs). Whereas the empirical potentials of lipids and water have been shown to work together, and while the same is also true for those of RTILs and water, all three have never been tested together. In a simplified way, we can conclude that the agreement between the MD simulations and the neutron reflectivity profiles (compare Figs. 3 and 4) is proof of validity of the empirical potential used.
Fig. 4.
Density distribution profiles as a function of height z obtained from our full-atom classical MD trajectories (Benedetto et al. 2015) for neat POPC bilayers (a) and bilayers doped with two RTILs (b). The computed profiles agreed with those measured by neutron reflectivity reported in Fig. 3: RTIL cations are absorbed in the lipid region, whereas the anions remain in the water in contact with the bilayers
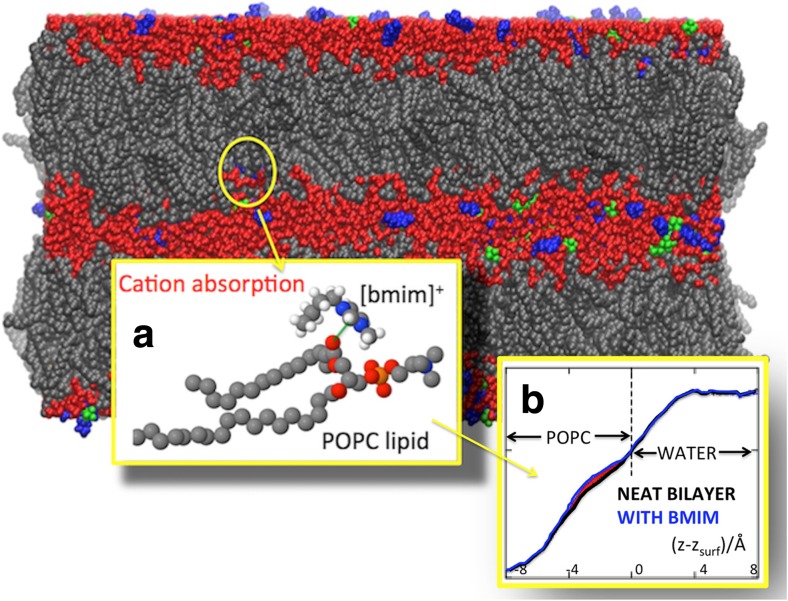
The MD simulations were able to determine or at least suggest the microscopic mechanism of the RTIL–biomembrane interaction, consisting of the following steps: (i) cations enter the phospholipid bilayer after 1–2 ns of simulations, (ii) the penetration of cations into the bilayer is driven by the Coulombic attraction between the positive charge of the cation itself with the negative charged groups in the lipid head region (e.g., the negative oxygens in the carbonyl group at the matching point of the hydrocarbon tails), and (iii) is stabilized by substantial dispersion forces between the cation and phospholipid tails, (iv) the absorption of the cations drives the penetration of a small amount of water into the polar portion of the phospholipid bilayer, and (v) stabilizes the hydrogen bonds at the lipid-water interface. Figure 5 presents some of the MD results.
Fig. 5.
Schematic view of one of the sample configurations used in our MD simulations (Benedetto et al. 2015). POPC domains in gray, water layers in red, [C4mim]+ in blue, and [PF6]− in green. Inset (a): Representative configuration of POPC and [C4mim]+. Inset (b): water density profiles: the difference (area in red) points to a water excess in the POPC doped with [C4mim]+
In the above MD study, the systems comprised 1,360 POPC molecules, and 26,000 water molecules, with a computational “production” time of about 100–150 ns. The simulation box had a length of about 200 × 100 × 120 Å along x, y, and z axes respectively. For classical full-atom MD simulations, these are “big numbers”, and to the best of our knowledge this is the biggest computer simulation study of phospholipids and RTILs. Both longer timescales and bigger systems are important to capture several features that otherwise cannot be detected. For example, only undulations of the bilayer surface whose wavelength is lower than twice the length of the simulation box can be detected, and only dynamical relaxations whose characteristic time is few times shorter that the total computational time can be properly identified. Needless to say, the simulation box sizes and computational “production” time affect also the error bars of all the observables extracted from the MD trajectories. Our MD simulations allow us to identify several trends due to the absorption of the RTIL cations in the lipid phase, from the shrinking of the bilayer thickness, to the variation in diffusion coefficients and of elastic properties. We found that both the isothermal (volume) compressibility and the surface compressibility moduli increase upon the addition of RTIL. It also changes the relaxation dynamics of hydration water and lipids by reducing the diffusion coefficient of water molecules, by increasing (in [bmim][Cl]) or decreasing (in [bmim][BF6]) the diffusion coefficient of the lipids. Neutron scattering can be used to probe most of these observables. Elastic and quasielastic neutron scattering can access the pico- to nanosecond dynamics (Benedetto and Kearley 2016; Magazù et al. 2008, 2009, 2010, 2012; Bee 1988; Volino 1978). Neutron spin-echo can be used to investigate slower relaxation processes (Mezei 1972) and to probe structural fluctuations (Nagao 2009; Woodka et al. 2012). Finally, small-angle neutron scattering and diffraction can be used for further structural characterization.
The joint experimental–simulation study presented above has its strength in combining two powerful approaches into one study. It allows a very detailed description of the microscopic mechanisms of RTIL–bilayer interaction. In the following, we will present recent results that either confirm or extend the scenario outlined above. Apart from a few cases, all the results are in agreement to each others, and what emerges is the extreme importance of the chemistry of lipids and RTILs. This suggests that case-by-case studies are needed. However, maintaining almost fixed chemistry, some general trends emerge, such as the correlation between RTILs chain lengths and concentration with cytotoxicity.
What matters in the microscopic world of RTILs-biomembranes interaction? From chain length and RTIL concentration to chemistry: a huge playground of challenges and opportunities.
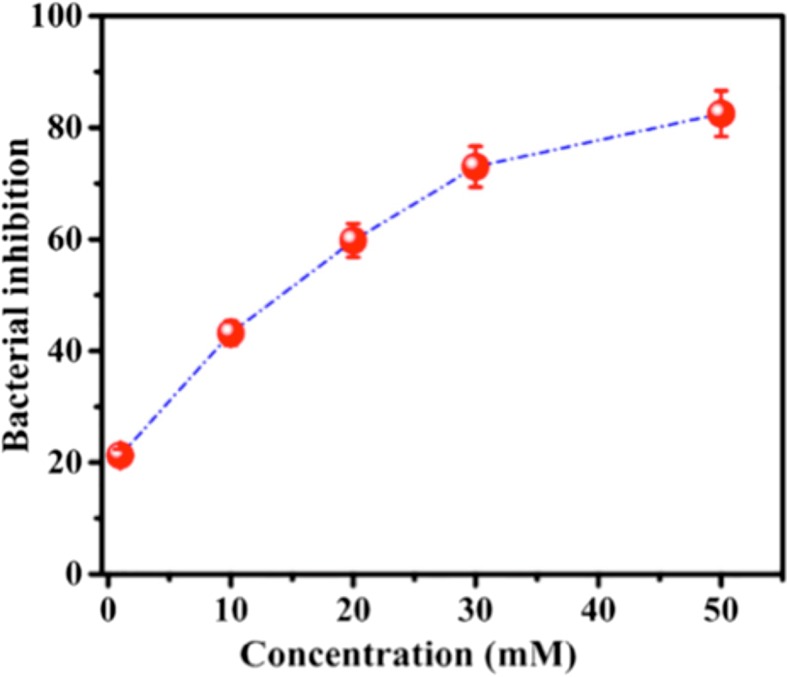
Shrinkage of the phospholipid bilayer thickness upon interaction with RTILs with water has been also confirmed by X-ray experiments (Bhattacharya et al. 2017, Kontro et al. 2016). In one case (Bhattacharya et al. 2017), single supported bilayers of DPPC (a zwitterionic lipid) interacting with aqueous solutions of [bmim][BF4] have been measured by means of X-ray reflectivity at several RTIL concentrations. As an additional result, the bilayer thickness seems to decrease faster with increasing RTIL concentration, although better error bars are needed to reach a final conclusion (see Table 1 of Bhattacharya et al. 2017). The effect of concentration has also been studied by monitoring the survival percentage of E. coli versus the concentration of [bmim][BF4], which shows an inverse correlation relationship (Fig. 6). In the same paper, Ghosh and co-workers (Bhattacharya et al. 2017) reported that the in-plane elasticity of supported monolayers decreases upon addition of the RTIL. This suggested that the rigid structure of the well-packed lipid monolayer relaxes in the presence of the RTIL. The in-plane elasticity has been determined by the pressure-area isotherm profiles on monolayers, with the water solution of RTIL added to the monolayer-forming lipid solution. Another way to probe changes in the mechano-elastic properties that are modified by the addition of RTILs is offered by AFM. Both the Young’s moduli and the rupture forces can be determined for single supported phospholipid bilayers and vesicles (Monocles et al. 2010; Garcia-Manyes and Sanz 2010; Roa et al. 2011; Ferenc et al. 2012; Stetter et al. 2016; Ding et al. 2017). To the best of our knowledge, there are no AFM studies of this kind. The elastic properties of phospholipid bilayers with and without RTILs have been studied by MD simulations (see below). AFM can also image the surface of the bilayers and tell us something about the degree of homogeneity of the surface and/or the presence of pores and defects.
Fig. 6.
Inhibition (%) of E. coli versus the concentration of the RTIL [BMIM][BF4] taken from (Bhattacharya et al. 2017). Figure reproduced with permission from the publisher
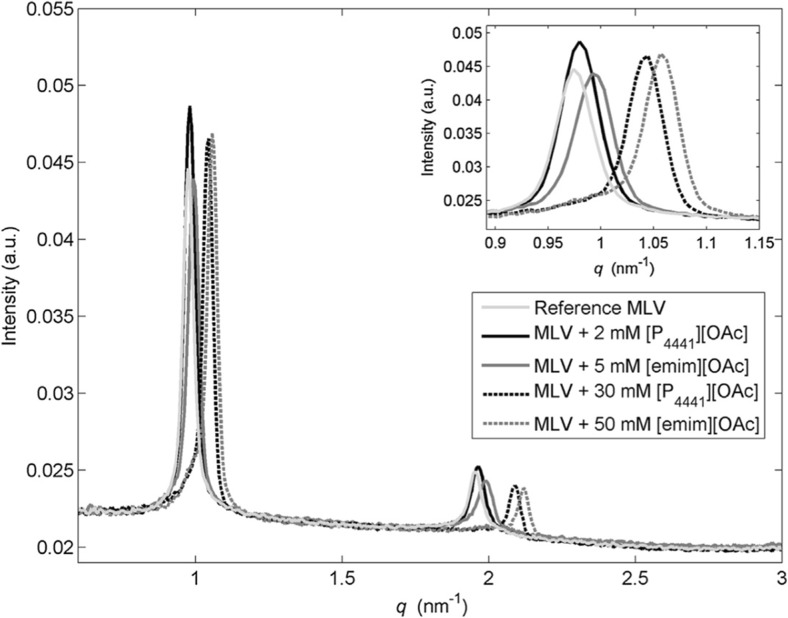
The second X-ray study cited above (Kontro et al. 2016) used small-angle X-ray scattering (SAXS) to study multi-lamellar liposomes of eggPC and eggPG (80:20 mol%) — and with cholesterol (60:20:20 mol%) — in interaction with water solutions of phosphonium-based RTILs. The results were compared with those of the more common imidazolium-based RTILs. Interest of this study is increased by the fact that some phosphonium-based RTILs have antibacterial activity (O’Toole et al. 2012). The lamellar spacing of liposomes decreases with increasing RTIL concentration (Fig. 7), so it is clear that in this case RTILs pass through the phospholipids multilayer superstructures. In the same study, Wiedmer and co-workers (Kontro et al. 2016) probed the effect of RTILs using dynamic light scattering (DLS) and zeta potential measurements on large uni-lamellar vesicles. They concluded that the ability of RTILs to affect liposomes is related to the length of the hydrocarbon chains of their cations. They also concluded that disruption of the phospholipid membrane is due to the disorder induced by the cation absorption. This result was confirmed by a more recent work again by Wiedmer et al. (Witos et al. 2017) using a nanoplasmonic sensing (NPS) measurement technique. They characterized the interaction between supported phospholipid uni-lamellar vesicles with amidinium- and phosphonium-based RTILs. NPS is a label-free optical technique that allows the study of surfaces and interfaces of metals, relying on surface plasmons, with a penetration depth of ~10 nm (the quartz crystal microbalance has a penetration depth of about 250 nm). Wiedmer et al. have also studied how the addition of cholesterol in the lipid phase changes the RTIL–biomembrane interaction (Kontro et al. 2016). This is quite important, since it is well known that cholesterol is present in several biomembranes and stabilizes them. As a result, vesicles without cholesterol ruptured at lower concentrations; however, above the rupture-concentration, the effects on the cholesterol-containing liposomes were more severe. The ruptured liposomes reassembled into organized lamellae, so under certain conditions, phosphonium-based ionic liquids have the ability to create new self-assembled structures from phospholipids.
Fig. 7.
SAXS pattern of multilamellar POPC vesicles in interaction with RTILs from (Kontro et al. 2016). Reference MLV (light gray), MLV treated with [P4441][OAc] (black), and MLV treated with [emim][OAc] (dark gray). Figure reproduced with permission from the publisher
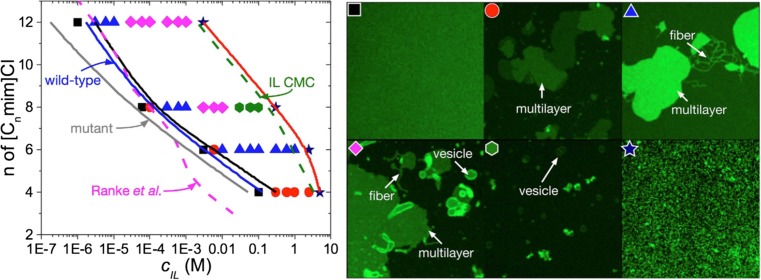
Further investigations into the connection between cytotoxicity, cation chain length, and RTIL concentration have been done by Maginn and co-workers by combining several experimental techniques with computer simulations (Yoo et al. 2014, 2016a, b). Their main result was the concentration-dependence study of RTILs on biomembranes. They found that RTIL cations nucleate morphological defects on biomembranes at concentrations near the half maximal effective concentration (EC50) of several microorganisms, and that RTILs destroy biomembranes at the RTIL critical micelle concentration (Fig. 8). The results suggest that the molecular mechanism of RTIL cytotoxicity may be linked to the RTIL-induced morphological reorganization of cell membranes initially caused by the insertion of RTILs into the membrane. In their studies, they have also shown that cytotoxicity increases with increasing alkyl chain length of the cation (Fig. 8), and relate this observation to the higher ability of alkyl chain to penetrate, and ultimately disrupt, cell membranes.
Fig. 8.
Phase diagram of [Cnmim][Cl] ionic liquid induced morphological changes to a supported α-PC bilayer taken from ref. (Yoo et al. 2016a). The EC50 toxicity line (in magenta) for IPC-cell shows a negative correlation between the toxic concentration and the RTIL cation chain length (Ranke et al. 2007a). The blue and gray lines are the predicted EC50 lines for wild-type (with cell wall) and mutant (without cell wall) strains of Chlamydomoas reinhardtii, respectively. The green line corresponds to the RTIL critical micelle concentration (CMC) of (Blesic et al. 2007). The symbols correspond to the specific morphologies as in the right image. Black square: neat bilayer; red circle: multilayer; blue triangle: multilayer and fiber/tube; pink diamond: multilayer, fiber/tube and vesicle; green hexagon: vesicle; navy star: disrupted bilayer. The solid black and red lines correspond to the onset of supported lipid bilayer disruption and the total disruption of the supported lipid bilayer respectively. Figure reproduced with permission from the publisher
Maginn et al. also performed all-atom and coarse-grained MD simulations. By sacrificing some atomic-level details, they were able to explore longer time-scales and larger length scales than atomistic MD simulations. This was important for studying both the absorption of RTILs and the changes in the bilayers structures. All-atom simulations usually can access a time scale of 100 ns and a spatial scale of tens of nanometers, whereas with coarse-grained simulations it is possible to access time scales of microseconds and spatial scales of micrometers. As a result, their coarse-grained MD simulations show that the short-tail RTIL [C4mim] cation spontaneously inserted into the lipid bilayer with the same orientation as that in the atomistic simulations, whereas the long-tail RTIL [C10mim] cation self-assembled into micelles that were eventually absorbed onto the upper bilayer leaflet, forming a RTIL monolayer. In the case of the short-tail cation, the number of inserted cations into the upper bilayer leaflet saturates at about 0.6 cations per lipid, after which the bilayer bends in response to the asymmetric distribution of inserted cations. Moreover, when RTIL cations are absorbed, the bending modules of the bilayer drops from 22.6 ± 1.7 × 10−20 J to 9.3 ± 0.9 × 10−20 J. Since this asymmetric situation does not occur when cations are absorbed in both the two leaflets, Maginn et al. (Yoo et al. 2016b) commented on the inability of cations to diffuse from one leaflet to the other, i.e., at origin of the bending fluctuations of the bilayer (Fig. 9).
Fig. 9.
Coarse-grained MD simulations of Ref. (Yoo et al. 2016b) suggest that it is the inability of some RTILs to diffuse from the outer leaflet to the inner leaflet of the phospholipid bilayer which is at the origin of the bilayer disruption. Figure reproduced with permission from the publisher
The inability of the RTILs to diffuse into the whole bilayer after inserted into the closest leaflet seems in contrast, however, with the SAXS data of (Kontro et al. 2016), and in our opinion is something that requires more investigations. Having a closer look at our neutron reflectivity data of Fig. 3, for example, we can conclude that it seems to depend on the phospholipids and RTILs used. It seems, moreover, that it depends on the difference between the lipid and the cation chain lengths. Since [bmim][Cl] diffuses in the inner leaflet of DMPC (Fig. 3c), but does not with POPC (Fig. 3b), we could conclude that the shorter the lipid chain, the higher the ability of RTILs to diffuse from one leaflet to the other. Since. Maginn and co-workers did not observe inter-leaflet diffusion of [bmim][Cl] in POPC, and S.K. Wiedmer and co-workers did observe inter-leaflet diffusion of different RTILs in POPC, we can conclude that the disagreement is due to the important role played by the RTIL chemistry, in line with our neutron reflectivity results (Benedetto et al. 2014b).
In our all-atom MD trajectories (Benedetto et al. 2015) done on POPC in interaction with [bmim][Cl], moreover, we did not observe any diffusion of cations from one layer to the other (result not yet published). However, longer simulation times may be needed to properly assess this inter-leaflet diffusion of RTILs, and its implication in RTILs cytotoxicity. AFM could also be a good technique for indirectly double-checking this in-bilayer diffusion of RTILs by measuring the distance between the bilayer surface and the support; since we know that RTILs shrink the bilayer thickness, any increment of such distance could be related to the diffusion of RTILs into the inner bilayer leaflet and the water interlayer on the top of the support. The ability of RTILs to diffuse in multi-layer geometries can be useful not only in setting-up experiments (such as diffraction experiments in which multi-layer lipid structures can be used) but also in applications. In our opinion, this subject deserves further investigations.
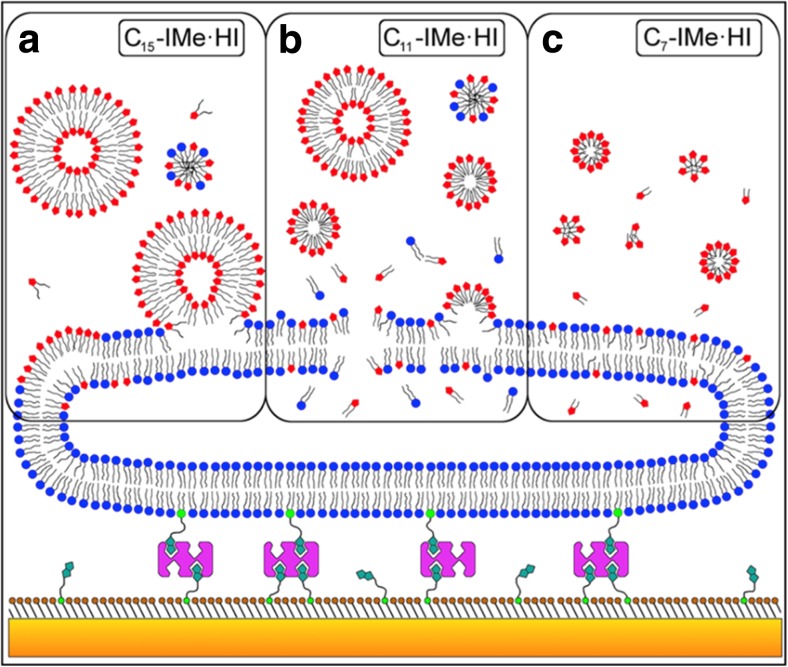
An important contribution to the saga has been made by Galla and coworkers (Wang et al. 2015a, b, 2016; Drücker et al. 2017, Rühling et al. 2017). They designed and synthesized a series of backbone-alkylated imidazolium-based RTILs composed of a hydrophilic N,N′-dimethylated imidazolium headgroup and two hydrophobic alkyl chains located at the 4- and 5-positions of the imidazole ring (Figs. 1d-e). The structure of these two-tail imidazolium-based RTILs (CnIMe·HI) is similar to that of phospholipids (Figs. 2a and b), explaining the similarity of their physicochemical properties. The interest in this new class of RTILs is increased by their significant anti-tumor activity and cellular toxicity. In comparison with the more common one-tail imidazolium RTILs, they show approximately three orders of magnitude higher anti-tumor activity. Having a global picture in mind, the most intriguing aspect is that their toxicity is negatively correlated with their chain lengths. The C7IMe·HI has the highest toxicity and anti-tumor activity, whereas the C15IMe·HI has the lowest toxicity. This circumstance contrasts with the “general picture” presented above, and highlights how the chemistry of the molecules plays an extremely important role. Moreover, the biological activity of these RTILs is inversely correlated with their lipophilicity (Fig. 10), something that also plays against the emerging picture where lipophilicity and toxicity are directly correlated. From Fig. 10a, it emerges that C15IMe·HI has the higher membrane activity, and by increasing its concentration the RTIL-lipid system shows a reorganization at 0.3 M fraction (see more details in the referred paper). Fig. 10b shows that C11IMe·HI can inhibit the supramolecular reorganization of the phospholipids, whereas Fig. 10c shows that the membrane activity of C7IMe·HI is negligible. These three different scenarios of RTIL-biomembrane interaction are summarized in Fig. 11.
Fig. 10.
Epifluorescence images of mixed monolayers of DPPC with double-tail imidazolium-based RTILs of different chain-length at different molar fractions at the air–water interface at room temperature taken from Wang et al. 2016. The membrane activity shows different behaviors depending on the chain length of the RTILs: the bilayers became more rigid for n = 15 (a), get inhibited for n = 11 (b), but almost unaffected for n = 7 (c). On the contrary, the biological activity of these RTILs goes the other way around, since the shortest one is the more toxic and the longest one stabilize the bilayer phase. Figure reproduced with permission from the publisher
Fig. 11.
A model for membrane interaction and structure formation of double-tail imidazolium-based RTILs taken from Drücker et al. 2017. Liposomes (blue) are tethered via biotin linkers (green) and streptavidin (purple) on a self-assembled monolayer (brown), which itself is chemisorbed on a gold-coated sensor surface (orange). a C15-IMe·HI is able to form vesicles in solution that can then associate, fuse, and intercalate into bilayer membranes. b C11-IMe·HI is able to form both vesicles and micelles while in water, which can then intercalate and lyse bilayer membranes. Bilayer disintegration is accompanied by the formation of micelles and mixed micelles. c C7-IMe·HI dissolves to micelles and single molecules and can pass though the membrane without disintegration. Figure reproduced with permission from the publisher
Galla and co-workers (Wang et al. 2015a, b, 2016; Drücker et al. 2017, Rühling et al. 2017) combined several experimental techniques such as film balance, quartz crystal microbalance, confocal laser scanning microscopy, calorimetry, epifluorescence microscopic measurements, with MD simulations. They have also measured the interaction of RTILs with biomembranes enriched with cholesterol (Fig. 12).
Fig. 12.
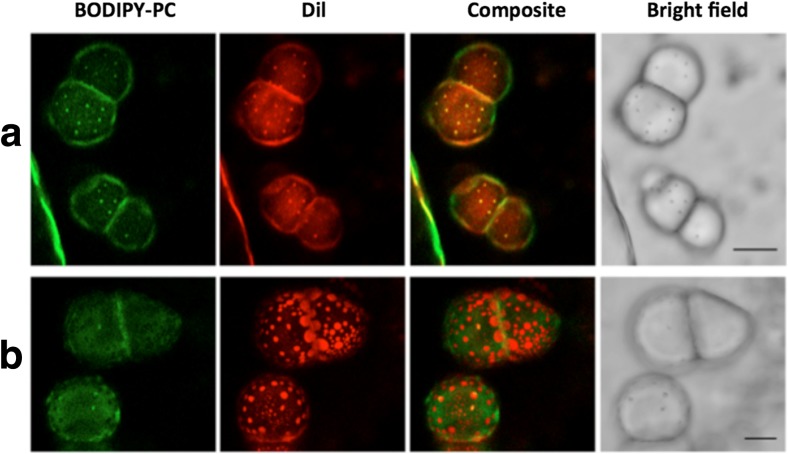
Bilayer domain fluidization of small bulged domains to flat large domains with enhanced dye specificity in the presence of 10% the double-tail imidazolium-based RTIL C15IMe·HI from Drücker et al. (2017). Giant uni-lamellar vesicles of a DOPC/SSM/Chol (33:33:33), and b DOPC/SSM/Chol/ C15IMe·HI (33:23:33:10) at 38 °C, scale 20 μm. Figure reproduced with permission from the publisher
Several other studies were done on this new and prom-ising subject, which more or less agreed with the results presented above. Namely: (i) positive correlation between RTILs chain lengths, RTILs concentration, and cytotoxicity (Witos et al. 2017; Losada-Pérez et al. 2016; Dusa et al. 2015; Galletti et al. 2015; Galluzzi et al. 2013; Kulacki and Lamberti 2008; Jeong et al. 2012; Mikkola et al. 2015; Jing et al. 2016), (ii) shrinkage of the bilayer thickness (Lim et al. 2014; Lim et al. 2015) and (iii) variation of its elasticity (Dusa et al. 2015) upon the absorption of RTIL cations, and (iv) Importance of the chemistry of the molecules (Weaver et al. 2013; Gal et al. 2012; Lee et al. 2015). The effects of RTILs on the thermotropic behavior of biomembranes were also probed in several reports (Weaver et al. 2013; Wang et al. 2016; Jeong et al. 2012.) At low concentrations of RTILs, the variation of the main phase transition of phospholipids bilayers is negligible, and at high concentration the variation is of the order of 5 to 10 degrees, sometimes reaching 20 degrees, but this just before the collapse of the bilayers. Several other MD simulation studies have also been done on this subject (e.g., Bingham and Ballone 2012; Cromie et al. 2009; Lim et al. 2015), which give access to the detailed mechanisms of interactions. In Lim et al., for example, the mutual interaction between RTIL cations once absorbed into the membrane has been studied, and the measured increment of permeability due to the absorption of RTILs has been related to their antibacterial activity. Investigations focused on specific cases and more complex systems, moreover, have enriched even more the broad and diverse panorama presented above (Patel et al. 2016; Modi et al. 2011; Jeong et al. 2012; Ryu et al. 2015; Lee et al. 2015). An interesting example is the study of Jeon, Lee and co-workers on the effect of RTILs on the ion channel function of Gramicidin A embedded in a phospholipid bilayer (Jeong et al. 2012; Ryu et al. 2015; Lee et al. 2015). Their results show for the first time how the changes of the physical properties of the biomembrane (e.g., thickness) induced by the absorption of RTIL cations can influence the activities of membrane proteins (Fig. 13). These effects are more significant with RTILs with longer alkyl chains, and at higher RTIL concentration. Interestingly, they figured out how the concentration of inorganic salts (i.e., NaCl) can play a major role. The picture that emerges is the following: RTILs disorder phospholipids and shrink the bilayer, which yields less membrane curvature around gramicidin A, thus stabilizes it, and leads to the increased ion permeability. However, this effect occurs at 1 M of NaCl, where RTILs only slightly increase the phospholipids dynamics because of the strong electrostatic interactions between NaCl and lipids. On the other hand, at 0.15 M of NaCl, RTILs also significantly increase the lateral mobility of both phospholipids and gramicidin A, which leads to the decreased ion permeability.
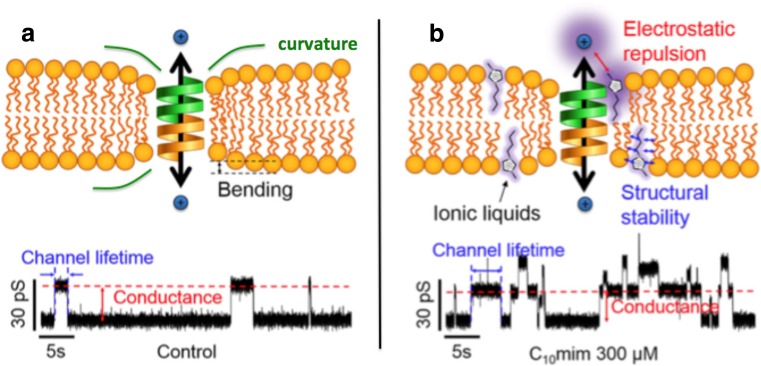
Fig. 13.
Effect of RTILs on gramicidin A ion channel from Ryu et al. (2015). a Neat system. b System doped with RTIL. The RTIL cation C10min (i) stabilizes the membrane–channel interaction by reducing the bilayer thickness and, in turn, its curvature closer to the channel location, and (ii) reduce the channel activity by electronic repulsion as sketched in a. The function of the channel seems also affected by the amount of the inorganic salt NaCl in the solution: the higher the amount, the higher the ion permeability. Figure reproduced with permission from the publisher
Summary and outlook for the future
The analysis of the current literature on the interaction between RTILs and biomembranes shows that the first target of these studies has been to determine the microscopic mechanism behind the cytotoxicity of RTILs, with the aim of designing greener RTILs for (industrial) applications. Whereas a positive correlation exists between RTILs chain lengths, RTILs concentration, and cytotoxicity, there are several exceptions that highlight how the effect of RTILs on biomembranes is a complex balance of different interactions where the chemistry of each molecule is playing a non-negligible role. On the one hand, this observation implies that the effect of RTILs on biomembranes has to be assessed on a case-by-case basis. On the other hand, the same observation opens a huge playground of new opportunities for applications in bio-nanotechnology. In our opinion, these opportunities are more important than the difficulties preventing an a priori assessment of the potential danger. Just a few nano-biotechnology studies have been done so far, and we are just at the beginning of this promising field of research.
Acknowledgements
A.B. thanks Prof. Pietro Ballone for fruitful discussions, and acknowledges support from (i) The European Community under the Marie-Curie Fellowship Grants HYDRA (No. 301463) and PSI-FELLOW (No. 290605), and (ii) The Science Foundation Ireland (SFI) under the Start Investigator Research Grant 15-SIRG-3538, with additional support provided by the School of Physics, University College Dublin, Ireland, and the Laboratory for Neutron Scattering, Paul Scherrer Institute (PSI), Switzerland.
Compliance with ethical standards
Conflict of interest
Antonio Benedetto declares that he has no conflicts of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Footnotes
This article is part of a Special Issue on ‘IUPAB Edinburgh Congress’ edited by Damien Hall
References
- Aroti A, Leontidis E, Dubois M, Zemb T. Effects of monovalent anions of the Hofmeister series on DPPC lipid bilayers part II: modeling the perpendicular and lateral equation-of-state. Biophys J. 2007;93:1580–1590. doi: 10.1529/biophysj.106.094482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bee M. Principles and applications in solid state chemistry, biology and materials science. Bristol PA: Adam Hilger; 1988. [Google Scholar]
- Benedetto A, Ballone P. Room temperature ionic liquids meet bio-molecules: a microscopic view of structure and dynamics. ACS Sustain Chem Eng. 2016;4:392–412. doi: 10.1021/acssuschemeng.5b01385. [DOI] [Google Scholar]
- Benedetto A, Ballone P. Room temperature ionic liquids meet bio-molecules: a microscopic view of structure and dynamics. Phi Mag. 2016;96:870–894. doi: 10.1080/14786435.2015.1119323. [DOI] [Google Scholar]
- Benedetto A, Kearley G. Elastic scattering spectroscopy (ESS): An instrument-concept for dynamics of complex (bio-)systems from elastic neutron scattering. Sci Rep. 2016;6:34266. doi: 10.1038/srep34266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedetto A, Heinrich F, Gonzalez MA, Fragneto G, Watkins E, Ballone P. Structure and stability of phospholipid bilayers hydrated by a room-temperature ionic liquid/water solution: A neutron reflectometry study. J Phys Chem B. 2014;118:12192–12206. doi: 10.1021/jp507631h. [DOI] [PubMed] [Google Scholar]
- Benedetto A, Bodo E, Gontrani L, Ballone P, Caminiti R. Amino acid anions in organic ionic compounds. An ab initio study of selected ion pairs. J Phys Chem. 2014;118:2471–2486. doi: 10.1021/jp412281n. [DOI] [PubMed] [Google Scholar]
- Benedetto A, Bingham RJ, Ballone P. Structure and dynamics of POPC bilayers in water solutions of room temperature ionic liquids. J Chem Phys. 2015;142:124706. doi: 10.1063/1.4915918. [DOI] [PubMed] [Google Scholar]
- Berkowitz ML, Vaćha R. Aqueous solutions at the interface with phospholipid bilayers. Acc Chem Res. 2012;45:74–82. doi: 10.1021/ar200079x. [DOI] [PubMed] [Google Scholar]
- Bernot R, Brueseke MA, Evans-White MA, Lamberti GA. Acute and chronic toxicity of imidazolium-based ionic liquids on daphnia magna. Envirom Toxic Chem. 2005;24:87–92. doi: 10.1897/03-635.1. [DOI] [PubMed] [Google Scholar]
- Bhattacharya G, Giri RP, Saxena H, Agrawal VV, Gupta A, Mukhopadhyay MK, Ghosh SK. X-ray reflectivity study of the interaction of an imidazolium-based ionic liquid with a soft supported lipid membrane. Langmuir. 2017;33:1295–1304. doi: 10.1021/acs.langmuir.6b03192. [DOI] [PubMed] [Google Scholar]
- Bingham RJ, Ballone P. Computational study of room-temperature ionic liquids interacting with a POPC phospholipid bilayer. J Phys Chem B. 2012;116:11205–11216. doi: 10.1021/jp306126q. [DOI] [PubMed] [Google Scholar]
- Blesic M, Marques MH, Plechkova NV, Seddon KR, Rebelo LPN, Lopes A. Self-aggregation of ionic liquids: micelle formation in aqueous solution. Green Chem. 2007;9:481–490. doi: 10.1039/b615406a. [DOI] [Google Scholar]
- Böckmann RA, Hac A, Heimburg T, Grubmüller H. Effect of sodium chloride on a lipid bilayer. Biophys J. 2003;85:1647–1655. doi: 10.1016/S0006-3495(03)74594-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne N, Angell CA. Protein unfolding, and the “tuning in” of reversible intermediate states, in protic ionic liquid media. J Mol Biol. 2008;378:707–714. doi: 10.1016/j.jmb.2008.02.050. [DOI] [PubMed] [Google Scholar]
- Byrne N, Angell CA. Formation and dissolution of hen egg white lysozyme amyloid fibrils in protic ionic liquids. Chem Commun. 2009;9:1046–1048. doi: 10.1039/b817590j. [DOI] [PubMed] [Google Scholar]
- Byrne N, Wang L-M, Belieres J-P, Angell CA. Reversible folding–unfolding, aggregation protection, and multi-year stabilization, in high concentration protein solutions, using ionic liquids. Chem Commun. 2007;26:2714–2716. doi: 10.1039/B618943A. [DOI] [PubMed] [Google Scholar]
- Cheng G, Varanasi P, Li C, Liu H, Melnichenko YB, Simmons BA, Kent MS, Singh S. Transition of cellulose crystalline structure and surface morphology of biomass as a function of ionic liquid pretreatment and its relation to enzymatic hydrolysis. Biomacromolecules. 2011;12:9344–9941. doi: 10.1021/bm101240z. [DOI] [PubMed] [Google Scholar]
- Clark KD, Nacham O, Yu H, Li T, Yamsek MM, Ronning DR, Anderson JL. Extraction of DNA by magnetic ionic liquids: tunable solvents for rapid and selective DNA analysis. Anal Chem. 2015;87:1552–1559. doi: 10.1021/ac504260t. [DOI] [PubMed] [Google Scholar]
- Clark KD, Sorensen M, Nacham O, Anderson JL. Preservation of DNA in nuclease-rich samples using magnetic ionic liquids. RSC Adv. 2016;6:39846–39851. doi: 10.1039/C6RA05932E. [DOI] [Google Scholar]
- Cromie SRT, Del Poṕolo MG, Ballone P. Interaction of room temperature ionic liquid solutions with a cholesterol bilayer. J Phys Chem B. 2009;113:11642–11648. doi: 10.1021/jp904060y. [DOI] [PubMed] [Google Scholar]
- Ding Y, Xu G-K, Wang G-F. On the determination of elastic moduli of cells by AFM based indentation. Sci Rep. 2017;7:45575. doi: 10.1038/srep45575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drücker P, Rühling A, Grill D, Wang D, Draeger A, Gerke V, Glorius F, Galla H-J. Imidazolium salts mimicking the structure of natural lipids exploit remarkable properties forming lamellar phases and giant vesicles. Langmuir. 2017;33:1333–1342. doi: 10.1021/acs.langmuir.6b03182. [DOI] [PubMed] [Google Scholar]
- Dusa F, Ruokonen S-V, Petrovaj J, Viitala T, Wiedmer SK. Ionic liquids affect the adsorption of liposomes onto cationic polyelectrolyte coated silica evidenced by quartz crystal microbalance. Colloids Surf B: Biointerfaces. 2015;136:496–505. doi: 10.1016/j.colsurfb.2015.09.059. [DOI] [PubMed] [Google Scholar]
- Earle MJ, Seddon KR. Ionic liquids. Green solvents for the future. Pure Appl Chem. 2000;72:1391–1398. doi: 10.1351/pac200072071391. [DOI] [Google Scholar]
- Evans KO. Supported phospholipid membrane interactions with 1-butyl-3-methylimidazolium chloride. J Phys Chem B. 2008;112:8558–8562. doi: 10.1021/jp7116592. [DOI] [PubMed] [Google Scholar]
- Ferenc R, Sanchez J, Blumich B, Herrmann W. AFM nanoindentation to determine Young’s modulus for different EPDM elastomers. Polym Test. 2012;31:425–432. doi: 10.1016/j.polymertesting.2012.01.003. [DOI] [Google Scholar]
- Gal N, Malferarri D, Kolusheva S, Galletti P, Tagliavini E, Jelinek R. Membrane interactions of ionic liquids: possible determinants for biological activity and toxicity. Biochim Biophys Acta. 2012;1818:2967–2974. doi: 10.1016/j.bbamem.2012.07.025. [DOI] [PubMed] [Google Scholar]
- Galletti P, Malferrari D, Samori C, Sartor G, Tagliavini E. Effects of ionic liquids on membrane fusion and lipid aggregation of egg-PC liposomes. Colloids Surf B: Biointerfaces. 2015;125:142–150. doi: 10.1016/j.colsurfb.2014.11.021. [DOI] [PubMed] [Google Scholar]
- Galluzzi M, Zhang S, Mohamadi S, Vakurov A, Podestà A, Nelson A. Interaction of imidazolium-based room-temperature ionic liquids with DOPC phospholipid monolayers: Electrochemical study. Langmuir. 2013;29:6573–6581. doi: 10.1021/la400923d. [DOI] [PubMed] [Google Scholar]
- Garcia-Manyes S, Sanz F. Nanomechanics of lipid bilayers by force spectroscopy with AFM: A perspective. Biochip Biophys Acta. 2010;1798:741–749. doi: 10.1016/j.bbamem.2009.12.019. [DOI] [PubMed] [Google Scholar]
- Ghandi K. A review of ionic liquids, their limits and applications. Green and Sustainable Chemistry. 2014;4:44–53. doi: 10.4236/gsc.2014.41008. [DOI] [Google Scholar]
- Hough WL, Smiglak M, Rodriguez H, Swatloski RP, Spear SK, Daly DY, Pernak J, Grisel JE, Carliss RD, Soutullo MD, Davis JH, Rogers RD. The third evolution of ionic liquids: Active pharmaceutical ingredients. New J Chem. 2007;31:1429–1436. doi: 10.1039/b706677p. [DOI] [Google Scholar]
- Jeong S, Ha SH, Han S-H, Lim M-C, Kim SM, Kim Y-R, Koo Y-M, So J-S, Jeon T-J. Elucidation of molecular interactions between lipid membranes and ionic liquids using model cell membranes. Soft Matter. 2012;8:5501–5506. doi: 10.1039/c2sm25223f. [DOI] [Google Scholar]
- Jing B, Lan N, Qiu J, Zhu Y. Interaction of ionic liquids with a lipid bilayer: A biophysical study of ionic liquid cytotoxicity. J Phys Chem B. 2016;120:2781–2789. doi: 10.1021/acs.jpcb.6b00362. [DOI] [PubMed] [Google Scholar]
- Kontro I, Svedström K, Duša F, Ahvenainen P, Ruokonen S-K, Witos J, Wiedmer SK. Effects of phosphonium-based ionic liquids on phospholipid membranes studied by small-angle X-ray scattering. Chem Phys Lipids. 2016;201:59–66. doi: 10.1016/j.chemphyslip.2016.11.003. [DOI] [PubMed] [Google Scholar]
- Kulacki KJ, Lamberti GA. Toxicity of imidazolium ionic liquids to freshwater algae. Green Chem. 2008;10:104–110. doi: 10.1039/B709289J. [DOI] [Google Scholar]
- Kumar A, Venkatesu P. A comparative study of myoglobin stability in the presence of Hofmeister anions of ionic liquids and ionic salts. Process Biochem. 2014;49:2158–2169. doi: 10.1016/j.procbio.2014.09.014. [DOI] [Google Scholar]
- Kumar A, Bisht M, Venkatesu P. Biocompatibility of ionic liquids towards protein stability: a comprehensive overview on the current understanding and their implications. Int J Bio Macromol. 2017;69:611–651. doi: 10.1016/j.ijbiomac.2016.12.005. [DOI] [PubMed] [Google Scholar]
- Lee H, Kim SM, Jeon T-J. Effects of imidazolium-based ionic liquids on the stability and dynamics of gramicidin a and lipid bilayers at different salt concentrations. J Mol Graph Model. 2015;61:53–60. doi: 10.1016/j.jmgm.2015.06.016. [DOI] [PubMed] [Google Scholar]
- Li Y, Liu X, Zhang S, Yao Y, Yao X, Xu J, Lu X. Dissolving process of a cellulose bunch in ionic liquids: a molecular dynamics study. Phys Chem Chem Phys. 2015;17:17894–17905. doi: 10.1039/C5CP02009C. [DOI] [PubMed] [Google Scholar]
- Lim GS, Zidar J, Cheong DW, Jaenicke S, Klähn M. Impact of ionic liquids in aqueous solution on bacterial plasma membranes studied with molecular dynamics simulations. J Phys Chem B. 2014;118:10444–10459. doi: 10.1021/jp5060952. [DOI] [PubMed] [Google Scholar]
- Lim GS, Jaenicke S, Klähn M. How the spontaneous insertion of amphiphilic imidazolium-based cations changes biological membranes: a molecular simulation study. Phys Chem Chem Phys. 2015;17:29171–29183. doi: 10.1039/C5CP04806K. [DOI] [PubMed] [Google Scholar]
- Liu H, Sale KL, Holmes BM, Simmons BA, Singh S. Understanding the interactions of cellulose with ionic liquids: a molecular dynamics study. J Phys Chem B. 2010;114:4293–4301. doi: 10.1021/jp9117437. [DOI] [PubMed] [Google Scholar]
- Losada-Pérez P, Khorshid M, Renner FU. Interactions of aqueous imidazolium-based ionic liquid mixtures with solid-supported phospholipid vesicles. PLoS One. 2016;11:9. doi: 10.1371/journal.pone.0163518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magazù S, Maisano G, Migliardo F, Benedetto A. Mean square displacement from self- distribution function evaluation by elastic incoherent neutron scattering. J Mol Struct. 2008;882:140–145. doi: 10.1016/j.molstruc.2007.09.022. [DOI] [Google Scholar]
- Magazù S, Maisano G, Migliardo F, Benedetto A. Biomolecular motion characterization by a self-distribution-function procedure in elastic incoherent neutron scattering. Phys Rev E. 2009;79:041915. doi: 10.1103/PhysRevE.79.041915. [DOI] [PubMed] [Google Scholar]
- Magazù S, Maisano G, Migliardo F, Benedetto A. Motion characterization by self-distribution–function procedure. Biochim Biophys Acta. 2010;1804:49–55. doi: 10.1016/j.bbapap.2009.09.017. [DOI] [PubMed] [Google Scholar]
- Magazù S, Migliardo F, Benedetto A, La Torre R, Hennet L. Bio-protective effects of homologous disaccharides on biological macromolecules. Eur Biophys J. 2012;41:361–367. doi: 10.1007/s00249-011-0760-x. [DOI] [PubMed] [Google Scholar]
- Mezei F. Neutron spin echo: A new concept in polarized thermal neutron techniques. Z Physik. 1972;255:146–160. doi: 10.1007/BF01394523. [DOI] [Google Scholar]
- Mikkola S-K, Robciuc A, Lokajova J, Holding AJ, Lämmmerhofer M, Kilpelaïnen I, Holopainen JM, AWT K, Wiedmer SK. Impact of amphiphilic biomass-dissolving ionic liquids on biological cells and liposomes. Environ Sci Technol. 2015;49:1870–1878. doi: 10.1021/es505725g. [DOI] [PubMed] [Google Scholar]
- Modi N, et al. Probing the transport of ionic liquids in aqueous solution through nanopores. J Phys Chem Lett. 2011;2:2331–2336. doi: 10.1021/jz201006b. [DOI] [Google Scholar]
- Monocles MA, Young TJ, Di Maio D. AFM indentation method used for elastic modulus characterization of interfaces and thin layers. J Mater Sci. 2010;45:3190–3197. doi: 10.1007/s10853-010-4326-6. [DOI] [Google Scholar]
- Nagao M. Observation of local thickness fluctuations in surfactant membranes using neutron spin echo. Phys Rev E. 2009;80:031606. doi: 10.1103/PhysRevE.80.031606. [DOI] [PubMed] [Google Scholar]
- O’Toole G, Wathier M, Zegans ME, Shanks RMQ, Kowalski R, Grinstaff MW. Diphosphonium ionic liquids as broad-spectrum antimicrobial agents. Cornea. 2012;31:810–816. doi: 10.1097/ICO.0b013e31823f0a86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabst G, Hodzic A, Štrancar J, Danner S, Rappolt M, Laggner P. Rigidification of neutral lipid bilayers in the presence of salts. Biophys J. 2007;93:2688–2696. doi: 10.1529/biophysj.107.112615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel R, et al. Effect of 1,4-bis(3-dodecylimidazolium-1-yl) butane bromide on channel form of gramicidin vesicles. Colloids Surf A Physicochem Eng Asp. 2016;508:150–158. doi: 10.1016/j.colsurfa.2016.08.058. [DOI] [Google Scholar]
- Petkovic M, Seddon KR, Rebelo LPN, Pereira CS. Ionic liquids: A pathway to environmental acceptability. Chem Soc Rev. 2011;40:1383–1403. doi: 10.1039/C004968A. [DOI] [PubMed] [Google Scholar]
- Plechkova NV, Rogers RD, Seddon KR (2009) Ionic liquids: from knowledge to applications. ACS Symposium Series no. 1030. American Chemical Society, Washington DC
- Pretti C, Chiappe C, Pieraccini D, Gregori M, Abramo F, Monni G, Intorre L. Acute toxicity of ionic liquids to the zebrafish (Danio Rerio) Green Chem. 2006;8:238–240. doi: 10.1039/B511554J. [DOI] [Google Scholar]
- Ranke J, Cox M, Müller A, Schmidt C, Beyersmann D. Sorption, cellular distribution, and cytotoxicity of imidazolium ionic liquids in mammalian cells — influence of lipophilicity. Toxicol Environ Chem. 2006;88:273–285. doi: 10.1080/02772240600589505. [DOI] [Google Scholar]
- Ranke J, Müller A, Bottin-Weber U, Stock F, Stolte S, Arning J, Störmann R, Jastorff B. Lipophilicity parameters for ionic liquid cations and their correlation to in vitro cytotoxicity. Ecotoxicol Environ Saf. 2007;67:430–438. doi: 10.1016/j.ecoenv.2006.08.008. [DOI] [PubMed] [Google Scholar]
- Ranke J, Stolte S, Stormann R, Arning J, Jastorff B. Design of sustainable chemical products—the example of ionic liquids. Chem Rev. 2007;107:2183–2206. doi: 10.1021/cr050942s. [DOI] [PubMed] [Google Scholar]
- Roa JJ, Oncins G, Diaz J, Sanz F, Zegarra M. Calculation of Young’s modulus value by means of AFM. Recent Pat Nanotechnol. 2011;5:27–36. doi: 10.2174/187221011794474985. [DOI] [PubMed] [Google Scholar]
- Rühling A, Wang D, Ernest JB, Wulff S, Honeker R, Richter C, Ferry A, Galla H-J, Glorius F. Influence of the headgroup of azolium-based lipids on their biophysical properties and cytotoxicity. Chem Eur J. 2017;23:5920–5924. doi: 10.1002/chem.201604182. [DOI] [PubMed] [Google Scholar]
- Ryu H, Lee H, Iwata S, Choi S, Kim MK, Kim Y-R, Maruta S, Kim SM, Jeon T-J. Investigation of ion channel activities of gramicidin a in the presence of ionic liquids using model cell membranes. Sci Rep. 2015;5 doi: 10.1038/srep11935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stetter FWS, Hyun S-H, Brander S, Urban JM, Thompson DH, Hugel T. Nanomechanical characterization of lipid bilayers with AFM-based methods. Polymer. 2016;102:326–332. doi: 10.1016/j.polymer.2015.11.038. [DOI] [Google Scholar]
- Stoimenovski J, Dean PM, Izgorodina EI, MacFarlane DR. Protic pharmaceutical ionic liquids and solids: aspects of protonics. Faraday Discuss. 2012;154:335–352. doi: 10.1039/C1FD00071C. [DOI] [PubMed] [Google Scholar]
- Stolte S, Arning J, Bottin-Weber U, Müller A, Pitner W-R, Welz-Biermann U, Jastorff B, Ranke J. Effects of different head groups and functionalised side chains on the cytotoxicity of ionic liquids. Green Chem. 2007;9:760–767. doi: 10.1039/B615326G. [DOI] [Google Scholar]
- Volino F. Spectroscopic methods for the study of local dynamics in polyatomic fluids. New York: Plenum; 1978. [Google Scholar]
- Wang D, Richter C, Rühling A, Drcker P, Siegmund D, Metzler-Nolte N, Glorius F, Galla H-J. A remarkably simple class of imidazolium-based lipids and their biological properties. Chem Eur J. 2015;21:1–5. doi: 10.1002/chem.201490218. [DOI] [PubMed] [Google Scholar]
- Wang D, Richter C, Rühling A, Hüwel S, Glorius F, Galla H-J. Anti-tumor activity and cytotoxicity in vitro of novel 4,5-dialkylimidazolium surfactants. Biochem Biophys Res Commun. 2015;467:1033–1038. doi: 10.1016/j.bbrc.2015.10.015. [DOI] [PubMed] [Google Scholar]
- Wang D, Jong DH, Rühling A, Lesch V, Shimizu K, Wulff S, Heuer A, Glorius F, Galla H-J. Imidazolium-based lipid analogues and their interaction with phosphatidylcholine membranes. Langmuir. 2016;32:12579–12592. doi: 10.1021/acs.langmuir.6b02496. [DOI] [PubMed] [Google Scholar]
- Weaver KD, Vorst MPV, Vijayaraghavan R, MacFarlane DR, Elliott GD. Interaction of choline salts with artificial biological membranes: DSC studies elucidating cellular interactions. Biochim Biophys Acta. 2013;1828:1856–1862. doi: 10.1016/j.bbamem.2013.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welton FT. Room-temperature ionic liquids. Solvents for synthesis and catalysis. Chem Rev. 1999;99:2071–2084. doi: 10.1021/cr980032t. [DOI] [PubMed] [Google Scholar]
- Witos J, Russo G, Ruokonen S-K, Wiedmer SK. Unraveling interactions between ionic liquids and phospholipid vesicles using nanoplasmonic sensing. Langmuir. 2017;33:1066–1076. doi: 10.1021/acs.langmuir.6b04359. [DOI] [PubMed] [Google Scholar]
- Woodka AC, Butler PD, Porcar L, Farago B, Nagao M. Lipid bilayers and membrane dynamics: Insight into thickness fluctuations. Phys Rev Lett. 2012;109:058102. doi: 10.1103/PhysRevLett.109.058102. [DOI] [PubMed] [Google Scholar]
- Yoo B, Shah JK, Zhu Y, Maginn EJ. Amphiphilic interactions of ionic liquids with lipid biomembranes: A molecular simulation study. Soft Matter. 2014;10:8641–8651. doi: 10.1039/C4SM01528B. [DOI] [PubMed] [Google Scholar]
- Yoo B, Jing B, Jones SE, Lamberti GA, Zhu Y, Shah JK, Maginn EJ. Molecular mechanisms of ionic liquid cytotoxicity probed by an integrated experimental and computational approach. Sci Rep. 2016;6:19889. doi: 10.1038/srep19889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo B, Zhu Y, Maginn EJ. Molecular mechanism of ionic-liquid-induced membrane disruption: Morphological changes to bilayers, multilayers, and vesicles. Langmuir. 2016;32:5403–5411. doi: 10.1021/acs.langmuir.6b00768. [DOI] [PubMed] [Google Scholar]
- Youngs TGA, Holbrey JD, Mullan CL, Norman SE, Lagunas MC, D’Agostino C, Mantle MD, Gladden LF, Bowron DT, Hardacre C. Neutron diffraction, NMR and molecular dynamics study of glucose dissolved in the ionic liquid 1-ethyl-3-methylimidazolium acetate. Chem Sci. 2011;2:1594–1605. doi: 10.1039/c1sc00241d. [DOI] [Google Scholar]
- Zhang J, Wu J, Yu J, Zhang X, Hea J, Zhang J. Application of ionic liquids for dissolving cellulose and fabricating cellulose-based materials: State of the art and future trends. Mater Chem Front. 2017;1:1273–1290. doi: 10.1039/C6QM00348F. [DOI] [Google Scholar]