Abstract
We describe a two-dimensional agarose gel electrophoresis procedure that improves the resolution of knotted DNA molecules. The first gel dimension is run at low voltage, and DNA knots migrate according to their compactness. The second gel dimension is run at high voltage, and DNA knots migrate according to other physical parameters such as shape and flexibility. In comparison with one-dimensional gel electrophoresis, this procedure segregates the knotted DNA molecules from other unknotted forms of DNA, and partially resolves populations of knots that have the same number of crossings. The two-dimensional display may allow quantitative and qualitative characterization of different types of DNA knots simply by gel velocity.
INTRODUCTION
The appearance of knotted DNA is frequent both in nature and in the laboratory (1–3). DNA recombinases (4,5) and topoisomerases (6,7) produce distinct types of knots when acting on circular DNA. Depending on the reaction environment, ligation of DNA free-ends also results in different degrees of knotting (8,9). Analysis of the knots produced in all these processes has become a useful tool to infer physical properties of the duplex, and to reconstruct the architecture of the complexes that enzymes or subcellular structures form with DNA. Currently, laborious electron microscopy is necessary to identify and quantify types of DNA knots (10). Here we describe an electrophoretic procedure that distinguishes individual populations of knots within complex mixtures of knotted, unknotted and linear DNA. As a source of knots we used DNA isolated from bacteriophage P4 capsids (11). Most of these DNA molecules are knotted circles that result from cohesive end-joining of the 11.6 kb phage genome. The P4 phage DNA is highly suitable for this study for two reasons. First, these DNA knots are complex and heterogeneous (11). Secondly, these knotted molecules are not covalently closed and therefore contain no confounding supercoiling, so they can be analyzed directly by gel electrophoresis without requiring enzymatic nicking. The two-dimensional gel electrophoresis described here readily resolved linear, unknotted and at least 10 populations of knots, including apparently two sets of 6- and 8-crossing knots, among the P4 phage DNA molecules.
MATERIALS AND METHODS
Phages and bacterial strains
Phages and bacterial strains used to obtain knotted DNA were kindly provided by Richard Calendar (University of California, Berkeley). Phage P4 vir1 del22 results from a 1.6 kb deletion of the P4 vir1 coliphage (12), which is a helper-dependent phage that does not lysogenize. Escherichia coli C-1895 is lysogenic for P2, a helper prophage, and it is used to grow stocks of P4 phages in liquid medium. Escherichia coli C-8001 is lysogenic for P2 amH13. As gene H encodes part of the phage tail, this strain is used to obtain tailless P4 capsids (13).
Isolation of knotted DNA from P4 tailless capsids
Knotted DNA was obtained by infecting the E.coli strain C-8001 with P4 vir1 del22 phages, following the procedure described by Isaksen et al. (13) with minor modifications. After bacterial lysis the released capsids were precipitated with PEG 8000, and then solubilized in P buffer (10 mM MgCl2, 10 mM Tris–HCl pH 7.2, 130 mM ammonium acetate). P4 tailless capsids were banded by cesium chloride centrifugation in an NVT65 rotor for 14 h at 45 000 r.p.m. The banded particles were dialyzed against P buffer. DNA was extracted twice with phenol, once with phenol/chloroform, precipitated with ethanol, and resuspended in TE buffer (10 mM Tris–HCl pH 8, 1 mM EDTA) to a concentration of ∼1 mg/ml.
Unknotting of DNA by yeast topoisomerase II
Yeast topoisomerase II was purified from Saccharomyces cerevisiae strain BCY123 harboring the topoisomerase II expression plasmid YEpTOP2GAL1 as previously described (14). DNA unknotting was carried out in 50 µl reaction volumes, containing 50 mM Tris–HCl pH 8, 1 mM EDTA, 150 mM KCl, 8 mM MgCl2, 7 mM 2-mercaptoethanol, 100 µg/ml bovine serum albumin, 5 µg of knotted DNA and 50 ng of topoisomerase II. Reactions started by the addition of ATP to 1 mM. Following incubation at 30°C, reactions were stopped by adding EDTA to 25 mM.
Electrophoretic analysis of knotted DNA
All gel electrophoresis was done using regular agarose equilibrated with TBE buffer (100 mM Tris–borate pH 8.3, 2 mM EDTA). To prepare two-dimensional gel slabs, a 2% agarose bed (20 × 20 × 0.2 cm) was first cast on the gel tray. On top of this bed, a 0.5 cm thick resolving slab containing 0.4% agarose was cast. In the two-dimensional analysis of the samples, this slab had only one well (2 × 2 mm section) close to the upper-left corner. The electrophoresis was done in 2 l gel tanks that had 30 cm of effective distance between the electrodes. In the first dimension, electrophoresis of DNA samples (usually of 8 µl volume containing up to 2 µg of DNA) was run at 0.8 V/cm for 40 h at room temperature. After a 90° rotation of the gel, the second dimension was run in the same electrophoresis buffer, applying 3.4 V/cm for 4 h at room temperature. Gels were stained with ethidium bromide and destained with water before photography.
RESULTS AND DISCUSSION
Effect of the applied voltage on the electrophoretic mobility of knotted DNA molecules
DNA from P4 vir1 del22 phage capsids was prepared as described in Materials and Methods. One-dimensional gel electrophoresis was used to identify the knotted DNA molecules, and to examine the effect of the applied voltage on the gel velocity of the knotted and the unknotted DNA. As shown in Figure 1A, P4 phage DNA generates a broad and complex distribution of knotted molecules (K, in lane 1). These knotted circles become easily untied by the transient denaturation of their non-covalent end-joins with heat (lane 2). They can also be linearized by a restriction endonuclease (lane 3), or converted into the unknotted circle by topoisomerase II (lane 4). Whereas the samples in the gel shown in Figure 1A were run at low voltage, equivalent samples were run at high voltage in the gel shown in Figure 1B. Comparison of both gels illustrates how migration of knotted DNA is affected by the applied voltage during electrophoresis. At low voltage (Fig. 1A), most knotted molecules (K) migrate faster than unknotted (C) or linear DNA (L). In these conditions, knot migration is known to be related to the average crossing number of the ideal representation of the corresponding knot (15–18). In general, the higher the crossing number, the more compact the knot structure and the faster it migrates in the gel. However, at high voltage (Fig. 1B), most knotted molecules (K) migrate much more slowly than the unknotted (C) or linear DNA (L). In these conditions, DNA migration depends on how fast the molecules can change shape to adapt to pore size while moving through the gel fibers (19). Accordingly, knotted molecules become retarded relative to unknotted molecules because they are less flexible than linear or unknotted ones.
Figure 1.
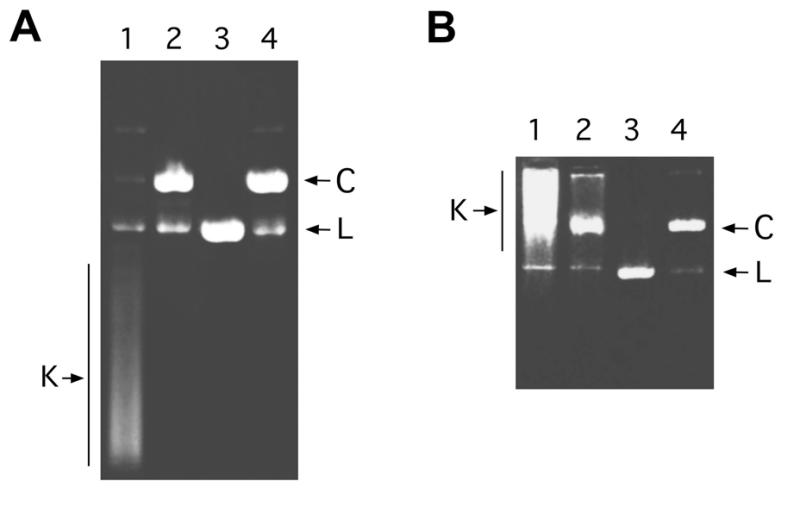
Effect of the applied voltage on the electrophoretic migration of knotted DNA. A sample of DNA obtained from bacteriophage P4 capsids was divided into four fractions. One fraction was not further treated (sample 1); a second fraction was heated to 75°C for 5 min (sample 2); a third fraction was digested with XbaI restriction endonuclease (sample 3); and a final fraction was treated with DNA topoisomerase II (sample 4). These four DNA samples were examined by electrophoresis (lanes 1–4, respectively) in a 0.4% agarose gel, which ran for 20 h at 25 V (A); and also in a 0.8% agarose gel, which ran for 1 h at 100 V (B). Both gels were stained with ethidium bromide and destained with water before photography. The positions of knotted DNA circles (K), unknotted DNA circles (C) and linearized DNA circles (L) are indicated.
Relative migration of knotted versus unknotted DNA by two-dimensional gel electrophoresis
The identification of knotted DNA by one-dimensional agarose gel electrophoresis is confounded when the gel position of some knotted molecules overlaps with that of linearized DNA or supercoiled DNA topoisomers, which often results from an excessive or an incomplete nicking of the DNA sample, respectively. This circumstance is depicted in Figure 2A, where the migration of linear (L) and covalently closed circular DNA topoisomers (CC) is shown to coincide with the migration of knotted DNA molecules (K). To improve the resolution of the knotted molecules and to segregate them from the unknotted ones, we combined the capacity of gel electrophoresis to discriminate either molecular compactness or molecular flexibility on a two-dimensional gel, in which the first and the second dimensions are run at different voltages. In the experiment shown in Figure 2B, a DNA sample containing different molecular forms of P4 phage DNA was prepared by mixing knotted molecules, linearized molecules and covalently closed unknotted topoisomers. This sample was loaded in a two-dimensional gel slab made as described in Materials and Methods, and run at low voltage (0.8 V/cm) in the first dimension and at high voltage (3.4 V/cm) in the second dimension. The two-dimensional gel clearly segregated the different molecular forms of the sample. In agreement with that observed in Figure 2A, in the first dimension (I) the migration of some knot populations overlapped with that of the covalently closed topoisomers or linear DNA. In the second dimension (II), however, the relative migration of linear DNA molecules (colored in green) markedly increased and became conveniently separated ahead of other molecular forms. The knotted molecules (colored in red), instead, generated a long arched distribution that migrated clearly apart from the straight array of covalently closed unknotted topoisomers (colored in blue) and linear DNA. As the two-dimensional gel had a lower concentration of agarose than the gel shown in Figure 1B, most knots still ran faster than the unknotted and linear DNA during the second (high voltage) dimension.
Figure 2.
Migration of knotted and unknotted DNA by two-dimensional gel electrophoresis. (A) Different molecular forms of P4 phage DNA were loaded in a 0.4% agarose gel that ran for 25 h at 30 V. Lane 1 contained a mixture of knotted molecules, linearized molecules and covalently closed unknotted molecules. Lane 2 mostly contained topoisomerase II unknotted molecules. Lane 3 mostly contained covalently closed unknotted molecules, which were obtained by treating the sample in lane 2 with T4-ligase. The gel positions of knotted molecules (K), linearized molecules (L), nicked unknotted molecules (C) and covalently closed unknotted molecules (CC) are indicated. (B) A sample containing different molecular forms of P4 phage DNA, similar to that described above, was loaded in a two-dimensional gel slab containing 0.4% agarose. The first dimension (I) ran at 25 V for 40 h. The second dimension (II) ran at 100 V for 4 h. The color scheme indicates the gel positions of linear molecules (green), nicked unknotted molecules (black), covalently closed unknotted molecules (blue) and knotted molecules (red).
Resolution of distinct DNA knot populations by two-dimensional gel electrophoresis
Further analysis of the migration of knotted DNA molecules by two-dimensional electrophoresis is depicted in Figure 3, in which native DNA from P4 phage capsids was run in the same gel conditions described in the above section. Individual populations of knot types were clearly distinguishable on the upper part of the arched distribution of knotted molecules. In the first dimension (I), the expected correlation between knot compactness (crossing number) and knot migration is observed (15–18). The positions of knots up to 10 crossings are clearly resolved as individual spots on the gel (colored in red). Knots with more crossings are embedded in the long tail, in which the faster species may have more than 30 crossings. In the second dimension (II), the migration of knots with a larger radius of gyration is no longer proportional to molecular compactness. The unknotted circle (colored in black) migrates faster than the 3-crossing knot, which in turn migrates faster than knot populations containing 4-, 5-, 6- and 7-crossings. Interestingly, whereas the knot populations containing 3-, 4-, 5- and 7-crossings run as single spots, the second dimension reveals two sets of knots at the level of 6- and 8-crossings. In these particular conditions, one set moves slower than the 3-crossing knot and the other faster. Therefore, at least two distinct types of knots likely containing 6- and 8-crossings are present in this DNA sample.
Figure 3.
Resolution of different populations of knotted DNA by two-dimensional gel electrophoresis. A sample containing 2 µg of P4 phage DNA was analyzed by two-dimensional gel electrophoresis in the same conditions described in Figure 2B. A magnification of this gel picture (middle panel) allows us to distinguish individual populations of knotted molecules. The color scheme indicates the gel positions of linear molecules (green), nicked unknotted molecules (black) and the knotted molecules (red). According to their migration in the first dimension, the most probable number of knot crossings of the resolved populations of knotted molecules is indicated. The identification of the 3-crossing knot (trefoil) was corroborated by comparison with the electrophoreitc mobility of trefoils obtained by random cyclization of linear P4 DNA. The assignment of the 4-, 5-, 6-, 7-, 8-, 9- and 10-crossing knots was reinforced, upon appreciable fitting of the electrophoretic display, by the nearby linear correlation previously determined between gel velocity at low voltage and the number of knot crossings (15–18).
CONCLUSION
The two-dimensional gel electrophoresis of DNA described here improves the possibility of identifying distinct DNA knot populations simply by gel velocity. This procedure segregates the knotted DNA molecules from linear and unknotted forms of DNA that could interfere with knot analysis. More importantly, it is also able to resolve different families of knots with the same number of crossings. This simple procedure may also advance our understanding of the electrophoretic separation of DNA molecules.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr R. Calendar for providing materials to prepare P4 phages. This work was supported by grant PB98-0487 of the Ministry of Science of Spain (to J.R.). S.T. was the recipient of a predoctoral fellowship of the Ministry of Science of Spain. J.A., M.E.V. and D.W.S. were supported by the Program in Mathematics and Molecular Biology (PMMB) through a Burroughs Wellcome Fund Interfaces Grant.
References
- 1.Liu L.F., Perkocha,L., Calendar,R. and Wang,J.C. (1981) Knotted DNA from bacteriophage capsids. Proc. Natl Acad. Sci. USA, 78, 5498–5502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Menissier J., de Murcia,G., Lebeurier,G. and Hirth,L. (1983) Electron microscopic studies of the different topological forms of the cauliflower mosaic virus DNA: knotted encapsidated DNA and nuclear minichromosome. EMBO J., 2, 1067–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ryan K.A., Shapiro,T.A., Rauch,C.A., Griffith,J.D. and Englund,P.T. (1988) A knotted free minicircle in kinetoplast DNA. Proc. Natl Acad. Sci. USA, 85, 5844–5848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wasserman S.A., Dungan,J.M. and Cozzarelli,N.R. (1985) Discovery of a predicted DNA knot substantiates a model for site-specific recombination. Science, 229, 171–174. [DOI] [PubMed] [Google Scholar]
- 5.Spengler S.J., Stasiak,A. and Cozzarelli,N.R. (1985) The stereostructure of knots and catenanes produced by phage lambda integrative recombination: implications for mechanism and DNA structure. Cell, 42, 325–334. [DOI] [PubMed] [Google Scholar]
- 6.Wasserman S.A. and Cozzarelli,N.R. (1991) Supercoiled DNA-directed knotting by T4 topoisomerase. J. Biol. Chem., 266, 20567–20573. [PubMed] [Google Scholar]
- 7.Roca J., Berger,J.M. and Wang,J.C. (1993) On the simultaneous binding of eukaryotic DNA topoisomerase II to a pair of double-stranded DNA helices. J. Biol. Chem., 268, 14250–14255. [PubMed] [Google Scholar]
- 8.Rybenkov V.V., Cozzarelli,N.R. and Vologodskii,A.V. (1993) Probability of DNA knotting and the effective diameter of the DNA double helix. Proc. Natl Acad. Sci. USA, 90, 5307–5311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shaw S.Y. and Wang,J.C. (1993) Knotting of a DNA chain during ring closure. Science, 260, 533–536. [DOI] [PubMed] [Google Scholar]
- 10.Zechiedrich E.L. and Crisona,N.J. (1989) Coating DNA with RecA protein to distinguish DNA path by electron microscopy. In Bjornsti,M. and Osheroff,N. (eds), Methods in Molecular Biology: DNA Topoisomerase Protocols. Humana Press, Totowa, NJ, Vol. 1, pp. 99–108. [DOI] [PubMed]
- 11.Liu L.F., Davis,J.L. and Calendar,R. (1981) Novel topologically knotted DNA from bacteriophage P4 capsids: studies with DNA topoisomerases. Nucleic Acids Res., 9, 3979–3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Raimondi A., Dunghi,R., Montaguti,A., Pessina,A. and Deho,G. (1985) Analysis of spontaneous deletion mutants of satellite bacteriophage P4. J. Virol., 54, 233–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Isaksen M., Julien,B., Calendar,R. and Lindqvist,B.H. (1999) Isolation of knotted DNA from coliphage P4. In Bjornsti,M. and Osheroff,N. (eds), Methods in Molecular Biology: DNA Topoisomerase Protocols. Humana Press, Totowa, NJ, Vol. 1, pp. 69–74. [DOI] [PubMed]
- 14.Worland S.T. and Wang,J.C. (1989) Inducible overexpression, purification and active site mapping of DNA topoisomerase II from the yeast Saccharomyces cerevisiae. J. Biol. Chem., 264, 4412–4416. [PubMed] [Google Scholar]
- 15.Stasiak A., Katritch,V., Bednar,J., Michoud,D. and Dubochet,J. (1996) Electrophoretic mobility of DNA knots. Nature, 384, 122. [DOI] [PubMed] [Google Scholar]
- 16.Vologodskii A.V., Crisona,N.J., Laurie,B., Pieranski,P., Katritch,V., Dubochet,J. and Stasiak,A. (1998) Sedimentation and electrophoretic migration of DNA knots and catenanes. J. Mol. Biol., 278, 1–3. [DOI] [PubMed] [Google Scholar]
- 17.Cantarella J., Kusner,R.B. and Sullivan,J.M. (1998) Tight knots deviate from linear relations. Nature, 392, 237–238. [Google Scholar]
- 18.Buck G. (1998) Four-thirds power law for knots and links. Nature, 392, 238–239. [Google Scholar]
- 19.Levene S.D. and Tsen,H. (1999) Analysis of DNA knots and catenanes by agarose-gel electrophoresis. In Bjornsti,M. and Osheroff,N. (eds), Methods in Molecular Biology: DNA Topoisomerase Protocols. Humana Press, Totowa, NJ, Vol. 1, pp. 75–86. [DOI] [PubMed]




