Abstract
The increasing interest in the evolution of human language has led several fields of research to focus on primate vocal communication. The ‘singing primates’, which produce elaborated and complex sequences of vocalizations, are of particular interest for this topic. Indris (Indri indri) are the only singing lemurs and emit songs whose most distinctive portions are “descending phrases” consisting of 2-5 units. We examined how the structure of the indris’ phrases varied with genetic relatedness among individuals. We tested whether the acoustic structure could provide conspecifics with information about individual identity and group membership. When analyzing phrase dissimilarity and genetic distance of both sexes, we found significant results for males but not for females. We found that similarity of male song-phrases correlates with kin in both time and frequency parameters, while, for females, this information is encoded only in the frequency of a single type. Song phrases have consistent individual-specific features, but we did not find any potential for advertising group membership. We emphasize the fact that genetic and social factors may play a role in the acoustic plasticity of female indris. Altogether, these findings open a new perspective for future research on the possibility of vocal production learning in these primates.
Introduction
Vocal signals often play a critical role in animal communication1. While many species make a conspicuous use of vocalizations, a limited number of taxa communicate using a sequence of vocal emissions, usually termed songs2. A song is a combination of different components that can be described hierarchically. Individual sounds are referred to as ‘units’, ‘elements’ or ‘notes’. One or more units that occur together can be called song ‘syllables’, and a sequence of one or more syllables is described as either a ‘phrase’ or ‘motif’. Since Darwin, scholars have been interested in understanding whether the complexity of singing is genetically determined, impacted by social experience, or the result of a learning process3, 4. The processes leading to song diversity have been widely investigated in birds. Endogenous and exogenous factors modulate the interplay between genetic characteristics, social experience, and learning5. For instance, early findings on song development in the zebra finches (Taeniogypta guttata) suggested that, while male birds develop their song during a sensitive period for vocal production learning6, song culture appears as a multi-generational phenotype, partially encoded in the genes of an isolated founding population. In this species, juvenile birds, that imitate isolated tutors, changed particular characteristics of the songs. These alterations can be accumulated over learning generations until a new natural song emerges7.
Although birds are the only animals in which vocal production learning has been rigorously associated with the modification of the cerebral connections, previous research showed that learning in vocal communication is not unique to them. Some terrestrial and marine mammals may possess the ability to learn the production of particular emissions8, and individuals of other species can learn the context in which to produce a particular call or how to modify their response to others’ vocalizations9. Communication using songs is widespread in different groups of birds and marine mammals, but is rare in primates10. A key question is to what extent primate vocalizations can be shaped by vocal production learning processes (hereafter, vocal learning) and social factors, and whether they may possess information useful for kin recognition. The current evidence is contradictory, with data suggesting both vocal learning and genetic relatedness as forces in shaping primate vocal signals. For instance, Marshall and colleagues11 suggested that genetic relatedness had a limited effect on the acoustic similarity in chimpanzee’s vocalizations (Pan troglodytes), since unrelated males showed a similar acoustic structure in their pant hoots. Further studies on chimpanzees consolidated the idea that vocal learning and convergence may play a role in the acoustic structure of food grunts shared within two groups12. The influence of social factors in shaping vocal signals has also been suggested for monkeys. Lemasson and colleagues found that social bonding better explained the acoustic similarity in the vocalizations of Campbell’s monkeys (Cercopithecus campbelli) when compared to genetic relatedness13. Although evidence seems to suggest that primate calls are not completely genetically determined, the question whether information about genetic relatedness, crucial for kin recognition, is retained in vocalization is still open. A study of mandrill’s vocalizations (Mandrillus sphinx)14 showed that the acoustic structure of contact calls was more similar between relatives than among unrelated individuals, suggesting that mandrill acoustic signals contain kin-specific information.
Because vocal learning has been typically associated with singing, studying the effects of genetics and social factors on the vocal output of primates that communicate using songs (i.e., tarsiers, gibbons, indris, titi monkeys15) is of great interest. Unfortunately, limited knowledge about the ontogenesis of primate singing behavior prevents a proper comparison with studies on birdsongs. However, previous research on gibbons reported that juveniles and young females produce immature vocalizations (Hylobates lar and H. agilis 16) and that the structure of the songs may reach a mature form at about six years old (Nomascus gabriellae 17). Moreover, the co-singing of mothers and daughters in Hylobates agilis has been interpreted as a possible form of tutoring to switch from an immature to a mature female great call18. On the other hand, concordance between song and genetic diversity across the crested gibbons19 suggests that genes may play a major role in shaping song structure. Additionally, both hybrid males and females showed intermediate song structure compared to the songs of the parental species (Hylobates lar x H. muelleri 20; Hylobates lar x H. pileatus 21). Interestingly, female songs tended to diverge more from their parents’ songs, while males’ appeared to resemble those of their father.
To further explore the processes shaping primate songs, we investigated the relationship between genetic distance and acoustic similarity in the indris. We also aimed to understand whether the acoustic structure of phrases differed when analyzed within closely (e.g.; father-offspring, mother-offspring) and distantly related indris in the population of Maromizaha.
The indri (Indri indri 22) is the only lemur that communicates through songs. The indris’ songs are long sequences of vocal units that are organized in phrases23, 24. They have the form of a chorus in which all the adults and the subadults of a group utter their contribution in a precise and coordinated manner25. Songs have various functions depending on the context in which are emitted and they are used for both inter and intra-group communication26, 27. Furthermore, songs are likely to provide information about the group composition and mediate the formation of new groups25, 28, 29. Because this species lives in familiar groups (Bonadonna, unpublished data) and the song has a rich repertoire of units25, 30, 31, the indris can be an excellent model to investigate the relationship between genetic relatedness and song similarity. Recent studies by Gamba and colleagues25 showed that the acoustic structure of phrases did not significantly change between age classes, suggesting that a limited variation may occur during ontogeny.
The inheritance of song characteristics has been inferred from the studies of hybrid gibbons. Geissmann21 studied the song of a male and a female hybrid (Hylobates pileatus x H. lar) finding that their songs differed markedly from the song characteristics of the parent species. However, studies on ground squirrels (Spermophilus suslicus) found a weak correlation between acoustic similarity and kinship, showing that other factors, such as the need for an individually distinctive acoustic structure, may play a critical role in vocal communication32–34.
In this study, we hypothesize that, if genetics strongly determines song characteristics, vocal learning may not play an important role in shaping the indris’ songs. We predicted that if song traits are mainly inherited, a high genetic distance will correspond to a reduced song similarity and that this reduction would be consistent within and between sexes. But it is also possible that emitters possess the potential for modifying their utterances and use songs to advertise their individuality and their belonging to a group35–37. In this second scenario, measures of genetic relatedness are not associated with song traits and we predicted that genetic distance and song similarity should not covary, but individuality and group membership would instead explain most of the acoustic variation.
Results
Extraction of the principal components
Four principal components accounted for 83.2% of the total variance of the temporal variables of the descending phrases of two units (DP2, see Supplementary Table S1), and five components accounted for 92.5% of the variance of frequency parameters (see Supplementary Table S2). We found six components for the temporal variables of the descending phrases consisting of three elements (DP3s) explaining 83% of the observed variance. We then found six components for the frequency variables of DP3s explaining 89.5% of the observed variance (see Supplementary Tables S3 and S4). The acoustic parameters showing the highest loadings on the principal components PC1 of each PCA were the first inter-onset interval (IOI1) and the total duration of unit 1 (Dur_unit1) for the temporal parameters of DP2. For frequency parameters of DP2, they were the average fundamental frequency of unit 2 (f0mean_unit2) and unit 1 (f0mean_unit1). For the temporal parameters of DP3, they were the second inter-onset interval (IOI2) and the duration of the second interval (Dur_int2). For the frequency parameters of DP3, they were the average fundamental frequency of unit 2 (f0mean_unit2), the frequency at the upper limit of the second quartiles of energy (Q50_unit2). A complete list of PCs and the loadings of the acoustic parameters are listed in the Supplementary Information (see Supplementary Tables S1–S4).
Acoustic similarity and genetic relatedness
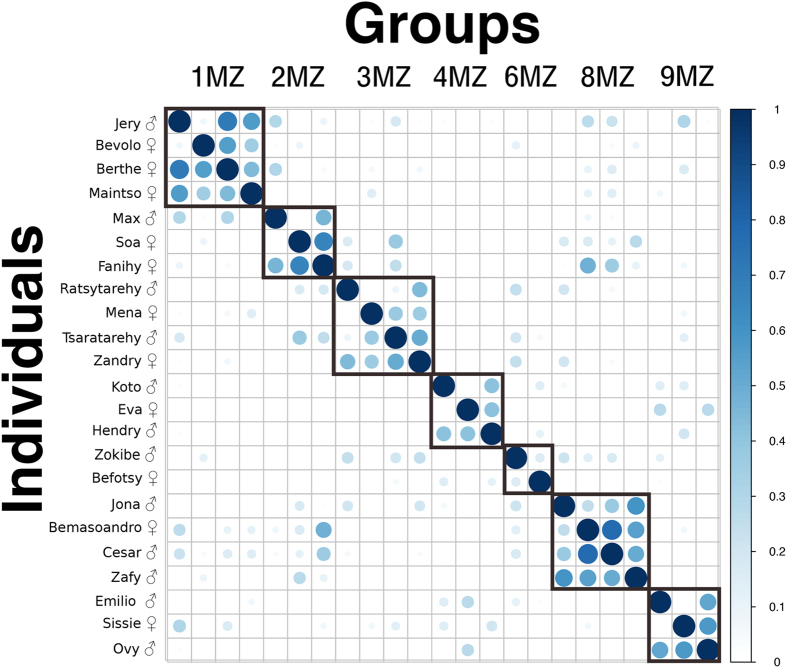
We found a variable degree of genetic variation within the groups of indris we sampled, confirming that each social group consisted of a male, an unrelated female, and their offspring (Fig. 1, Supplementary Table S5). When analyzing whether genetic distances had an effect on the temporal and frequency characteristics of DP2 at the population level, Mantel tests revealed a significant positive effect for temporal but not for frequency parameters (Table 1). For DP3, we did not find a significant effect either for temporal or frequency parameters (Table 1). Since male and female phrases are sexually dimorphic25, we replicated the Mantel tests separately for each sex. We found that combining the sexes together was underestimating the effect of genetic distance on male song dissimilarity, which showed a highly significant value for the temporal characteristics of DP2 (Table 1). We did not find any significant p-values for DP3 temporal and frequency features (Table 1). While there was a tendency for frequency parameters to be related to genetic relatedness in males, Mantel tests revealed a non-significant correlation between acoustics and genetics among females for DP2 and DP3 (Table 1).
Figure 1.
Plot of the trioml estimator75 showing the genetic relatedness among the individuals in the study groups of the indris of Maromizaha (see also Supplementary Table 3). Individual names and sexes are shown on the vertical axis; group composition is shown on the horizontal axis. Dot size and color refer to genetic relatedness: the darker and bigger the dot, the more genetically related are the individuals. The correlation plot was generated using the R package corrplot 82.
Table 1.
Results of the Mantel tests analyzing the correlation between acoustic similarity for the temporal (temp.) and frequency (freq.) parameters of descending phrases DP2 and DP3 and genetic relatedness.
| MALES | FEMALES | OVERALL | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| N | R | P-VALUE | N | R | P-VALUE | N | R | P-VALUE | ||
| DP2 | Temp. | 12 | 0.306 | 0.004 | 11 | 0.119 | 0.201 | 23 | 0.119 | 0.034 |
| Freq. | 0.058 | 0.309 | 0.077 | 0.309 | 0.072 | 0.136 | ||||
| DP3 | Temp. | 12 | 0.164 | 0.080 | 10 | 0.006 | 0.473 | 22 | 0.062 | 0.171 |
| Freq. | 0.172 | 0.055 | 0.015 | 0.234 | 0.094 | 0.074 | ||||
The genetic similarity of adults ranged between 0-0.30, 0.09-0.77 for parent-offspring, and 0.11-0.50 for siblings. When we compared the pairwise acoustic indices of the categories father-daughter, father-son, mother-daughter, and mother-son to those of unrelated adults-offspring pairs, we found significant correlations of fathers and sons for the temporal and frequency parameters of DP2 and DP3 (Table 2), showing that similarity was higher within phrases emitted by related than unrelated male indris. We have also found that similarity was higher within phrases emitted by mothers and daughters than unrelated females, but only for the frequency parameters of DP2.
Table 2.
Results of the Mantel tests analyzing the correlation between acoustic similarity for the temporal (temp.) and frequency (freq.) parameters of descending phrases DP2 and DP3 and kin information.
| SONS | DAUGHTERS | |||||||
|---|---|---|---|---|---|---|---|---|
| N | R | p | N | R | p | |||
| DP2 | Temp. | FATHERS | 7 | 0.397 | 0.010 | 7 | 0.033 | 0.410 |
| MOTHERS | 7 | 0.126 | 0.295 | 7 | 0.227 | 0.114 | ||
| Freq. | FATHERS | 7 | 0.281 | 0.019 | 7 | 0.026 | 0.533 | |
| MOTHERS | 7 | 0.152 | 0.191 | 7 | 0.311 | 0.038 | ||
| DP3 | Temp. | FATHERS | 7 | 0.509 | 0.010 | 5 | 0.221 | 0.200 |
| MOTHERS | 7 | −0.001 | 0.486 | 5 | 0.115 | 0.200 | ||
| Freq. | FATHERS | 7 | 0.342 | 0.010 | 5 | 0.059 | 0.500 | |
| MOTHERS | 7 | 0.183 | 0.171 | 5 | 0.001 | 0.500 | ||
Individual signature and group membership
We investigated whether the variation of the DPs was consistent with the individual ID and the group membership. Despite an overall within-sex structural similarity between the DPs recorded, we found a remarkable similarity at the individual level, which reflected in the high rates of correct classification of the permuted Discriminant Function Analysis (pDFA) (Table 3). DP2s and DP3s could be assigned to the individual with accuracy greater than chance using both temporal and frequency parameters (Table 3). These results were confirmed when we repeated the analyses separating sexes (Table 3).
Table 3.
Percentage of correctly assigned descending phrases DP2 and DP3 to the individuals, overall and by sex (N = 23 for DP2, of which N = 12 for males and N = 11 for females, N = 22 for DP3, of which N = 12 for males and N = 10 for females), and groups (group memb., N = 7) for the indris of Maromizaha.
| TRAINING | P-VALUE | TESTING | P-VALUE | ||||
|---|---|---|---|---|---|---|---|
| DP2 | INDIVIDUALITY | Temporal | Overall | 44.03 | 0.001 | 24.07 | 0.001 |
| Males | 51.64 | 0.024 | 29.23 | 0.001 | |||
| Females | 50.50 | 0.003 | 37.30 | 0.001 | |||
| Frequency | Overall | 46.62 | 0.001 | 19.59 | 0.001 | ||
| Males | 53.56 | 0.046 | 18.15 | 0.002 | |||
| Females | 56.39 | 0.001 | 32.13 | 0.001 | |||
| GROUP MEMB. | Temporal | 41.47 | 0.475 | 31.09 | 0.258 | ||
| Frequency | 45.13 | 0.380 | 29.98 | 0.225 | |||
| DP3 | INDIVIDUALITY | Temporal | Overall | 81.02 | 0.001 | 28.73 | 0.001 |
| Males | 91.83 | 0.009 | 35.92 | 0.001 | |||
| Females | 71.40 | 0.001 | 45.91 | 0.001 | |||
| Frequency | Overall | 73.50 | 0.001 | 27.79 | 0.001 | ||
| Males | 83.00 | 0.009 | 23.04 | 0.001 | |||
| Females | 70.47 | 0.001 | 44.60 | 0.001 | |||
| GROUP MEMB. | Temporal | 56.04 | 0.443 | 35.56 | 0.125 | ||
| Frequency | 56.89 | 0.325 | 33.72 | 0.269 |
In contrast, indris produced phrases that did not signal group membership accurately. In fact, pDFAs using the temporal and frequency parameters of DP2 and DP3 showed statistically significant p-values neither during the training, nor during the testing (Table 3).
Discussion
The results presented here come from the first intra-population analysis of indris comparing the acoustic characteristics of song phrases and genetic relatedness. Using this approach, we demonstrated that the structure of males’ phrases transmits information about relatedness more consistently than females’ ones, and that song phrases possess the potential to provide conspecifics with a cue to individual identity of the emitter in both sexes. Male song-phrases DP2 transmit information about relatedness in the form of time parameters, but this information is not encoded in female calls. We found a tendency for encoding of relatedness information in the frequency structure of male song-phrases DP3. We also found that similarity of both temporal and frequency parameters in male phrases DP2 and DP3 correlate significantly correlate with genetic distance in the ‘father-son’ category. A result that for female phrases is limited only to the frequency parameters of DP2. We did not find significant correlations across sexes. Overall, we can confirm our prediction that the indris’ song phrases contain information about genetic relatedness as it has been found in other primate species (e.g.: Mandrillus sphinx 14). It is interesting to notice that the encoding of this information is strong in the temporal characteristics of the descending phrases when analyzed using genetic similarity indices, but significantly stronger in both temporal and frequency parameters of DP2 and DP3 when compared between related and unrelated males. The correlation between temporal patterns and genetic relatedness is especially interesting in the light of those studies that investigated how hybrids differed from their parental species in the acoustic characteristics of their utterances38, 39. In gibbons and lemurs, pulse structure and rhythmic characteristics have a particular relevance in the discrimination between hybrids and parental species21, 38. Our results are in agreement with analyses of intra-population variation on ground squirrels (Urocitellus beldingi) showing that individuals produce calls more similar to their relatives than to unrelated individuals40. We extended these findings showing that, in indri, relatedness is also encoded by frequency parameters of both DP2 and DP3 in males, and DP2 in females.
While the vast amount of data about primate kin recognition is devoted to the highly complex social groups of African monkeys41, 42, Kessler and colleagues43 demonstrated that the advertisement calls for the grey mouse lemur (Microcebus murinus) possess patrilineal signatures that mediate paternal kin recognition. Male vocal signatures have been indicated as an important mechanism for inbreeding avoidance43. This mechanism may have sense in the light of the long-term pair bonding of indris44, but may also play a role in a scenario in which extra-pair copulation can potentially contribute to increasing levels of genetic diversity within a population45. Moreover, even though physical fights between individuals of neighboring groups are rare in indris46, they involve primarily males and always include choral vocal displays47. Thus, song similarity between related males may mediate kin recognition and de-escalate aggressions. From a different perspective, the descending phrases are paradigmatic examples of vocal emissions with remarkable frequency modulation23. It is therefore possible that those phrases are acoustically more flexible and less genetically-determined. This interpretation can explain only part of our results, but it is interesting to notice that in the study by Lemasson and colleagues48 the genetic similarity between Campbell’s monkey females (Cercopithecus campbelli campbelli) did not explain the acoustic similarities of their contact calls. A higher degree of acoustic plasticity within females is supported by previous research on baboons (Papio cynocephalus ursinus 49), Campbell’s monkeys (Cercopithecus campbelli campbelli 50), and Japanese macaques (Macaca fuscata 51). Whether social factors may play a role in the acoustic plasticity of female indris need further investigation, but there is evidence that the contribution of the temporal structure to the song is less genetically determined in females than in males. Previous studies have found that females change the duration of their song to partially overlap with males’ singing29. Females may also adjust duration according to the number of males in their social group, while males tend to avoid overlapping each other25, 26.
The lack of knowledge about dispersal patterns in the indris prevents further speculation regarding the relatively higher genetic signature in male calls. In socially monogamous species there is a tendency towards female-biased dispersal52 that data from the field do not support for the indris. Data collected over 14 years and in different forests suggest that both male and female indris disperse (Giacoma, unpublished data), although dispersal frequency and distance are currently not available.
Individual variation in the vocal signals is a precondition for individual recognition, which can result in both affiliative and aggressive situations53. Previous research has demonstrated that sex differences may override individual differences25, but the results of the present study complement those findings showing that strong individuality is nevertheless encoded in the indris’ phrases. The presence of an individual signature is confirmed by both the training and testing phase of the permuted Discriminant analysis and it is valid for both temporal and frequency parameters. Our findings are in agreement with previous studies that have reported a strong individual signature in the acoustic signals of social mammals (e.g.; yellow-bellied marmots54; Speckled ground squirrels32, 33). The individuality encoding was also found in the Cao Vit gibbon male phrases55 and in the Bornean gibbon female great calls56, two species that emit songs like the indris. Our analyses suggest that individuality is encoded in both males and females, tracing an interesting path for future research in other singing primates. While most of the previous research on the indris’ song suggests that the temporal parameters play a major role in the sex-specific encoding of the vocal emissions25, 29, we found that also frequency parameters have potential for individual recognition. This evidence expands the recent findings of Gamba and colleagues25 about a sex-specific difference in pitch patterns during the song. In general, both time and frequency variables appeared to play a role in encoding individuality as they probably do for sex-specificity.
Behavioral observations support our results that singing in indris may facilitate the exchange of identity information in the context of distant communication26, 46. This idea is in agreement with previous studies on other primates, where long distance vocalizations were found to be useful for identifying individuals55 and estimating male fighting ability57. Although we do not have data in support of the hypothesis that song may be useful to estimate individuals’ fighting ability, our behavioral observations suggest that they indris can vocally discriminate individuals. Two lines of evidence support this idea. Torti and colleagues26 showed that songs elicited regrouping of particular individuals within a group. Bonadonna and colleagues45 observed that a female that has just been involved in extra-pair copulation did not join the song of her group mates, which were singing at a distance. We found support for the hypothesis that the use of song phrases to broadcast individuality may be essential during pair formation at distance26 and for the regulation of territorial spacing, where other communicative signals may be ineffective58.
When we analyzed the dissimilarity of phrases emitted by members of a social group, we found that pDFA could assign neither DP2 nor DP3 to the group with a classification rate higher that those predicted by chance. This result is in contrast to the findings of Knörnschild and colleagues on greater sac-winged bats (Saccopteryx bilineata 59) and Vester and colleagues on pilot whales (Globicephala melas 60), where differences of vocalizations within social groups were significantly lower than intergroup differences. The work on the greater sac-winged bats is particularly interesting because the authors found that pups modified their emissions during ontogeny and learned their songs through vocal imitation of their harem males, independently of their genetic relatedness59. Studies on apes and monkeys suggested a consistent degree of acoustic plasticity in nonhuman primate calls, which was observed in particular after social changes61, 62 or during vocal interactions63.
A previous investigation on the rhythmic structure of the indris’ song showed that the structure of the descending phrases did not change significantly during ontogeny25. Combined with our finding that group membership cues do not appear to be encoded in the phrase structure, these two elements seem to demonstrate that the indris’ song has limited flexibility when compared to other animals’ utterances and that learning may play a secondary role in song acquisition. Our results disagree with previous findings by Baker-Medard and colleagues27 that found significant differences in chorus songs between three groups in the Analamazaotra Reserve. There can be multiple reasons for these different findings. First, the number of songs and the number of groups considered by Baker-Medard and colleagues27 is smaller than those we used in the present study, possibly reducing the variation of the acoustic measurements and leading to an intergroup dissimilarity that we were not able to find. Second, it is possible that the fact that Baker-Medard and colleagues27 considered all the units in the songs for their analysis contributed some essential trait for group discrimination that is lacking when only the DPs are considered. However, we think that considering the most common DP types occurring in the songs should allow a proper evaluation of the acoustic variability exhibited by this species. As for the individual discrimination, we lack any evidence that the indris make similar discriminations regarding groups. However, according to our data, the potential for group recognition appears weaker than that for individual recognition. We can hypothesize that the song may play a role in the numerical assessment of group size, as McComb and colleagues64 have demonstrated on the African lions (Panthera leo). Future playbacks of the indris’ songs may improve our knowledge about the amount of information encoded by the songs.
Overall, our results confirm that vocal signals can be shaped by both genetic factors and social experience in the indris. Even in primates that emit songs with complex temporal and frequency patterns, phrases contain information about genetic relatedness and subtle variation in the acoustic structure may play a role in providing conspecifics with cues for kin identification and individual discrimination.
Methods
Observations and recordings
We studied seven groups living in the Maromizaha Forest (18°56′49′′S, 48°27′53′′E). We collected data in the field from 2011 to 2016, for a total of 24 months. We observed a social group per day, approximately from 6 AM to 1 PM. We identified the indris individually thanks to natural marks. Group composition ranged from 2 to 4 indris (Fig. 2; details are provided in Table 4). Recordings were made using solid-state recorders (SoundDevices 702, Olympus S100 and LS05, and Tascam DR-100, DR-40, and DR-05) equipped with Sennheiser (ME 66 and ME 67) or AKG (CK 98) shotgun microphones. The microphone signal was recorded at a sampling rate of 44.1 kHz, 16 bit. We recorded all the songs at a distance comprised between 2 and 20 m, keeping visual contact with the vocalizing animals (Fig. 3). We made all efforts to orientate the microphone toward the focal uttering individuals. All recordings were carried out without the use of playback stimuli, and nothing was done to modify the behavior of the indris. When in the field, we had one observer per individual in a group. Using the focal animal sampling technique65, we were able to attribute each vocalization to a signaler. From the individual song contributions, we extracted 1066 descending phrases consisting of two units (hereafter, DP2), and 1259 descending phrases consisting of three units (DP3; Fig. 4). We focused on DP2s and DP3s because they are the most common phrase types in the indris’ song25. The sampling included phrases emitted by seven males (of which six sired at least one offspring), seven siring females, 9 offspring (five males and four females, Table 4). We included in our analyses only those individuals contributing at least four DPs. Our final sample included 23 individuals for the DP2s and 22 individuals for the DP3s.
Figure 2.
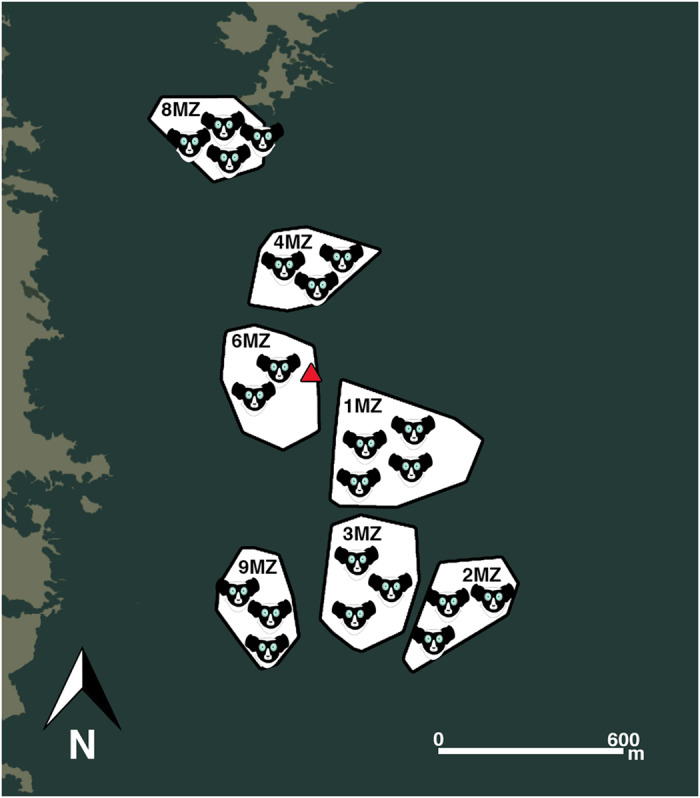
Map of the study area in the Maromizaha Forest. Minimum Convex Polygons (MCP) generated with ArcGIS 9.1 (ESRI) correspond to the 2016 home range of the study groups. Group ID is reported onto each MCP, and the indris’ face icons indicate the number of animals per group. The red shape indicates the geographical location of the Maromizaha Research Center (18°58′34.06″S 48°27′53.88″E). Drawings by Dr. Valeria Torti.
Table 4.
Summary of group, ID, sex and year of birth of offspring.
| Group | ID | Sex/YOB |
|---|---|---|
| 1MZ | Jery | ♂ |
| Bevolo | ♀ | |
| Maintso | ♀ 2010 | |
| Berthe | ♀ 2012 | |
| 2MZ | Max | ♂ |
| Soa | ♀ | |
| Fanihy | ♀ 2012 | |
| 3MZ | Ratsytarehy | ♂ |
| Mena | ♀ | |
| Tsaratarehy | ♂ before 2009 | |
| Zandry | ♀ 2010 | |
| 4MZ | Koto | ♂ |
| Eva | ♀ | |
| Hendry | ♂ before 2009 | |
| 6MZ | Zokibe | ♂ |
| Befotsy | ♀ | |
| 8MZ | Jona | ♂ |
| Bemasoandro | ♀ | |
| Cesar | ♂ before 2009 | |
| Zafy | ♂ 2012 | |
| 9MZ | Emilio | ♂ |
| Sissie | ♀ | |
| Ovy | ♂ 2013 |
For each group, the reproductive pair is listed first. The year of birth is not reported for the reproductive pair.
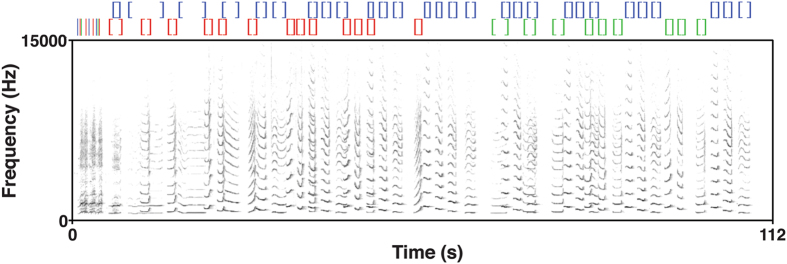
Figure 3.
Spectrogram of the indris’ song generated using PRAAT. In this song recorded in the Maromizaha Forest, a reproductive pair is singing with a female offspring (Group 2MZ). At the top of the spectrogram, the color brackets indicate the start (“[”) and the end (“]”) of each male’s units (in blue), reproductive female’s (in red), and female offspring’s (in green).
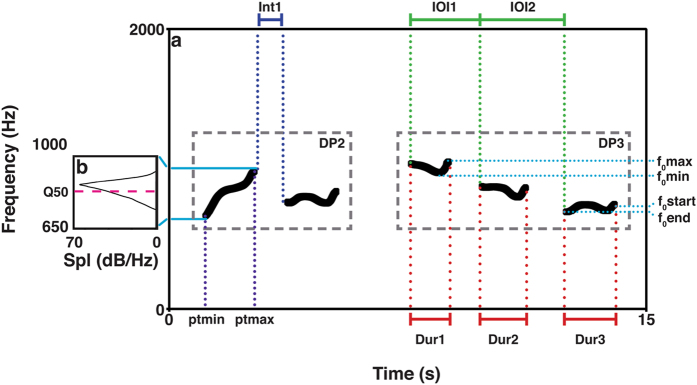
Figure 4.
Schematic representation of the spectrogram of the isolated fundamental frequency of two descending phrases, a DP2 and a DP3 (a). The sound spectrogram displays time (seconds) on the x-axis, frequency (Hz) on the vertical axis. We describe acoustic parameter collection of unit duration (in red); intervals (in blue); inter-onset intervals (in green); percentage of time to the minimum (ptmin) and maximum of pitch (ptmax, in purple); maximum and minimum pitch (f0max and f0min), fundamental frequency at the beginning and at the end of a unit (f0start, f0end, in light blue). In the spectrum (b), the fuchsia dotted line marks the frequency corresponding to the upper limit of the second quartile of energy in the spectrum (Q50). The sound spectrum displays sound pressure level (Spl/dB) on the x-axis, frequency on the vertical axis.
Genetic analyses
The genetic analysis confirmed the identification of the reproductive pairs we independently derived from our behavioral observations25, 26. We collected fecal samples from 23 individuals and we stored all the samples in RNAlater® Ambion66 at room temperature in the field and at 4 °C in the laboratory.
DNA extraction
We extracted genomic DNA from feces using the QIAamp DNA® Stool Mini Kit (Qiagen®, Hilden, Germany) with the following changes from the manufacturer’s protocol (QIAamp DNA Stoll Handbook 04/2010). We used 300 mg stool; we added 35 μl of proteinase K and incubated at 70° Celsius for 30 minutes during the purification phase. We applied 75 μl Buffer AE on the QIAamp membrane for the first DNA elution and incubated the spin column with Buffer AE at room temperature for 15 minutes.
For the samples collected in 2014, we used the automated robotic workstation QIAcube HT supported by the software QIAxtractor 4.17.1 (Qiagen®) to conduct DNA purification. We set the protocol for QXT Liquid DNA V1. First, we bathed the 2.0 mL tubes containing 300 mg of smashed feces and 1.6 mL of Buffer ASL at 70 °C for at least 5 minutes. After that, tubes were centrifuged at 13000 RPM for 10 minutes. We transferred 200 μl of supernatant to separate wells of the QIAextractor lysis plate and we started the run. At the end of the process, we obtained 70 µl of DNA elution for each sample. We stored the extracted DNA at 4 °C for immediate use.
DNA genotyping
We selected a set of 6 microsatellite marker loci identified as potentially variable in indri that provided good quality amplification products for multiplex PCRs67 (Table 5). A fluorescent dye (FAM, HEX) labeled the 5′ end forward primer of each locus to analyze simultaneously loci of similar allelic size. PCR amplification was carried out in 10 µL reaction volume containing: 5 µL Multiplex PCR Master Mix (Qiagen®), 0.1 µm of each primer, 2 µL DNA template, 2 µL RNase-free water. We set the cycle conditions as follows: a pre-incubation step at 95 °C for 15 min; 50 cycles with denaturation at 94 °C for 30 s, annealing at 54 °C or 60 °C (depending on the locus, see Table 5) for 90 s. The first extension phase was at 72 °C for 60 s; the final extension phase at 60 °C for 30 min.
Table 5.
Microsatellite loci used in this study, with respective primers, and annealing temperatures. The number of PCR cycles is 50.
| Locus | Forward primer | Reverse Primer | Repeat motif | Annealing temp. (°C) | Size range (bp) |
|---|---|---|---|---|---|
| 67HDZ25 | GGACCCTAATTCAAATATCACCTC | GGCATTTCTACTCCAGGTTGG | (CA)16 | 54 | 218-253 |
| 67HDZ62 | AGCCCTTTCTCTCAATGCC | CCTTCTTTGTTATCTTTCTGCATC | (GT)21 | 54 | 203-217 |
| 67HDZ18 | GGACTGGTAGATTTCTGGGTTTAG | CAGCCACTCCAATGCAAAG | (CA)7C(CA)15 | 60 | 164-190 |
| 67HDZ55 | TCAGGAGTTGGGACCAGGG | ATGAAGGGATGGAGGTGGG | (GT)18 | 60 | 312-334 |
| 67HDZ180 | TCCCCTCCTCAGTCATTTCTC | CGTGAAGCTCGTGTGTATGG | (CA)17 | 60 | 113-136 |
| 67HDZ39 | CAGAGCCAGGGTTCAAATTC | TTGTCTTTTCTGCCACTGTAGG | (CA)11 | 60 | 148-162 |
We estimated the allele size by electrophoresis using a 48-capillary ABI 3730 DNA Analyzer (Applied Biosystems). We mixed 1 µL of PCR product with 6.85 µL HiDi formamide (Applied Biosystems) and 0.15 µL Genescan 500-ROX size standard (Applied Biosystems). We conducted automated allele calling using the software GENEMAPPER 3.7 (Applied Biosystems). We then confirmed by eye all the allele sizes and checked for consistency across replicate PCRs of the same sample or from the same individual for a certain locus (minimum three replicates for heterozygotes and five replicates for homozygotes).
Relatedness analysis
We estimated relatedness among individuals using the R package related 68. First, we compared seven different relatedness estimators commonly used in the literature, five moment estimators69–73 and two likelihood-based estimators, the dyadic likelihood estimator – dyadml 74 and the triadic likelihood estimator – trioml 75. Using the allele frequencies observed in our dataset, we simulated datasets of 100 pairs for four known relatedness categories (parent–offspring, full-sibling, half-sibling, and unrelated). We chose the trioml 75 estimator to calculate relatedness for all possible dyadic combinations because it showed the highest consistency and obtained a matrix in which the more positive the index, the more two individuals are genetically related. Since indri lives in family group28, 47, the historical record of group composition since 2009 allowed us to infer parental relationships among individuals based on behavioral observations, especially between mother and offspring. We were able to assign a social father to each of the offspring included in the study. To define parental information for the comparison of acoustic distances, we run paternity analyses including as potential fathers all the adult males sampled (Bonadonna, unpublished data) and could assign paternity for nine offspring (out of 10).
Acoustic and statistical analyses
Because the singers’ phrases could overlap each in the temporal and frequency domain, we first extracted the fundamental frequency using a manual procedure and then obtained the pitch contour using a semi-automatic process in Praat76. We then added 0.5 s of silence at the beginning and the end of each phrase. Because each unit within a phrase went through the same set of measurements, we collected a minimum of 10 measurements in the temporal domain and a minimum of 20 measurements of pitch variability for each DP. The complete list of variables we measured is in Supplementary Table S6, while some parameters are presented in Fig. 4. Further details about the methodology used can be found in Gamba and colleagues25.
We used principal components analysis (Factor analysis in IBM SPSS 24.0.0.1) to reduce the data to uncorrelated principal components (PCs) using separately temporal and frequency measurements of DP2s and DP3s. We obtained four PCs exceeding eigenvalue 1 for the temporal measurements and five for frequency variables of DP2s. We obtained six PCs exceeding eigenvalue 1 for each of the two sets of variables of DP3s. To understand whether genetic relatedness could explain acoustic similarities, we transformed the PCs obtained for each of these sets in a Euclidean distance matrix (function dist in R 3.2.3) and then calculated the average individual means. We then run the Mantel tests (9999 randomizations77, 78) on the average individual means against the matrix of genetic indices (package vegan in R79). All matrix indices were normalized to have a value between 0 and 1 before entering the analyses.
We used the Mantel test to evaluate whether the acoustic distance differed among individuals paired by categories (‘father-daughter’; ‘mother-daughter’, ‘father-son’, ‘mother-son’). We assessed the correlation between the acoustic similarity matrix and a binary matrix indicating the category (e.g.; ‘father-daughter’; refs 77 and 80) for each pair of song phrases. A significant correlation would indicate a difference in the similarity of phrases given by one of the pairs mentioned above (e.g.; father-daughter) when compared with phrases emitted by unrelated reproductive adults (e.g.; other ‘fathers’ in the sample) and offspring (e.g.; ‘daughters’ of other pairs).
To understand whether we could identify a potential for individual recognition or group membership we submitted the component scores to permutated linear discriminant function analysis81 in R (using a custom script by Roger Mundry). When testing for individual differences, we used the individual as test factor and the song from which the DPs were extracted as a control factor. We also ran the analyses split by sex. When testing for group membership, we used the group as test factor and the individual identity as control factor. We split all the analyses into two phases, a training phase and a testing phase (R. Mundry, personal communication) for which we collected the correct classification rate and the p-value.
Data availability
Data and programs used for the analyses presented in the paper are available to the Editorial board members and the referees upon request or already included in the Supporting Information.
Electronic supplementary material
Acknowledgements
This research was supported by Università degli Studi di Torino and the African, Caribbean, and Pacific (ACP) Science and Technology Programme of the ACP Group of States, with the financial assistance of the European Union, through the Projects BIRD (Biodiversity Integration and Rural Development; No. FED/2009/217077) and SCORE (Supporting Cooperation for Research and Education; Contract No. ACP RPR 118 # 36) and by grants from the Parco Natura Viva—Centro Tutela Specie Minacciate. We are grateful to GERP (Groupe d’Etudes et des Recherche sur les Primates) and Dr Jonah Ratsimbazafy. We thank Dr Cesare Avesani Zaborra and Dr Caterina Spiezio for helping us with the organization of the field station in Maromizaha. We are grateful to the researchers and the international guides, to Lanto and Mamatin, for their help and logistical support. We also thank San Diego Zoo Global, LDVI and Dr Chia L. Tan. The contents of this document are the sole responsibility of the authors and can under no circumstances be regarded as reflecting the position of the European Union. We have received permits for this research, each year, from “Direction des Eaux et Forêts” and “Madagascar National Parks” (formerly ANGAP) [(2004 (N° 190/MINENV.EF/SG/DGEF/DPB/SCBLF/RECH) 2005 (N° 197/MINENV.EF/SG/DGEF/DPB/SCBLF/RECH), 2006 (N° 172/06/MINENV.EF/SG/DGEF/DPB/SCBLF), 2007 (N° 0220/07/MINE NV.EF/SG/DGEF/DPSAP/SSE), 2008 (N° 258/08/MEFT/SG/DG EF/DSAP/SSE), 2009 (N° 243/09/MEF/SG/DGF/DCB.SAP/SLRSE), 2010 (N° 118/10/MEF/SG/DGF/DCB.SAP/SCBSE; N° 293/10/MEF/SG/DGF/DCB.SAP/SCB), 2011 (N° 274/11/MEF/SG/D GF/DCB.SAP/SCB), 2012 (N°245/12/MEF/SG/DGF/DCB.SAP/SCB), 2014 (N°066/14/MEF/SG/DGF/DCB.SAP/SCB)]. Data collection did not require a permit for 2013 because it has been performed by Malagasy citizens only.
Author Contributions
V.T., G.B., and C.G. contributed to study design, analyzed the data, performed literature review, helped to interpret the results, and helped to draft the manuscript. C.D.G., D.V., L.P., and O.F. analyzed the data, performed literature review, helped to interpret the results and to draft the manuscript. R.M.R. oversaw field studies and data collection, performed data collection in 2013 and helped collate the data. M.G. contributed to study design, collated and analyzed the data, interpreted results, and drafted the manuscript. All authors contributed to writing the final manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Valeria Torti and Giovanna Bonadonna contributed equally to this work.
Marco Gamba and Cristina Giacoma jointly supervised this work.
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-10656-9
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Bradbury, J. W. and Vehrencamp, S. L. Principles of animal communication. (Sinauer Associates, 1998).
- 2.Clarke E, Reichard UH, Zuberbühler K. The Syntax and Meaning of Wild Gibbon Songs. PLoS ONE. 2006;1(1) doi: 10.1371/journal.pone.0000073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marler P. Three models of song learning: Evidence from behavior. J. Neurobiol. 1997;33:501–516. doi: 10.1002/(SICI)1097-4695(19971105)33:5<501::AID-NEU2>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 4.Warren WC, et al. The genome of a songbird. Nature. 2010;464:757–762. doi: 10.1038/nature08819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marler P. Innateness and the instinct to learn. An. Acad. Bras. Cienc. 2004;76:189–200. doi: 10.1590/S0001-37652004000200002. [DOI] [PubMed] [Google Scholar]
- 6.Immelmann, K. Song development in the zebra finch and other estrildid finches in Bird vocalizations (ed. Hinde, R. A.) 64–74 (Cambridge University Press,1969).
- 7.Fehér O, Wang H, Saar S, Mitra PP, Tchernichovski O. De novo establishment of wild-type song culture in the zebra finch. Nature. 2009;459:564–568. doi: 10.1038/nature07994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tyack PL. Convergence of calls as animals form social bonds, active compensation for noisy communication channels, and the evolution of vocal learning in mammals. J. Comp. Psychol. 2008;122(3):319–331. doi: 10.1037/a0013087. [DOI] [PubMed] [Google Scholar]
- 9.Nowicki S, Searcy WA. The evolution of vocal learning. Curr. Opin. Neurobiol. 2014;28:48–53. doi: 10.1016/j.conb.2014.06.007. [DOI] [PubMed] [Google Scholar]
- 10.Haimoff EH. The organization of song in the agile gibbon (Hylobates agilis) Folia Primatol. 1984;42:42–61. doi: 10.1159/000156143. [DOI] [Google Scholar]
- 11.Marshall AJ, Wrangham RW, Arcadi AC. Does learning affect the structure of vocalizations in chimpanzees? Anim. Behav. 1999;58:825–830. doi: 10.1006/anbe.1999.1219. [DOI] [PubMed] [Google Scholar]
- 12.Slocombe K, Watson SK, Townsend SW. Vocal Learning in the functionally referential food grunts of chimpanzees. Curr. Biol. 2015;25(4):495–499. doi: 10.1016/j.cub.2014.12.032. [DOI] [PubMed] [Google Scholar]
- 13.Lemasson A, Ouattara K, Petit EJ, Zuberbühler K. Social learning of vocal structure in a nonhuman primate? BMC Evol. Biol. 2011;11 doi: 10.1186/1471-2148-11-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levréro, F. et al. Social shaping of voices does not impair phenotype matching of kinship in mandrills. Nat. Commun. 6, 7609, doi:1038/ncomms8609 (2015). [DOI] [PubMed]
- 15.Geissmann, T. Gibbon songs and human music from an evolutionary perspective in The origins of music (ed. Wallin, N., Merker, B. and Brown, S.) 103–123 (MIT Press, 2000).
- 16.Koda H, et al. Immature male gibbons produce female-specific songs. Primates. 2013;55:13–7. doi: 10.1007/s10329-013-0390-2. [DOI] [PubMed] [Google Scholar]
- 17.Merker B, Cox C. Development of the female great call in Hylobates gabriellae: A case study. Folia Primatol. 1999;70:97–106. doi: 10.1159/000021680. [DOI] [PubMed] [Google Scholar]
- 18.Koda H, Lemasson A, Oyakawa C, Rizaldi D. Possible role of mother-daughter vocal interactions on the development of species-specific song in gibbons. PLoS ONE. 2013;8(8) doi: 10.1371/journal.pone.0071432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thin, V., Hallam, C., Roos, C., Hammerschmidt, K. Concordance between vocal and genetic diversity in crested gibbons. BMC Evol. Biol. 11–36 (2011). [DOI] [PMC free article] [PubMed]
- 20.Tenaza R. Songs of hybrid gibbons (Hylobates lar × H. muelleri) Am. J. Primatol. 1985;8(3):249–253. doi: 10.1002/ajp.1350080307. [DOI] [PubMed] [Google Scholar]
- 21.Geissmann T. Inheritance of song parameters in the gibbon song, analyzed in 2 hybrid gibbons (Hylobates pileatus x Hylobates lar) Folia Primatol. 1984;42:216–235. doi: 10.1159/000156165. [DOI] [Google Scholar]
- 22.Gmelin, J. F. Caroli a Linné systema naturae per regna tria naturae, secundum classes, ordines, genera, species, cum characteribus, differentiis, synonymis, locis. Tomus I. Editio decima tertia, aucta, reformata. Lipsiae. 1–500 (1788).
- 23.Gamba M, Favaro L, Torti V, Sorrentino V, Giacoma C. Vocal tract Flexibility and variation in the vocal output in wild indris. Bioacoustics. 2011;20(3):251–265. doi: 10.1080/09524622.2011.9753649. [DOI] [Google Scholar]
- 24.Thalmann U, Geissman T, Simona A, Mutschler T. The Indris of Anjanaharibe-Sud, Northeastern Madagascar. Int. J. Primatol. 1993;14:357–381. doi: 10.1007/BF02192772. [DOI] [Google Scholar]
- 25.Gamba M, et al. The Indris Have Got Rhythm! Timing and Pitch Variation of a Primate Song Examined between Sexes and Age Classes. Front. Neurosci. 2016;10 doi: 10.3389/fnins.2016.00249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Torti V, Gamba M, Rabemananjara ZH, Giacoma C. The songs of the indris (Mammalia: Primates: Indridae): contextual variation in the long-distance calls of a lemur. Ital. J. Zool. 2013;80(4):596–607. doi: 10.1080/11250003.2013.845261. [DOI] [Google Scholar]
- 27.Baker-Médard, M. S. A., Baker, M. C. and Logue, D. M. Chorus Song of the Indri (Indri indri: Primates, Lemuridae): Group Differences and Analysis of Within-group Vocal Interactions. Int. J. Comp. Psychol. 26, 241–255 (2013).
- 28.Pollock JI. The song of the Indris (Indri indri; Primates: Lemuroidea): natural history, form and function. Int. J. Primatol. 1986;7:225–267. doi: 10.1007/BF02736391. [DOI] [Google Scholar]
- 29.Giacoma C, Sorrentino V, Rabarivola C, Gamba M. Sex differences in the song of Indri indri. Int. J. Primatol. 2010;31:539–551. doi: 10.1007/s10764-010-9412-8. [DOI] [Google Scholar]
- 30.Maretti G, Sorrentino V, Finomana A, Gamba M, Giacoma C. Not just a pretty song: An overview of the vocal repertoire of Indri indri. JASs. 2010;88:151–165. [PubMed] [Google Scholar]
- 31.Sorrentino, V., Gamba, M., Giacoma, C. A quantitative description of the vocal types emitted in the indri’s song. Leaping ahead: advances in prosimian biology, 315–322 (2013).
- 32.Matrosova VA, Volodin IA, Volodina EV, Vasilieva NA, Kochetkova AA. Between-year stability of individual alarm calls in the yellow ground squirrel Spermophilus fulvus. J. Mammal. 2010;91:620–627. doi: 10.1644/09-MAMM-A-143.1. [DOI] [Google Scholar]
- 33.Matrosova VA, Blumstein DT, Volodin IA, Volodina EV. The potential to encode sex, age, and individual identity in the alarm calls of three species of Marmotinae. Naturwissenschaften. 2011;98:181–192. doi: 10.1007/s00114-010-0757-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matrosova VA, et al. Genetic and alarm call diversity across scattered populations of speckled ground squirrels (Spermophilus suslicus) Mamm. Biol. 2016;81:255–265. doi: 10.1016/j.mambio.2016.01.001. [DOI] [Google Scholar]
- 35.Volodin IA, Lapshina EN, Volodina EV, Frey R, Soldatova NV. Nasal and oral calls in juvenile goitred gazelles (Gazella subgutturosa) and their potential to encode sex and identity. Ethology. 2011;117:294–308. doi: 10.1111/j.1439-0310.2011.01874.x. [DOI] [Google Scholar]
- 36.Volodin IA, Volodina EV, Lapshina EN, Efremova KO, Soldatova NV. Anim. Cogn. 2014;17 doi: 10.1007/s10071-013-0666-3. [DOI] [PubMed] [Google Scholar]
- 37.Knörnschild M, Jung K, Nagy M, Metz M, Kalko EKV. Bat echolocation calls facilitate social communication. Proc. R. Soc. B. 2012;279(1748):4827–4835. doi: 10.1098/rspb.2012.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brockelman Y. R. & Gittins S. P. Natural hybridization in the Hylobates lar species group: Implications for speciation in gibbons in The Lesser Apes: Evolutionary and Behavioural Biology, (ed. Preuschoft, H., Chivers, D. J., Brockelman, W. Y. & Creel N.) 498–532, (Edinburgh University Press, 1984).
- 39.Macedonia JM, Taylor LL. Subspecific divergence in a loud call of the ruffed lemur (Varecia variegata) Am. J. Primatol. 1985;9:295–304. doi: 10.1002/ajp.1350090406. [DOI] [PubMed] [Google Scholar]
- 40.Kastely, C. R. Belding’s Ground Squirrels (Urocitellus beldingi) are more acoustically similar to Relatives than to Unrelated Individuals. http//nature.berkeley.edu/classes/es196/projects/2012final/KastelyC_2012.pdf (2012).
- 41.Cheney DL, Seyfarth RM. Recognition of other individuals’ social relationships by female baboons. Anim. Behav. 1999;58:67–75. doi: 10.1006/anbe.1999.1131. [DOI] [PubMed] [Google Scholar]
- 42.Rendall, D. Recognizing kin: mechanisms, media, minds, modules, and muddles in Kinship and Behavior in Primates (ed. Chapais, B., Berman, C. M.) 295–316 (Oxford University Press, 2004).
- 43.Kessler, S. E., Scheumann, M., Nash, L. T. and Zimmermann, E. Paternal kin recognition in the high frequency/ultrasonic range in a solitary foraging mammal. BMC Ecol. 12(26) (2012). [DOI] [PMC free article] [PubMed]
- 44.Pollock, J. I. The ecology and sociology of feeding in Indri indri In Primate Ecology. Feeding and Ranging Behaviour of Lemurs, Monkeys and Apes (ed. Clutton-Brock, T. H.) 37–69 (London: Academic Press, 1977).
- 45.Bonadonna G, et al. Behavioral correlates of extra-pair copulation in Indri indri. Primates. 2014;55(1):119–123. doi: 10.1007/s10329-013-0376-0. [DOI] [PubMed] [Google Scholar]
- 46.Bonadonna, G. et al. Territory exclusivity and intergroup encounters in Indri indri upon methodological tuning. Eur. Zool. J. 84(1) (2017).
- 47.Pollock. J. I. The social behaviour and ecology of Indri indri. Ph.D. dissertation, University of London (1975).
- 48.Lemasson A, Hausberger M. Acoustic variability and social significance of calls in female Campbell’s monkeys (Cercopithecus campbelli campbelli) J. Acoust. Soc. Am. 2011;129:3341–3352. doi: 10.1121/1.3569704. [DOI] [PubMed] [Google Scholar]
- 49.Lemasson A, Ouattara K, Petit EJ, Züberbühler K. Social learning of vocal structure in a nonhuman primate? BMC Evol. Biol. 2011;11 doi: 10.1186/1471-2148-11-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lemasson A, Hausberger M, Züberbühler K. Socially meaningful vocal plasticity in adult Campbell’s monkeys (Cercopithecus campbelli) J. Comp. Psychol. 2005;119:220–229. doi: 10.1037/0735-7036.119.2.220. [DOI] [PubMed] [Google Scholar]
- 51.Lemasson, A., Jubin, R., Masataka, N., & Arlet, M. Copying hierarchical leaders’ voices? Acoustic plasticity in female Japanese macaques. Sci. Rep. 6, 21289, doi:10.138/srep21289 (2016). [DOI] [PMC free article] [PubMed]
- 52.Mabry KE, Shelley EL, Davis KE, Blumstein DT, Van Vuren DH. Social Mating System and Sex-Biased Dispersal in Mammals and Birds: A Phylogenetic Analysis. PLoS ONE. 2013;8(3) doi: 10.1371/journal.pone.0057980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shapiro, A. D. Recognition of individuals within the social group: signature vocalizations in Handbook of Mammalian Vocalization (eds. Stefan M. Brudzynski) 495–504 (Oxford Academic Press, 2009).
- 54.Blumstein DT, Munos O. Individual, age and sex-specific information is contained in yellow-bellied marmot alarm calls? Anim. Behav. 2005;69:353–361. doi: 10.1016/j.anbehav.2004.10.001. [DOI] [Google Scholar]
- 55.Feng JJ, Cui LW, Ma CY, Fei HL, Fan PF. Individuality and Stability in Male Songs of Cao Vit Gibbons (Nomascus nasutus) with Potential to Monitor Population Dynamics. PLoS ONE. 2014;9(5) doi: 10.1371/journal.pone.0096317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Clink, D. J., Bernard, H., Crofoot, M. C., Marshall, A. J. Investigating Individual Vocal Signatures and Small-Scale Patterns of Geographic Variation in Female Bornean Gibbon (Hylobates muelleri) Great Calls. Int. J. Primatol. 1–16; 10.1007/s10764-017-9972-y (2017).
- 57.Barelli C, Mundry R, Heistermann M, Hammerschmidt K. Cues to Androgens and Quality in Male Gibbon Songs. PLoS ONE. 2013;8(12) doi: 10.1371/journal.pone.0082748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Marler, P. On the origin of speech from animal sounds in The Role of Speech In Language (ed. Kavanagh, J. F. & Cutting, J.) 11–37 (MIT Press 1975).
- 59.Knörnschild M, Nagy M, Metz M, Mayer F, von Helversen O. Complex vocal imitation during ontogeny in a bat. Biol. Lett. 2010;6(2):156-–159. doi: 10.1098/rsbl.2009.0685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vester H, Hammerschmidt K, Timme M, Hallerberg S. Quantifying group specificity of animal vocalizations without specific sender information. Phys. Rev. E. 2016;93 doi: 10.1103/PhysRevE.93.022138. [DOI] [PubMed] [Google Scholar]
- 61.Snowdon CT, Elowson AM. Pygmy Marmosets Modify Call Structure When Paired. Ethology. 1999;105:893–908. doi: 10.1046/j.1439-0310.1999.00483.x. [DOI] [Google Scholar]
- 62.Watson SK, et al. Vocal learning in the functionally referential food grunts of chimpanzees. Curr. Biol. 2015;25(4):495–499. doi: 10.1016/j.cub.2014.12.032. [DOI] [PubMed] [Google Scholar]
- 63.Candiotti A, Zuberbühler K, Lemasson A. Convergence and divergence in Diana monkey vocalizations. Biol. Lett. 2012;8(3):382–385. doi: 10.1098/rsbl.2011.1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.McComb K, Packer C, Pusey A. Roaring and numerical assessment in contests between groups of female lions. Panthera leo. Anim Behav. 1994;47:379–387. doi: 10.1006/anbe.1994.1052. [DOI] [Google Scholar]
- 65.Altmann J. Observational study of behavior: sampling methods. Behaviour. 1974;49:227–267. doi: 10.1163/156853974X00534. [DOI] [PubMed] [Google Scholar]
- 66.Nsubuga AM, et al. Factors affecting the amount of genomic DNA extracted from ape faeces and the identification of an improved sample storage method. Mol. Ecol. 2004;13:2089–2094. doi: 10.1111/j.1365-294X.2004.02207.x. [DOI] [PubMed] [Google Scholar]
- 67.Zaonarivelo JR, et al. Morphometric data for Indri (Indri indri) collected from ten forest fragments in eastern Madagascar. Lemur News. 2007;12:19–23. [Google Scholar]
- 68.Pew J, Muir PH, Wang J, Frasier TR. Related: an R package for analysing pairwise relatedness from codominant molecular markers. Mol. Ecol. Resour. 2015;15:557–561. doi: 10.1111/1755-0998.12323. [DOI] [PubMed] [Google Scholar]
- 69.Queller DC, Goodnight KF. Estimating Relatedness Using Genetic Markers. Evolution. 1989;43(2):258–275. doi: 10.1111/j.1558-5646.1989.tb04226.x. [DOI] [PubMed] [Google Scholar]
- 70.Li C, Weeks D, Chakravarti A. Similarity of DNA fingerprints due to chance and relatedness. Hum. Hered. 1993;43:45–52. doi: 10.1159/000154113. [DOI] [PubMed] [Google Scholar]
- 71.Ritland K. Estimators for pairwise relatedness and individual inbreeding coefficients. Genet. Res. 1996;67:175–185. doi: 10.1017/S0016672300033620. [DOI] [Google Scholar]
- 72.Lynch M, Ritland K. Estimation of pairwise relatedness with molecular markers. Genetics. 1999;152:1753–1766. doi: 10.1093/genetics/152.4.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang J. An estimator for pairwise relatedness using molecular markers. Genetics. 2002;160:1203–1215. doi: 10.1093/genetics/160.3.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Milligan B. Maximum-likelihood estimation of relatedness. Genetics. 2003;163:1153–116. doi: 10.1093/genetics/163.3.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang J. Triadic IBD coefficients and applications to estimating pairwise relatedness. Genet. Res. 2007;89:135–153. doi: 10.1017/S0016672307008798. [DOI] [PubMed] [Google Scholar]
- 76.Boersma, P. & Weenink, D. Praat: doing phonetics by computer [Computer program]. Version 6.0.14; http://www.praat.org/ (2016).
- 77.Sokal, R. R., & Rohlf, F. J. Biometry: the principles and practice of statistics in biological research. Third edition (ed. Freeman, W. H.) 887 (1995).
- 78.Manly, B. J. F. Randomization, Bootstrap and Monte Carlo Methods in Biology. Second edition. (ed. Chapman & Hall) 480 (1997).
- 79.Oksanen, J. et al. Vegan: Community Ecology Package. R package version 2.3-3. http://CRAN.R-project.org/package=vegan (2016).
- 80.Krull CR, et al. Analyses of sex and individual differences in vocalizations of Australasian gannets using a dynamic time warping algorithm. J. Acoust. Soc. Am. 2012;132(2):1189–98. doi: 10.1121/1.4734237. [DOI] [PubMed] [Google Scholar]
- 81.Mundry S, Sommer C. Discriminant function analysis with nonindependent data: consequences and an alternative. Anim. Behav. 2007;74:965–976. doi: 10.1016/j.anbehav.2006.12.028. [DOI] [Google Scholar]
- 82.Wei, T. & Simko, V. Corrplot: Visualization of a Correlation Matrix. R package version 0.77. https://CRAN.R-project.org/package=corrplot (2016).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data and programs used for the analyses presented in the paper are available to the Editorial board members and the referees upon request or already included in the Supporting Information.