Abstract
TFEB is a master regulator for transcription of genes involved in autophagy and lysosome biogenesis. Activity of TFEB is inhibited upon its serine phosphorylation by mTOR. The overall mechanisms by which TFEB activity in the cell is regulated are not well elucidated. Specifically, the mechanisms of TFEB turnover and how they might influence its activity remain unknown. Here, we show that STUB1, a chaperone‐dependent E3 ubiquitin ligase, modulates TFEB activity by preferentially targeting inactive phosphorylated TFEB for degradation by the ubiquitin–proteasome pathway. Phosphorylated TFEB accumulated in STUB1‐deficient cells and in tissues of STUB1‐deficient mice resulting in reduced TFEB activity. Conversely, cellular overexpression of STUB1 resulted in reduced phosphorylated TFEB and increased TFEB activity. STUB1 preferentially interacted with and ubiqutinated phosphorylated TFEB, targeting it to proteasomal degradation. Consistent with reduced TFEB activity, accumulation of phosphorylated TFEB in STUB1‐deficient cells resulted in reduced autophagy and reduced mitochondrial biogenesis. These studies reveal that the ubiquitin–proteasome pathway participates in regulating autophagy and lysosomal functions by regulating the activity of TFEB.
Keywords: autophagy, CHIP (C terminus of HSC70‐Interacting Protein), lysosomes, STUB1, TFEB
Subject Categories: Autophagy & Cell Death; Metabolism; Post-translational Modifications, Proteolysis & Proteomics
Introduction
TFEB (transcription factor EB) is a master regulator for transcription of genes involved in autophagy and lysosomal biogenesis (Sardiello et al, 2009; Settembre et al, 2011). Activity of TFEB is inhibited upon its serine phosphorylation by the mammalian target of rapamycin (mTOR). The phosphorylated TFEB is the inactive form and is sequestered in the cytosol. Upon inhibition of mTOR1, for example, by starvation or under conditions of lysosomal stress, TFEB is activated by dephosphorylation and is translocated to the nucleus where it upregulates the transcription of its target genes (Martina et al, 2012; Roczniak‐Ferguson et al, 2012; Settembre et al, 2012).
Although the regulation of TFEB activity by phosphorylation has been established, the overall mechanisms by which TFEB levels in the cell are regulated remain to be elucidated. Specifically, the mechanisms of TFEB turnover and how they might influence its activity remain unknown. STIP1 homology and U‐Box containing protein 1 (STUB1), formerly known as CHIP (Cterminus of HSC70‐Interacting Protein), is a chaperone‐dependent E3 ubiquitin ligase (Meacham et al, 2001). Here, we show that STUB1 modulates TFEB activity by preferentially targeting inactive phosphorylated TFEB for degradation by the ubiquitin–proteasome pathway. Thus, the ubiquitin–proteasome pathway participates in regulating autophagy and lysosomal functions by regulating the activity of TFEB.
Results and Discussion
STUB1 regulates steady state of phosphorylated TFEB
We have previously generated STUB1 knockout (−/−) mice (Wei et al, 2014). In this study, additional characterization of these mice suggested reduced autophagy. In an attempt to explain these findings, we evaluated mouse embryonic fibroblasts (MEFs) from these mice. We observed increased steady‐state TFEB in STUB1‐deficient (−/−) MEFs (Fig 1A). More importantly, the increase in TFEB, observed in STUB1−/− MEFs, was mostly observed in the phosphorylated S142 form of TFEB, which represents the inactive form (Settembre et al, 2011). To investigate these observations in human cells, we evaluated HeLa cells made deficient in STUB1 using specific shRNA. Similar to the observations in MEFs, HeLa cells deficient in STUB1 had increased levels of TFEB caused largely by increase in phosphorylated S142‐TFEB (Fig 1B). These data were further confirmed by exogenously expressing FLAG‐tagged TFEB in HeLa cells and observing marked increase in phosphorylated TFEB in cells deficient in STUB1 (Fig 1C). Because TFEB activity is regulated by mTOR, we determined the effect of STUB1 deficiency on mTOR activity by evaluating the level of its downstream substrate phosphorylated S6. There was no significant difference in mTOR activity or its inhibition by starvation, between STUB1‐deficient MEFs or HeLa cells and their wild‐type counterparts (Appendix Fig S1).
Figure 1. STUB1 deficiency leads to accumulation of phosphorylated TFEB.
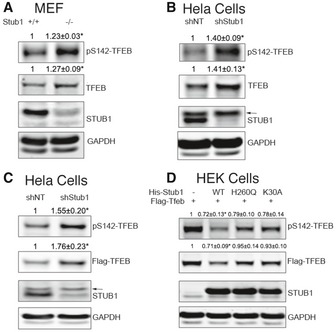
- Wild‐type (+/+) or STUB1 knockout (−/−) mouse embryonic fibroblasts (MEFs) were analyzed by Western blot using indicated antibodies.
- HeLa cells, stably expressing control shRNA (shNT) or STUB1‐specific shRNA (shStub1), were analyzed by Western blot.
- HeLa cells stably expressing shNT or shStub1 were transfected for 48 h with Flag‐TFEB, and cell lysates were analyzed by Western blot.
- HEK293 cells were cotransfected for 48 h with Flag‐TFEB together with His‐STUB1, H260Q‐STUB1, or K30A‐STUB1. Cell lysates were then analyzed by Western blot.
To confirm that STUB1 can affect steady‐state level of TFEB, we co‐expressed FLAG‐TFEB together with either His‐tagged STUB1 or His‐STUB1 mutants into HEK293 cells. Co‐expression of His‐STUB1, but not His‐STUB1 mutants, led to reduction in phosphorylated S142‐TFEB (Fig 1D). His‐STUB1 mutants used were His‐STUB1‐K30A or His‐STUB1‐H260Q. STUB1‐K30A has a point mutation in the TPR chaperone‐binding domain and therefore is unable to bind chaperones and thus its ability to bind substrates is greatly reduced. STUB1‐H260Q has a point mutation in the U‐box domain of STUB1 and therefore is deficient in E3 ubiquitin ligase activity, and thus, its ability to ubiquitinate the substrate to mark it for degradation is diminished (Xu et al, 2002). To determine if the effects of STUB1 on TFEB can be observed in vivo, we evaluated tissues obtained from STUB1−/− mice and their wild‐type +/+ controls. TFEB and phosphorylated TFEB were markedly increased in livers and brains of STUB1−/− mice, compared to controls (Fig EV1). These data suggested that STUB1 has an important in vivo role in regulating TFEB activity.
Figure EV1. Accumulation of TFEB in STUB1 knockout mice.
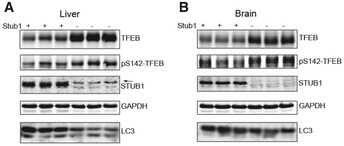
-
A, BLiver (A) and brain (B) tissues from wild‐type (+/+) or STUB1−/− mice were analyzed by Western blot analysis using indicated antibodies. Arrow denotes a previously described non‐specific band (Sha et al, 2009).
STUB1 targets TFEB to the ubiquitin–proteasome pathway
Taken together, the data above suggested that STUB1 could reduce the steady‐state level of TFEB by reducing the level of the inactive phosphorylated TFEB. STUB1 functions as a specialized ubiquitin ligase, which interacts with chaperone‐bound substrates and targets them for proteasomal degradation (Meacham et al, 2001; Xu et al, 2002; Sha et al, 2009; Wei et al, 2014). Therefore, we hypothesized that STUB1 interacted with TFEB and that interaction was mediated by molecular chaperones. To test this hypothesis, we carried out co‐immunoprecipitation analysis following cotransfection of TFEB with wild‐type STUB1 or STUB1 mutants. TFEB was detected in immune complexes with HSP70 and with either wild‐type STUB1 or STUB1‐H260Q (Fig 2A). However, the interaction of STUB1 with TFEB and HSP70 was abrogated by mutation in the TPR chaperone‐binding domain in STUB1‐K30A mutant. These data suggested that STUB1 interacted with TFEB and such interaction was mediated by chaperones including HSP70. TFEB ubiquitination was increased by cotransfection of wild‐type STUB1 in HEK293 cells but to a much lesser extent by STUB1‐H260Q or STUB1‐K30A mutants (Fig 2B). These data suggested that STUB1 interacted with and ubiquitinated TFEB.
Figure 2. STUB1 interacts with TFEB and ubiquitinates it.
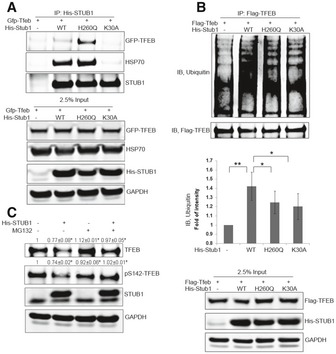
-
A, BHEK293 cells were cotransfected for 48 h with GFP‐TFEB (A) or Flag‐TFEB (B) together with His‐STUB1, H260Q‐STUB1, or K30A‐STUB1. Cell lysates were then subjected to co‐immunoprecipitation using His (A) or Flag (B) antibodies. The immunocomplexes and input were analyzed by Western blot.
-
CHEK293 cells, cotransfected for 48 h with His‐STUB1 and GFP‐TFEB, were treated for 4 h with 50 μM MG132 or vehicle only. Cell lysates were analyzed by Western blot.
Ubiquitination can target substrates for degradation via either ubiquitin–proteasome system or macroautophagy (hereafter called autophagy)–lysosome system (Zhao et al, 2007; Korolchuk et al, 2010; Schreiber & Peter, 2014). We investigated if TFEB was predominately degraded by proteasome pathway or autophagy–lysosome system. Inhibition of the proteasome with MG132 led to accumulation of TFEB in HeLa cells (Fig EV2A). Furthermore, we analyzed the effect of proteasomal or lysosomal inhibition on TFEB half‐life using pulse‐chase strategy. S35‐labeled TFEB half‐life was measured in HeLa cells in the presence of proteasome inhibitor PS341 or lysosome inhibitor bafilomycin A. TFEB half‐life increased significantly in the presence of proteasome inhibitor PS341, but not with the presence of lysosome inhibitor bafilomycin A (Fig EV2B and C). Taken together these data suggested that TFEB was degraded predominantly through ubiquitin–proteasome system.
Figure EV2. TFEB is predominantly degraded by proteasome.
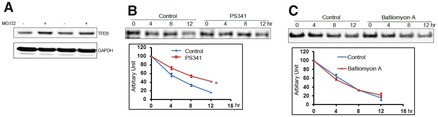
-
AHeLa cells were treated for 2 h with proteasome inhibitor MG132 (50 μM), and cell lysates were analyzed by Western blot.
-
B, CHeLa cells were mock‐treated or treated with proteasome inhibitor PS341 (10 μM; B), or the lysosome inhibitor bafilomycin (10 μM; C), and TFEB half‐life was measured using pulse‐chase analysis. Data are mean ± SD, n = 3. * denotes P < 0.05 using two‐way ANOVA.
We then reasoned that STUB1‐mediated reduction in TFEB levels was caused by degradation of phosphorylated TFEB by the proteasome, and therefore, proteasomal inhibition should abrogate the effect of STUB1 on TFEB. We evaluated the effect of proteasomal inhibition on TFEB in HeLa cells, following overexpression of STUB1. The reduction in TFEB by STUB1 was markedly attenuated in the presence of MG132 (Fig 2C). These data suggested STUB1 ubiquitinated TFEB and targeted it for degradation through proteasome pathway.
STUB1 preferentially targets phosphorylated TFEB for degradation
Activity of TFEB and subcellular localization of TFEB is regulated by phosphorylation and dephosphorylation on residues S142 and S211 (Settembre et al, 2011, 2012, 2013a,b; Martina et al, 2012; Roczniak‐Ferguson et al, 2012). Our data above suggested that STUB1 targeted phosphorylated TFEB for degradation. Co‐immunoprecipitation analysis of STUB1 with wild‐type TFEB, S142A‐TFEB, or S211A TFEB in HEK293 cells showed that the interaction of STUB1 with TFEB was reduced when STUB1 was cotransfected with S142A‐TFEB or S211‐TFEB compared with wild‐type TFEB (Fig 3A). These data suggested that STUB1 preferentially interacted with phosphorylated TFEB.
Figure 3. STUB1 preferentially targets phosphorylated TFEB for degradation.
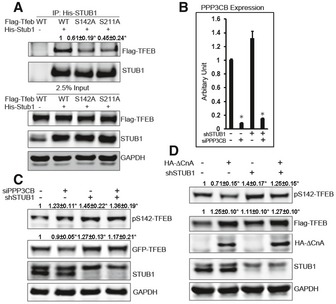
-
AHEK293 cells were cotransfected for 48 h with His‐STUB1 together with Flag‐TFEB, S142A‐TFEB, or S211A‐TFEB. Cell lysates were subjected to co‐immunoprecipitation using His antibodies. The immunocomplexes and input were analyzed by Western blot.
-
B, CHeLa cells stably expressing control shRNA or STUB1‐specific shRNA were transfected for 72 h with non‐target siRNA or PPP3CB siRNA. Cell lysates were then analyzed by real‐time (RT)‐PCR using PPP3CB‐specific primers (B) or by Western blot (C).
-
DFlag‐TFEB expressing HeLa cells were stably transfected with control shRNA or STUB1‐specific shRNA. Cells were transfected for 48 h with vector only or ∆CnA (constitutively active calcineurin), and cell lysates were analyzed by Western blot.
Prior studies have shown that phosphorylated TFEB is sequestered in cytosol (Settembre et al, 2011, 2012, 2013a,b; Roczniak‐Ferguson et al, 2012). However, the fate of the phosphorylated TFEB is not known. TFEB remains phosphorylated at baseline and is dephosphorylated by the phosphatase calcineurin under conditions leading to TFEB activation such as starvation or lysosomal stress (Medina et al, 2015). We hypothesized that phosphorylated TFEB is prone to STUB1‐mediated degradation. We reasoned that enhancing phosphorylation of TFEB would lead to reduction in its steady state, as it would be more prone for degradation. We, therefore, investigated steady‐state levels of TFEB following modulation of its phosphatase calcineurin.
Consistent with recent studies (Medina et al, 2015), phosphorylation of TFEB was increased in cells made deficient in calcineurin catalytic subunit PPP3CB by siRNA knockdown, whereas phosphorylation of TFEB was decreased in cells expressing a constitutive active calcineurin A subunit (∆CnA) (Fig 3B–D). More importantly, TFEB levels were reduced in cells made deficient in calcineurin compared to cells transfected with non‐target siRNA. Further, the reduction in TFEB by calcineurin knockdown was abrogated in cells deficient in STUB1 (Fig 3C).
In contrast, TFEB levels were increased in cells transfected with constitutive active calcineurin A subunit (∆CnA), compared to cells transfected with vector only. Further, levels of TFEB increased more in cells with overexpression of ∆CnA that were also deficient in STUB1 (Fig 3D). These data suggested that phosphorylated TFEB is preferentially targeted for STUB1‐mediated degradation.
STUB1 deficiency reduced TFEB activity
Because phosphorylated TFEB is inactive (Settembre et al, 2011, 2012, 2013a,b; Roczniak‐Ferguson et al, 2012), and our data suggested that phosphorylated TFEB was preferentially degraded by STUB1, we speculated that STUB1 could influence TFEB activity. Upon activation, TFEB binds to CLEAR (coordinated lysosomal expression and regulation) element in promoter region of its target. TFEB activity is often measured by its ability to upregulate peroxisome proliferator‐activated receptor coactivator 1α (PGC1α; also known as Ppargc1a) and promotes the transcription of its target genes (Settembre et al, 2013a,b). We evaluated TFEB activity, in STUB1‐deficient cells and their wild‐type counterpart, using PGC1α promoter‐driven luciferase activity. We speculated that accumulation of phosphorylated TFEB in STUB1‐deficient cells would result in reduction in overall TFEB activity, detected by reduced PGC1α transcription. Confirming such speculation, we found that PGC1α luciferase activity was significantly reduced in HeLa cells made deficient in STUB1 by specific shRNA, compared to cells expressing control shRNA. In contrast, there was no significant difference in luciferase activity between control cells and STUB1‐deficient cells when a CLEAR element mutated PGC1α promoter was used (Fig 4A). There was no difference in CMV promoter‐driven Renilla luciferase activity between HeLa cells expressing STUB1 shRNA or cells expressing control shRNA (Appendix Fig S2A), confirming that the difference of PGC1α‐luciferase activity was not caused by shRNA transfection. PGC1α promoter activity was also lower in STUB1−/− MEFs, compared to +/+ MEFs (Fig 4B).
Figure 4. STUB1 modulates TFEB activity.
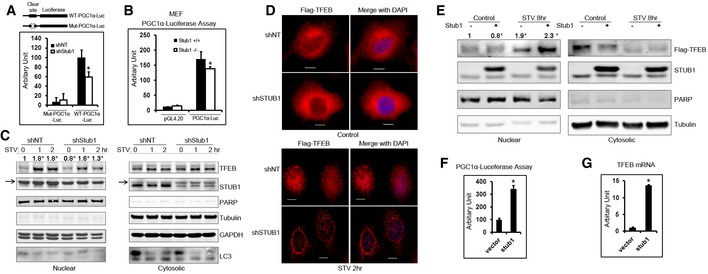
- HeLa cells, stably expressing control shRNA (shNT) or STUB1‐specific shRNA (shStub1), were transfected with wild‐type PGC1α promoter–luciferase vector or Clear site mutated PGC1α promoter–luciferase vector and luciferase activity was assayed 24 h post‐transfection.
- Wild‐type (+/+) or STUB1−/− MEFs were transfected with vector only or PGC1α promoter–luciferase vector, and luciferase activities were assayed 24 h post‐transfection.
- HeLa cells stably expression shNT or shSTUB1 were mock‐treated, starved for 1 or 2 h, and cell lysates were fractionated into nuclear and cytoplasmic fractions and analyzed by Western blot. Arrows denote a previously described non‐specific band (Sha et al, 2009).
- HeLa cells stably expressing Flag‐TFEB were transfected for 72 h with shNT or shSTUB1. Cells were mock‐treated or starved for 2 h and analyzed by immunofluorescence using Flag antibodies. Scale bar, 10 μm.
- HeLa cells, stably expressing Flag‐TFEB, were transfected with vector only or STUB1. Cells were mock‐treated or starved for 8 or 12 h, and cell lysates (nuclear and cytoplasmic fractions) were analyzed by Western blot.
- HeLa cells were transfected for 24 h with vector only or STUB1. Cells were then transfected, for another 24 h, with PGC1α‐luciferase, and luciferase activity assay was performed.
- HeLa cells were transfected for 48 h with vector only or STUB1, and TFEB mRNA expression was assayed by RT–PCR.
The activity of TFEB can also be evaluated by translocation of non‐phosphorylated TFEB to the nucleus in response to activating stimulus such as starvation (Settembre et al, 2011, 2012, 2013a,b; Roczniak‐Ferguson et al, 2012). Therefore, we used starvation to determine the effect of STUB1 on TFEB activity. In STUB1‐deficient HeLa cells, TFEB nuclear translocation in response to starvation was markedly reduced compared to that observed in control cells, as shown by biochemical cellular fractionation and by immunofluorescence analysis (Fig 4C and D, respectively). There was increased nuclear non‐phosphorylated active TFEB caused by starvation, (Appendix Fig S2B). In contrast, HeLa cells overexpressing STUB1 had increased TFEB activity, as evidenced by increased nuclear TFEB (Fig 4E) and by increased PGC1α‐luciferase transcription (Fig 4F). Whereas TFEB moved to the nucleus in response to starvation, there was no observed nuclear translocation of STUB1, which remained mostly cytosolic and colocalized with cytosolic “inactive” TFEB (Fig EV3A).
Figure EV3. STUB1 regulates TFEB and mitochonrial respiration.
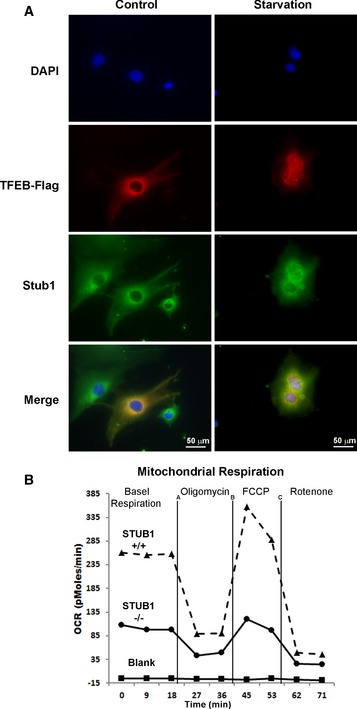
- TFEB colocalizes with STUB1 in cytosol. HeLa cells stably expressing Flag‐TFEB were mock‐treated or starved for 2 h and analyzed by immunofluorescence using Flag or Stub1 antibodies. Scale bar, 20 μm.
- STUB1 deficiency reduced mitochondrial biogenesis. Wild‐type (+/+) or STUB1−/− MEFs were subjected to mitochondrial respiration analysis.
It has been shown that TFEB activates its own transcription through multiple CLEAR elements in TFEB promoter (Settembre et al, 2013a,b). Thus, increased TFEB activity would generally lead to increased TFEB mRNA. Consistent with this notion, TFEB mRNA was higher in HeLa cells transfected with STUB1, compared to cells transfected with vector only (Fig 4G). These data suggested that STUB1 modulates TFEB activity by preferentially degrading phosphorylated TFEB and thereby regulating the level of inactive phosphorylated TFEB in the cytosol.
The above data indicated that TFEB activity, executed by non‐phosphorylated TFEB, inversely correlated with the cellular level of phosphorylated TFEB. Therefore, these data implied that phosphorylated TFEB could exert a dominant negative effect by interfering with non‐phosphorylated TFEB translocation to the nucleus. In exerting its activity, TFEB binds DNA as a homodimer or as heterodimer with closely related b‐HLH‐LZ protein family members (Fisher et al, 1991). We hypothesized that phosphorylated TFEB might exert its dominant negative effect by interacting with non‐phosphorylated TFEB. To test this hypothesis, we constructed a phosphorylation‐defective and phosphomimetic Flag‐tagged TFEB mutants, S142A/S211A‐TFEB and S142D/S211D‐TFEB, respectively. Flag‐tagged S142A/S211A‐TFEB mutant was cotransfected, in HEK293 cells, with a GFP‐tagged TFEB construct (wild‐type, S142A/S211A, or S142D/S211D). Co‐immunoprecipitation analysis, using Flag antibody, suggested that Flag‐tagged S142A/S211A‐TFEB mutant interacted with GFP‐tagged TFEB independent of its phosphorylation status (Fig 5A). These data further implied that binding of accumulated phosphorylated TFEB to non‐phosphorylated TFEB is the underlying mechanism for reduced TFEB activity in STUB1‐deficient cells.
Figure 5. Interaction of TFEB with STUB1 is induced by starvation or mTOR inhibition.
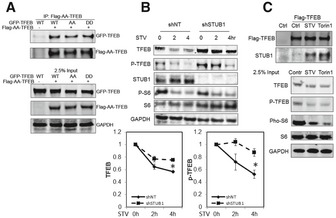
- Flag‐AA‐TFEB (TFEB‐S142A/S211A) was cotransfected with GFP‐TFEB, GFP‐AA‐TFEB, or GFP‐DD‐TFEB (TFEB‐S142D/S211D) into HEK293 cells. Cell lysates were subjected to immunoprecipitation using Flag antibodies. Immunocomplexes or 2.5% input were analyzed by Western blot.
- HeLa cells stably expressing Flag‐TFEB were transfected for 72 h with shNT or shSTUB1. Cells were mock‐treated or starved for 2 or 4 h and analyzed by Western blot. Graphs show quantitative analysis of TFEB and P‐TFEB.
- HeLa cells stably expressing Flag‐TFEB were subjected for 2 h to mock treatment, starvation, or 250 nM Torin1 treatment. Cell lysates were subjected to immunoprecipitation analysis using Flag antibodies.
The above findings suggested that degradation of inactive phosphorylated TFEB would constitute an inherent mechanism for increasing TFEB activity. Therefore, we reasoned that activation of TFEB, by stimuli such as starvation, would be accompanied by enhanced degradation of phosphorylated TFEB and thus reduction in overall steady‐state level of TFEB. We found that, in HeLa cells, starvation induced time‐dependent reduction in steady‐state TFEB. Importantly, such reduction was attenuated in cells deficient in STUB1 (Fig 5B). These data suggested that targeting phosphorylated TFEB for degradation is an important cellular mechanism to increase TFEB activity upon induction by conditions such as starvation. We therefore reasoned that upon activation of TFEB, there could be enhanced interaction between TFEB and STUB1 and conducted experiments to test such possibility. We found that induction of TFEB activity, by starvation or mTOR inhibition, led to marked increased in the interaction between TFEB and STUB1 (Fig 5C). These data confirmed that modulation of the level of phosphorylated TFEB by STUB1 is an integral cellular mechanism of regulation of TFEB activity. They further implied that under stress conditions, phosphorylated TFEB, which is the major TFEB pool in the cytosol, was degraded in order to free more of the non‐phosphorylated TFEB to translocate to the nucleus. The non‐phosphorylated TFEB would then activate target genes including TFEB itself, leading to increasing the overall cellular pool of active TFEB. These data also explained the paradoxical nature of increased levels of TFEB (phosphorylated form) and decreased TFEB activity observed in STUB1‐deficient cells.
Accumulation of phosphorylated TFEB in STUB1‐deficient cells resulted in reduced autophagy
TFEB is the master transcription factor for the expression of autophagy and lysosomal genes (Sardiello et al, 2009; Settembre et al, 2011). Therefore, we reasoned that reduced TFEB activity in STUB1‐deficient cells would result in reduced autophagy. During autophagy, a cytosolic form of LC3 (LC3‐I) is conjugated to phosphatidylethanolamine to form LC3‐phosphatidylethanolamine conjugate (LC3‐II), which is recruited to autophagosomal membranes (Kabeya et al, 2000; Tanida et al, 2008). LC3‐II serves as an autophagosomal marker and widely used to monitor autophagy. LC3‐II formation can be detected by its enhanced mobility in immunoblots and by forming cytoplasmic puncta detected by fluorescence microscopy. Sequestosome 1 (SQSTM1), also called p62, is a receptor for cargo destined to be degraded by autophagy and itself is degraded by autophagy. p62 accumulates when autophagy is inhibited, and is reduced by autophagy induction (Bjørkøy et al, 2009; Rusten & Stenmark, 2010). Under baseline conditions, autophagy was reduced in STUB1−/− MEFs, compared to +/+ MEFs, as shown by reduced LC3‐II formation and increased SQSTM1 (Fig 6A). Upon induction of autophagy by starvation, LC3‐II increased and SQSTM1 was reduced in STUB1+/+ MEFs but these responses were greatly attenuated in STUB1−/− MEFs (Fig 6B). Wild‐type and ATG7−/− MEFs were used as controls for autophagy flux analysis. To further evaluate autophagic flux, we used GFP‐LC3 to evaluate autophagy by fluorescence microscopy in MEFs that were treated with the lysosomal inhibitor chloroquine. GFP‐LC3 autophagosomes were markedly decreased in STUB1−/− MEFs, compared with wild‐type MEFs (Fig 6C). These data suggested that autophagy was reduced in STUB1‐deficient MEFs and that reduction was likely due to reduced initiation of autophagosomes rather than reduced lysosomal degradation. To confirm these findings, we evaluated expression of genes in the autophagy–lysosomal pathway by RT–PCR‐based array analysis. The expression of these genes was reduced in STUB1‐deficient cells (Fig 6D).
Figure 6. STUB1‐deficient cells exhibit reduced autophagy.
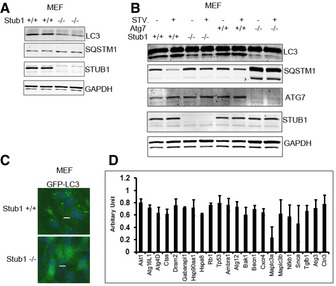
- MEFs were analyzed by Western blot.
- MEFs were mock‐treated or starved for 2 h (STV) and then analyzed by Western blot. Wild‐type or ATG7−/− MEFs were used as control for autophagy flux analysis.
- MEFs were transfected with GFP‐LC3 for 48 h and then treated with chloroquine (50 μm) for 2 h before they were fixed in 4% PFA for fluorescence microscopy. Scale bar, 10 μm.
- HeLa cells, stably expressing control shRNA (shNT) or STUB1‐specific shRNA (shStub1), were subjected to RT–PCR‐based gene expression array analysis. Genes that were significantly reduced (P < 0.05 in Student's t‐test analysis) in STUB1 knockdown cells are shown. Data are mean ± SD, n = 3.
These data suggested that autophagy was reduced in STUB1‐deficient cells because of reduction in transcription of autophagy genes, secondary to accumulation of phosphorylated TFEB leading to reduction in TFEB activity.
Reduced mitochondrial biogenesis in STUB1‐deficient cells
TFEB directly controls PGC1α induction during stress response (Settembre et al, 2013a,b). PGC1a is a master regulator of mitochondrial biogenesis and function (Scarpulla, 2011; Austin & St‐Pierre, 2012). We reasoned that reduced TFEB activity in STUB1−/− cells would result in reduction in PGC1α and consequently decreased mitochondrial biogenesis. We found that PGC1α mRNA expression was significantly reduced in STUB1−/− MEFs (Fig 7A). Mitochondrial copy number was also reduced in STUB1−/− MEFs, compared to wild‐type MEFs (Fig 7B). The majority of cellular ATP is produced by mitochondria (Dahout‐Gonzalez et al, 2006). ATP content was significantly reduced in STUB1−/− MEFs, compared with wild‐type MEFs (Fig 7C). Furthermore, we investigated mitochondrial respiration in STUB1−/− and +/+ MEFs using Seahorse XF analyzer as previously described (Scurtu et al, 2015; van der Windt GJ, 2016). Mitochondrial oxygen consumption rate (OCR) in STUB1−/− MEFs was lower than in wild‐type MEFs (Figs 7D and EV3B). Consistent with the above findings in MEFs, HeLa cells with shRNA‐mediated knockdown of STUB1 expressed lower PGC1α mRNA and had lower ATP content, compared with cells treated with non‐target shRNA (Fig 7E and F). Taken together, these data suggested that mitochondrial biogenesis was compromised in STUB1‐deficient cells, and mitochondrial‐dependent ATP generation was reduced. These data further implied that deficiency of STUB1 and consequently of TFEB activity would reduce the ability of cells to adapt to stress conditions due to failure to increase energy stores or to induce autophagic–lysosomal stress responses. Consistent with this notion, we found that STUB1−/− mice exhibited near complete neonatal lethality (Fig EV4A–C) and the surviving STUB1−/− mice were smaller in size and less active than wild‐type sibling mice (Fig EV4D). Cell proliferation assay using XTT assay showed that STUB1−/− MEF cells proliferate slower than that of wild‐type MEFs (Fig EV4E).
Figure 7. Reduced mitochondrial biogenesis in STUB1‐deficient cells.
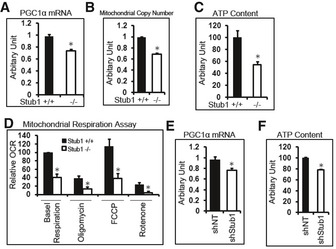
-
A–DWild‐type (+/+) or STUB1−/− MEFs were subjected to RT–PCR analysis of PGC1α (A), mitochondrial copy number analysis (B), ATP content analysis (C), or mitochondrial respiration analysis (D).
-
E, FHeLa cells, stably expressing control shRNA (shNT) or STUB1‐specific shRNA (shStub1), were subjected to RT–PCR analysis of PGC1α (E) or ATP content analysis (F).
Figure EV4. STUB1 knockout mice exhibit partial neonatal lethality.
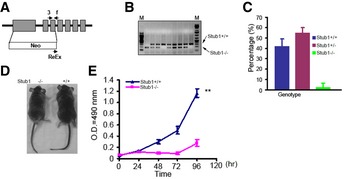
- Schematic diagram of generating STUB1 knockout (−/−) mice. Arrows denotes the primers used for genotyping the transgenic mice.
- Genotyping of STUB1−/− mice using PCR.
- Homozygous STUB1 knockout mice are significantly lower than the expected Mendelian inheritance.
- STUB1−/− mice are smaller than wild‐type (+/+) mice.
- Proliferation assay of MEFs from STUB1−/− and +/+ mice.
Constitutively active TFEB mutant rescued autophagy and mitochondrial biogenesis in STUB1‐deficient cells
We then wanted to confirm that reduced autophagy and mitochondrial biogenesis, observed in STUB1‐deficient cells, was due to reduced TFEB activity. To enhance TFEB activity in STUB1‐deficient HeLa cells, we transfected these cells with S142A/S211A‐TFEB, which is a constitutively active version of TFEB (Settembre et al, 2012). Exogenous expression of S142A/S211A‐TFEB in HeLa cells resulted in increase in TFEB activity, estimated by measuring PGC1α promoter luciferase activity and rescued TFEB activity in STUB1‐deficient cells (Fig 8A and Appendix Fig S3). Furthermore, expression of S142A/S211A‐TFEB mutant into STUB1‐deficient HeLa cells rescued autophagy deficiency, as evidenced by increased formation of LC3‐II, and ATP (Fig 8B and C, respectively). Similar results were obtained using MEFs from wild‐type and STUB1−/− mice (Fig EV5). These data indicated that lower TFEB activity in STUB1‐deficient cells was responsible for reduced autophagy and mitochondrial biogenesis in these cells.
Figure 8. Overexpression of the non‐phosphorylatable TFEB mutant rescues autophagy and mitochondrial biogenesis in STUB1‐deficient cells.
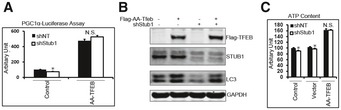
- Cells were transfected for 24 h with vector only (control) or TFEB‐S142A/S211A mutant (Flag‐AA‐TFEB) and then transfected for another 24 h with PGC1α promoter–luciferase before luciferase activity was analyzed.
- Cells were transfected for 48 h with vector only or Flag‐AA‐TFEB, and cell lysates were analyzed by Western blot.
- Cells were treated with transfection reagent only (control) or transfected with vector only or Flag‐AA‐TFEB. ATP content in cell lysates was analyzed 48 h post‐transfection.
Figure EV5. Overexpression of the non‐phosphorylatable TFEB mutant rescues autophagy and mitochondrial biogenesis in STUB1 knockout MEFs.
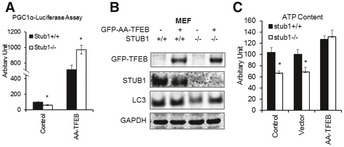
- MEFs were transfected for 24 h with vector only (control) or TFEB‐S142A/S211A mutant (GFP‐AA‐TFEB) and then transfected for another 24 h with PGC1α promoter–luciferase before luciferase activity was analyzed.
- MEFs were transfected for 48 h with vector only or GFP‐AA‐TFEB, and cell lysates were analyzed by Western blot.
- MEFs were treated with transfection reagent only (control) or transfected with vector only or GFP‐AA‐TFEB. ATP content in cell lysates was analyzed 48 h post‐transfection.
In this study, we elucidated that STUB1 plays a critical role in maintaining the homeostasis of the autophagy–lysosome pathway and mitochondrial biogenesis by controlling the steady state of phosphorylated TFEB. The model depicted in Fig 9 illustrates how STUB1 regulates TFEB activity. At nutrient‐rich environment, activated mTOR phosphorylates TFEB, resulting in a low baseline TFEB activity. Upon stress, such as in starvation and/or mTOR inhibition, there is enhanced interaction between STUB1 and phosphorylated TFEB. The latter is ubiqutinated by STUB1 and is targeted for proteasomal degradation. Non‐phosphorylated TFEB translocates to the nucleus exerting its transcriptional activity by promoting genes of the autophagy–lysosomal and mitochondrial pathways as well as TFEB itself. The results of this study suggested that targeting phosphorylated TFEB for degradation was an important mechanism to enhance TFEB activity. TFEB is active as a dimer (Fisher et al, 1991). In the absence of phosphorylated TFEB, there will be increased formation of active non‐phosphorylated homodimers of TFEB. One consequence of this cellular strategy of degrading inactive TFEB would be the need to resynthesize new TFEB. Consistent with this notion, it has been shown that TFEB promotes its own transcription. In STUB1‐deficient cells, phosphorylated TFEB is not efficiently degraded. Accumulating phosphorylated TFEB is inactive and it further reduces TFEB activity by forming heterodimers with non‐phosphorylated TFEB, leading to reduced TFEB translocation to the nucleus. Reduced TFEB activity, in STUB1‐deficient cells, leads to inhibition of autophagy–lysosome pathway and mitochondrial biogenesis. Taken together, our study suggests that the mechanism of degrading inactive phosphorylated TFEB and resynthesizing new TFEB is an important cellular mechanism to cope with cellular stress conditions requiring increased autophagic–lysosomal and mitochondrial biogenesis.
Figure 9. Proposed model for the regulation of TFEB activity by STUB1.
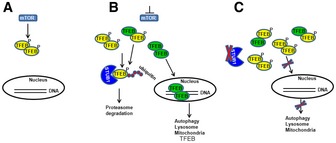
- In a nutrient‐rich environment, activated mTOR phosphorylates TFEB, resulting in reduced TFEB activity.
- Under conditions of mTOR inhibition, such as during starvation, STUB1 targets phosphorylated TFEB for proteasomal degradation. Non‐phosphorylated TFEB translocates to the nucleus exerting its transcriptional activity promoting genes of the autophagy–lysosomal and mitochondrial pathways as well as TFEB itself.
- In STUB1‐deficient cells, phosphorylated TFEB is not efficiently degraded. Accumulating phosphorylated TFEB is inactive and it further reduces TFEB activity by forming heterodimers with non‐phosphorylated TFEB, leading to reduced TFEB translocation to the nucleus. Reduced TFEB activity, in STUB1‐deficient cells, leads to inhibition of autophagy–lysosome pathway and mitochondrial biogenesis.
Lysosomal storage disorders are caused by lysosomal dysfunction as a consequence of enzyme deficiencies and abnormal buildup of various toxic materials in cells (Winchester et al, 2000). TFEB upregulates the expression of key genes involved in folding, proteostasis, and lysosomal trafficking pathways (Song et al, 2013; Spampanato et al, 2013). Further, activation of TFEB attenuates lysosomal storage pathology both in vitro and in vivo. (Medina et al, 2011; Song et al, 2013; Spampanato et al, 2013). Therefore, TFEB is considered as a therapeutic target in diseases associated with defects in autophagy–lysosomal pathways. Methods to induce TFEB activation to enhance autophagy–lysosome‐mediated degradation of misfolded proteins have been suggested for treatment of diseases associated with misfolded proteins such as neurodegenerative diseases (Decressac & Björklund, 2013; Martini‐Stoica et al, 2016). Our study showed that STUB1 plays a critical role in modulating TFEB activity and reveals STUB1 as a novel potential therapeutic target in these diseases.
Materials and Methods
Reagents
N‐Carbobenzoxyl‐L‐leucinyl‐L‐leucinyl‐L‐norleucinal (MG132), N‐ethylmaleimide (NEM), Chloroquine (CQ), and Triton X‐100 were from Sigma. mTOR inhibitor Torin1 and Tween 20 were from Calbiochem. TFEB and PARP antibodies were from Cell Signaling. STUB1 antibody (pc711) was from Calbiochem. Phospho‐S142 TFEB antibody was a gift from Dr. Gerard Karsenty. GAPDH antibody was from Advanced ImmunoChemical. α‐tubulin antibody was from Abcam. GFP antibody was from Thermo Scientific. Ubiquitin antibody (u5379) and HA antibody (HA‐7) were from Sigma. LC3B antibody was from Novus. p62 antibody was from American Research Products. ATG7 antibody was from Abcam. STUB1, STUB1‐H260Q, and STUB1‐K30A expressing vectors are kindly provided by Dr. Len Neckers. TFEB mutants of TFEB‐S142A, TFEB‐S211A, TFEB‐S142A/S211A were generated by site‐directed point mutagenesis. PGC1α‐luciferase reporter vectors were previously described (Settembre et al, 2013a,b). HA‐tagged‐∆CaN vector was kindly provided by Dr. Beverly Rothermel at University of Texas Southwestern Medical Center. PPP3CB siRNA was purchased from Dharmacon. STUB1+/− mice were from Texas A&M Institute for Genomic Medicine. Mice were maintained in a pathogen‐free animal facility at Baylor College of Medicine. Mouse embryo fibroblasts (MEFs) were generated from Day 13.5 embryos. ATG7−/− MEFs were obtained from Dr. Masaaki Komatsu. All experimental protocols were approved by the Institutional Animal Care and Use Committee.
Cell proliferation assay
XTT [3‐(4,5‐Dimethylthiazol‐2‐yl)‐2,5‐Diphenyltetrazolium Bromide] assay was performed according to the user's manual (Roche). Briefly, cells were plated in 96‐well plates at a density of 1.3 × 103 cells per well in 0.1 ml culture medium. At the times specified, 50 μl XTT labeling mixture per well was added and incubated for 4 h at 37°C and 5% CO2. The spectrophotometrical absorbance of samples was measured at wavelength of 490 nm. The reference wavelength was 690 nm.
Nuclear and cytoplasmic fractionation
Nuclear and cytoplasmic fraction was extracted using NE‐PER™ Nuclear and Cytoplasmic Extraction Reagents. Briefly, cells were washed twice with PBS and resuspended in cytoplasmic extraction reagents I and incubated on ice for 10 min. Cytoplasmic extraction reagents II were added and incubated on ice for 1 min and centrifuged at 4°C, 16,000 g, for 5 min. Supernatant was saved as cytoplasmic fraction. The pellet were washed once with PBS and suspended in nuclear extraction reagent and incubated on ice for 40 min with vortexing for 15 s every 10 min. The supernatant nuclear fraction was harvested by centrifuge at 4°C, 16,000 g, for 10 min.
Autophagy induction
Autophagy was induced by starvation by incubating cells in EBSS for 2 h or by incubating cells for 2 h in regular medium with 250 nM of the mTOR inhibitor Torin1.
Immunoprecipitation
Immunoprecipitation was carried out by lysing cells in RIPA buffer and 1 mg of cell lysates was incubated at 4°C with primary antibody for 2 h before Dynabeads protein G (Thermo Scientific) were added to the samples. After further incubation for 1 h at 4°C, beads were washed three times in ice‐cold lysis buffer. Immunoprecipitated proteins were eluted by heating at 95°C for 5 min in 2× LDS buffer. Co‐immunoprecipitation assay was carried out by lysing cells in NETN buffer (20 mM Tris–Cl, pH 8.0, 100 mM NaCl, 1 mM EDTA, and 0.5% NP‐40), and 1 mg of cell lysates was precleared by Dynabeads protein G and incubated with primary antibody for 2 h before Dynabeads protein G (Thermo Scientific) were added to the samples. Immunoprecipitated proteins were eluted by heating at 95°C for 5 min in 2× LDS buffer.
Fluorescence microscopy
Cells were grown on glass coverslips coated with poly‐D‐lysine, fixed in 4% paraformaldehyde, permeabilized by 0.2% Triton X‐100, and blocked in 10% normal goat serum. Primary antibody incubation was done at room temperature for 1 h, followed by a 30‐min incubation with Alexa Fluor‐labeled secondary antibodies (Molecular Probes, Eugene, OR). Coverslips were mounted by the blue nuclear chromatin stain 4, 6‐diamidino‐2‐phenylindole (DAPI; Molecular Probes), and images were acquired by a Zeiss Axiovert microscope deconvolution microscopy system.
Real‐time PCR
Total RNA was reverse‐transcribed into cDNA and reaction was performed using TaqMan‐specific primer sets (AB, Applied Biosystems). Relative expressions of specific genes were calculated by ∆∆CT method by normalizing to the expression of glyceraldehyde phosphate dehydrogenase (GAPDH) or 18s rRNA. Quantitative analysis of autophagy gene expression was carried out using RT² Profiler™ PCR Array Human Autophagy (Qiagen). Fold changes were calculated using ΔΔCT method with normalization of raw data to housekeeping genes.
Mitochondrial copy number analysis
Total DNA was extracted using DNeasy Blood & Tissue Kit (Qiagen). TaqMan probe sets specific to human or mouse mitochondrial cytochrome c oxidase subunit I (mtCOXI), and TaqMan probe sets specific to human or mouse genomic GAPDH were used for quantitative PCR‐based analysis of relative mitochondrial copy number assay in 50 ng of total DNA.
Mitochondrial respiration assay
MEFs from wild‐type or STUB1 knockout mice were seeded in XF24 cell culture plates (Seahorse Bioscience, North Billerica, MA). After 24 h, OCR was measured using the Seahorse Bioscience XF24 Extracellular Flux Analyzer (Seahorse Bioscience). OCR of cells was measured in sequence to basal levels (with no additives), with 1 μg/ml oligomycin, with 4 μM FCCP and 1 μM rotenone. Cells were subjected in sequence to the following additions: (i) Basal levels were measured with no additives; (ii) 1 μM OLIGO, which reversibly inhibits ATP synthase and OXPHOS, was added to show glycolysis alone; (iii) 0.3 μM FCCP, a mitochondrial uncoupler, was added to induce maximal respiration; (iv) 0.1 μM ROT, a complex I inhibitor and mitochondrial poison, was added to end the reaction. Triplicate experimental wells were examined, and the results were plotted by Seahorse software.
Luciferase assay
Luciferase activity was measured using Steady‐Glo® Luciferase Assay System (Promega Corporation). Briefly, cells were cultured in 100 μl medium using 96‐well white plate, and 100 μl of CCLR (cell culture lysis reagent) with luciferase substrate was added to each well. Luminescence was measured using SpectroMax M3 multi‐mode microplate reader (Molecular Devices).
Statistical analyses
Data are presented as the mean ± standard deviation of at least three independent experiments. Differences between samples were analyzed using Student's t‐test or as otherwise indicated in the text. Statistical significance was set at P < 0.05.
Author contributions
YS performed experiments, analyzed data, and wrote manuscript. LR performed experiments and analyzed data. CS and AB provided critical reagents, analyzed data, and edited manuscript. NTE supervised research, analyzed data, and wrote manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Review Process File
Source Data for Figure 1
Acknowledgements
We acknowledge current and former members of the Eissa laboratory for stimulating discussions and technical assistance. This study was supported by National Heart, Lung and Blood Institute.
The EMBO Journal (2017) 36: 2544–2552
References
- Austin S, St‐Pierre J (2012) PGC1α and mitochondrial metabolism–emerging concepts and relevance in ageing and neurodegenerative disorders. J Cell Sci 125(Pt 21): 4963–4971 [DOI] [PubMed] [Google Scholar]
- Bjørkøy G, Lamark T, Pankiv S, Øvervatn A, Brech A, Johansen T (2009) Monitoring autophagic degradation of p62/SQSTM1. Methods Enzymol 452: 181–197 [DOI] [PubMed] [Google Scholar]
- Dahout‐Gonzalez C, Nury H, Trézéguet V, Lauquin GJ, Pebay‐Peyroula E, Brandolin G (2006) Molecular, functional, and pathological aspects of the mitochondrial ADP/ATP carrier. Physiology 21: 242–249 [DOI] [PubMed] [Google Scholar]
- Decressac M, Björklund A (2013) TFEB: pathogenic role and therapeutic target in Parkinson disease. Autophagy 9: 1244–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher DE, Carr CS, Parent LA, Sharp PA (1991) TFEB has DNA‐binding and oligomerization properties of a unique helix‐loop‐helix/leucine‐zipper family. Genes Dev 5: 2342–2352 [DOI] [PubMed] [Google Scholar]
- Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T (2000) LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J 19: 5720–5728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korolchuk VI, Menzies FM, Rubinsztein DC (2010) Mechanisms of cross‐talk between the ubiquitin‐proteasome and autophagy‐lysosome systems. FEBS Lett 584: 1393–1398 [DOI] [PubMed] [Google Scholar]
- Martina JA, Chen Y, Gucek M, Puertollano R (2012) MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy 8: 903–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martini‐Stoica H, Xu Y, Ballabio A, Zheng H (2016) The autophagy‐lysosomal pathway in neurodegeneration: a TFEB perspective. Trends Neurosci 39: 221–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meacham GC, Patterson C, Zhang W, Younger JM, Cyr DM (2001) The Hsc70 co‐chaperone CHIP targets immature CFTR for proteasomal degradation. Nat Cell Biol 3: 100–105 [DOI] [PubMed] [Google Scholar]
- Medina DL, Fraldi A, Bouche V, Annunziata F, Mansueto G, Spampanato C, Puri C, Pignata A, Martina JA, Sardiello M, Palmieri M, Polishchuk R, Puertollano R, Ballabio A (2011) Transcriptional activation of lysosomal exocytosis promotes cellular clearance. Dev Cell 21: 421–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina DL, Di Paola S, Peluso I, Armani A, De Stefani D, Venditti R, Montefusco S, Scotto‐Rosato A, Prezioso C, Forrester A, Settembre C, Wang W, Gao Q, Xu H, Sandri M, Rizzuto R, De Matteis MA, Ballabio A (2015) Lysosomal calcium signalling regulates autophagy through calcineurin and TFEB. Nat Cell Biol 17: 288–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roczniak‐Ferguson A, Petit CS, Froehlich F, Qian S, Ky J, Angarola B, Walther TC, Ferguson SM (2012) The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci Signal 5: ra42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusten TE, Stenmark H (2010) p62, an autophagy hero or culprit? Nat Cell Biol 12: 207–209 [DOI] [PubMed] [Google Scholar]
- Sardiello M, Palmieri M, di Ronza A, Medina DL, Valenza M, Gennarino VA, Di Malta C, Donaudy F, Embrione V, Polishchuk RS, Banfi S, Parenti G, Cattaneo E, Ballabio A (2009) A gene network regulating lysosomal biogenesis and function. Science 325: 473–477 [DOI] [PubMed] [Google Scholar]
- Scarpulla RC (2011) Metabolic control of mitochondrial biogenesis through the PGC‐1 family regulatory network. Biochim Biophys Acta 1813: 1269–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber A, Peter M (2014) Substrate recognition in selective autophagy and the ubiquitin‐proteasome system. Biochim Biophys Acta 1843: 163–181 [DOI] [PubMed] [Google Scholar]
- Scurtu I, Sturza A, Pavel IZ, Popescu R, Privistirescu A, Duicu OM, Muntean DM (2015) Bioenergetic characterization of h9c2 cells using the extracellular flux analyzer. Rev Med Chir Soc Med Nat Iasi 119: 491–495 [PubMed] [Google Scholar]
- Settembre C, Di Malta C, Polito VA, Garcia Arencibia M, Vetrini F, Erdin S, Erdin SU, Huynh T, Medina D, Colella P, Sardiello M, Rubinsztein DC, Ballabio A (2011) TFEB links autophagy to lysosomal biogenesis. Science 332: 1429–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C, Zoncu R, Medina DL, Vetrini F, Erdin S, Erdin S, Huynh T, Ferron M, Karsenty G, Vellard MC, Facchinetti V, Sabatini DM, Ballabio A (2012) A lysosome‐to‐nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J 31: 1095–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C, De Cegli R, Mansueto G, Saha PK, Vetrini F, Visvikis O, Huynh T, Carissimo A, Palmer D, Klisch TJ, Wollenberg AC, Di Bernardo D, Chan L, Irazoqui JE, Ballabio A (2013a) TFEB controls cellular lipid metabolism through a starvation‐induced autoregulatory loop. Nat Cell Biol 15: 647–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C, Fraldi A, Medina DL, Ballabio A (2013b) Signals from the lysosome: a control centre for cellular clearance and energy metabolism. Nat Rev Mol Cell Biol 14: 283–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha Y, Pandit L, Zeng S, Eissa NT (2009) A critical role for CHIP in the aggresome pathway. Mol Cell Biol 29: 116–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W, Wang F, Savini M, Ake A, di Ronza A, Sardiello M, Segatori L (2013) TFEB regulates lysosomal proteostasis. Hum Mol Genet 22: 1994–2009 [DOI] [PubMed] [Google Scholar]
- Spampanato C, Feeney E, Li L, Cardone M, Lim JA, Annunziata F, Zare H, Polishchuk R, Puertollano R, Parenti G, Ballabio A, Raben N (2013) Transcription factor EB (TFEB) is a new therapeutic target for Pompe disease. EMBO Mol Med 5: 691–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanida I, Ueno T, Kominami E (2008) LC3 and autophagy. Methods Mol Biol 445: 77–88 [DOI] [PubMed] [Google Scholar]
- Wei Q, Sha Y, Bhattacharya A, Abdel Fattah E, Bonilla D, Jyothula SS, Pandit L, Khurana Hershey GK, Eissa NT (2014) Regulation of IL‐4 receptor signaling by STUB1 in lung inflammation. Am J Respir Crit Care Med 189: 16–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winchester B, Vellodi A, Young E (2000) The molecular basis of lysosomal storage diseases and their treatment. Biochem Soc Trans 28: 150–154 [DOI] [PubMed] [Google Scholar]
- Xu W, Marcu M, Yuan X, Mimnaugh E, Patterson C, Neckers L (2002) Chaperone‐dependent E3 ubiquitin ligase CHIP mediates a degradative pathway for c‐ErbB2/Neu. Proc Natl Acad Sci USA 99: 12847–12852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Brault JJ, Schild A, Cao P, Sandri M, Schiaffino S, Lecker SH, Goldberg AL (2007) FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab 6: 472–483 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Review Process File
Source Data for Figure 1


