Abstract
An old question about regeneration is whether it is an ancestral character which is a general property of living matter, or whether it represents a set of specific adaptations to the different circumstances faced by different types of animal. In this review, some recent results on regeneration are assessed to see if they can throw any new light on this question. Evidence in favour of an ancestral character comes from the role of Wnt and bone morphogenetic protein signalling in controlling the pattern of whole‐body regeneration in acoels, which are a basal group of bilaterian animals. On the other hand, there is some evidence for adaptive acquisition or maintenance of the regeneration of appendages based on the occurrence of severe non‐lethal predation, the existence of some novel genes in regenerating organisms, and differences at the molecular level between apparently similar forms of regeneration. It is tentatively concluded that whole‐body regeneration is an ancestral character although has been lost from most animal lineages. Appendage regeneration is more likely to represent a derived character resulting from many specific adaptations.
Keywords: adaptation, bone morphogenetic protein, regeneration, Wnt
Subject Categories: Development & Differentiation
Glossary
- ADMP
Anti‐dorsalising morphogenetic protein—a BMP (q.v.)‐like signalling molecule expressed on the dorsal side of early vertebrate embryos.
- AP
Anterior–posterior—the anterior is the head end of an animal, or the end having a concentration of sensory organs, and the posterior is the tail end. In human anatomy, anterior means the same as ventral (q.v.)
- Bilateria
Animals having bilateral symmetry, typically with a head, a tail, and dorsal and ventral sides. The bilateria comprise all metazoa (q.v.) other than the phyla Cnidaria (hydroids, anemones, jellyfish), Ctenophora (comb jellies) and Porifera (sponges), along with the enigmatic Trichoplax adhaerens.
- Blastema
A mass of undifferentiated cells which grow and differentiate to produce a regenerated structure. In planarians, the region called the blastema is visible as a depigmented area, but it consists of post‐mitotic cells, the mitotic cells contributing to the regenerate being located more proximally. In situations such as the vertebrate tail where there is no dedifferentiation and the regenerate is formed from several different cell populations, it is preferable to use the term “regeneration bud” rather than “blastema”.
- BMP
Bone morphogenetic protein—one of the key groups of signalling molecules controlling development and regeneration.
- CFP
Cyan fluorescent protein—one of several fluorescent proteins used as reporters in transgenic experiments.
- Cre‐lox
A system derived from bacteriophage P1, which is widely used for making genetic modifications in animals. Cre is a recombinase enzyme, which cuts DNA at specific sites called loxP sites. In the context of this article, the Cre‐lox system is used to label cells derived from those expressing a specific gene. The promoter of interest is used to drive Cre, and this excises an inhibitory sequence enabling a reporter gene to be switched on permanently. This reporter subsequently remains active in all progeny cells regardless of their state of differentiation. In the CreER version of the system, activity of the Cre enzyme can be regulated by oestrogen analogues such as tamoxifen. In this article, the promoters used for Cre labelling experiments are those of the following genes: K14 (keratin 14, expressed in the basal layer of the epidermis), Prx1 (encodes a transcription factor expressed in limb mesenchyme), Scleraxis (encodes a transcription factor expressed in tendons), Sox9 (encodes a transcription factor expressed in bone, and many other tissues), Tie2 (encodes an angiopoietin receptor expressed in endothelial cells), VE‐cadherin (encodes a component of adherens junctions present in endothelial cells).
- Dedifferentiation
Functional cell types look different down the microscope because of high concentrations of specific proteins concerned with their function. Dedifferentiation means the loss of the visible differentiated characteristics of a cell. Dedifferentiated cells may be, but are not necessarily, either stem (q.v.) or progenitor cells.
- Deuterostomia
The super‐phylum of animals including the phyla Chordata (including the vertebrates), Echinodermata (sea urchins, starfish, crinoids) and Hemichordata (acorn worms).
- DV
Dorsal–ventral—the dorsal side of an animal is normally that away from the substratum and the ventral side is towards the substratum.
- FACS
Fluorescence‐activated cell sorting—a method for separating cell types based on the binding of specific fluorescent antibodies to their surfaces.
- Hypostome
The region containing the oral opening of Hydra or another species of the hydrozoan class within the Cnidaria. The term is also used to refer to a mouthpart of arachnids.
- K14‐CreER
See Cre‐lox.
- Metazoa
Synonym for multicellular animals.
- mTmG
A reporter mouse strain used in Cre‐lox (q.v.) cell lineage labelling experiments. When recombination is catalysed by Cre, expression of the fluorescent protein tdTomato is replaced by that of GFP (green fluorescent protein).
- Planarian
A free‐living flatworm of the phylum Platyhelminthes. The term “planarian” sometimes refers to all of class Turbellaria, and sometimes just to the order Tricladida.
- Pluripotency
The ability to form all cell types in the body under suitable culture conditions. In the case of mammals, the definition excludes the trophectoderm. The mammalian zygote is defined as totipotent and the inner cell mass and epiblast as pluripotent.
- Prod1
A glycerophosphatidyl‐anchored cell surface protein found only in salamanders.
- Prx1‐Cre
See Cre‐lox.
- Scleraxis‐Cre
See Cre‐lox.
- Sox9‐Cre
See Cre‐lox.
- Stem cell
A cell type that, as a population, divides to reproduce itself and to generate differentiated progeny. Normally stem cells persist for the life of the animal and exist within a specific niche of non‐stem cells. Mammalian pluripotent cells, which exist in tissue culture, comprise embryonic stem cells and induced pluripotent stem cells.
- Tie2‐Cre
See Cre‐lox.
- VE‐cadherin‐CreER
See Cre‐lox.
- Wnt
One of the key groups of signalling molecules controlling development and regeneration. The name derives from “wingless‐related MMTV integration site 1”, wingless being a Drosophila gene and MMTV being murine mammary tumour virus.
The regeneration of missing parts by animals remains one of the more mysterious branches of developmental biology. Nonetheless, there is rapid research progress and some specific mechanisms are now quite well understood 1, 2, 3, 4, 5. In this article, I am going to ask whether certain recent discoveries enable us to say any more about the old question raised by the title. To say that regenerative ability is ancestral in evolutionary terms implies that it may be an intrinsic or pristine property of living matter that is always present but can become masked in various circumstances. On the other hand, if regenerative capacity is an evolutionary novelty, this implies that there is no general ability of living matter to regenerate, but that the ability to do so has evolved many times as an adaptation to specific circumstances. Although the issue is a very old one, it still has an obvious practical interest. If regeneration is really an intrinsic property of living matter, then there may be a simple way to unmask it in humans by restoring a component that has been switched off or lost in the recent evolutionary past. But if every instance of regeneration is an evolutionary novelty, then each one may involve the generation of new mechanisms and perhaps the involvement of new genes. To introduce such a mechanism to a non‐regenerating organism would require many new or modified components and many new connections between them. It may of course turn out that both views are correct to some degree and that some regeneration behaviours are ancestral while others have originated recently.
Bely and Nyberg 6 provide a useful classification of regenerative processes into five types (Fig 1). They distinguish cellular regeneration, for example regrowth of severed nerve axons; tissue regeneration, for example growth of epidermis covering a wound; organ regeneration of the heart, liver or lens, which tends to restore organ mass but without internal pattern. They refer to the distal regeneration of appendages in vertebrates and arthropods as structural regeneration. Most dramatic is whole‐body regeneration, also called bidirectional regeneration 7, in which either heads or tails can regenerate from the same body level depending on the orientation of the cut surface. These different levels of performance clearly involve different mechanisms. Internal organ regeneration has to be able to sense total organ size and to stop growing when this is restored 8. Structural regeneration of appendages needs to be able to do this and also to generate spatial pattern, including often a pattern of structures not present at the amputation surface. Whole‐body regeneration needs to be able to do both these things and also to decide on polarity (head versus tail?) before doing anything else.
Figure 1. The five types of regeneration found in animals, according to Bely and Nyberg 6 .
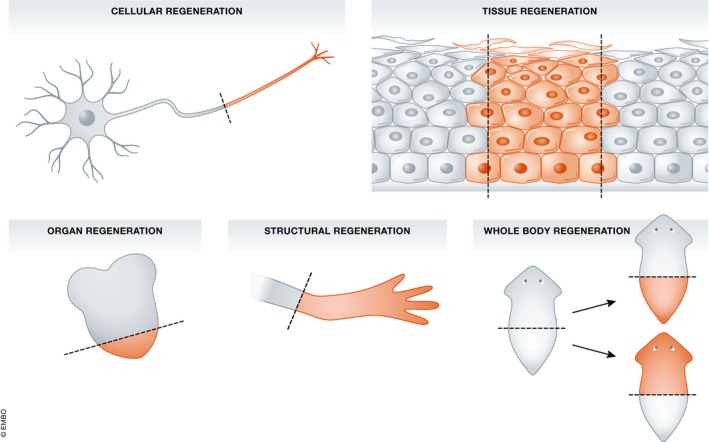
Cellular regeneration is mostly apparent as the regrowth of severed nerve axons. Tissue regeneration indicates the closure of gaps in a homogeneous cell population. Organ regeneration indicates the restoration of size of an organ, often comprising multiple cell types. Structural regeneration refers to regeneration of appendages in arthropods and vertebrates. This involves regeneration of pattern, but always in a distalwards direction. Whole‐body regeneration indicates the ability to regenerate both heads and tails, often from small body fragments.
The survey of regenerative ability among animal phyla and classes by Bely and Nyberg 6 reminds us that regenerative ability is scattered among animal groups with no obvious pattern. It is not the case, as often supposed, that regeneration is found only in more basal phyla. Regeneration of limbs and tails in urodele amphibians and of tails in lizards have been familiar for a long time. We now know that regeneration of the head can occur in the hemichordate Ptychodera flava 9. Hemichordates are considered to be a sister group to echinoderms and so members of the Deuterostomia group of phyla that also includes ourselves. Even more remarkable, in the colonial urochordate Botrylloides leachi, entire fully functional zooids can regenerate from small blood vessel fragments 10. Morphological evidence suggests that it is cells from the blood, possibly pluripotent stem cells, that undergo this regeneration. Urochordates are now considered a sister group of the vertebrates within the phylum Chordata, so this is currently the “highest” (=most derived) animal type known to undergo whole‐body regeneration.
This article will focus on the top two types of regeneration, structural (appendage) regeneration and whole‐body regeneration, which both involve the formation of new pattern as well as the expansion of existing structures. I shall also examine the recently discovered case of the spiny mouse, which has unusually good ability for a mammal at the level of tissue regeneration. I shall start by suggesting some criteria for deciding whether regeneration mechanism is ancestral or derived, and then consider whether recent findings can lead to any new conclusions. In the later Discussion, some further conceptual issues and caveats will be outlined.
General arguments
The overall patchy distribution of regenerative ability across the phylogenetic tree of animals has caused many to surmise that regeneration is indeed an ancestral property of living matter but that its maintenance involves some cost. In other words, whole‐body regeneration may be maintained as a by‐product of asexual reproduction by fission, or structural regeneration of appendages may be retained to cope with high levels of non‐lethal predation, but otherwise maintenance of regenerative capacity confers some selective disadvantage and becomes lost 11. The alternative view is that every instance of regeneration is a fresh adaptation to loss of body parts in normal life, especially the occurrence of non‐lethal predation affecting the regenerating parts. It might seem that the occurrence of non‐lethal predication is consistent with either view. But to support the idea of ancestral regeneration, it also needs to be shown that the entire ancestral line of animals has had a similar regenerative ability, something which is very difficult to prove, although some examples of regenerating vertebrate appendages have been found in the fossil record 12.
A regenerative ability which has persisted continuously since the origin of Bilateria might be expected to occur via a common mechanism, which is likely to involve reuse of components of the mechanisms for embryonic development. A novel regenerative process might also reuse a developmental mechanism, but need not do so, and could be completely different both from that species’ own developmental mechanism and from regenerative mechanisms in other animals. So different mechanisms from those used in development, or different mechanisms from those found in otherwise similar animals, could both be considered evidence for evolutionary novelty.
The persistence of all the genes in all the cells of most animals would seem to make regeneration possible in principle. However, some recent results on gene loss indicate that this is not as universal a feature as is often supposed, as even vertebrates can lose genes during development, so it should not be assumed that all genes are always present 13. The ability to force several cell types to become pluripotent by introduction of selected transcription factors 14, 15 might also be considered an argument for a universal ancestral regenerative ability. Pluripotent stem cells are cells that can grow without limit in vitro and can also, following appropriate treatment, form any of the cell types in the normal body. At least some of the neoblasts, or stem cells, found in planarians are pluripotent 16, and feed the continuous replacement of the differentiated cells and structures both in homeostasis and in regeneration. However, mammals do not seem to have pluripotent cells later in the life cycle than early embryogenesis 17, and pluripotent cells injected into animals generate teratomas rather than regenerating structures 18, 19. Also, recent results (see below) have indicated that the degree of dedifferentiation in vertebrate regeneration is rather less than had formerly been supposed and never extends to full pluripotency of regenerating cells. Some regenerating structures, such as the vertebrate tail, undergo excess division of those tissues that are still mitotic, but probably no dedifferentiation at all 20, 21. Cell differentiation is generally inimical to regeneration, particularly when differentiated cells become post‐mitotic, as is found, for example, for skeletal muscle cells and neurons in mammals. Current understanding of chromatin biology indicates that differentiated cells have large areas of chromatin in a closed compacted state, difficult of access for transcription factors 22.
Evidence for the ancestral nature of whole‐body regeneration
Conservation of signalling systems
In recent years, there has been considerable progress in elucidating the signalling systems used to control regeneration. Space precludes consideration of most of these data, but some results are particularly relevant to the issue of ancestral versus adaptive regeneration. Perhaps the main theme that has emerged is a consensus that the establishment of all bilaterian body plans is controlled during development by gradients of Wnt (anteroposterior, AP) and bone morphogenetic protein (BMP) (dorsal–ventral, DV) 23 (Fig 2).
Figure 2. A phylogenetic tree of some groups of animals familiar to regeneration research.
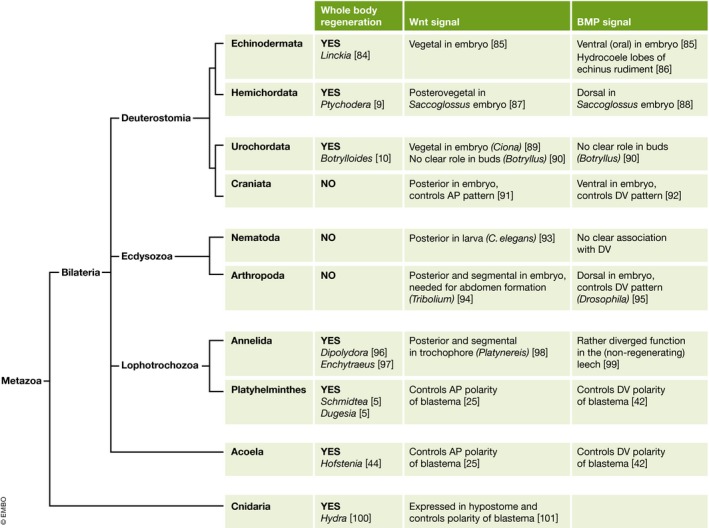
The first column on the right shows the occurrence of whole‐body regeneration in at least one species. The second indicates the disposition of Wnt signalling in relation to anteroposterior pattern and the third of BMP signalling in relation to dorsoventral pattern.
Planarians contain several Wnt genes and the components of the canonical Wnt signalling pathway via β‐catenin. The Wnt genes are expressed in a posterior high‐anterior low gradient. Blockage of Wnt signalling with RNAi causes posterior‐facing cuts to regenerate heads, and treatment with Wnt agonists causes anterior‐facing cuts to regenerate tails 24, 25. Wnt factors are also among the signals controlling the anteroposterior pattern of vertebrate embryos and are expressed in the posterior of the gastrulating embryo 26. It has been argued that the role of Wnt in the control of polarity in planarian regeneration represents an embryonic signalling process that is found in all bilaterian animals and is therefore ancestral 26. In fact, not all free‐living flatworms are capable of regeneration 27, but investigation of some species incapable of head regeneration has shown that this can be restored by inhibition of the Wnt system, indicating that other aspects of the anterior regeneration mechanism are still present 28, 29, 30.
Is it possible to subsume the posterior (tail) regeneration of vertebrates under the heading of whole‐body regeneration requiring Wnt signalling? Wnt signalling is indeed necessary for regeneration of the Xenopus tadpole 31 and the zebrafish caudal fin 32. Following amputation, Wnt pathway genes are upregulated in the regenerating lizard tail but not in the non‐regenerating lizard limb 33. However, in these cases there is no dedifferentiation and each tissue type provides the cells for its own regeneration. This, together with the fact that tail regeneration is always distal, even from a proximal‐facing cut surface 34, makes it doubtful whether there is a true homology with whole‐body regeneration. The whole‐body regeneration of Hydra also involves the Wnt system. Here, most Wnt factors are expressed around the hypostome and are needed for “head” regeneration 35, 36. The “head” of hydroids is considered by some to be homologous to the posterior of bilaterians 37, although there remains considerable uncertainty about whether there are any homologies of body plan at all between the basal metazoa (cnidarians, ctenophores and sponges) and the bilaterian animals 38.
A comparable story has emerged about BMP signalling and the control of dorsoventral pattern. In development, probably all bilaterian animals have a system of BMP signalling antagonised by BMP inhibitors 39. In chordates, BMP is expressed on the future ventral side, in protostome invertebrates on the future dorsal side. In Xenopus embryos it has been shown that an interaction between BMP and ADMP (anti‐dorsalising morphogenetic protein) can explain the proportion regulation which occurs after parts of the embryo are excised 40. Adult planarians also have a BMP‐ADMP system controlling dorsoventral pattern. BMP is expressed in subepidermal cells along the dorsal midline and ADMP along the ventral midline and lateral regions. Terminal regeneration does occur in the absence of BMP signalling, provoked by appropriate RNAi treatment, but the new structures are ventralised. Lateral regeneration of thin parasagittal strips (i.e. the thin part of a worm transected longitudinally to one side of the midline) requires the re‐establishment of BMP secreting dorsal cells and this does need BMP and ADMP, suggesting a similar relationship between these factors as is found during the regulation of Xenopus early embryos 41, 42, 43.
Recently, a group of worms called acoels have been studied 44. These superficially resemble planaria, but molecular phylogeny studies show that they do not fall within the phylum Platyhelminthes, and are probably basal Bilateria equally related to protostomes and deuterostomes. They also have a BMP‐ADMP system very similar to that of planaria, and in view of their phylogenetic position, this system was most likely already established in the first ancestors of the modern bilaterian animals.
In Fig 2 is a phylogenetic tree of metazoa showing some of the recently studied instances of whole‐body regeneration, together with the role of the Wnt and BMP systems in controlling pattern. Data are necessarily scanty, and often the Wnt and BMP studies relate to different species from the analysis of regeneration behaviour. Although the Wnt system controlling AP pattern and the BMP system controlling DV pattern are certainly of evolutionary antiquity, it is not currently clear whether they maintain these roles in the stages of the life cycle of those echinoderms and urochordates, which are capable of whole‐body regeneration.
If persistent gradients of Wnt and BMP are needed for whole‐body regeneration, then the process may be incompatible with the hierarchical type of development typical of vertebrates or insects. In these well‐studied groups, there is a succession of developmental decisions in response to signalling molecules 45. During each of these, a cell population becomes divided into more than one population having different states of commitment. For the next subdivision, the competences of the cell populations change such that a new set of states of commitment can be accessed. There are usually about six developmental decisions between the pluripotent stage (blastula, blastoderm, epiblast) and the terminally differentiated cell types. If, for example, a graded Wnt signal controls the patterning of the posterior to anterior axis, then the competence to respond to Wnt in this way is normally lost after the primary body plan has been determined. Wnt signals are reused to pattern specific organ rudiments such as the heart or limbs, but now the competence is altered and the target genes are different from those responsible for whole‐body patterning. Under these circumstances even if a Wnt gradient were re‐established across the whole adult body, it would not elicit the correct responses for whole‐body regeneration. To do this, we should require also that the cells near the wound were dedifferentiated right back to an epiblast or pluripotent state. This degree of dedifferentiation has not been seen in vertebrates except when forced by the methods used to make iPS cells.
Pluripotent stem cells and regeneration
In a survey of regenerative ability by different groups of planarian, there is a very good correlation between those groups capable of whole‐body regeneration and those which normally undergo asexual regeneration by fission 27. This supports the view that whole‐body regeneration has been maintained as a by‐product of asexual reproduction. In planarians, the only dividing cells are the neoblasts, which maintain the body under normal conditions as well as providing cells for regeneration following transection. There is evidence that at least some of the neoblasts are pluripotent, at least under certain experimental conditions 16. Two methods were used to establish this. The first is an adaptation of older work done on the mammalian intestine. Worms were X‐irradiated to a level that destroys almost all their neoblasts. This means that worms that recover must do so by regeneration from one or a few surviving neoblasts. The regenerating clones repopulate all the cell types so presumably the originating cell for each clone was pluripotent, at least under the conditions of extreme radiation stress. The other method involves transplantation of single cells to genetically distinguishable lethally irradiated hosts. A small proportion of transplants enable successful regeneration and the formation of a complete worm composed of donor cells, which is itself capable of further growth and of asexual reproduction. Since neoblasts comprise all the dividing cells of a planarian, it seems unlikely that all of them could be pluripotent and more likely that they comprise some sort of stem cell hierarchy. A recent study of neoblasts using single‐cell RNAi shows that there are at least two identifiable classes called ζ, which are lineage‐committed, and σ which have a wider potency 46. Many neoblasts express some genes indicative of early differentiation 47, suggesting that the proportion of truly pluripotent cells is small.
In vertebrate development, there are probably no pluripotent cells remaining after the formation of the general body plan from the epiblast/blastoderm/blastula. Analysis of the requirement for Oct4 activity in the mouse indicates that there is none after this stage, except in developing germ cells 48. Reports of the presence of pluripotent cells in adult mammals have been hard to reproduce, and intensive study has indicated that tissue‐specific stem cells are, indeed, tissue specific and not pluripotent 49, 50. It has often been suggested that the persistence of pluripotent cells would be dangerous, as their artificial generation in vivo leads to tumours 18.
Are pluripotent stem cells needed for whole‐body regeneration? It is tempting to draw an association but so far the evidence only relates to planaria, which do have pluripotent cells, and mammals, which do not. Curiously, the example of Hydra suggests that pluripotent cells may not be necessary. Here, the stem cell‐like interstitial cells are not pluripotent and the organism consists of three distinct cell lineages, whose proportions are maintained during growth and which do not seem to interconvert 51.
Evidence for specific adaptations in structural and tissue regeneration
Non‐lethal predation of regenerating body parts
In many types of animal, there is substantial evidence of non‐lethal predation. Lizard tails, brittle star arms, polychaete heads, insect legs, bivalve siphons and many others may often be found in the process of regeneration 6. Sometimes, the predation pressure is extreme. For example, ref. 52 reported that bivalve siphons were cropped 25 times each season by juvenile fish; or, at any one time, 97% of an Antarctic brittle star were found to have lost parts of their arms 53. Surprisingly, considering the vast amount of research on regeneration of the urodele limb, there seem to be remarkably few studies of predation of urodele amphibians in the wild. But some recent reports from Eastern Europe indicate that significant levels of limb and tail predation are indeed found in wild newts, finding, respectively, 24% of injuries 54, 9% 55 and 21% 56. These various studies do support the idea of sustained selective pressure to maintain, or evoke, regenerative ability in numerous animal groups, including the well‐studied urodeles. However, quantitative confirmation of selective pressure is still lacking. It remains necessary to show that the loss of reproductive fitness due to the injury exceeds that due to the regeneration process itself, for example the extra energy requirement and the potential inconvenience of having a partly regenerated structure for a period of time. So although there are field data showing abundant predation, it is difficult at present to make a definitive statement about the extent to which regenerative abilities are actually maintained by selection due to predation. The existence of non‐lethal predation of regenerating parts is a necessary condition for de novo adaptation. It is also consistent with long‐term retention of an ancestral ability, but in this case evidence would also need to be provided for consistent retention right back to the origin of the lineage (e.g. the earliest limbed vertebrate in the case of limb regeneration).
New genes required for regeneration
Evidence for adaptation may be found through the existence of molecules essential to regeneration that are present only in a regenerating taxon, and not in its non‐regenerating relatives. New genes arise frequently in evolution, both by gene duplication and divergence, and from formerly non‐expressed DNA 57. An example is provided by the gene for the GPI‐anchored protein Prod1, which behaves like a proximalising factor in limb regeneration 58. One of the remarkable attributes of urodele limb regeneration is that the proximal–distal pattern of the regenerate starts at exactly the level of the amputation. The character of the blastema in terms of proximal–distal fate is known to be correlated with a gradient of cell adhesion in the limb 59. Prod1 is expressed in a gradient, high at the proximal end and low at the distal end of the limb. It antagonises cell adhesion, and experiments with blocking antibodies indicate that it is responsible for the gradient of adhesivity. When regenerating limbs are treated with retinoic acid, an increase in Prod1 is correlated with a proximalisation of the character of the blastema in terms of the structures subsequently regenerated. Furthermore, when overexpressed by electroporation, it can proximalise a blastema in terms both of cell movements and of the structures formed 58. All these properties indicate that Prod1 is intimately involved in the regeneration mechanism. But, although there are distant homologs of Prod1 in other animals, the Prod1 gene itself has been found to occur only in salamanders 60. Maybe its appearance was a key evolutionary novelty responsible for, or necessary for, limb regeneration in salamanders. Another, previously known, example of a regeneration‐specific gene product is the newt‐specific retinoic acid receptor δ2, needed for response of the blastema to the proximalising effect of retinoic acid 61, 62.
Modest degrees of dedifferentiation
It is often implied that all types of regeneration require extreme dedifferentiation from functional cell types to pluripotent cells. But recent results have tended to show that regeneration involves much less dedifferentiation than had previously been supposed. It also involves very little metaplasia: namely, the formation of one cell type from dedifferentiated cells derived from another cell type. This was first shown in the Xenopus tadpole tail, in which tadpoles were constructed by embryonic grafts to have a part of one specific tissue type genetically labelled. The results showed that regeneration following amputation occurred in all cases from the cell populations already present in the tail (specifically spinal cord, notochord, muscle satellite cells and melanophores) 20, 63, 64. Similar methods, along with some grafts of pure tissue explants, were used to study the regenerating limb of the axolotl 21 (Fig 3). Here it was found that epidermis, muscle, connective tissue and nerve all regenerated from the corresponding cell types in the stump. There is, however, some dedifferentiation in this case, as the connective tissues (fibroblasts, tendons, cartilage, ligaments) are capable of interconversion, as had been shown previously using triploid marked grafts of single tissue types 65.
Figure 3. How to make a blastema with cells of a specific tissue type labelled.
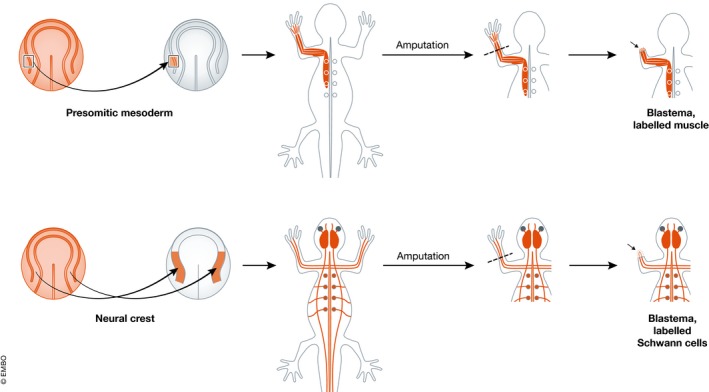
A graft is made at the neurula stage from a uniformly GFP‐labelled embryo to an unlabelled host. This graft populates only one tissue type. Amputation of the limb generates a blastema with a heritable label in the cells derived from that tissue type. See Kragl et al 21.
Even in the mouse digit tip, where a limited degree of regeneration is found in adult life, a heroic series of Cre‐lox procedures showed that the regenerated digit tip contained tissues derived from the respective precursors in the stump without metaplasia 66, 67. Epidermis and its derivatives were labelled using K14‐CreER. Connective tissues were labelled with Prx1‐Cre. Bone and cartilage within the mesoderm were labelled with Sox9‐Cre (this also labels epidermis). Tendons were labelled with Scleraxis‐Cre. Vessel endothelia were labelled with Tie2‐Cre (this also labels blood cells) and VE‐cadherin‐CreER. The reporter mouse strain used was mTmG, which switches from tdTomato to GFP expression when recombination is catalysed by Cre. Where CreER strains were used, they were activated by tamoxifen before amputation. In addition, the contribution of haematopoietic cells to the regenerate was investigated using two methods. First, haematopoietic stem cells were purified by FACS from a CFP‐expressing donor and grafted into conditioned hosts before amputation. Secondly, the experimental mice were parabiosed with GFP‐labelled partners before amputation. In neither case was there any contribution of labelled cells to the tissues of the regenerate other than blood cells.
Comparable results have also been obtained on other models. Two studies of the regeneration of the zebrafish caudal fin, one using transposon labels of different tissue types, and the other Cre‐lox labelling, both showed a lack of metaplasia in the regenerate 68, 69. In the amphipod crustacean Parhyale hawaiensis, specific cell lineages were labelled using a transposon injected into individual embryonic blastomeres populating ectoderm, mesoderm, endoderm or the germ line. Embryos with appropriate labelling patterns were grown up and subjected to appendage amputation. The regenerated appendages showed a complete germ line restriction of labelled cells indicating that the epidermis and muscles came from separate lineages 70.
So a wide range of recent studies using various labelling techniques on several vertebrate and one invertebrate model have all shown a very modest level of metaplasia or none at all. This suggests that the degree of dedifferentiation deployed in regeneration is the minimum required to provide cells for the regenerate. It indicates that a high dedifferentiation ability, such as complete reversion to pluripotency, has not been retained in evolution.
Differences between related taxa
One particular recent set of investigations of cell lineage in regeneration concerns the origin of new multinucleated muscle fibres. In mammals, multinucleate muscle fibres are completely post‐mitotic, not only not dedifferentiating, but not even undergoing DNA replication. New muscle fibres in mammals can arise from muscle satellite cells, which are specialised muscle stem cells that reside beneath the basement membrane of the muscle fibres and are responsible for fibre regeneration following muscle damage 71. In regeneration of the urodele limb, it was long believed that muscle fibres dedifferentiated into proliferating mononuclear cells. Originally the evidence for this was based on electron microscope studies 72. In the 1990s, the laboratory of Brockes carried out some lineage label experiments in axolotls and newts using retroviral labelling of implanted muscle fibres followed by amputation 73, 74. This indicated the reality of dedifferentiation and some formation of muscle in the regenerate by the labelled cells. But later it was found for the axolotl limb, and also the Xenopus tadpole tail, that the muscle of the regenerated limb was entirely derived from satellite cells and not from dedifferentiation 63, 75. In the amphipod P. hawaiensis, the identification of PAX3/7‐positive cells in the muscle, capable of generating muscle on regeneration, also suggested a close similarity to the axolotl 70.
Recently, this discordance of results has been investigated more thoroughly using Cre‐lox labelling and it turns out that, in the axolotl (Ambystoma mexicanum), muscle regeneration indeed occurs from satellite cells, while in the newt (Notophthalamus viridescens) it occurs by dedifferentiation of fibres to yield mononuclear myoblasts 76. This was a considerable surprise. Because the morphological events of limb regeneration are very similar across urodele amphibians, several species have been used in research in the past, and results have generally been supposed valid for all.
Can we deduce anything about the main question of this article from the muscle example? We have apparently a sharp difference of regeneration mechanism in two similar looking animals. One uses a normal method of muscle regeneration while the other uses a novel mechanism not found in normal development. This does suggest that in the case of the newt Notophthalamus, there has been evolution of a novel mechanism, even though Notophthalamus also has muscle satellite cells. If we did not know about Notophthalamus, from our knowledge of mammals, the axolotl and Parhyale we might easily have concluded that all muscle in all animals regenerates from satellite cells. Although Notophthalamus and Ambystoma look fairly similar to us, they have had plenty of time to diversify. The evolutionary divergence of the lines leading to the modern genera is estimated to have taken place about 100 million years ago 77 This is within the Cretaceous period, similar to the divergence time of armadillos and humans, and so does give plenty of time for diversification. We do not know what selective pressures drove the process but it could be that the capacity for limb regeneration arose independently in the two lineages, or was lost and reacquired with a different mechanism in the lineage leading to Notophthalamus.
The case of the spiny mouse
Most knowledge of mammalian regeneration, or non‐regeneration, comes from those very well‐known laboratory rodents: the rat and the mouse. We are here considering tissue regeneration, rather than the regeneration of new pattern, according to the classification of Bely and Nyberg 6, although it fits within the current section as a probable example of adaptation.
In rats and mice, wounds in the skin are repaired by growth of the epidermis but this is underlain by a collagen‐rich scar and fails to regenerate epidermal appendages such as hair follicles. There is a little more regeneration normally found in the rabbit, which can regenerate ear punch holes, an ability lacking in most strains of mouse. Recently, a study on two species of Acomys, the African spiny mouse, has shown a significantly greater regenerative ability than in the laboratory mouse 78, 79. Large wounds in the skin are rapidly healed, partly by contraction and partly by rapid epidermal growth. There is little scar formation in the dermis. Few collagen I‐secreting myofibroblasts are formed, and the extracellular matrix reformed in the wound area is soft, with a high content of tenascin and fibronectin and collagen III in place of collagen I. Most interesting, unlike in normal mice, the hair follicles and other epidermal appendages are present in the new skin. Ear punch holes can also close rapidly, with regeneration of cartilage, dermis, adipose tissue and epidermis with hair follicles.
The skin of the spiny mice is very fragile: tensile strength measurements showed that it is 20 times weaker than normal mouse skin. Because of this, the skin is easily detached by modest insults. It has been suggested that Acomys has evolved the ability to shed skin to enable escape from predators, and the rapid regeneration is an aspect of this adaptation. If this is correct, then this particular regenerative ability would support the evolutionary novelty viewpoint. Acomys also illustrates a general tendency for regeneration and scarring to be mutually exclusive 80. It has long been known that the skin of amphibians and of foetal mammals heals without scarring 81. This is associated with a much lower level of inflammation than is found in wounds in adult mice or humans. In scar‐free healing, macrophages are absent and there is a much lower level of proinflammatory cytokines, although at least some types of macrophage are apparently needed for urodele limb regeneration 82. It is generally supposed that both the inflammatory response and the scar formation in mammals are adaptations to avoid infection and to close wounds rapidly, even if full functionality is not restored. The adaptation for skin shedding and rapid regeneration shown by Acomys evidently involves a reduction in inflammation, so perhaps it has also evolved other methods for avoiding infection.
Discussion
This article has only considered a few of the interesting results uncovered by regeneration research in recent years. These were selected in the hope that they might have something to say about the essential dichotomy presented in the title. It is always difficult to arrive at firm conclusions on evolutionary matters but these results do seem to point to a difference between whole‐body regeneration, which is probably ancestral although has been lost in most animal lineages, and the structural regeneration of appendages, where there currently is more evidence for de novo adaptation.
Whole‐body regeneration and pluripotency
The occurrence of whole‐body regeneration in acoels as well as in some taxa of more derived metazoa (platyhelminths, annelids, echinoderms, urochordates) suggests that this capacity is ancestral although it has been lost from most animal lineages. Because of the good correlation between whole‐body regeneration and the occurrence of asexual reproduction by fission, it seems likely that the selective force for persistence arises from the need to maintain asexual reproduction. This constraint would not exist for asexual reproduction by parthenogenesis, which is found in many animal groups that do not regenerate.
But it is not quite the same to say that whole‐body regeneration is ancestral and to say that it is an inherent quality of living matter. To be inherent, regenerative behaviour has not only to be ancestral but also to be retained in all lineages, even though it may be masked in some way, and it is doubtful whether this is the case.
Although evidence only really exists for the planarians and acoels, the control of polarity and patterning in whole‐body regeneration seems to rely on the graded distributions of Wnt and BMP. These factors are involved during development in setting up the body axes of animals across the Bilateria, and their persistence into adult life may be necessary for whole‐body regeneration. In planarians and acoels, the “adult” worms retain embryonic properties in that the gradients are still active and cell renewal depends on continuous production of cells from a population of neoblasts including at least some pluripotent stem cells. But most groups of Bilateria do not show this persistent embryonic state, and it may be that in its absence, whole‐body regeneration is not possible. Development of the more complex animals typically involves a hierarchy of decisions. Each involves the subdivision of a field of cells in response to a graded factor, and factors such as Wnts, BMPs, fibroblast growth factors (FGFs) and hedgehogs (HHs) are reused for several decisions. As indicated above, the competence of cells changes at each stage, so even if there were gradients of Wnt and BMP present in adult life, they would not evoke the same responses as in the early embryo. There seems no obvious way in which a hierarchical mode of development would be compatible with whole‐body regeneration.
The other essential ingredient for whole‐body regeneration is cells with a high degree of dedifferentiation. In planarians, there are always pluripotent cells present, so no dedifferentiation is needed for regeneration. But in most metazoa, pluripotent cells will probably have disappeared after the early stages of embryonic development. Certainly in vertebrates, regeneration does not utilise pluripotent cells. If the tissue types of the missing part are the same as those found near the amputation plane, as in the tail, then there is no need for dedifferentiation at all. The dedifferentiation found in the regenerating urodele limb is just for the connective tissues and involves a one‐step reversion to something resembling a limb mesenchyme state, not a full reversion to pluripotency. For appendage regeneration in arthropods, regeneration occurs during a moult cycle and the tissues of epidermis and mesoderm remain separate.
In terms of the selective costs of regeneration, it is possible that regenerative ability itself is dangerous because it can lead to uncontrolled growth. In mammals, the relative stability of states of cell differentiation, and the restriction of persistent cell division to small cell populations in adult animals are often viewed as defences against cancer. In particular, we know that pluripotent stem cells are potentially dangerous. When pluripotent cells are grafted into adult mammals, they do not integrate into the host tissues but instead give rise to tumours called teratomas 19. Naturally occurring germ cell tumours are also believed to arise from pluripotent cells.
In conclusion, whole‐body regeneration may be ancestral but its maintenance in evolution is probably not compatible with the hierarchical nature of development, the absence of persistent pluripotent cells, or the absence of dedifferentiation to pluripotency. It would not therefore be correct to say that it was an inherent or essential attribute of animal life.
Structural and tissue regeneration
In the case of the urodeles, there is now evidence for significant non‐lethal predation of limbs and tails in the wild. While we do not know the true costs and benefits of limb or tail regeneration, this is at least consistent with distal regeneration in salamanders being an adaptation to predation which need not have been retained since the origin of limbs. In addition, at least one key molecular component responsible for proximal–distal patterning, Prod1, is unique to salamanders. The remarkable difference between types of salamander in the mechanism of muscle regeneration cannot be explained, but it does provide further evidence of evolutionary novelty in this area. Although limb regeneration reuses many of the molecular components responsible for limb development, there is another feature not found in development, which has been known for a very long time, namely nerve dependence. Some of the underlying mechanism for this is now known 83 and nerve dependence does seem like the sort of additional control that would minimise the possibility of neoplastic growths arising from dedifferentiation in situations other than a limb or tail amputation. The reuse of embryonic mechanisms may be tempered by the different scale of regeneration: a large salamander will have a limb maybe 1 cm across, whereas the embryonic limb bud is patterned by gradients operating across about 100 μm. The rate of diffusion goes down with the square of the distance, and large limbs do regenerate very slowly, although other modifications to mechanism may be needed as well.
The remarkable skin shedding ability of the spiny mouse looks like a specific adaptation to predation. The improved regenerative ability, as compared to normal mice, is achieved at the cost of less inflammation and scarring. So presumably there are additional, as yet unknown, adaptations that make this mouse resistant to infection after suffering very large wounds to the skin.
Conclusion
The more we get to know about regeneration, the more it appears to represent a large range of somewhat disparate processes, and the less there appear to be any general principles governing all of them together. So it does help to consider separately each of the types of regeneration. The recent results surveyed in this article do not allow very firm conclusions to be drawn, but there is perhaps a suggestion that whole‐body regeneration is genuinely ancestral, although becomes irretrievably lost in animals which show a hierarchical, multistep, type of embryonic development. By contrast, the structural regeneration of appendages seems more likely to be an adaptive response to non‐lethal predation, although it does tend to reuse many developmental mechanisms, which gives an appearance of ancestrality.
These tentative conclusions do not mean that it will not be possible one day to engineer replacement human organs using cellular reprogramming and tissue engineering techniques, which are currently advancing rapidly. But it does suggest that there will probably not be a simple elixir that can be sprinkled onto a human amputation stump to make it grow again.
Conflict of interest
The author declares that he has no conflict of interest.
Box 1: In need of answers.
Are pluripotent cells needed for whole‐body regeneration?
When in evolutionary time did the first animals lose the ability to undergo whole‐body regeneration and why?
Was it because of the acquisition of more than one level in the developmental hierarchy?
-
To what extent does stable inactive chromatin inhibit dedifferentiation and regeneration?
Research progress on these issues will require a better understanding of the molecular developmental biology of regeneration in understudied animal groups showing whole‐body regeneration, especially the annelids and basal chordates.
How often are new genes found involved in examples of structural (appendage) regeneration?
-
How often do mechanisms of appendage regeneration differ between similar taxa of animals?
These are both likely to be associated with recent adaptations. Research progress will require study at the molecular level of a wider range of urodele species showing limb regeneration, and of more groups of amphibians and reptiles which undergo tail regeneration.
EMBO Reports (2017) 18: 1497–1508
See the Glossary for abbreviations used in this article.
References
- 1. Brockes JP, Kumar A (2008) Comparative aspects of animal regeneration. Annu Rev Cell Dev Biol 24: 525–549 [DOI] [PubMed] [Google Scholar]
- 2. Nakamura T, Mito T, Bando T, Ohuchi H, Noji S (2008) Dissecting insect leg regeneration through RNA interference. Cell Mol Life Sci 65: 64–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nacu E, Tanaka EM (2011) Limb regeneration: a new development? Annu Rev Cell Dev Biol 27: 409–440 [DOI] [PubMed] [Google Scholar]
- 4. Simon A, Tanaka EM (2013) Limb regeneration. Wiley Interdiscip Rev Dev Biol 2: 291–300 [DOI] [PubMed] [Google Scholar]
- 5. Elliott SA, Alvarado AS (2013) The history and enduring contributions of planarians to the study of animal regeneration. Wiley Interdiscip Rev Dev Biol 2: 301–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bely AE, Nyberg KG (2010) Evolution of animal regeneration: re‐emergence of a field. Trends Ecol Evol 25: 161–170 [DOI] [PubMed] [Google Scholar]
- 7. Slack JMW (1980) A serial threshold theory of regeneration. J Theor Biol 82: 105–140 [DOI] [PubMed] [Google Scholar]
- 8. Berg DA, Kirkham M, Wang H, Frisen J, Simon A (2011) Dopamine controls neurogenesis in the adult salamander midbrain in homeostasis and during regeneration of dopamine neurons. Cell Stem Cell 8: 426–433 [DOI] [PubMed] [Google Scholar]
- 9. Rychel AL, Swalla BJ (2008) Anterior regeneration in the hemichordate Ptychodera flava . Dev Dyn 237: 3222–3232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rinkevich Y, Paz G, Rinkevich B, Reshef R (2007) Systemic bud induction and retinoic acid signaling underlie whole body regeneration in the urochordate Botrylloides leachi . PLoS Biol 5: 900–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bely AE (2010) Evolutionary loss of animal regeneration: pattern and process. Integr Comp Biol 50: 515–527 [DOI] [PubMed] [Google Scholar]
- 12. Tanaka EM (2016) The molecular and cellular choreography of appendage regeneration. Cell 165: 1598–1608 [DOI] [PubMed] [Google Scholar]
- 13. Wang J, Davis RE (2014) Programmed DNA elimination in multicellular organisms. Curr Opin Genet Dev 27: 26–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Maherali N, Hochedlinger K (2008) Guidelines and techniques for the generation of induced pluripotent stem cells. Cell Stem Cell 3: 595–605 [DOI] [PubMed] [Google Scholar]
- 15. Sánchez Alvarado A, Yamanaka S (2014) Rethinking differentiation: stem cells, regeneration, and plasticity. Cell 157: 110–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wagner DE, Wang IE, Reddien PW (2011) Clonogenic neoblasts are pluripotent adult stem cells that underlie planarian regeneration. Science 332: 811–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Slack JMW (2008) Origin of stem cells in organogenesis. Science 322: 1498–1501 [DOI] [PubMed] [Google Scholar]
- 18. Abad M, Mosteiro L, Pantoja C, Canamero M, Rayon T, Ors I, Grana O, Megias D, Dominguez O, Martinez D et al (2013) Reprogramming in vivo produces teratomas and iPS cells with totipotency features. Nature 502: 340–345 [DOI] [PubMed] [Google Scholar]
- 19. Bulic‐Jakus F, Katusic Bojanac A, Juric‐Lekic G, Vlahovic M, Sincic N (2016) Teratoma: from spontaneous tumors to the pluripotency/malignancy assay. Wiley Interdiscip Rev Dev Biol 5: 186–209 [DOI] [PubMed] [Google Scholar]
- 20. Gargioli C, Slack JMW (2004) Cell lineage tracing during Xenopus tail regeneration. Development 131: 2669–2679 [DOI] [PubMed] [Google Scholar]
- 21. Kragl M, Knapp D, Nacu E, Khattak S, Maden M, Epperlein HH, Tanaka EM (2009) Cells keep a memory of their tissue origin during axolotl limb regeneration. Nature 460: 60–65 [DOI] [PubMed] [Google Scholar]
- 22. Grewal SIS, Moazed D (2003) Heterochromatin and epigenetic control of gene expression. Science 301: 798–802 [DOI] [PubMed] [Google Scholar]
- 23. Niehrs C (2010) On growth and form: a Cartesian coordinate system of Wnt and BMP signaling specifies bilaterian body axes. Development 137: 845–857 [DOI] [PubMed] [Google Scholar]
- 24. Gurley KA, Rink JC, Alvarado AS (2008) {beta}‐Catenin defines head versus tail identity during planarian regeneration and homeostasis. Science 319: 323–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Petersen CP, Reddien PW (2008) Smed‐{beta}catenin‐1 is required for anteroposterior blastema polarity in planarian regeneration. Science 319: 327–330 [DOI] [PubMed] [Google Scholar]
- 26. Petersen CP, Reddien PW (2009) Wnt signaling and the polarity of the primary body axis. Cell 139: 1056–1068 [DOI] [PubMed] [Google Scholar]
- 27. Egger B, Gschwentner R, Rieger R (2007) Free‐living flatworms under the knife: past and present. Dev Genes Evol 217: 89–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sikes JM, Newmark PA (2013) Restoration of anterior regeneration in a planarian with limited regenerative ability. Nature 500: 77–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Umesono Y, Tasaki J, Nishimura Y, Hrouda M, Kawaguchi E, Yazawa S, Nishimura O, Hosoda K, Inoue T, Agata K (2013) The molecular logic for planarian regeneration along the anterior‐posterior axis. Nature 500: 73–76 [DOI] [PubMed] [Google Scholar]
- 30. Liu SY, Selck C, Friedrich B, Lutz R, Vila‐Farre M, Dahl A, Brandl H, Lakshmanaperumal N, Henry I, Rink JC (2013) Reactivating head regrowth in a regeneration‐deficient planarian species. Nature 500: 81–84 [DOI] [PubMed] [Google Scholar]
- 31. Lin G, Slack JMW (2008) Requirement for Wnt and FGF signaling in Xenopus tadpole tail regeneration. Dev Biol 316: 323–335 [DOI] [PubMed] [Google Scholar]
- 32. Stoick‐Cooper CL, Weidinger G, Riehle KJ, Hubbert C, Major MB, Fausto N, Moon RT (2007) Distinct Wnt signaling pathways have opposing roles in appendage regeneration. Development 134: 479–489 [DOI] [PubMed] [Google Scholar]
- 33. Vitulo N, Dalla Valle L, Skobo T, Valle G, Alibardi L (2017) Transcriptome analysis of the regenerating tail vs. the scarring limb in lizard reveals pathways leading to successful vs. unsuccessful organ regeneration in amniotes. Dev Dyn 246: 116–134 [DOI] [PubMed] [Google Scholar]
- 34. Stefanelli A, Thermes G, Poddie M (1950) La Rigenerazione del midollo spinale delle larve di anfibi anuri e sue relazioni con la corda dorsale. Riv Biol 42: 239–264 [PubMed] [Google Scholar]
- 35. Lengfeld T, Watanabe H, Simakov O, Lindgens D, Gee L, Law L, Schmidt HA, Özbek S, Bode H, Holstein TW (2009) Multiple Wnts are involved in Hydra organizer formation and regeneration. Dev Biol 330: 186–199 [DOI] [PubMed] [Google Scholar]
- 36. Gee L, Hartig J, Law L, Wittlieb J, Khalturin K, Bosch TCG, Bode HR (2010) [beta]‐catenin plays a central role in setting up the head organizer in Hydra. Dev Biol 340: 116–124 [DOI] [PubMed] [Google Scholar]
- 37. Meinhardt H (2004) Different strategies for midline formation in bilaterians. Nat Rev Neurosci 5: 502–510 [DOI] [PubMed] [Google Scholar]
- 38. Martindale MQ (2005) The evolution of metazoan axial properties. Nat Rev Genet 6: 917–927 [DOI] [PubMed] [Google Scholar]
- 39. DeRobertis EM, Sasai Y (1996) A common plan for dorsoventral patterning in Bilateria. Nature 380: 37–40 [DOI] [PubMed] [Google Scholar]
- 40. Reversade B, De Robertis EM (2005) Regulation of ADMP and BMP2/4/7 at opposite embryonic poles generates a self‐regulating morphogenetic field. Cell 123: 1147–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Reddien PW, Bermange AL, Kicza AM, Sanchez Alvarado A (2007) BMP signaling regulates the dorsal planarian midline and is needed for asymmetric regeneration. Development 134: 4043–4051 [DOI] [PubMed] [Google Scholar]
- 42. Gavino MA, Reddien PW (2011) A BMP/ADMP regulatory circuit controls maintenance and regeneration of dorsal‐ventral polarity in planarians. Curr Biol 21: 294–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Molina MD, Neto A, Maeso I, Gomez‐Skarmeta JL, Salo E, Cebria F (2011) Noggin and noggin‐like genes control dorsoventral axis regeneration in planarians. Curr Biol 21: 300–305 [DOI] [PubMed] [Google Scholar]
- 44. Srivastava M, Mazza‐Curll KL, van Wolfswinkel JC, Reddien PW (2014) Whole‐body acoel regeneration is controlled by Wnt and BMP‐ADMP signaling. Curr Biol 24: 1107–1113 [DOI] [PubMed] [Google Scholar]
- 45. Slack J (2014) Establishment of spatial pattern. Wiley Interdiscip Rev Dev Biol 3: 379–388 [DOI] [PubMed] [Google Scholar]
- 46. van Wolfswinkel Josien C, Wagner Daniel E, Reddien Peter W (2014) Single‐Cell analysis reveals functionally distinct classes within the planarian stem cell compartment. Cell Stem Cell 15: 326–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Scimone ML, Kravarik KM, Lapan SW, Reddien PW (2014) Neoblast specialization in regeneration of the planarian Schmidtea mediterranea . Stem Cell Rep 3: 339–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lengner CJ, Camargo FD, Hochedlinger K, Welstead GG, Zaidi S, Gokhale S, Scholer HR, Tomilin A, Jaenisch R (2007) Oct4 expression is not required for mouse somatic stem cell self‐renewal. Cell Stem Cell 1: 403–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wagers AJ, Weissman IL (2004) Plasticity of adult stem cells. Cell 116: 639–648 [DOI] [PubMed] [Google Scholar]
- 50. Phinney DG, Prockop DJ (2007) Concise review: mesenchymal stem/multipotent stromal cells: the state of transdifferentiation and modes of tissue repair current views. Stem Cells 25: 2896–2902 [DOI] [PubMed] [Google Scholar]
- 51. Hobmayer B, Jenewein M, Eder D, Eder MK, Glasauer S, Gufler S, Hartl M, Salvenmoser W (2012) Stemness in Hydra—a current perspective. Int J Dev Biol 56: 509–517 [DOI] [PubMed] [Google Scholar]
- 52. Sasaki K, Kudo M, Tomiyama T, Ito K, Omori M (2002) Predation pressure on the siphons of the bivalve Nuttallia olivacea by the juvenile stone flounder Platichthys bicoloratus in the Natori River estuary, north‐eastern Japan. Fish Sci 68: 104–116 [Google Scholar]
- 53. Clark MS, Dupont S, Rossetti H, Burns G, Thorndyke MC, Peck LS (2007) Delayed arm regeneration in the Antarctic brittle star Ophionotus victoriae . Aquat Biol 1: 45–53 [Google Scholar]
- 54. Nemes S (2002) Morphometry of metamorphosed smooth newts Triturus vulgaris (Amphibia : Salamandridae) with notes on limb, toe and tail injury frequency. Biologia 57: 615–619 [Google Scholar]
- 55. Kopecky O (2013) Predation‐induced injuries in wild populations of alpine newt. Pak J Zool 45: 417–422 [Google Scholar]
- 56. Macat Z, Jerabkova L, Reiter A, Rulik M, Jablonski D (2015) Malformations and body injuries in a hybrid zone of crested newts (Caudata: Salamandridae: Triturus cristatus superspecies). Acta Herpetol 10: 135–141 [Google Scholar]
- 57. Tautz D, Domazet‐Loso T (2011) The evolutionary origin of orphan genes. Nat Rev Genet 12: 692–702 [DOI] [PubMed] [Google Scholar]
- 58. da Silva SM, Gates PB, Brockes JP (2002) The newt ortholog of CD59 is implicated in proximodistal identity during amphibian limb regeneration. Dev Cell 3: 547–555 [DOI] [PubMed] [Google Scholar]
- 59. Nardi JB, Stocum DL (1984) Surface properties of regenerating limb cells: evidence for gradation along the proximodistal axis. Differentiation 25: 27–31 [Google Scholar]
- 60. Garza‐Garcia A, Harris R, Esposito D, Gates PB, Driscoll PC (2009) Solution structure and phylogenetics of Prod1, a member of the three‐finger protein superfamily implicated in salamander limb regeneration. PLoS One 4: e7123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ragsdale CW, Petkovich M, Gates PB, Chambon P, Brockes JP (1989) Identification of a novel retinoic acid receptor in regenerative tissues of the newt. Nature 341: 654–657 [DOI] [PubMed] [Google Scholar]
- 62. Pecorino LT, Entwistle A, Brockes JP (1996) Activation of a single retinoic acid receptor isoform mediates proximodistal respecification. Curr Biol 6: 563–569 [DOI] [PubMed] [Google Scholar]
- 63. Chen Y, Lin GF, Slack JMW (2006) Control of muscle regeneration in the Xenopus tadpole tail by Pax7. Development 133: 2303–2313 [DOI] [PubMed] [Google Scholar]
- 64. Lin G, Chen Y, Slack JMW (2007) Regeneration of melanophores and other neural crest derivatives in the Xenopus tadpole tail. BMC Dev Biol 7: 56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Namenwirth M (1974) Inheritance of cell‐differentiation during limb regeneration in axolotl. Dev Biol 41: 42–56 [DOI] [PubMed] [Google Scholar]
- 66. Rinkevich Y, Lindau P, Ueno H, Longaker MT, Weissman IL (2011) Germ‐layer and lineage‐restricted stem/progenitors regenerate the mouse digit tip. Nature 476: 409–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Lehoczky JA, Robert B, Tabin CJ (2011) Mouse digit tip regeneration is mediated by fate‐restricted progenitor cells. Proc Natl Acad Sci USA 108: 20609–20614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Tu S, Johnson Stephen L (2011) Fate restriction in the growing and regenerating zebrafish fin. Dev Cell 20: 725–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Stewart S, Stankunas K (2012) Limited dedifferentiation provides replacement tissue during zebrafish fin regeneration. Dev Biol 365: 339–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Konstantinides N, Averof M (2014) A common cellular basis for muscle regeneration in arthropods and vertebrates. Science 343: 788–791 [DOI] [PubMed] [Google Scholar]
- 71. Fan C‐M, Li L, Rozo ME, Lepper C (2012) Making skeletal muscle from progenitor and stem cells: development versus regeneration. Wiley Interdiscip Rev Dev Biol 1: 315–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Hay ED (1959) Electron microscopic observations of muscle dedifferentiation in regenerating Amblystoma (sic) limbs. Dev Biol 1: 555–585 [Google Scholar]
- 73. Lo DC, Allen F, Brockes JP (1993) Reversal of muscle differentiation during limb regeneration in the axolotl. Proc Natl Acad Sci USA 90: 7230–7234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Kumar A, Velloso CP, Imokawa Y, Brockes JP (2000) Plasticity of retrovirus‐labelled myotubes in the newt limb regeneration blastema. Dev Biol 218: 125–136 [DOI] [PubMed] [Google Scholar]
- 75. Morrison JI, Loof S, He P, Simon A (2006) Salamander limb regeneration involves the activation of a multipotent skeletal muscle satellite cell population. J Cell Biol 172: 433–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Sandoval‐Guzman T, Wang H, Khattak S, Schuez M, Roensch K, Nacu E, Tazaki A, Joven A, Tanaka EM, Simon A (2014) Fundamental differences in dedifferentiation and stem cell recruitment during skeletal muscle regeneration in two salamander species. Cell Stem Cell 14: 174–187 [DOI] [PubMed] [Google Scholar]
- 77. Steinfartz S, Vicario S, Arntzen JW, Caccone A (2007) A Bayesian approach on molecules and behavior: reconsidering phylogenetic and evolutionary patterns of the Salamandridae with emphasis on Triturus newts. J Exp Zool B Mol Dev Evol 308B: 139–162 [DOI] [PubMed] [Google Scholar]
- 78. Seifert AW, Kiama SG, Seifert MG, Goheen JR, Palmer TM, Maden M (2012) Skin shedding and tissue regeneration in African spiny mice (Acomys). Nature 489: 561–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Brant JO, Lopez MC, Baker HV, Barbazuk WB, Maden M (2015) A comparative analysis of gene expression profiles during skin regeneration in Mus and Acomys . PLoS One 10: e0142931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Mescher AL, Neff AW (2005) Regenerative capacity and the developing immune system. Adv Biochem Eng Biotechnol 93: 39–66 [DOI] [PubMed] [Google Scholar]
- 81. Eming SA, Martin P, Tomic‐Canic M (2014) Wound repair and regeneration: mechanisms, signaling, and translation. Sci Transl Med 6: 265sr6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Godwin JW, Pinto AR, Rosenthal NA (2013) Macrophages are required for adult salamander limb regeneration. Proc Natl Acad Sci USA 110: 9415–9420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Kumar A, Godwin JW, Gates PB, Garza‐Garcia AA, Brockes JP (2007) Molecular basis for the nerve dependence of limb regeneration in an adult vertebrate. Science 318: 772–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Rivera YC, Hernandez RI, del Angel PS, Meza EZ, Gonzalez RC (2016) Regenerative potential of the sea star Linckia guildinguii . Hidrobiologica 26: 103–108 [Google Scholar]
- 85. McClay DR (2011) Evolutionary crossroads in developmental biology: sea urchins. Development 138: 2639–2648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Koop D, Cisternas P, Morris VB, Strbenac D, Yang JYH, Wray GA, Byrne M (2017) Nodal and BMP expression during the transition to pentamery in the sea urchin Heliocidaris erythrogramma: insights into patterning the enigmatic echinoderm body plan. BMC Dev Biol 17: 4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Darras S, Gerhart J, Terasaki M, Kirschner M, Lowe CJ (2011) beta‐Catenin specifies the endomesoderm and defines the posterior organizer of the hemichordate Saccoglossus kowalevskii . Development 138: 959–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Lowe CJ, Terasaki M, Wu M, Freeman RM, Runft L, Kwan K, Haigo S, Aronowicz J, Lander E, Gruber C et al (2006) Dorsoventral patterning in hemichordates: insights into early chordate evolution. PLoS Biol 4: e291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Imai K, Takada N, Satoh N, Satou Y (2000) (beta)‐Catenin mediates the specification of endoderm cells in ascidian embryos. Development 127: 3009–3020 [DOI] [PubMed] [Google Scholar]
- 90. Rosner A, Alfassi G, Moiseeva E, Paz G, Rabinowitz C, Lapidot Z, Douek J, Haim A, Rinkevich B (2014) The involvement of three signal transduction pathways in botryllid ascidian astogeny, as revealed by expression patterns of representative genes. Int J Dev Biol 58: 677–692 [DOI] [PubMed] [Google Scholar]
- 91. Kiecker C, Niehrs C (2001) A morphogen gradient of Wnt/beta‐catenin signalling regulates anteroposterior neural patterning in Xenopus . Development 128: 4189–4201 [DOI] [PubMed] [Google Scholar]
- 92. Dosch R, Gawantka V, Delius H, Blumenstock C, Niehrs C (1997) BMP‐4 acts as a morphogen in dorsoventral mesoderm patterning in Xenopus . Development 124: 2325–2334 [DOI] [PubMed] [Google Scholar]
- 93. Coudreuse DYM, Roël G, Betist MC, Destrée O, Korswagen HC (2006) Wnt gradient formation requires retromer function in Wnt‐producing cells. Science 312: 921–924 [DOI] [PubMed] [Google Scholar]
- 94. Bolognesi R, Farzana L, Fischer TD, Brown SJ (2008) Multiple Wnt Genes are required for segmentation in the short‐germ embryo of Tribolium castaneum . Curr Biol 18: 1624–1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Ferguson EL, Anderson KV (1992) Decapentaplegic acts as a morphogen to organize dorsoventral pattern in the Drosophila embryo. Cell 71: 451–461 [DOI] [PubMed] [Google Scholar]
- 96. Lindsay SM, Jackson JL, He SQ (2006) Anterior regeneration in the spionid polychaetes Dipolydora quadrilobata and Pygospio elegans . Mar Biol 150: 1161–1172 [Google Scholar]
- 97. Myohara M (2004) Differential tissue development during embryogenesis and regeneration in an annelid. Dev Dyn 231: 349–358 [DOI] [PubMed] [Google Scholar]
- 98. Pruitt MM, Letcher EJ, Chou HC, Bastin BR, Schneider SQ (2014) Expression of the Wnt gene complement in a spiral‐cleaving embryo and trochophore larva. Int J Dev Biol 58: 563–573 [DOI] [PubMed] [Google Scholar]
- 99. Kuo DH, Weisblat DA (2011) A new molecular logic for BMP‐mediated dorsoventral patterning in the leech helobdella. Curr Biol 21: 1282–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Bosch TCG (2007) Why polyps regenerate and we don't: towards a cellular and molecular framework for Hydra regeneration. Dev Biol 303: 421–433 [DOI] [PubMed] [Google Scholar]
- 101. Hobmayer B, Rentzsch F, Kuhn K, Happel CM, von Laue CC, Snyder P, Rothbacher U, Holstein TW (2000) WNT signalling molecules act in axis formation in the diploblastic metazoan Hydra. Nature 407: 186–189 [DOI] [PubMed] [Google Scholar]


