Abstract
Long non‐coding RNAs (lncRNAs) regulating gene expression at the chromatin level are widespread among eukaryotes. However, their functions and the mechanisms by which they act are not fully understood. Here, we identify new fission yeast regulatory lncRNAs that are targeted, at their site of transcription, by the YTH domain of the RNA‐binding protein Mmi1 and degraded by the nuclear exosome. We uncover that one of them, nam1, regulates entry into sexual differentiation. Importantly, we demonstrate that Mmi1 binding to this lncRNA not only triggers its degradation but also mediates its transcription termination, thus preventing lncRNA transcription from invading and repressing the downstream gene encoding a mitogen‐activated protein kinase kinase kinase (MAPKKK) essential to sexual differentiation. In addition, we show that Mmi1‐mediated termination of lncRNA transcription also takes place at pericentromeric regions where it contributes to heterochromatin gene silencing together with RNA interference (RNAi). These findings reveal an important role for selective termination of lncRNA transcription in both euchromatic and heterochromatic lncRNA‐based gene silencing processes.
Keywords: heterochromatin, non‐coding RNA (ncRNA), sexual differentiation, transcription, YTH domain
Subject Categories: Development & Differentiation, RNA Biology, Transcription
Introduction
Long non‐coding RNAs (lncRNAs) are widespread regulators of gene transcription and chromatin modification among eukaryotes. LncRNAs control gene expression, in cis or in trans, by serving as decoy or scaffold that interact with chromatin modifiers and remodelers to regulate the chromatin state of specific genomic sites (Rinn & Chang, 2012; Morris & Mattick, 2014). Studies on nuclear RNA interference (RNAi)‐mediated gene silencing have further provided evidence that RNAi co‐transcriptionally eliminates regulatory lncRNAs and, in addition, mediates the function of these lncRNAs in forming heterochromatin or silent chromatin in the fission yeast Schizosaccharomyces pombe and other eukaryotes (Castel & Martienssen, 2013). However, most of the co‐transcriptional activity eliminating lncRNAs relies on the conserved exosome complex instead of RNAi (Kilchert et al, 2016) and, in this case, the functional and physical connections between exosome‐dependent co‐transcriptional elimination of lncRNAs and lncRNA‐based gene silencing processes are poorly characterized.
Among the extensively studied cases of regulatory lncRNAs co‐transcriptionally controlled by RNAi are the S. pombe pericentromeric lncRNAs, which play a central role in the formation of heterochromatin (Buhler & Moazed, 2007; Cam et al, 2009), characterized by the methylation of histone H3 on lysine 9 (H3K9me; Grewal & Jia, 2007). Schizosaccharomyces pombe pericentromeric regions are mainly composed of DNA repeats, named dg and dh, transcribed by the RNA polymerase II (RNAPII; Djupedal et al, 2005; Kato et al, 2005). Production of dg and dh sense and anti‐sense lncRNAs is believed to lead to the formation of double‐stranded RNAs (dsRNAs; Reinhart & Bartel, 2002). The RNAi protein Dicer (Dcr1) processes dsRNAs into small interfering RNAs (siRNAs) that load on the RNA‐induced transcriptional gene silencing (RITS) complex (Verdel et al, 2004). RITS uses the siRNAs as guides to co‐transcriptionally base‐pair with nascent and complementary lncRNAs (Motamedi et al, 2004; Buhler et al, 2006) that are then eliminated by a cis‐acting positive feedback loop. In this loop, the targeted nascent lncRNAs serve as matrixes to locally synthesize double‐stranded RNAs and produce more siRNAs and RITS that bind the nascent pericentromeric lncRNAs (Motamedi et al, 2004; Colmenares et al, 2007). In complement to RNAi, the exosome complex and its cofactor, the TRAMP complex, also contribute to the elimination of dg and dh lncRNAs (Buhler et al, 2007; Wang et al, 2008; Reyes‐Turcu et al, 2011). The exosome is a highly conserved nucleocytoplasmic complex that degrades RNA (Chlebowski et al, 2013; Kilchert et al, 2016), including the nascent lncRNAs issued from pervasive transcription (Jensen et al, 2013). Because both Rrp6, a 3′ → 5′ exonuclease only present in the nuclear form of the exosome complex, and Cid14, a subunit of the TRAMP complex, localize in the vicinity of pericentromeric heterochromatin (Keller et al, 2012; Oya et al, 2013), it has been proposed that the exosome and TRAMP reinforce heterochromatin gene silencing by a cis‐acting post‐transcriptional gene silencing that degrades lncRNAs produced from heterochromatin regions.
Like in other eukaryotes, a major function of the S. pombe nuclear exosome is to degrade co‐transcriptionally the lncRNAs produced by pervasive transcription (Zhou et al, 2015). In addition, the nuclear exosome is also part of an RNA surveillance machinery that targets meiotic pre‐mRNAs in a selective manner to prevent S. pombe cells from undergoing meiosis during vegetative growth (Harigaya et al, 2006; Yamanaka et al, 2010; Hiriart et al, 2012; Zofall et al, 2012; Tashiro et al, 2013). The selective targeting of the surveillance machinery is achieved by the YTH RNA‐binding domain of Mmi1 (Harigaya et al, 2006; meiotic mRNA interception protein 1), which recognizes the hexameric RNA motif UNAAAC (where N can be any nucleotide) present in several copies in these meiotic transcripts (Chen et al, 2011; Hiriart et al, 2012; Yamashita et al, 2012). In parallel, Mmi1 RNA surveillance machinery also eliminates specific nascent lncRNAs by recognizing the same hexameric motif (Hiriart et al, 2012; Ard et al, 2014; Shah et al, 2014; Chatterjee et al, 2016). Mmi1‐mediated elimination of the lncRNA meiRNA regulates meiosis (Hiriart & Verdel, 2013; Yamashita et al, 2016), while its elimination of the lncRNA prt1 regulates phosphate uptake (Shah et al, 2014; Chatterjee et al, 2016). Mmi1 binding to prt1 and to meiotic pre‐mRNAs triggers the recruitment of RNAi proteins (Hiriart et al, 2012; Shah et al, 2014) and the formation of facultative heterochromatin (Hiriart et al, 2012; Zofall et al, 2012; Tashiro et al, 2013; Shah et al, 2014). Mmi1 also promotes the transcription termination of its targeted meiotic and lncRNA genes (Shah et al, 2014; Chalamcharla et al, 2015). It has been proposed that Mmi1/exosome‐mediated transcription termination serves to prime the lncRNA targets for degradation (Shah et al, 2014), yet this remains to be tested. More broadly, the biological significance and the direct implication of this transcription termination in Mmi1‐mediated gene silencing have not been addressed.
According to its primary sequence, Mmi1 belongs to the large family of YTH (YT521‐B Homology) RNA‐binding proteins (Stoilov et al, 2002; Zhang et al, 2010; Wang & He, 2014). The recent structures of its YTH domain confirmed its overall similarity to the other YTH domains studied (Chatterjee et al, 2016; Wang et al, 2016). However, in contrast to budding yeast and mammalian YTH domains that bind to RNA by specifically recognizing the methylated adenine (m6A) RNA modification (Li et al, 2014; Theler et al, 2014; Xu et al, 2014; Zhu et al, 2014), Mmi1 YTH domain binds to RNA by recognizing the unmethylated UNAAAC motif (Chen et al, 2011; Yamashita et al, 2012; Wang et al, 2016). On the other hand, and similarly to Mmi1, other YTH domain‐containing proteins associate to both mRNAs and lncRNAs (Xu et al, 2014; Patil et al, 2016), and their association with mRNAs controls RNA decay (Wang et al, 2014) and splicing (Xiao et al, 2016), while the function of their association with lncRNAs is poorly characterized.
In this study, thanks to a combination of high‐throughput sequencing, computational prediction, and protein structure‐driven analyses, we identify new lncRNAs targeted by the YTH domain of Mmi1 and find that the co‐transcriptional binding of Mmi1 to some of these lncRNAs controls sexual differentiation and heterochromatin gene silencing. We uncover that Mmi1 binding to a unique nascent lncRNA, that we named non‐coding RNA associated to Mmi1 (nam1), is sufficient to control entry into sexual differentiation. Importantly, Mmi1 binding to nam1 not only promotes the recruitment of the exosome but also imposes a robust termination of transcription of nam1. We further demonstrate that, by doing so, Mmi1 prevents nam1 read‐through transcription from repressing the downstream mitogen‐activated protein kinase kinase kinase (MAPKKK) essential to entry into sexual differentiation. In addition, we also uncover that Mmi1 binding to pericentromeric lncRNAs mediates heterochromatin gene silencing, in particular by promoting transcription termination. Finally, we show that Mmi1‐mediated termination of lncRNA transcription may not act in parallel but rather alternate during the cell cycle with the RNAi‐mediated heterochromatin gene silencing. Altogether, these findings demonstrate that the selective transcription termination of lncRNA genes mediated by the YTH domain of Mmi1 regulates lncRNA‐based gene silencing processes implicated in important cellular processes such as cell differentiation and heterochromatin gene silencing.
Results
Extensive identification of RNAs targeted by Mmi1's YTH domain
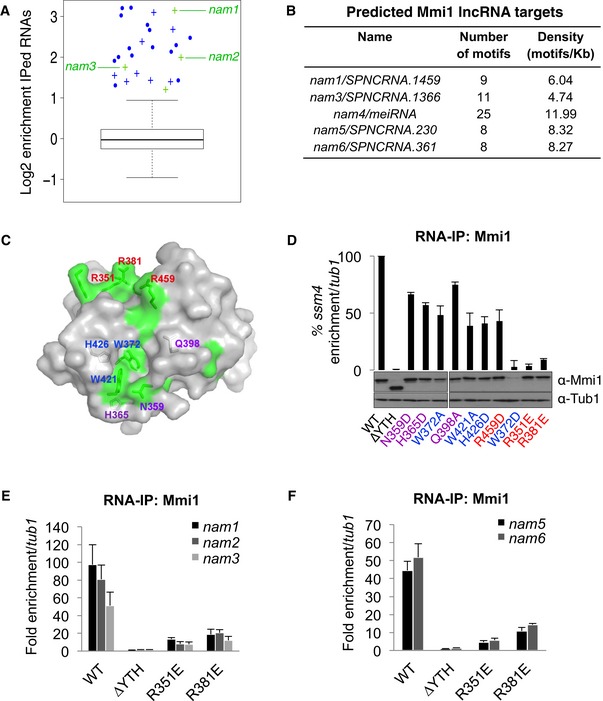
To better characterize the function of Mmi1 RNA‐binding protein, we searched for the RNAs targeted by Mmi1 on a genomewide scale. We first conducted Mmi1 RNA‐IPs coupled to high‐throughput sequencing. Thousands of RNAs were identified in both Mmi1 and control RNA‐IPs, but only 27 RNAs were enriched at least twofold in all Mmi1 RNA‐IPs (Fig 1A and Appendix Table S1); 15 of the 20 previously validated mRNA targets of Mmi1 (Harigaya et al, 2006; Hiriart et al, 2012) as well as eight new mRNAs were among them. As expected, the enriched mRNAs showed almost exclusively a meiotic expression profile (Appendix Table S1) and a high density of Mmi1 binding motifs UNAAAC in comparison with the complete set of S. pombe mRNAs (Fig EV1A). Interestingly, three new lncRNAs produced from different euchromatic regions and a snoRNA were also enriched in Mmi1 RNA‐IPs (Fig 1A). All three lncRNAs possess an overrepresentation of UNAAAC motifs in their sequence relative to the complete set of S. pombe‐annotated non‐coding RNAs (Fig EV1A), suggesting that they are also targets of Mmi1.
Figure 1. Extensive identification of Mmi1 RNA targets by combining RNA‐IP sequencing and computational approaches.
-
ABox plot of the enrichment of the RNAs identified by Mmi1 RNA‐IPs coupled to high‐throughput sequencing. The log of the average enrichments obtained from two independent RNA‐IPs is plotted. The enrichment is relative to the no antibody RNA‐IPs conducted in parallel with Mmi1 RNA‐IPs. The boxes represent the median and the upper and lower quartiles (25% and 75%). The whiskers extend to 1.5 times the interquartile range from the box. The mRNAs and lncRNAs enriched at least twofold in both Mmi1 RNA‐IPs are shown in blue and green, respectively. Crosses represent newly identified Mmi1 targets and dots known targets.
-
BLncRNAs predicted to be targets of Mmi1 with a high confidence by our computational approach.
-
CSurface representation of Mmi1 YTH domain. Area corresponding to conserved residues (according to the alignment shown in Fig EV2A) and located at the surface are highlighted in green. Mutated residues in Mmi1 YTH domain are indicated in blue for the ones located within the aromatic cage, in purple for the ones surrounding the cage, and in red for the rest.
-
DRNA‐IPs showing the impact of Mmi1 YTH domain point mutations on Mmi1 binding to ssm4 mRNA. The lower part shows a Western blot monitoring the protein level of WT and mutant Mmi1 proteins in the cells used for the RNA‐IPs. Loading was monitored using an anti‐Tub1 (tubulin) antibody.
-
E, FRNA‐IPs showing the YTH‐dependent association of Mmi1 with the lncRNAs identified in (A) and (B).
Figure EV1. Density of Mmi1 binding sites within mRNAs and lncRNAs, and computational approach developed to predict Mmi1 RNA targets.
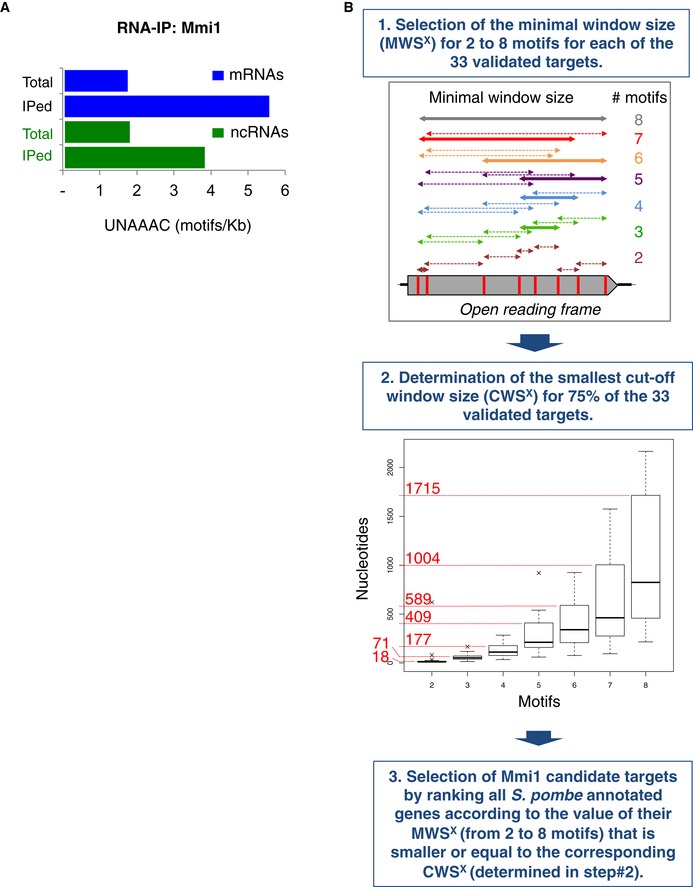
- Density of Mmi1 binding motif (UNAAAC) among all the annotated mRNAs and lncRNAs versus the mRNAs and lncRNAs enriched at least twofold in Mmi1 RNA‐IP Seq.
- Diagram of the genomewide computational approach used to predict Mmi1 targets. In step 1, a hypothetical open reading frame that has eight UNAAAC motifs in its sequence is shown as an example. The double‐headed arrows indicate all possible windows containing from two to eight motifs. The thick double‐headed arrows correspond to the windows yielding the minimal window size (MWS) from two to eight motifs. In step 2, each box plot represents the MWSx calculated for all of the validated targets. The boxplots were produced using the “boxplot” function in R with default parameters: the boxes represent the median and the upper and lower quartiles (25% and 75%); and the whiskers attempt to capture the extreme values, but extend to at most 1.5 times the interquartile range from the box, in which case any outliers beyond this range appear as crosses. The numbers in red represent the cutoff window size (CWS) for two to eight motifs. See the Materials and Methods for more details.
We conducted in parallel a computational approach to identify Mmi1 targets. Conversely, to the RNA‐IPs sequencing approach, the computational approach does not require a minimal level of expression of the target RNA to be identified. The computational approach considers the number and density of UNAAAC motifs per RNA as criteria to screen among all S. pombe‐annotated mRNAs and lncRNAs (Fig EV1B). Using stringent filtering conditions (see the Materials and Methods for more details), a total of 17 mRNAs mostly expressed during meiosis (Appendix Table S2) as well as five lncRNAs were identified as high‐confidence targets of Mmi1 (Fig 1B). Noticeably, the set of lncRNAs included two of the lncRNAs identified by the high‐throughput RNA‐IP approach, the meiRNA (Hiriart et al, 2012; Yamashita et al, 2012) and, intriguingly, two lncRNAs produced from pericentromeric regions embedded within heterochromatin, suggesting that Mmi1 binding to lncRNAs may not be restricted to euchromatic lncRNAs but also encompasses heterochromatic lncRNAs expressed at low levels.
In the process of characterizing Mmi1 binding to RNA, we obtained the structure of the Mmi1 YTH domain, at 1.5 Å resolution (Figs 1C and EV2A and B, and Appendix Table S3) and, based on this structure, we made a series of point mutants that may interfere with Mmi1 binding to RNA without impacting on the structure of the domain. Wild‐type and mutant Mmi1 proteins were expressed in mmi1∆ cells, and their in vivo binding to ssm4, spo5, and rec8 mRNAs, three previously validated targets of Mmi1 (Hiriart et al, 2012), was monitored. Out of the 10 mutations made, mutations R351E and R381E were found to cause a marked reduction of Mmi1 binding to the three target mRNAs without reducing the protein level of Mmi1 (Figs 1D and EV2C). In agreement with the possibility that these two mutations impact on Mmi1 YTH domain binding to RNA rather than on its structure, gel filtration experiments showed similar elution patterns for R351E, R381E, and wild‐type Mmi1 YTH domains (Fig EV2D), and RNA pull‐down experiments indicated that both mutations negatively impact on Mmi1 binding to RNA in vitro (Fig EV2E and F). Additionally, the analysis of the subcellular localization of Mmi1 R351E and R381E proteins by immunofluorescence showed that their localization is similar to the wild‐type Mmi1 protein (Fig EV2G). Importantly, the RNA‐IP of Mmi1 R351E and R381E point mutant coupled to PCR (which is more sensitive than the RNA‐IP Seq) confirmed that Mmi1 YTH domain specifically recognizes the five new lncRNAs identified by our RNA‐IP high‐throughput sequencing and computational approaches (Fig 1E and F). We named these lncRNAs non‐coding RNA associated to Mmi1 (nam). Hence, from our broad search of Mmi1 RNA targets based on the combination of different approaches, we discovered new RNAs, including euchromatic and heterochromatic lncRNAs.
Figure EV2. Characterization of Mmi1 YTH domain point mutants R351E and R381E.
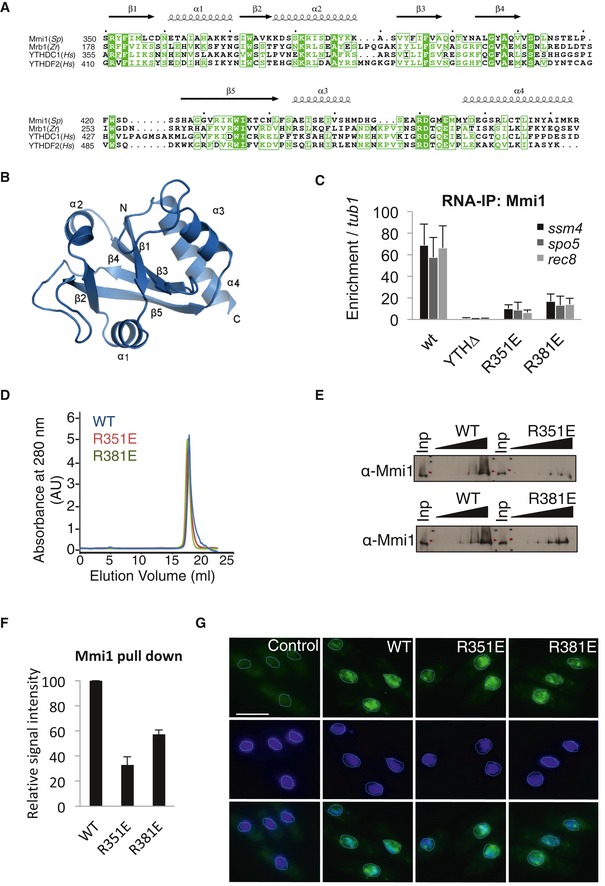
- Sequence alignment of YTH domains from the yeast Schizosaccharomyces pombe (Sp), the yeast Zygosaccharomyces rouxii (Zr), and Homo sapiens (Hs).
- Ribbon diagram of Mmi1 YTH domain structure in the same orientation as in Fig 2C.
- RNA‐IP showing that the in vivo binding of Mmi1 to three of its known targets (ssm4, rec8, and spo5 mRNAs) is strongly reduced for R351E and R381E Mmi1 mutants.
- Gel filtration showing similar elution behavior for WT, R351E, and R381E Mmi1 YTH domains.
- RNA pull‐down showing in vitro that mutations R351E and R381E impair Mmi1 binding to a RNA containing the UUAAAC motif.
- Relative enrichments of Mmi1 protein recovered after RNA pull‐downs done in (E). The quantification was estimated from three independent experiments. See the Materials and Methods for more details.
- Fluorescence microscopy images showing the cellular localization of Mmi1‐R351E, Mmi1‐R381E and Mmi1 wild‐type (WT) proteins. Nuclear DNA was stained with DAPI (blue). Scale bar, 10 μm.
Mmi1 association with the sole nam1 lncRNA regulates the MAPK‐mediated entry into sexual differentiation
Mmi1 is well known as an inhibitor of sexual differentiation progression that prevents entry into meiosis by targeting and triggering the degradation of meiotic mRNAs (Harigaya et al, 2006). In response to nutrient starvation (mainly nitrogen), S. pombe cells undergo sexual differentiation (Fig 2A), to allow them to adapt and resist to conditions not favorable to cell growth. At the onset of sexual differentiation, two cells of opposite mating type (h+ and h‐) mate to produce a zygote. The zygote then undergoes genome duplication followed by meiosis and the formation of four spores. Quite unexpectedly, we found that mmi1∆ cells poorly execute sexual differentiation, indicating that Mmi1 may be also required to promote sexual differentiation (Figs 2A and EV3A). Further analysis of mmi1∆ cells showed that they poorly mate, indicating that the onset of sexual differentiation is defective (Fig 2B). This defect occurs irrespective of the mating type identity of mmi1∆ cells (Fig EV3B and C). We then assessed the importance of Mmi1 binding to RNA for the control of entry into sexual differentiation by taking advantage of our Mmi1 R351E and R381E point mutants and analyzing whether their expression rescues the cell differentiation defect. While the expression of Mmi1 wild‐type protein completely rescues the sexual differentiation of mmi1∆ cells, the expression of Mmi1‐R351E or Mmi1‐R381E YTH mutant proteins does not (Figs 2A and B, and EV3A), indicating that the binding of Mmi1 YTH domain to one or more RNA is essential for the proper control of entry into sexual differentiation.
Figure 2. Mmi1 binding to nam1 lncRNA controls MAPK‐mediated entry into sexual differentiation.
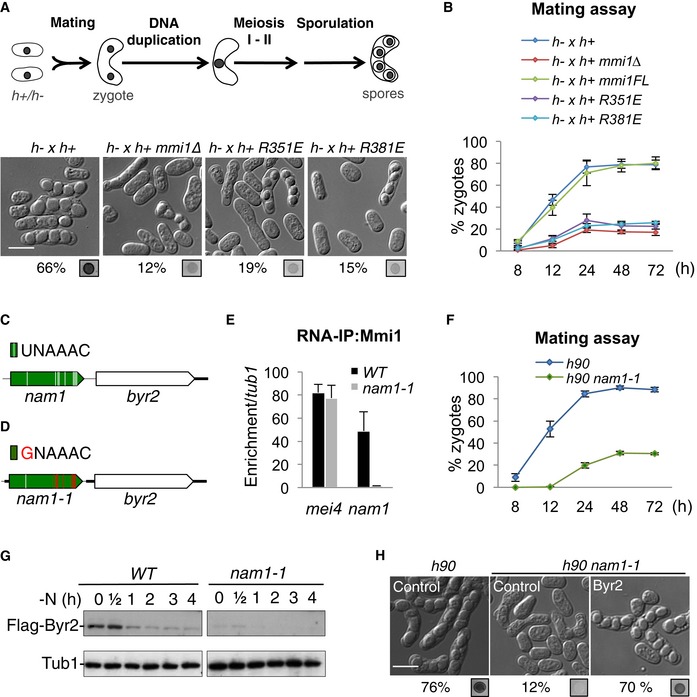
- Upper part, scheme of Schizosaccharomyces pombe sexual differentiation. Lower part, microscopy images showing WT and mmi1∆ cells, and mmi1∆ cells expressing Mmi1‐R351E or Mmi1‐R381E mutant proteins. Images were taken after 24 h of induction of sexual differentiation by growth on SPAS medium. The percentage of cells that underwent differentiation and iodine vapor assays, conducted on the corresponding patches, are shown at the bottom of each image. Scale bar, 10 μm.
- Mating assay showing the percentage of zygotes forming over time in the same cells as in (A).
- Scheme of nam1‐byr2 locus. Mmi1 UNAAAC binding motifs are depicted by white lines.
- Scheme of nam1‐byr2 locus in nam1‐1 cells highlighting the eight UNAAAC motifs mutated (red lines).
- Mmi1 RNA‐IPs showing the specific loss of binding of Mmi1 to nam1‐1 lncRNA but not to mei4 mRNA, another target of Mmi1.
- Mating assay showing the percentage of zygotes formed over time in WT and nam1‐1 cells.
- Western blots showing the level of Flag‐Byr2 protein over the first 4 h of sexual differentiation in WT and nam1‐1 cells. Tubulin (Tub1) level was used as a loading control.
- Microscopy images of WT (h90) cells transformed with an empty plasmid (Control) and nam1‐1 cells transformed with either an empty plasmid (Control) or a plasmid expressing Byr2 protein (Byr2), after 24 h of induction of sexual differentiation. Scale bar, 10 μm.
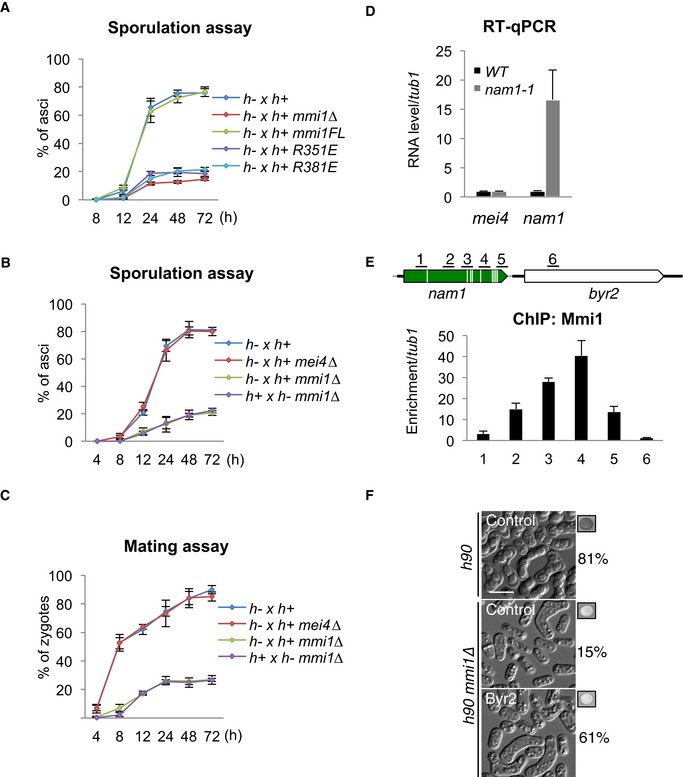
Figure EV3. Mmi1‐dependent entry into sexual differentiation (related to Fig 2).
-
ASporulation assay showing the percentage of asci forming over time in WT, mmi1∆, mmi1‐R351E, and mmi1‐R381E cells.
-
B, CGraphs showing the percentage of asci (B) and zygotes (C) forming over time in WT, mei4∆, and mmi1∆ mei4∆ cells.
-
DRT–qPCR showing the level of nam1 lncRNA and mei4 mRNA in WT and nam1‐1 cells.
-
EChIP monitoring Mmi1 localization over nam1‐byr2 locus in WT cells. Upper part of the panel highlights genomic regions (black lines).
-
FMicroscopy images showing the state of sexual differentiation of WT and mmi1∆ cells expressing or not expressing Byr2 from a plasmid, after 24 h of growth on SPAS. The percentage of cells that underwent differentiation and iodine vapor assays conducted on the corresponding patch are shown at the right side of each picture. Scale bar, 10 μm.
In relation to Mmi1's function in entry into sexual differentiation, we noticed that nam1 lncRNA, one of the most enriched RNAs in Mmi1 RNA‐IPs (Fig 1A), maps just upstream of byr2 (Fig 2C), a gene encoding a MAPKKK essential for entry into sexual differentiation (Wang et al, 1991; Styrkarsdottir et al, 1992). This prompted us to test whether Mmi1 targeting of nam1 plays a role in regulating MAPK‐mediated entry into sexual differentiation. We previously showed that a single mutation in Mmi1 RNA binding motif (UNAAAC), consisting in the replacement of the first ribonucleotide U with a G, compromises its binding to Mmi1 both in vitro and in vivo (Hiriart et al, 2012; Yamashita et al, 2012). We thus made recombinant cells expressing a mutant version of nam1 lncRNA from the endogenous locus, named nam1‐1, in which the first ribonucleotide for eight out of its nine UNAAAC motifs was swapped from U to G (Fig 2D). As expected, the specific binding of Mmi1 to nam1‐1 is lost and nam1‐1 accumulates, while it still binds to its other targets as illustrated by its preserved binding to mei4 mRNA another known target of Mmi1 (Figs 2E and EV3D). Remarkably, we found that nam1‐1 cells recapitulate the mating defect of mmi1∆ cells, when induced to differentiate (Fig 2F). In agreement with Mmi1 directly regulating byr2 gene expression, Mmi1 localizes to nam1 gene (Fig EV3E). Furthermore, Byr2 protein level is significantly reduced in nam1‐1 cells compared to wild‐type cells (Fig 2G), and, importantly, the expression of Byr2 from a plasmid rescues the mating defect of both nam1‐1 and mmi1∆ cells (Figs 2H and EV3F). From these findings, we conclude that the sole binding of Mmi1 to nam1 lncRNA plays a central role at the onset of sexual differentiation by regulating the expression of the byr2 MAPKKK gene.
Mmi1‐induced transcription termination of nam1 gene promotes expression of the downstream byr2 MAPKKK gene
We next investigated the mechanism by which Mmi1 binding to nam1 lncRNA favors expression of Byr2. The detection of nam1 lncRNA and byr2 mRNA by Northern blot showed that their levels are anti‐correlated, with the level of byr2 mRNA being low when the level of nam1 is high when comparing wild‐type cells to nam1‐1 cells (Fig 3A). We also noticed the accumulation of a second and longer form of nam1 lncRNA (that we named nam1‐L) and that was detected with a probe specific for either nam1 or the 5′ end of byr2. This latter result indicated that, in the absence of Mmi1 binding to nam1 lncRNA, the nam1 gene might be experiencing read‐through transcription. In agreement with this possibility, strand‐specific RT‐qPCR experiments showed a 13‐fold increase of read‐through transcripts in nam1‐1 cells (Fig EV4A). Importantly, while the occupancy of the overall population of RNAPII increases only modestly downstream of nam1 in nam1‐1 cells relative to wild‐type cells (Fig EV4B), the elongating RNAPII (RNAPII‐S2P), which rapidly decreases after the 3′ end of nam1 in wild‐type cells, stays at a high level in nam1‐1 cells (Fig 3B), as well as in mmi1∆ cells (Fig EV4C). Moreover, the occupancy of the initiating RNAPII (RNAPII‐S5P) at byr2 promoter strongly decreases in nam1‐1 cells (Fig EV4D). Collectively, these results show that in the absence of Mmi1 binding to nam1, the termination of transcription at nam1 gene is defective.
Figure 3. Mmi1 promotes transcription termination of nam1 non‐coding gene and prevents nam1 read‐through transcription from repressing the downstream MAPKKK gene byr2 .
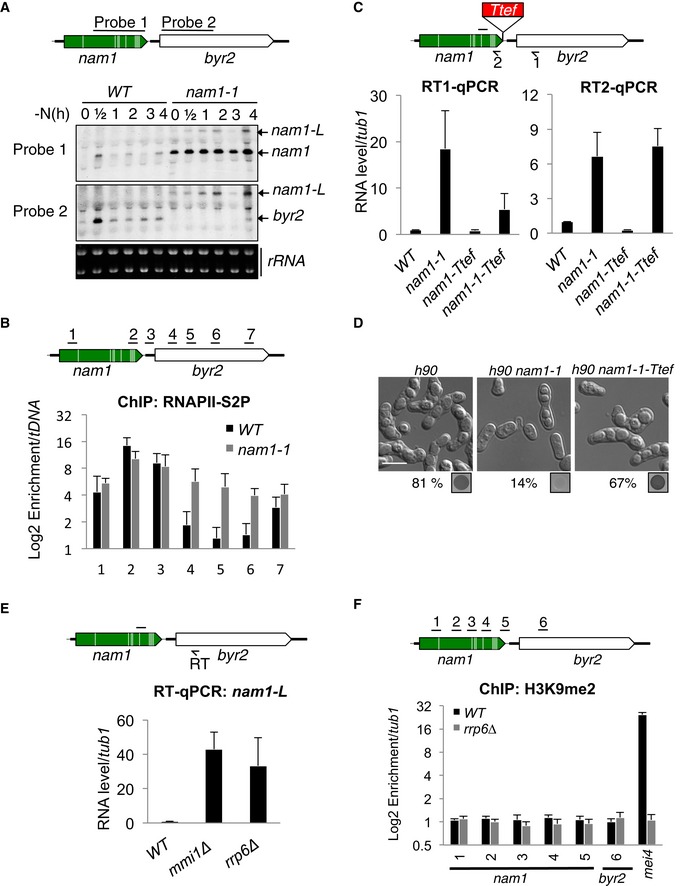
- Northern blots showing nam1 and byr2 RNA levels during the first 4 h of sexual differentiation. Ribosomal RNAs (rRNAs) stained with ethidium bromide were used as loading controls. Black lines indicate probes used to detect nam1 and byr2 RNAs.
- ChIPs showing the occupancy of the elongating RNAPII (RNAPII‐S2P) over nam1‐byr2 locus, in WT and nam1‐1 cells. RNAPII‐S2P was immunoprecipitated with an antibody recognizing the heptameric repeats (present in the C‐terminal domain of the polymerase) when it is phosphorylated on its serine 2. Black lines, genomic regions investigated.
- RT–qPCRs showing the accumulation of nam1 read‐through transcripts (RT1–qPCR) and nam1 lncRNAs (RT2–qPCR) in cells with or without the transcription terminator Ttef inserted at the 3′ end of nam1 (scheme). Black arrow, primer used for the strand‐specific reverse transcription (RT); black line, location of the region amplified by PCR.
- Microscopy images of, respectively, WT (h90), nam1‐1, and nam1‐1‐Ttef cells, after induction of sexual differentiation for 24 h. The percentage of sporulation and iodine vapor assays are shown at the bottom of the images. Scale bar, 10 μm.
- RT–qPCRs showing the accumulation of nam1 read‐through transcripts in mmi1∆ and rrp6∆ cells. Black arrow and line as in (C).
- ChIPs monitoring the enrichment of H3K9me2 over nam1‐byr2 locus and mei4 gene in WT and rrp6∆ cells.
Figure EV4. Mmi1 control of byr2 gene expression (related to Fig 3).
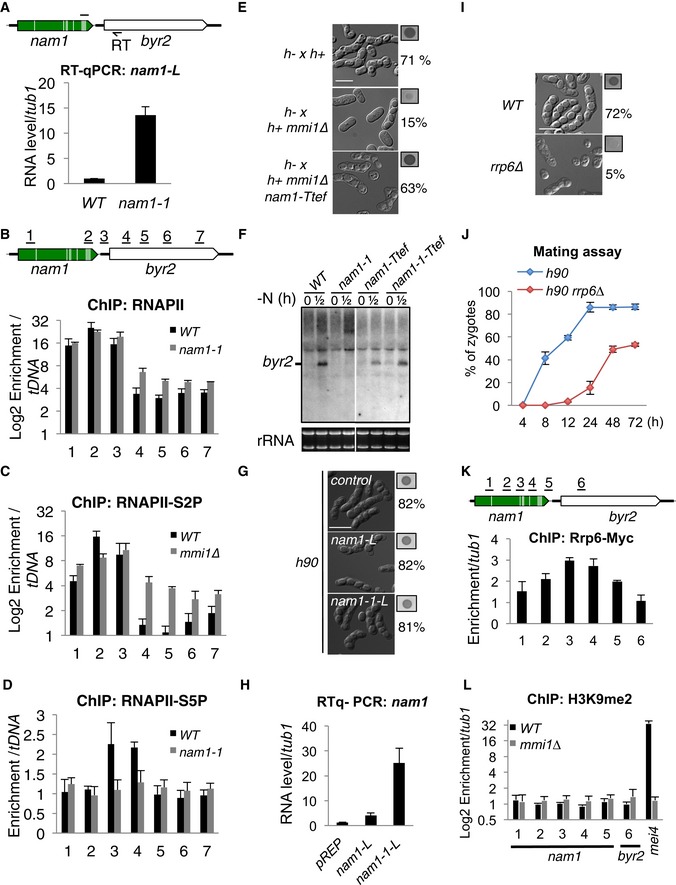
- RT–qPCR showing the accumulation of nam1 read‐through transcripts in nam1‐1 cells. Black arrow, primer used for the strand‐specific reverse transcription (RT); black line, location of the region amplified by PCR.
- ChIPs showing the occupancy of the whole population of RNAPII over nam1‐byr2 locus, in WT and nam1‐1 cells. Black lines above the scheme, genomic regions investigated.
- ChIPs showing the occupancy of the elongating RNAPII (RNAPII‐S2P) over nam1‐byr2 locus, in WT and mmi1∆ cells. The same genomic regions as in (B) were investigated.
- ChIPs showing the occupancy of the initiating RNAPII (RNAPII‐S5P) over nam1‐byr2 locus, in WT and nam1‐1 cells. The same genomic regions as in (B) were investigated.
- Microscopy images showing the sexual differentiation status in WT, mmi1∆, and mmi1∆ nam1‐Ttef cells, after 24 h of growth on SPAS. The percentage of cells that underwent differentiation and iodine vapor assays conducted on the corresponding patch are shown at the right side of each picture. Scale bar, 10 μm.
- Northern blot showing the level of byr2 mRNA before (0 h) and after (0.5 h) induction of sexual differentiation in WT, nam1‐1, nam1‐Ttef, and in nam1‐1‐Ttef. Note that the white separation indicates that lanes on the left and the right come from the same gel but that the images were cropped to place them next to each other.
- Microscopy images showing WT cells transformed with an empty plasmid or a pREP plasmid expressing nam1‐L or nam1‐1L lncRNA, after having induced sexual differentiation by growth on SPAS for 24 h. Scale bar, 10 μm.
- RT–qPCR showing the level of nam1‐L and nam1‐1‐L lncRNA expressed from the pREP plasmid.
- Microscopy images showing WT (h90) and rrp6∆ cells, after having induced sexual differentiation by growth on SPAS for 24 h. Scale bar, 10 μm.
- Mating assay showing the percentage of zygotes forming over a 72‐h period, from cells induced to undergo sexual differentiation by growth on SPAS.
- ChIP showing the enrichment of Rrp6 over nam1‐byr2 locus.
- ChIP monitoring the enrichment of H3K9me2 over nam1‐byr2 locus and mei4 gene in WT and mmi1∆ cells.
To test the importance of Mmi1‐dependent transcription termination of nam1 in regulating sexual differentiation, we introduced a potent terminator of transcription (Ttef) at the 3′ end of nam1 to prevent nam1 read‐through transcription. As expected, the insertion of Ttef significantly reduces the accumulation of nam1 read‐through transcripts in nam1‐1 cells (Fig 3C, left part). Strikingly, the insertion of Ttef rescues most of the defect in entry into sexual differentiation of both nam1‐1 and mmi1∆ cells (Figs 3D and EV4E), as well as the expression of byr2 (Fig EV4F). Of note, the rescue caused by the insertion of Ttef occurs despite the sevenfold increase of nam1 lncRNA level (Fig 3C, right part), suggesting that the accumulation of nam1 lncRNA by itself has little or no role in regulating byr2 expression and sexual differentiation. Additionally, and in agreement with nam1 transcripts acting only in cis, the production of nam1 read‐through transcripts from a plasmid did not interfere with sexual differentiation even with a 25‐fold accumulation of nam1‐L relative to wild‐type cells (Fig EV4G and H). Altogether, these findings demonstrate that Mmi1‐mediated transcription termination of nam1 prevents nam1 read‐through transcription from repressing the immediately downstream byr2 gene.
Rrp6, but not H3K9 methylation, contributes to Mmi1‐dependent control of nam1 expression
Given the tight functional connection between the exosome and Mmi1, we next examined whether the exosome is implicated in Mmi1‐mediated control of sexual differentiation. Similarly to mmi1∆ and nam1‐1 cells, rrp6∆ cells present a defect in entry into sexual differentiation (Fig EV4I), although the defect is less pronounced after 48 and 72 h of sexual differentiation induction (Fig EV4J). Additionally, nam1 read‐through transcripts accumulate in rrp6∆ cells (Fig 3E), and Rrp6 localizes to nam1 gene (Fig EV4K). Because facultative heterochromatin forms in an exosome‐dependent manner at some of Mmi1 targets (Hiriart et al, 2012; Zofall et al, 2012; Tashiro et al, 2013; Shah et al, 2014), we also examined whether this was the case at the nam1‐byr2 locus. However, no H3K9 methylation was detected at this locus in wild‐type, rrp6∆ or mmi1∆ cells (Figs 3F and EV4L). Thus, Rrp6, but not the deposition of the H3K9me mark, contributes to Mmi1‐mediated control of nam1 expression.
Mmi1 induces Rrp6‐dependent heterochromatin gene silencing at pericentromeric DNA
Following on our computational approach that revealed Mmi1 binding to pericentromeric heterochromatic lncRNAs (Fig 1B), we also investigated the function of Mmi1 in heterochromatin gene silencing at pericentromeric DNA repeats. The two heterochromatic lncRNAs identified, nam5 and 6, are produced from slightly divergent dh repeats (Fig 4A). According to the transcriptomic analysis of pericentromeric DNA repeats, an additional and non‐annotated lncRNA, sharing the same UNAAAC‐rich sequence with nam5 and 6, is expressed from centromere 3 repeats. By using RNA‐IPs, we found that this lncRNA is also bound to Mmi1, and named it nam7 (Fig EV5A). In contrast, dg‐specific lncRNAs were not enriched in Mmi1 RNA‐IPs (Fig EV5B).
Figure 4. Mmi1 drives Rrp6‐dependent heterochromatin gene silencing at pericentromeric regions.
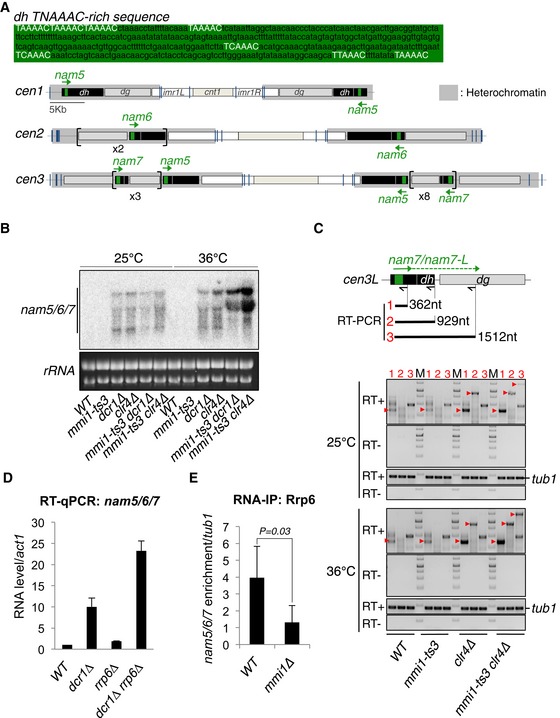
- Upper part, TNAAAC‐rich sequence present in nam5, 6, and 7 lncRNAs. Lower part, schematic representation of three S. pombe centromeres showing the different pericentromeric DNA repeats susceptible to produce nam5, 6, and 7 (green arrows).
- Northern blot showing the level of nam5/6/7 lncRNA population in mmi1‐ts3, dcr1∆, clr4∆ single mutant cells, and in mmi1‐ts3 dcr1∆ and mmi1‐ts3 clr4∆ double mutant cells, at the permissive (25°C) and restrictive (36°C) temperatures.
- RT–PCRs monitoring the accumulation of nam7 lncRNAs and nam7‐L read‐through transcripts, in the same cells and conditions as in (B). Red arrow heads point to the expected PCR products while the other bands correspond to non‐specific PCR products. From the scheme and the agarose gels: black arrows, primers used for the three different reverse transcriptions; black lines and red numbers, expected PCR products of the three different RT–PCRs; M, DNA ladder markers; tub1, tubulin control.
- RT–qPCRs showing the levels of nam5/6/7 lncRNA population in the double mutant rrp6∆ dcr1∆ cells, relative to the single mutant rrp6∆ and dcr1∆ cells.
- RNA‐IPs showing that Rrp6‐Myc13 binds to nam5/6/7 lncRNAs in a Mmi1‐dependent manner. P‐value was calculated using a two‐tailed Student's t‐test.
Figure EV5. Mmi1‐directed heterochromatin gene silencing (related to Fig 4).
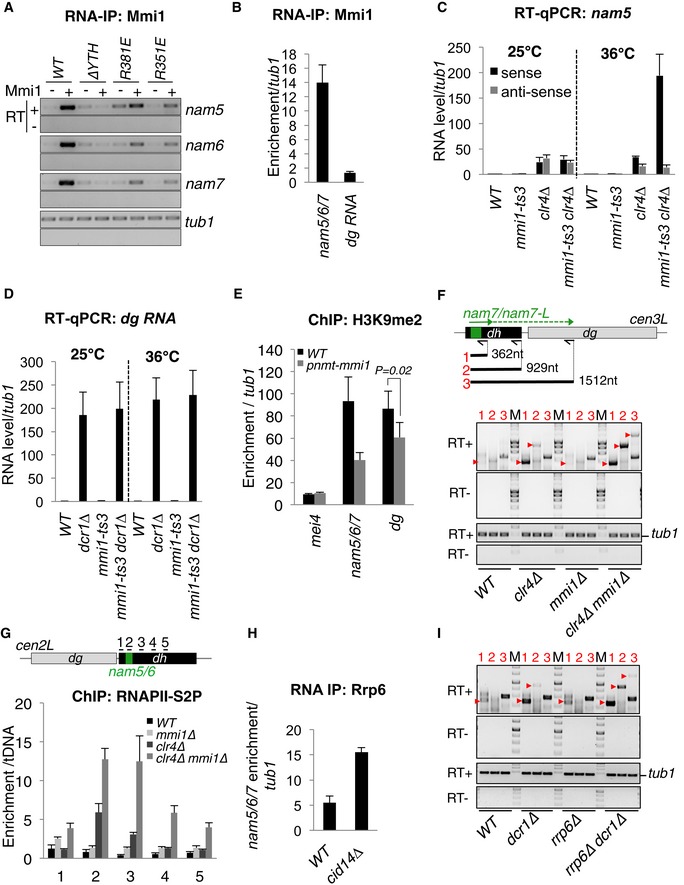
- RNA‐IP showing that in vivo, the binding of Mmi1 to nam5/6/7 lncRNAs is strongly reduced by Mmi1 mutations R351E and R381E expressed from a plasmid.
- RNA‐IP showing that Mmi1 binds to nam5/6/7 lncRNAs but not to dg lncRNAs.
- RT–qPCR showing the synergy of accumulation of nam5/6/7 lncRNAs in mmi1‐ts3 clr4∆ double mutant cells at restrictive temperature (36°C).
- RT–qPCR showing no additional accumulation of dg lncRNAs in mmi1‐ts3 dcr1∆ double mutant cells at restrictive temperature (36°C).
- ChIP showing a reduction of the level of H3K9me2 at the nam5/6/7 and dg pericentromeric regions but not at mei4 gene, in cells overexpressing Mmi1 (pnmt1‐mmi1) compared to WT cells. P‐value was calculated from four independent experiments using a two‐tailed Student's t‐test.
- RT–PCRs monitoring the accumulation of nam7‐L in WT, clr4∆, and mmi1∆ and mmi1∆ clr4∆ cells. In the upper part of the panel, the black arrows indicate the location of the primers used for reverse transcription (RT), and the black lines indicate the expected PCR products.
- ChIPs showing the occupancy of the elongating RNAPII (RNAPII‐S2P) over dh repeats in WT, mmi1∆, clr4∆, and mmi1∆ clr4∆ cells. Black lines above the scheme, genomic regions investigated.
- RNA‐IP showing that Cid14 is not required for the association of Rrp6‐Myc13 to nam5/6/7 lncRNAs.
- RT–PCRs monitoring the accumulation of nam7‐L in rrp6∆, dcr1∆, and rrp6∆ and dcr1∆ cells. RT–PCRs were conducted using the same conditions as in (F).
To test whether Mmi1 could play a role in pericentromeric heterochromatin gene silencing, we examined the levels of heterochromatic lncRNAs in mmi1∆ and mmi1‐ts3 thermosensitive cells. Northern blot and RT‐qPCR experiments showed no significant accumulation of nam5, 6, and 7 lncRNAs in mmi1‐deficient cells (Figs 4B and EV5C). However, since RNAi and the methylation of H3K9, catalyzed by the methyltransferase Clr4, play a major role in pericentromeric heterochromatin gene silencing, we also examined the level of the nam5/6/7 lncRNA population in cells deficient for both Mmi1 and Dcr1 or Mmi1 and Clr4. Importantly, Northern blot and RT‐qPCR experiments showed a synergy of accumulation of nam5/6/7 lncRNA population in mmi1‐ts3 dcr1∆ and mmi1‐ts3 clr4∆ double mutant cells at restrictive temperature (36°C; Figs 4B and EV5C). This requirement of Mmi1 is specific to nam5/6/7 lncRNAs, since no additional accumulation was observed for nam5/6/7 anti‐sense lncRNAs or for dg‐specific lncRNAs in the same double mutant cells (Fig EV5C and D). Interestingly, overexpression of Mmi1 causes a reduction of H3K9 methylation at nam5/6/7 repeats as well as dg repeats (Fig EV5E), indicating that Mmi1 may have a general impact on pericentromeric heterochromatin. In addition, strand‐specific RT–PCR experiments showed that nam7 read‐through transcripts also accumulate in mmi1‐ts3 clr4∆ but not in wild‐type cells (Fig 4C), suggesting that Mmi1 contributes to heterochromatin gene silencing especially by promoting termination of transcription. Similar results were obtained in mmi1∆ clr4∆ cells (Fig EV5F). Accordingly, ChIP experiments showed an increase of the elongating RNAPII downstream of nam5/6 repeats which is dependent on Mmi1 (Fig EV5G). Furthermore, in agreement with the fact that Mmi1 and Rrp6 act together, RT–qPCR experiments showed that nam5/6/7 lncRNAs accumulate more in dcr1∆ rrp6∆ double mutant cells compared to the single mutant cells (Fig 4D). Moreover, RNA‐IPs showed that Rrp6 is recruited to nam5/6/7 lncRNAs in a Mmi1‐dependent and Cid14‐independent manner (Figs 4E and EV5H), and RT‐qPCRs showed that nam7 read‐through transcripts further accumulate in rrp6∆ dcr1∆ double mutant compared to the single mutants (Fig EV5I). From these findings, we conclude that Mmi1 binding to pericentromeric lncRNAs mediates Rrp6‐dependent heterochromatin gene silencing and the termination of lncRNA transcription within heterochromatin.
Mmi1 silences pericentromeric DNA transcription preferentially in early S phase
By conducting ChIP experiments, we noticed that the modest association of Mmi1 with pericentromeric DNA in wild‐type cells increases significantly in clr4∆ cells (Fig 5A). Because the level of pericentromeric H3K9 methylation as well as RNAi‐mediated gene silencing vary during the cell cycle progression (Chen et al, 2008; Kloc et al, 2008), we reasoned that Mmi1‐mediated gene silencing at pericentromeric heterochromatin might also vary. Using synchronized cells, we found that Mmi1 localization to heterochromatin reaches a peak in the G1/S transition phase, when H3K9 methylation is minimal (Fig 5B). Within the same time window, the level of nam5/6/7 lncRNA population reaches a low point in a Mmi1‐dependent manner (Fig 5C). Moreover, Mmi1‐dependent nam7 read‐through transcripts accumulate preferentially within the same time window (Fig 5D). Hence, these findings reveal that, during the cell cycle, Mmi1‐mediated heterochromatin gene silencing does not continuously act in parallel of RNAi but acts preferentially in early S phase when pericentromeric DNA is being replicated and RNAi‐mediated heterochromatin formation is idling.
Figure 5. Mmi1‐ and RNAi‐mediated silencing of pericentromeric DNA transcription alternate during the progression of the cell cycle.
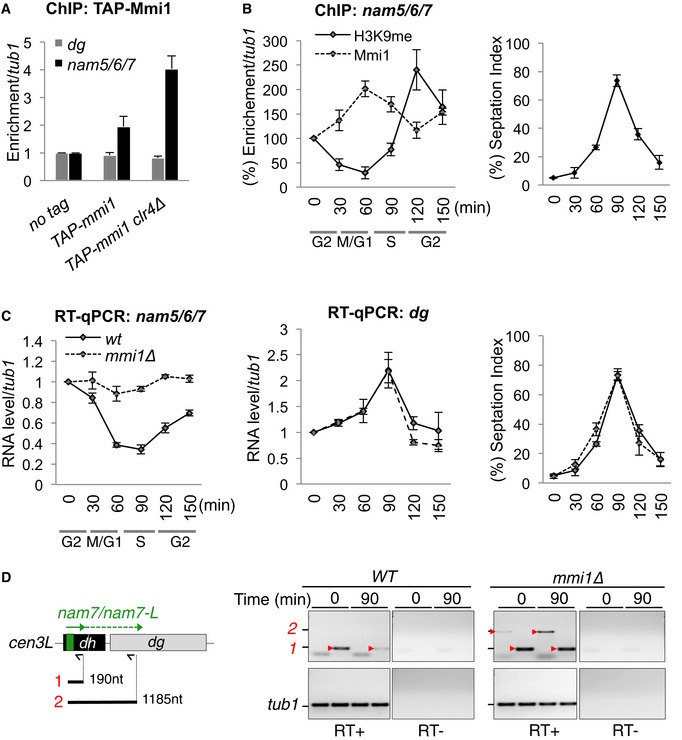
- ChIPs assessing the localization of Mmi1 to pericentromeric DNA in WT and clr4∆ cells.
- ChIPs showing the localization of Mmi1 (dashed line) and H3K9me2 (black line) to the pericentromeric nam5/6/7 DNA regions during the progression of the cell cycle. Cell synchronization was achieved by using cdc25‐ts cells (see the Materials and Methods for more details). Cell synchronization was monitored by measuring the percentage of cells with a septum (right part).
- RT–qPCRs monitoring the levels of nam5/6/7 (left part) and dg (middle part) lncRNA populations during the cell cycle.
- RT–PCRs monitoring the accumulation of nam7 lncRNAs and nam7‐L read‐through transcripts in G2/M and G1/S phases from synchronized cells as in (B). Red arrow heads point to the expected PCR products. The other bands are non‐specific PCR products. From the scheme: black arrows, primers used for the reverse transcriptions; black lines and red numbers, regions amplified by PCR.
Discussion
Although the coupling of non‐coding transcription to the elimination of its nascent lncRNA occurs at many sites in eukaryotic genomes, its potential to contribute to gene regulation is only emerging. Here, we report a multiscale study that provides insights into how exosome‐dependent co‐transcriptional elimination of lncRNAs is linked to the function of lncRNAs acting as regulators of gene expression. First, this study discovers new regulatory lncRNAs recognized by the YTH domain of Mmi1 and degraded in cis by the exosome. Second, it reveals that the co‐transcriptional degradation of these lncRNAs by Mmi1/exosome machinery is linked to the control of sexual differentiation and heterochromatin gene silencing. Third, it provides evidence that transcription termination mediated by the binding of Mmi1 to a unique nascent lncRNA (nam1) plays a key role in the control of sexual differentiation. Fourth, it uncovers that Mmi1‐mediated termination of lncRNA transcription also takes place at pericentromeric DNA repeats where it acts together with RNAi and in a cell‐cycle‐regulated fashion, to silence transcription within heterochromatin. Below, we discuss the implication of these findings for lncRNA‐based gene regulation.
LncRNA‐mediated control of cell differentiation and protein‐coding gene expression
Our extensive search for RNAs targeted by Mmi1 led to the identification of several regulatory lncRNAs, including nam1, which regulates the MAPK‐mediated entry into sexual differentiation. Additionally, we demonstrate that Mmi1 co‐transcriptional binding to nam1 promotes its transcription termination and this plays a central role in promoting the expression of the downstream MAPKKK gene byr2, which is essential for entry into sexual differentiation (Styrkarsdottir et al, 1992). The regulation of protein‐coding gene expression by the transcription of an adjacent non‐coding gene has emerged as a widespread regulatory process among eukaryotes known as transcription interference (Guil & Esteller, 2012; Hiriart et al, 2012; Jensen et al, 2013; Kornienko et al, 2013; Yamashita et al, 2016). Non‐coding transcription positively or negatively impacts on gene expression, and occurs in sense or anti‐sense orientation relative to the regulated gene. Transcription interference may be mediated by either the lncRNA under synthesis, recruiting repressive or activating factors, or by the elongating polymerase itself, which can interfere with the binding of transcription factors or of other RNA polymerases on the adjacent protein‐coding gene. Importantly, in all these cases, it is the switch ON or OFF of the non‐coding transcription that was found, or proposed, to be the key step for regulating the adjacent protein‐coding gene. Here, we provide evidence for the existence of another type of switch acting at the step of transcription termination. In the case of byr2 MAPKKK gene regulation, the binding of Mmi1 to nam1 nascent transcript promotes robust termination of RNAPII transcription of nam1 (Fig 6, left part). By doing so, Mmi1 prevents nam1 read‐through transcription from invading and interfering with the transcription of byr2. The exact mechanism by which Mmi1 binding to a nascent lncRNA promotes efficient transcription termination is unclear. However, our finding that read‐through transcripts from nam1 accumulate in rrp6∆ cells, together with the recent finding that Rrp6 could directly contribute to the termination of transcription (Lemay et al, 2014), suggest that Mmi1 may promote transcription termination of nam1 by recruiting the exosome. In agreement with this possibility, Mmi1 and Rrp6 were both reported to impose early transcription termination at meiotic genes (Shah et al, 2014; Chalamcharla et al, 2015). We note that, although our results suggest that nam1 lncRNA has no function by itself in silencing byr2 gene (Fig 3C), it is possible that nam1 read‐through transcripts have such a function by, for example, interfering with transcription factors, forming double‐stranded RNA with potential byr2 anti‐sense RNAs or recruiting histone modifiers, as previously reported (Guil & Esteller, 2012; Kornienko et al, 2013; Wery et al, 2016).
Figure 6. Model for Mmi1/exosome YTH‐mediated control of cell differentiation and heterochromatin gene silencing mediated by its targeting of nascent lncRNA and the induction of their transcription termination.
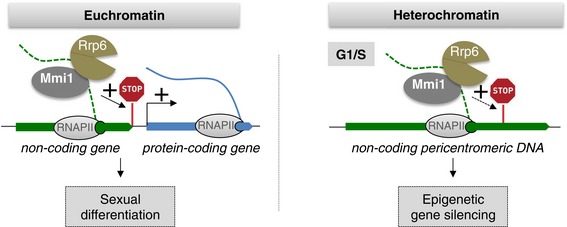
- The YTH domain of Mmi1 co‐transcriptionally binds to specific lncRNAs expressed from either euchromatin or heterochromatin regions. We propose that Mmi1 binding to a euchromatic nascent lncRNA (in green) induces the recruitment of the exosome and together they degrade the lncRNA and promote robust termination of the lncRNA transcription, which otherwise will inhibit the expression of the downstream protein‐coding gene (in blue). In the case of nam1‐byr2 locus, the efficient transcription termination prevents the occurrence of read‐through transcription from nam1 gene, which represses byr2 MAPKKK gene, a critical regulator of the entry into sexual differentiation. In parallel, Mmi1 co‐transcriptional binding to the heterochromatic pericentromeric lncRNAs also recruits the exosome and contributes to heterochromatin gene silencing by degrading the nascent lncRNAs as well as by inducing precocious termination of their transcription. STOP sign, site of Mmi1‐dependent lncRNA transcription termination.
Our finding that Mmi1 also silences non‐coding read‐through transcription at pericentromeric DNA regions shows that this process is not limited to the nam1‐byr2 locus. Accordingly, our computational approach identified several other non‐coding genes located upstream of protein‐coding genes that are potentially regulated by Mmi1 (Table EV1). Thus, several other protein‐coding genes may be regulated by Mmi1‐mediated surveillance of lncRNA transcription. More broadly, in human cells, a new class of unstable transcripts, termed short intergenic ncRNAs (sincRNAs), that localize upstream of many protein‐coding genes was identified very recently (Schwalb et al, 2016). The role of these unstable lncRNAs remains unknown, but from our findings, it is possible that, in a similar way to the termination of nam1 transcription that regulates the expression byr2 gene, the control of sincRNAs transcription termination might regulate the expression of diverse protein‐coding genes.
LncRNA‐based heterochromatin gene silencing
Heterochromatin gene silencing at S. pombe centromeres relies on the processing and elimination of pericentromeric nascent lncRNAs by RNAi (Motamedi et al, 2004; Colmenares et al, 2007) and the exosome connected to the TRAMP complex (Buhler et al, 2007; Wang et al, 2008; Reyes‐Turcu et al, 2011). Our findings further reveal that Mmi1 recruits the exosome subunit Rrp6 to specific pericentromeric lncRNAs, in a TRAMP‐independent manner, indicating that Mmi1 also regulates expression of heterochromatic genes and that the exosome can be recruited to heterochromatic lncRNAs by different co‐factors. Additionally, our finding that read‐through transcripts accumulate in a Mmi1‐ and Rrp6‐dependent manner suggests that Mmi1 RNA surveillance machinery mediates heterochromatin gene silencing especially by inducing precocious transcription termination of RNAPII as it does at nam1 gene (Fig 6, right part). Interestingly, transcription termination also contributes to heterochromatin gene silencing in the evolutionary distant budding yeast (Vasiljeva et al, 2008), indicating that precocious transcription termination is a mechanism of heterochromatin gene silencing potentially shared by many eukaryotes.
We also report that Mmi1/exosome and RNAi machineries eliminate the same heterochromatic lncRNAs. What could be the advantage of having these two RNA surveillance machineries acting at the same genomic loci? One obvious possibility is that by implicating different RNA elimination machineries, the overall robustness of heterochromatin gene silencing is improved, as it has been proposed for the meiotic mRNAs targeted by Mmi1 RNA surveillance machinery and the RNAi effector complex RITS (Hiriart et al, 2012). However, our findings also suggest that the Mmi1‐ and RNAi‐mediated gene silencing processes may mostly alternate during the cell cycle progression, indicating that the implication of both machineries may rather insure a continuous heterochromatin gene silencing throughout the cell cycle. Another and non‐mutually exclusive possibility is that Mmi1/exosome and RNAi machineries compete for the same RNA substrate. This possibility is supported by our finding that overexpression of Mmi1 reduces the level of pericentromeric H3K9 methylation (Fig EV5E). Such a competition has already been described at S. pombe retrotransposons (Yamanaka et al, 2013). In wild‐type cells, retrotransposons are silenced in an exosome‐dependent fashion, while in rrp6‐deficient cells, retrotransposons are silenced by RNAi‐mediated formation of heterochromatin. At pericentromeric heterochromatin, when cells are not replicating, RNAi‐mediated heterochromatin formation is favored. Elimination of lncRNAs by RNAi would then contribute to the efficient formation and maintenance of heterochromatin, via the positive action of the RNAi‐dependent positive feedback loops. In early S phase, we propose that Mmi1/exosome RNA surveillance takes over and the elimination of the nascent transcripts may inhibit the RNAi amplification loops. This possibility is supported by the fact that Mmi1/exosome silencing activity is predominant when the amount of pericentromeric siRNAs is the lowest during the cell cycle (Kloc et al, 2008). Intriguingly, in mmi1∆ cells although the nam heterochromatic lncRNAs are no more degraded by the Mmi1/exosome machinery, they do not accumulate in early S phase, conversely to the dg heterochromatic lncRNAs. The reason for this apparent discrepancy is at the moment unclear. Importantly, regardless of what may be the exact reason for having both RNAi and the exosome acting on the same lncRNAs, our findings provide insights on the mechanism and function of the elimination of nascent heterochromatic lncRNAs by the exosome. Knowing that the exosome was found to contribute to heterochromatin gene silencing in plants (Shin et al, 2013) and drosophila (Eberle et al, 2015), and that in mammals pericentromeric lncRNAs accumulate in a cell‐cycle‐regulated manner (Lu & Gilbert, 2007), our findings on Mmi1‐mediated heterochromatin gene silencing have the potential to shed light on the mechanism and function of the exosome‐dependent heterochromatin gene silencing acting in other eukaryotes. In addition, our study demonstrates the direct implication of Mmi1, a member of the family of YTH domain‐containing proteins, in heterochromatin gene silencing. Remarkably, another member of the YTH family has been recently implicated in heterochromatin gene silencing at the inactive X chromosome in mammalian female cells (Patil et al, 2016). Thus, the function of YTH‐mediated heterochromatin gene silencing is conserved between fission yeast and mammals, and future studies shall determine whether they implicate similar lncRNA‐based mechanisms.
Materials and Methods
Strains, media, and plasmids
Genotypes of strains used in this study are listed in Appendix Table S4. Mating and sporulation assays were done using exponentially growing cells in MM(+N) media, washed three times with water, and transferred to MM media without ammonium chloride MM(‐N) for nitrogen starvation in liquid culture or on SPAS plates for the indicated times (with 105 cells used at each time point). Mating and sporulation efficiency were monitored from three independent experiments and by counting under the microscope at least 500 cells for each experiment. New S. pombe strains were made using the PCR‐based gene targeting method (Forsburg & Rhind, 2006). Positive transformants were selected by growth on YEA medium containing the appropriate antibiotic and confirmed by genomic PCR. Point mutants of Mmi1 protein were made using the Quick‐Change mutagenesis protocol (Agilent) and the plasmid pJRL81 (Moreno et al, 2000) containing wild‐type mmi1 coding sequence as the template. Mutations were confirmed by DNA sequencing before introduction of the plasmids in mmi1∆ mei4∆ cells. Note that, as previously reported (Hiriart et al, 2012), we used mmi1∆ mei4∆ cells because mmi1 deletion causes a strong growth defect due to the expression of Mei4 transcription factor. mei4∆ cells were thus used to control that the effects are indeed caused by the deletion of mmi1. A unique copy of WT mmi1, mmi1‐R351E, and mmi1‐R381E was integrated at ars1 genomic site in mmi1∆ mei4∆ cells, and grown on MM‐LEU. The nam1‐1, nam1‐1‐Ttef, and nam1‐Ttef cells were generated in two steps. First, ura4 gene was integrated into nam1 locus and positive transformants were selected by growth on –URA and genomic PCR. Second, nam1::ura4+ cells were transformed with synthesized DNA fragments of nam1 (Shine gene) containing either the single mutation of the first nucleotide (T to G) in eight of the nine TNAAAC motifs present in nam1, alone or together with the addition of the transcription terminator (Ttef) to generate, respectively, nam1‐1 and nam1‐1‐Ttef. Terminator Ttef has been integrated at the 3′ end of nam1 in cells named nam1‐Ttef or nam1‐1‐Ttef. Positive transformants were validated by sequencing the recombined genomic regions. Generation of cells expressing Flag‐Byr2 from the endogenous gene was realized following the same strategy as described above and using a synthetic DNA including the sequence of three Flag upstream and in frame with the coding sequence of byr2 (Shine gene). Ectopic expression of Byr2‐HA protein from pREP41 and pJRU41 plasmids (Moreno et al, 2000) was achieved by cloning byr2 coding sequence between NdeI and BglII restriction sites. Expression of nam1‐L and nam1‐1‐L from pREP3 plasmids was achieved by cloning, between PstI and SacI sites, a fragment of nam1‐byr2 genomic DNA sequence, which encompasses 160 nt upstream and 1,700 nt downstream of nam1, from WT and nam1‐1 cells, respectively.
RT‐qPCR
Total RNA was isolated using phenol/chloroform from 25 ml of log‐phase cell cultures; 1 μg of total RNA was reverse‐transcribed using Transcriptor reverse transcriptase (Roche). Strand‐specific RT–qPCRs were performed using specific primers. PCR and qPCR were done using the BioMixTM Red and the MESA BLUE qPCR MasterMix for SYBR® (Bioline, Eurogentec), respectively. Briefly, for the detection of read‐through transcription of nam1, the cDNA was synthesized using RT primers located in byr2 sequence, and amplified using primers located in nam1. For the detection of read‐through transcription of nam7, three different RT (1, 2 and 3) were performed, RT1 and RT2 using primers located in nam7 sequence, RT3 using a primer downstream of nam7 sequence. Primers are listed in Appendix Table S5.
RNA‐IP and chromatin‐IP
ChIP and RNA‐IP coupled to PCR analysis were performed as described previously (Hiriart et al, 2012). RNA‐IP experiments coupled to high‐throughput sequencing were conducted in duplicates. Immunoprecipitated RNA was fragmented 200–300nt long using RNA Fragmentation Reagent Kit from (Ambion). Fragmented RNA was 5′ phosphorylated using T4 Polynucleotide Kinase (Fermentas) and ligated to the 5′ adaptor using the T4 RNA ligase (Fermentas). After incubation overnight at 20°C, ligated RNA was purified (Absolutely RNA kit, Stratagene) and reverse‐transcribed (Superscript III Reverse Transcriptase, Invitrogen). PCR amplification of the cDNA library was performed using Phusion polymerase (NEB). 5′ adaptors and primers used for the RT and the PCR are listed in Appendix Table S5. The antibodies used for the IPs are anti‐Mmi1 (Hiriart et al, 2012), anti‐dimethylated H3K9 (Abcam, ab1220), anti‐Myc (9E10, Santa Cruz, sc‐40), anti‐TAP (Thermo Fisher, CAB 1001), anti‐RNAPII 8WG16 (Abcam, ab817), anti‐RNAPII‐Ser‐2P (Millipore, 04‐1571), and anti‐RNAPII‐Ser‐5P (Covance, MMS‐134R).
High‐throughput sequencing
DNA libraries from the RNA‐IPs were deep‐sequenced (Solexa, Illumina). Bioinformatic analysis was performed as described previously (Xiol et al, 2012). Briefly, the sequencing reads were mapped to the genome and associated with the annotated region at that genomic locus. For each experiment, the number of unique reads associated to each gene was divided by the total number of unique reads from that experiment, to obtain a normalized count in reads per million (RPM). For each gene in each duplicate RNA‐IP experiment, the control and RNA‐IP RPMs were adjusted by adding a fixed pseudo‐count of 10, and an enrichment, defined as the ratio between the adjusted RNA‐IP and control RPMs, was calculated. Our list of candidate Mmi1 targets contains only RNAs whose enrichment exceeds 2 in both RNA‐IP experiments. The high‐throughput sequencing files are available at the GEO database under the accession number GSE90688.
Computational prediction of Mmi1 targets
The computational method comprises three main steps (Fig EV1B) and relies on the following definition: for any given annotated sequence of RNA, we denote MWSX (minimal window size) as the smallest number of nucleotides that encompasses X occurrences of the UNAAAC motif in the RNA. For example, the MWS3 of a given RNA is the size of the smallest subsequence of the RNA that contains three UNAAAC motifs. In a first step, we focused on our 33 validated Mmi1 RNA targets [21 from (Hiriart et al, 2012), and 12 from this study]. For each value of X between 2 and 8, we determined the smallest possible cutoff window size CWSX (such that MWSX ≤ CWSX) for 75% of the 33 targets. X did not go above eight motifs since < 75% of the validated Mmi1 targets possess more than eight motifs. In a second step, we calculated the MWSX for each individual annotated gene in S. pombe, represented by its unspliced transcript sequence. Finally, in a third step, the CWSX cutoffs were used to rank all the genes. In this study, we only considered as strong candidates the ones that satisfy the following: MWSX ≤ CWSX for every X from 2 to 8 (Appendix Table S2).
Protein expression and purification
A His‐GST fusion of S. pombe Mmi1 YTH domain (residues 347–488) was expressed in E. coli BL21Star (DE3) from pPETM‐30 vector. The protein was first purified by affinity chromatography using Ni2+ resin. After His‐tag cleavage with TEV protease, the protein was further purified using a second Ni2+ column followed by a size‐exclusion chromatography. Purified Mmi1 YTH domain was concentrated to 8.5 mg/ml in a buffer containing 20 mM Tris, pH 7.0, 150 mM NaCl, and 10 mM β‐mercaptoethanol. The best‐diffracting crystals grew within 3 days at 20°C in a solution containing 0.2 M ammonium sulfate, 0.1 M Tris pH 8.5, and 25 % PEG3350. Selenomethionine (SeMet)‐substituted Mmi1 was produced using E. coli BL21Star (DE3) in a defined medium containing 60 mg/l of SeMet. Purification and crystallization of the SeMet‐substituted Mmi1 were done as for the native protein. For data collection at 100 K, crystals were snap‐frozen in liquid nitrogen with a solution containing mother liquor and 30% (v/v) glycerol.
Crystallization, data collection, and structure determination
Crystals of Mmi1 YTH domain (347–488) belong to the space group P21 with unit cell dimensions a = 57 Å, b = 58 Å, c = 94.1 Å, β = 106.6°. The asymmetric unit contains four Mmi1 molecules and has a solvent content of 47%. A complete native data set was collected to a resolution of 1.45 Å on beamline ID23EH1 at the European Synchrotron Radiation Facility (ESRF, Grenoble, France; Appendix Table S3). The data were processed using XDS (Kabsch, 2010). The structure of Mmi1 was determined by multiple‐wavelength anomalous dispersion (MAD) phasing method using a SeMet‐substituted crystal. Data sets with a resolution of 1.7–1.8 Å were collected at wavelengths corresponding to the peak and inflection point wavelength of the Se K‐edge (0.979012 and 0.979314 Å, respectively). The positions of selenium sites were identified, refined, and used for phasing in autoSHARP (Bricogne et al, 2003). COOT (Emsley & Cowtan, 2004) was used for model building. Structure was refined with REFMAC5 (Murshudov et al, 1997) to final R‐factor of 15.7% and Rfree of 17.9% with all residues in allowed (99% in favored) regions of the Ramachandran plot (Davis et al, 2004). Crystal diffraction data and refinement statistics for the structure are displayed in Appendix Table S3. Coordinates of the Mmi1 YTH domain have been deposited to the Protein Data Bank and assigned the accession number PDB ID 5O8M.
Immunofluorescent microscopy
Schizosaccharomyces pombe mid‐log‐phase cultures were fixed with 3.8% paraformaldehyde at room temperature for 30 min. After digestion of the cell wall with 0.25 mg/ml of Novozym and 0.25 mg/ml of Zymolyase (Sigma), cells were permeabilized 2 min with 1% Triton X‐100 and blocked with 3% BSA for 30 min. Detection of Mmi1 was obtained using a Mmi1 primary antibody (1/100 dilution) and a secondary antibody coupled to a fluorescent dye (Alexa 448 1/400 dilution). DNA was stained with 4′,6‐diamidino‐2‐phenylindole (DAPI) 1 ng/ml for 5 min. Fluorescence microscopy and differential interference contrast (DIC) imaging were done using a Zeiss Apotome microscope (Carl Zeiss MicroImaging). Raw images were processed using the AxioVision software (Carl Zeiss MicroImaging).
RNA pull‐down assay
3 × 1010 E. coli BL21Star (DE3) cells expressing either His‐Mmi1 WT, His‐Mmi1R351E, or His‐Mmi1R381E mutant proteins were lysed by sonication in 10 ml lysis buffer (50 mM HEPES, 150 mM NaCl, 1 mM EDTA, 1% Triton X‐100, 5 mM DTT, 1 mM PMSF); 100 ng of the biotinylated RNA (5′Biot‐GGAUCCUUAAACAGAUCU) was denaturated 2 min at 90°C and incubated 20 min at room temperature in RNA structure buffer (10 mM Tris pH 7, 0.1 M KCl, 10 mM MgCl2). The biotinylated RNA was added to a series of 100 μl of lysis extracts diluted 10‐fold from 1 to 1,000, and incubated 1 h at room temperature; 15 μl of 10 mg/ml Dynabeads™ M280 Streptavidin (Invitrogen) was added to the mixture and incubated for an additional 30 min at room temperature. The beads were then washed three times with 1 ml of lysis buffer and then eluted by boiling 5 min in SDS Laemmli buffer. The efficiency of Mmi1 binding to the RNA was analyzed by Western blot using anti‐Mmi1 antibody. Relative enrichments of Mmi1 wild‐type and mutant proteins after RNA pull‐down were calculated from three independent experiments as follows: for each experiment and each Mmi1 proteins, the signals obtained with the dilutions 1/1, 1/10, and 1/100 of the extracts were quantified using ImageJ. The average quantification of the three signals was then normalized to the input signal to obtain the enrichment.
Northern blot
Northern blot experiments were conducted following the usual procedure. 10 μg of total RNA was used for each sample. Probes were labeled radioactively by 5‐min labeling using T4 Polynucleotide Kinase (Fermentas) for the strand‐specific nam5/6/7 probes, and by random priming for byr2 and nam1 probes. Hybridization step was done overnight at 65°C in ULTRAHyb Buffer (Thermo Fisher). The membranes were exposed to a PhosphorImager screen (PMI—Bio‐Rad).
Cell cycle synchronization
Cell cycle synchronization experiments using the cdc25‐22 temperature‐sensitive mutant strain were performed as described previously (Forsburg & Rhind, 2006). Briefly, cell synchronization was achieved by a block and release of cell proliferation, consisting in shifting an early log‐phase cell culture from 25 to 36°C for 3 h to block the cells in G2 phase. The temperature of the culture was rapidly lowered to 25°C by cooling it in cold water. At 25°C, cells restart their growth in a synchronized manner. To evaluate the level of cell synchronization, the septation index (which corresponds to the proportion of cells with a septum) was determined after fixation of the cells in 70% ethanol and staining with DAPI (1 μg/ml) and Calcofluor (10 μg/ml).
Author contributions
LT‐T and AV planned the study. LT‐T, YS, AY, MY, and AV analyzed the data. LT‐T and AV wrote the original draft and all of the authors refined the manuscript. LT‐T and MR performed RNA‐IPs. BG, NT‐M, RS, RP, and AV performed the analyses on massive sequencing data. NT‐M and AV designed the computational screen. JB and JK obtained the YTH domain crystal structure. LT‐T, MD, and YS performed mating and sexual differentiation assays. YS and LT‐T performed the Northern blots, and YS, AY, and MY analyzed them. LT‐T performed all other experiments with help from MD, EL, and EH.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Table EV1
Source Data for Expanded View
Review Process File
Source Data for Figure 1
Source Data for Figure 2
Source Data for Figure 3
Source Data for Figure 4
Source Data for Figure 5
Acknowledgements
We thank R. Allshire, F. Bachand, P. Bernard, D. Moazed for strains and reagents. We thank El C. Ibrahim, D. Libri, S. Rousseaux, and members of the A.V. laboratory for their helpful comments and critical reading of the manuscript, and X. Ronot for his support in the initial stage of the study. We thank the microscope facility from IAB and the Center for Radioisotope Facilities from Okazaki Research Facilities NINS for technical support. M.D. was supported in part by a PhD fellowship from the Association pour la Recherche sur le Cancer (ARC). This study was supported by JSPS KAKENHI (Grant Number 15H04333), a grant from The Naito Foundation to A.Y. and a grant for Basic Science Research Projects from The Sumitomo Foundation (Grand Number 140283) to A.Y., and by the Institut National de la Santé Et de la Recherche Médicale (INSERM) Avenir program, a European Research Council (ERC) Starting Grant (ERC‐StG‐RNAiEpiMod‐210896) and the “RNAgermSilence” ANR grant to A.V.
The EMBO Journal (2017) 36: 2626–2641
References
- Ard R, Tong P, Allshire RC (2014) Long non‐coding RNA‐mediated transcriptional interference of a permease gene confers drug tolerance in fission yeast. Nat Commun 5: 5576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bricogne G, Vonrhein C, Flensburg C, Schiltz M, Paciorek W (2003) Generation, representation and flow of phase information in structure determination: recent developments in and around SHARP 2.0. Acta Crystallogr D Biol Crystallogr 59: 2023–2030 [DOI] [PubMed] [Google Scholar]
- Buhler M, Verdel A, Moazed D (2006) Tethering RITS to a nascent transcript initiates RNAi‐ and heterochromatin‐dependent gene silencing. Cell 125: 873–886 [DOI] [PubMed] [Google Scholar]
- Buhler M, Moazed D (2007) Transcription and RNAi in heterochromatic gene silencing. Nat Struct Mol Biol 14: 1041–1048 [DOI] [PubMed] [Google Scholar]
- Buhler M, Haas W, Gygi SP, Moazed D (2007) RNAi‐dependent and ‐independent RNA turnover mechanisms contribute to heterochromatic gene silencing. Cell 129: 707–721 [DOI] [PubMed] [Google Scholar]
- Cam HP, Chen ES, Grewal SI (2009) Transcriptional scaffolds for heterochromatin assembly. Cell 136: 610–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castel SE, Martienssen RA (2013) RNA interference in the nucleus: roles for small RNAs in transcription, epigenetics and beyond. Nat Rev Genet 14: 100–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalamcharla VR, Folco HD, Dhakshnamoorthy J, Grewal SI (2015) Conserved factor Dhp1/Rat1/Xrn2 triggers premature transcription termination and nucleates heterochromatin to promote gene silencing. Proc Natl Acad Sci USA 112: 15548–15555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee D, Sanchez AM, Goldgur Y, Shuman S, Schwer B (2016) Transcription of lncRNA prt, clustered prt RNA sites for Mmi1 binding, and RNA polymerase II CTD phospho‐sites govern the repression of pho1 gene expression under phosphate‐replete conditions in fission yeast. RNA 22: 1011–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ES, Zhang K, Nicolas E, Cam HP, Zofall M, Grewal SI (2008) Cell cycle control of centromeric repeat transcription and heterochromatin assembly. Nature 451: 734–737 [DOI] [PubMed] [Google Scholar]
- Chen HM, Futcher B, Leatherwood J (2011) The fission yeast RNA binding protein Mmi1 regulates meiotic genes by controlling intron specific splicing and polyadenylation coupled RNA turnover. PLoS ONE 6: e26804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chlebowski A, Lubas M, Jensen TH, Dziembowski A (2013) RNA decay machines: the exosome. Biochim Biophys Acta 1829: 552–560 [DOI] [PubMed] [Google Scholar]
- Colmenares SU, Buker SM, Buhler M, Dlakic M, Moazed D (2007) Coupling of double‐stranded RNA synthesis and siRNA generation in fission yeast RNAi. Mol Cell 27: 449–461 [DOI] [PubMed] [Google Scholar]
- Davis IW, Murray LW, Richardson JS, Richardson DC (2004) MOLPROBITY: structure validation and all‐atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res 32: W615–W619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djupedal I, Portoso M, Spahr H, Bonilla C, Gustafsson CM, Allshire RC, Ekwall K (2005) RNA Pol II subunit Rpb7 promotes centromeric transcription and RNAi‐directed chromatin silencing. Genes Dev 19: 2301–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberle AB, Jordan‐Pla A, Ganez‐Zapater A, Hessle V, Silberberg G, von Euler A, Silverstein RA, Visa N (2015) An interaction between RRP6 and SU(VAR)3‐9 targets RRP6 to heterochromatin and contributes to heterochromatin maintenance in Drosophila melanogaster . PLoS Genet 11: e1005523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K (2004) Coot: model‐building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Forsburg SL, Rhind N (2006) Basic methods for fission yeast. Yeast 23: 173–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal SI, Jia S (2007) Heterochromatin revisited. Nat Rev Genet 8: 35–46 [DOI] [PubMed] [Google Scholar]
- Guil S, Esteller M (2012) Cis‐acting noncoding RNAs: friends and foes. Nat Struct Mol Biol 19: 1068–1075 [DOI] [PubMed] [Google Scholar]
- Harigaya Y, Tanaka H, Yamanaka S, Tanaka K, Watanabe Y, Tsutsumi C, Chikashige Y, Hiraoka Y, Yamashita A, Yamamoto M (2006) Selective elimination of messenger RNA prevents an incidence of untimely meiosis. Nature 442: 45–50 [DOI] [PubMed] [Google Scholar]
- Hiriart E, Vavasseur A, Touat‐Todeschini L, Yamashita A, Gilquin B, Lambert E, Perot J, Shichino Y, Nazaret N, Boyault C, Lachuer J, Perazza D, Yamamoto M, Verdel A (2012) Mmi1 RNA surveillance machinery directs RNAi complex RITS to specific meiotic genes in fission yeast. EMBO J 31: 2296–2308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiriart E, Verdel A (2013) Long noncoding RNA‐based chromatin control of germ cell differentiation: a yeast perspective. Chromosome Res 21: 653–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen TH, Jacquier A, Libri D (2013) Dealing with pervasive transcription. Mol Cell 52: 473–484 [DOI] [PubMed] [Google Scholar]
- Kabsch W (2010) Integration, scaling, space‐group assignment and post‐refinement. Acta Crystallogr D Biol Crystallogr 66: 133–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, Goto DB, Martienssen RA, Urano T, Furukawa K, Murakami Y (2005) RNA polymerase II is required for RNAi‐dependent heterochromatin assembly. Science 309: 467–469 [DOI] [PubMed] [Google Scholar]
- Keller C, Adaixo R, Stunnenberg R, Woolcock KJ, Hiller S, Buhler M (2012) HP1(Swi6) mediates the recognition and destruction of heterochromatic RNA transcripts. Mol Cell 47: 215–227 [DOI] [PubMed] [Google Scholar]
- Kilchert C, Wittmann S, Vasiljeva L (2016) The regulation and functions of the nuclear RNA exosome complex. Nat Rev Mol Cell Biol 17: 227–239 [DOI] [PubMed] [Google Scholar]
- Kloc A, Zaratiegui M, Nora E, Martienssen R (2008) RNA interference guides histone modification during the S phase of chromosomal replication. Curr Biol 18: 490–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornienko AE, Guenzl PM, Barlow DP, Pauler FM (2013) Gene regulation by the act of long non‐coding RNA transcription. BMC Biol 11: 59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemay JF, Larochelle M, Marguerat S, Atkinson S, Bahler J, Bachand F (2014) The RNA exosome promotes transcription termination of backtracked RNA polymerase II. Nat Struct Mol Biol 21: 919–926 [DOI] [PubMed] [Google Scholar]
- Li F, Zhao D, Wu J, Shi Y (2014) Structure of the YTH domain of human YTHDF2 in complex with an m(6)A mononucleotide reveals an aromatic cage for m(6)A recognition. Cell Res 24: 1490–1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Gilbert DM (2007) Proliferation‐dependent and cell cycle regulated transcription of mouse pericentric heterochromatin. J Cell Biol 179: 411–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno MB, Duran A, Ribas JC (2000) A family of multifunctional thiamine‐repressible expression vectors for fission yeast. Yeast 16: 861–872 [DOI] [PubMed] [Google Scholar]
- Morris KV, Mattick JS (2014) The rise of regulatory RNA. Nat Rev Genet 15: 423–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motamedi MR, Verdel A, Colmenares SU, Gerber SA, Gygi SP, Moazed D (2004) Two RNAi complexes, RITS and RDRC, physically interact and localize to noncoding centromeric RNAs. Cell 119: 789–802 [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ (1997) Refinement of macromolecular structures by the maximum‐likelihood method. Acta Crystallogr D Biol Crystallogr 53: 240–255 [DOI] [PubMed] [Google Scholar]
- Oya E, Kato H, Chikashige Y, Tsutsumi C, Hiraoka Y, Murakami Y (2013) Mediator directs co‐transcriptional heterochromatin assembly by RNA interference‐dependent and ‐independent pathways. PLoS Genet 9: e1003677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil DP, Chen CK, Pickering BF, Chow A, Jackson C, Guttman M, Jaffrey SR (2016) m6A RNA methylation promotes XIST‐mediated transcriptional repression. Nature 537: 369–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart BJ, Bartel DP (2002) Small RNAs correspond to centromere heterochromatic repeats. Science 297: 1831 [DOI] [PubMed] [Google Scholar]
- Reyes‐Turcu FE, Zhang K, Zofall M, Chen E, Grewal SI (2011) Defects in RNA quality control factors reveal RNAi‐independent nucleation of heterochromatin. Nat Struct Mol Biol 18: 1132–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Chang HY (2012) Genome regulation by long noncoding RNAs. Annu Rev Biochem 81: 145–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwalb B, Michel M, Zacher B, Fruhauf K, Demel C, Tresch A, Gagneur J, Cramer P (2016) TT‐seq maps the human transient transcriptome. Science 352: 1225–1228 [DOI] [PubMed] [Google Scholar]
- Shah S, Wittmann S, Kilchert C, Vasiljeva L (2014) lncRNA recruits RNAi and the exosome to dynamically regulate pho1 expression in response to phosphate levels in fission yeast. Genes Dev 28: 231–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin JH, Wang HL, Lee J, Dinwiddie BL, Belostotsky DA, Chekanova JA (2013) The role of the Arabidopsis exosome in siRNA‐independent silencing of heterochromatic loci. PLoS Genet 9: e1003411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoilov P, Rafalska I, Stamm S (2002) YTH: a new domain in nuclear proteins. Trends Biochem Sci 27: 495–497 [DOI] [PubMed] [Google Scholar]
- Styrkarsdottir U, Egel R, Nielsen O (1992) Functional conservation between Schizosaccharomyces pombe ste8 and Saccharomyces cerevisiae STE11 protein kinases in yeast signal transduction. Mol Gen Genet 235: 122–130 [DOI] [PubMed] [Google Scholar]
- Tashiro S, Asano T, Kanoh J, Ishikawa F (2013) Transcription‐induced chromatin association of RNA surveillance factors mediates facultative heterochromatin formation in fission yeast. Genes Cells 18: 327–339 [DOI] [PubMed] [Google Scholar]
- Theler D, Dominguez C, Blatter M, Boudet J, Allain FH (2014) Solution structure of the YTH domain in complex with N6‐methyladenosine RNA: a reader of methylated RNA. Nucleic Acids Res 42: 13911–13919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasiljeva L, Kim M, Terzi N, Soares LM, Buratowski S (2008) Transcription termination and RNA degradation contribute to silencing of RNA polymerase II transcription within heterochromatin. Mol Cell 29: 313–323 [DOI] [PubMed] [Google Scholar]
- Verdel A, Jia S, Gerber S, Sugiyama T, Gygi S, Grewal SI, Moazed D (2004) RNAi‐mediated targeting of heterochromatin by the RITS complex. Science 303: 672–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Xu HP, Riggs M, Rodgers L, Wigler M (1991) byr2, a Schizosaccharomyces pombe gene encoding a protein kinase capable of partial suppression of the ras1 mutant phenotype. Mol Cell Biol 11: 3554–3563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SW, Stevenson AL, Kearsey SE, Watt S, Bahler J (2008) Global role for polyadenylation‐assisted nuclear RNA degradation in posttranscriptional gene silencing. Mol Cell Biol 28: 656–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, He C (2014) Reading RNA methylation codes through methyl‐specific binding proteins. RNA Biol 11: 669–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, Fu Y, Parisien M, Dai Q, Jia G, Ren B, Pan T, He C (2014) N6‐methyladenosine‐dependent regulation of messenger RNA stability. Nature 505: 117–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Zhu Y, Bao H, Jiang Y, Xu C, Wu J, Shi Y (2016) A novel RNA‐binding mode of the YTH domain reveals the mechanism for recognition of determinant of selective removal by Mmi1. Nucleic Acids Res 44: 969–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wery M, Descrimes M, Vogt N, Dallongeville AS, Gautheret D, Morillon A (2016) Nonsense‐mediated decay restricts LncRNA levels in yeast unless blocked by double‐stranded RNA structure. Mol Cell 61: 379–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao W, Adhikari S, Dahal U, Chen YS, Hao YJ, Sun BF, Sun HY, Li A, Ping XL, Lai WY, Wang X, Ma HL, Huang CM, Yang Y, Huang N, Jiang GB, Wang HL, Zhou Q, Wang XJ, Zhao YL et al (2016) Nuclear m(6)A reader YTHDC1 regulates mRNA splicing. Mol Cell 61: 507–519 [DOI] [PubMed] [Google Scholar]
- Xiol J, Cora E, Koglgruber R, Chuma S, Subramanian S, Hosokawa M, Reuter M, Yang Z, Berninger P, Palencia A, Benes V, Penninger J, Sachidanandam R, Pillai RS (2012) A role for Fkbp6 and the chaperone machinery in piRNA amplification and transposon silencing. Mol Cell 47: 970–979 [DOI] [PubMed] [Google Scholar]
- Xu C, Wang X, Liu K, Roundtree IA, Tempel W, Li Y, Lu Z, He C, Min J (2014) Structural basis for selective binding of m6A RNA by the YTHDC1 YTH domain. Nat Chem Biol 10: 927–929 [DOI] [PubMed] [Google Scholar]
- Yamanaka S, Yamashita A, Harigaya Y, Iwata R, Yamamoto M (2010) Importance of polyadenylation in the selective elimination of meiotic mRNAs in growing Schizosaccharomyces pombe cells. EMBO J 29: 2173–2181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka S, Mehta S, Reyes‐Turcu FE, Zhuang F, Fuchs RT, Rong Y, Robb GB, Grewal SI (2013) RNAi triggered by specialized machinery silences developmental genes and retrotransposons. Nature 493: 557–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita A, Shichino Y, Tanaka H, Hiriart E, Touat‐Todeschini L, Vavasseur A, Ding DQ, Hiraoka Y, Verdel A, Yamamoto M (2012) Hexanucleotide motifs mediate recruitment of the RNA elimination machinery to silent meiotic genes. Open Biol 2: 120014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita A, Shichino Y, Yamamoto M (2016) The long non‐coding RNA world in yeasts. Biochim Biophys Acta 1859: 147–154 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Theler D, Kaminska KH, Hiller M, de la Grange P, Pudimat R, Rafalska I, Heinrich B, Bujnicki JM, Allain FH, Stamm S (2010) The YTH domain is a novel RNA binding domain. J Biol Chem 285: 14701–14710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Zhu J, Schermann G, Ohle C, Bendrin K, Sugioka‐Sugiyama R, Sugiyama T, Fischer T (2015) The fission yeast MTREC complex targets CUTs and unspliced pre‐mRNAs to the nuclear exosome. Nat Commun 6: 7050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu T, Roundtree IA, Wang P, Wang X, Wang L, Sun C, Tian Y, Li J, He C, Xu Y (2014) Crystal structure of the YTH domain of YTHDF2 reveals mechanism for recognition of N6‐methyladenosine. Cell Res 24: 1493–1496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zofall M, Yamanaka S, Reyes‐Turcu FE, Zhang K, Rubin C, Grewal SI (2012) RNA elimination machinery targeting meiotic mRNAs promotes facultative heterochromatin formation. Science 335: 96–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Table EV1
Source Data for Expanded View
Review Process File
Source Data for Figure 1
Source Data for Figure 2
Source Data for Figure 3
Source Data for Figure 4
Source Data for Figure 5


