Abstract
A universal response to elevated temperature and other forms of physiological stress is the induction of heat shock proteins (HSPs). Hsp16 in Schizosaccharomyces pombe encodes a polypeptide of predicted molecular weight 16 kDa that belongs to the HSP20/α-crystallin family whose members range in size from 12 to 43 kDa. Heat shock treatment increases expression of the hsp16 gene by 64-fold in wild-type cells and 141-fold in cdc22-M45 (ribonucleotide reductase) mutant cells. Hsp16 expression is mediated by the spc1 MAPK signaling pathway through the transcription factor atf1 and in addition through the HSF pathway. Nucleotide depletion or DNA damage as occurs in cdc22-M45 mutant cells, or during hydroxyurea or camptothecin treatment, is sufficient to activate hsp16 expression through atf1. Our findings suggest a novel role for small HSPs in the stress response following nucleotide depletion and DNA damage. This extends the types of damage that are sensed by the spc1 MAPK pathway via atf1.
INTRODUCTION
A universal response to elevated temperature and other forms of stress is the induction of heat shock proteins (HSPs). HSPs are classified into seven major families according to their size, structure and function. These include HSP100, HSP90, HSP70, HSP60, HSP40, HSP33 and the small HSPs related to α-crystallins (12–43 kDa). Large HSPs are highly conserved in species as diverse as bacteria, yeast and mammals. In contrast, for small HSPs the main region of homology is a hydrophobic stretch of only ∼80–100 amino acids showing sequence similarity to vertebrate α-crystallin (1). The conserved α-crystallin domain is located in the C-terminus (2–5; Pfam database) and is preceded by an N-terminal domain of variable length and sequence (2).
In Schizosaccharomyces pombe, three small HSPs have been identified. Two of these, hsp16 and hsp20, are characterized as members of the Hsp20/α-crystallin family (6; NCBI), while the other, hsp9, has homology to HSP12 of Saccharomyces cerevisiae and hsp26 of Drosophila melanogaster (7). In other organisms, the number of small HSPs is variable. For example, there are at least 20 in Arabidopsis thaliana, (Arabidopsis database), 28 in human (NCBI), four in D.melanogaster (Drosophila database), five in mouse (NCBI), 16 in Caenorhabditis elegans (NCBI) and five in S.cerevisiae (8–10; NCBI). The Pfam database currently contains 364 members of this family from all species (11). Some characterized members of the Hsp20/α-crystallin family include: vertebrate hsp27 (hsp25), induced by a variety of environmental stresses; D.melanogaster hsp22, hsp23, hsp26, hsp27; the C.elegans hsp16 multigene family; in fungi, HSP26 (S.cerevisiae) and hsp30 (Neurospora crassa and Aspergillus nidulans); and in plants four classes of hsp20, plus α-crystallin A and B chains.
The large HSPs have been implicated in major physiological processes such as cell division, transcription, protein folding, transport and membrane function (12–15). To date, however, there is no experimental evidence that small HSPs are essential for normal cellular function. The Hsp20/α-crystallin family act as molecular chaperones in vitro protecting other proteins against heat-induced denaturation and aggregation (15–20). They can form large oligomeric complexes (17,21–23) and have a role in thermotolerance in mammalian cells and Drosophila (24–26) but not in yeast cells (8). In mammalian cells, small HSPs bind specifically to cytoskeletal elements such as actin and to intermediate filaments such as desmin, vimentin and glial fibrillary acidic protein (24,27–29). It has also been reported that small HSPs modulate apoptosis through the Fas/Apo1 receptor (30) and are involved in cell growth and differentiation (31).
Hsp26 in S.cerevisiae functions as a molecular chaperone in vivo (21). It accumulates to high levels after heat shock, during the transition to sporulation and even under other stresses such as increased salt concentration and starvation (32,33). However, Hsp26 does not appear to be required for cell viability under these conditions (8,34). This suggests that the function of Hsp26 in stress response overlaps the functions of other HSPs. To date the regulation and function of small HSPs in yeast remains elusive.
In this paper, we report the isolation of S.pombe hsp16, a member of the small HSP family. We show that hsp16 expression is induced by a number of environmental stimuli including heat shock. In addition, expression of hsp16 is responsive to deoxyribonucleotide depletion or DNA damage and this response is dependent on the spc1 MAPK pathway and the atf1 transcription factor.
MATERIALS AND METHODS
Strains and media
Schizosaccharomyces pombe strains were derived from wild-type 972 h– or 975 h+ (35) (Table 1). Strains were grown in YEA complex medium (yeast extract medium containing adenine) or Edinburgh minimal medium containing nutritional supplements as necessary (36).
Table 1. Schizosaccharomyces pombe strains used in this study.
| Strains |
Genotype |
Source |
| Q250 | h– wild-type | Laboratory collection |
| Q1411 | h– ura4-D18 | Laboratory collection |
| Q1825 | h– hsp16::ura4+ ura4-D18 | This study |
| Q243 | h90 cdc22-M45 | Laboratory collection |
| Q1832 | h– hsp16::ura4+ cdc22-M45 ura4-D18 | This study |
| Q1833 | h– hsp16::ura4+ spc1::ura4+ ura4-D18 | This study |
| Q1834 | h+ hsp16::ura4+ atf1::ura4+ ura4-D18 | This study |
| Q815 | h– rad1-1 ade6– | Laboratory collection |
| Q956 | h– rad1-1 cdc22-M45 | Laboratory collection |
| Q1622 | h– atf1::ura4+leu1-32 ura4-D18 his3-D1 | W. Wahls |
| Q1692 | h– atf1::ura4+ cdc22-M45 leu1-32 his3-D1 ura4-D18 | This study |
| Q1510 | h– spc1::ura4+ leu1-32 | Shiozaki and Russell |
| Q1676 | h+ spc1::ura4+ cdc22-M45 ura4-D18 | This study |
| Q910 | h– pyp1::ura4+ ura4-D18 | S. Ottilie |
| Q1820 | h+ pyp1::ura4+ cdc22-M45 ura4-D18 | This study |
| Q225 | h90 ras1::leu1+ leu1-32 ade6-210 | Nadin-Davis and Nasim |
| Q227 | h90 rasval17 leu1-32 ade6-210 | Nadin-Davis and Nasim |
| Q1814 | h90 hsp16–GFP(S65T)-ura4+ ura4-D18 | This study |
| Q1636 | h+ cdc22-M45 hsp16–GFP(S65T)-ura4+ ura4-D18 | This study |
| Q1662 | h– spc1::ura4+ hsp16–GFP(S65T)-ura4+ ura4-D18 leu1-32 | This study |
| Q1681 | h+ spc1::ura4+ cdc22-M45 hsp16–GFP(S65T)-ura4+ ura4-D18 | This study |
| Q1683 | h– atf1::ura4+ hsp16–GFP(S65T)-ura4+ ura4-D18 leu1-32 his3-D1 | This study |
| Q1694 | h+ atf1::ura4+ cdc22-M45 hsp16–GFP(S65T)-ura4+ ura4-D18 his3-D1 | This study |
| Q1708 | h– pcr1::his3+ hsp16–GFP(S65T)-ura4+ ura4-D18 leu1-32 his3-D1 | This study |
| Q1730 | h+ pcr1::his3+ cdc22-M45 hsp16–GFP(S65T)-ura4+ ura4-D18 his3-D1 | This study |
| Q1721 | h– wis1::his1+ hsp16–GFP(S65T)-ura4+ ura4-D18 leu1-32 his1-102 | This study |
| Q1728 | h+ wis1::his1+ cdc22-M45 hsp16–GFP(S65T)-ura4+ ura4-D18 leu1-32 his1-102 | This study |
| Q1639 | h– rad1-1 hsp16–GFP(S65T)-ura4+ ura4-D18 | This study |
| Q1644 | h+ rad1-1 cdc22-M45 hsp16–GFP(S65T)-ura4+ ura4-D18 | This study |
| Q1740 | h– rad1-1 spc1:: ura4+ hsp16–GFP(S65T)-ura4+ ura4-D18 | This study |
| Q1792 | h90 ras::leu1+ hsp16–GFP(S65T)-ura4+ ura4-D18 leu1-32 ade6-210 | This study |
| Q1858 | h– hsp20::ura4+ ura4-D18 | This study |
| Q1859 | h– hsp16::ura4+ hsp20::ura4+ ura4-D18 | This study |
Cloning of hsp16 gene
Standard molecular biological and genetic techniques were used (36,37). In an attempt to identify genes whose expression was dependent on cell cycle stage, a differential hybridization screen was mounted using a fission yeast λ genomic library which was replica-blotted and probed with radioactive cDNA probes. The probes were reverse transcribed from RNA isolated from various cdc mutant strains following arrest at 36°C for 4 h (cdc10-129, G1; cdc22-M45, S; cdc25-22, G2). One λ clone, 15-66, was found to give a very strong signal with the cdc22-M45 probe but not with the others. The hybridizing region was subcloned and sequenced yielding hsp16 (38,39). hsp16 has been independently sequenced by the fission yeast genome project (Sanger database) and has also recently been characterized by Danjoh and Fujiyama (6).
Construction of Δhsp16
The entire hsp16 open reading frame (ORF) was replaced with the ura4+ gene by one-step gene replacement (37). Stable ura4+ haploids were selected and exact gene replacement was confirmed by PCR, northern blot hybridization and western blotting. The strain was extensively out-crossed to ensure that no background mutations were present.
Construction of hsp20::ura4+
Hsp20 was identified by similarity to hsp16 within the fission yeast genome project. The hsp20 gene from –172 to +591 bp relative to the 420 bp ORF was amplified by PCR from S.pombe genomic DNA using high fidelity Taq polymerase (Roche Molecular Biochemicals). The PCR product was subcloned into pCR2.1-Topo (Invitrogen) and the ura4+ gene subcloned into the blunt ended BstEII site at +52 bp in the hsp20 ORF to generate pCR2.1-Topo-hsp20::ura4+. The hsp20::ura4+ cassette in the recombinant vector was then PCR amplified with high fidelity Taq polymerase (Roche Molecular Biochemicals) and this PCR product used to replace hsp20 in a haploid strain (ura4-D18 h–) (37). Stable ura4+ haploids were selected and exact gene replacement was confirmed by PCR.
Expression of hsp16
Total RNA was prepared as described (37) and 5 µg of each sample was resolved on a formaldehyde gel. Hybridization probes were labeled using the Rediprime II random prime labeling system ([α-32P]dCTP; Amersham Pharmacia Biotech). An NdeI–SalI hsp16 fragment was used to detect the hsp16 mRNA and a BamHI fragment of rDNA (plasmid provided by M. Yanagida, Kyoto University, Japan) was used to detect ribosomal RNA as a control. Signals were quantitated directly using a PhosphorImager (Molecular Dynamics) and expressed relative to 18S rDNA levels, which served as an internal loading control. Two independent RNA extractions were prepared and analyzed.
Quantitation of green fluorescent protein (GFP)
A very strong correlation exists between the amount of GFP in a cell and the total fluorescence (40,41). This was therefore used to quantitate protein expression.
Wild-type and mutant yeast strains were grown with shaking at 25°C in YEA to a density of 2–5 × 106 cells/ml and then shifted to the restrictive temperature of 36°C for 4 h. At various times 10 ml of liquid culture was collected into ice-cold water to a final cell density of 1 × 107 cells/ml and kept on ice. Hsp16–GFP levels were quantitated using a Luminescence Spectrometer LS50B (Perkin Elmer) and normalized relative to fluorescence levels in a wild-type cell not expressing GFP. Assays were performed in triplicate and independently repeated three times. Control experiments showed that the GFP fluorescence was stable for at least 6 h while samples were kept at 0°C. The washing and analysis procedure alone did not induce expression of this gene.
Production of GST–hsp16 fusion protein
To prepare the protein product of the hsp16 gene, the coding region of hsp16 was fused to the IPTG-inducible glutathione S-transferase (GST) in pGEX-2T (Amersham Pharmacia Biotech). Following expression in Escherichia coli with 1 mM IPTG at 37°C the GST–hsp16 fusion protein was purified on a glutathione–agarose column and eluted with 10 mM reduced glutathione in 50 mM Tris–HCl pH 8.0 according to the manufacturer’s instructions (Amersham Pharmacia Biotech).
Immunochemical analysis
To generate polyclonal antibodies against the hsp16 protein, GST–hsp16 protein was separated on a 12% SDS–polyacrylamide gel, excised, eluted and mixed with Titre Max Gold Adjuvant (Cedarlane) according to the manufacturer’s instruction. Following a second injection at day 28, serum was collected on day 40 and used as a source of antibody for western blot analysis and immunofluorescence experiments.
Fluorescence microscopy
All fluorescent images were taken with a Leica fluorescence microscope equipped with a high performance CCD camera (Sensicam) and Slidebook software (Intelligent Imaging System). Cells were collected using Whatman 934-AH glass micofibre filters (Fisher Scientific) and fixed in 100% methanol at –20°C for at least 20 min. Immunofluorescence was carried out as described in Sawin and Nurse (42). The primary antibody used was the rabbit polyclonal GST–hsp16 antiserum generated in the laboratory (1:5000) and the secondary antibody used was Alexa™ goat anti-rabbit IgG (H + L) conjugate (1:250) (Molecular Probes). Stained cells were counterstained with 1 µg/ml DAPI.
Protein extraction
For native protein extracts, cells were harvested by centrifugation, washed once with ice-cold stop buffer (150 mM NaCl, 50 mM NaF, 10 mM EDTA, 1 mM NaN3 pH 8.0) and immediately frozen at –70°C. The cell pellet was resuspended in 200 µl lysis buffer (50 mM Tris–HCl pH 8.0, 150 mM NaCl, 5 mM EDTA, 1% Nonidet P-40, 1 mM DTT, 10% glycerol, 50 mM NaF, 1 mM Na3VO4, 1 mM PMSF and 20 µg/ml each leupeptin, pepstatin and aprotinin) and glass beads added to the meniscus. Cells were broken open by vortexing with glass beads and centrifuged to prepare a cleared whole-cell extract. Protein concentration was determined using the Bio-Rad protein assay. Extracts (20 µg) were separated by 12% SDS–PAGE, electroblotted to a PVDF membrane (Santa Cruz) and detected by immunoblotting with polyclonal anti-GST–hsp16 antibody (1:1000). Immunoreactive bands were revealed with HRP-conjugated secondary goat anti-rabbit IgG antibody (1:2000) (Santa Cruz) and the luminol-based ECL detection kit (Santa Cruz).
Yeast two-hybrid screen
The full-length hsp16 orf was fused to the 3′ end of the lexA DNA-binding domain in pEG202 (43) by generating a BamHI/NotI hsp16 fragment by PCR and cloning into the BamHI/NotI sites, to yield the bait plasmid. The λ ACT S.pombe cDNA library (obtained from ATCC 87289) was fused to the Gal4 transcriptional activation domain and cloned into the XhoI site with subsequent conversion to plasmid form using cre-lox site-specific recombination (44). The two-hybrid experiments were performed with S.cerevisiae strain Y1003 (MATa/MATαURA3::lexAop-lacZ/8lexA-ADE2::URA3 ura3-1/ura3-1 leu2-2/leu2-3 his3-11/his3-11 trp1-1/trp1-1 ade2-1/ade2 can1-100/can1-100) (45). Approximately 750 000 cDNA clones were screened. Following confirmation of the assay, positive clones were sequenced.
RESULTS
hsp16 expression is strongly induced in a cdc22-M45 mutant background
A λ plaque was identified that displays a very strong signal when hybridized to labeled cDNA from cdc22-M45 arrested cells but not from probes made from cdc10-129 or cdc25-22 arrested cells. The λ clone (designated 15–66) was nick-translated and used to hybridize to northern blots of RNA derived from the same three arrested cell populations. A mRNA of ∼750 bp was found to account for the differential signal (39, data not shown). The region coding for this transcript was subcloned, sequenced and found to be hsp16. The clone was of interest since it was clearly responsive to more than just heat shock because that had been kept constant for all strains. It was subsequently found to be somewhat elevated, relative to heat shock response alone, in cdc17-117 (DNA ligase). This response was lower than seen for cdc22-M45 (38,39).
To examine this quantitatively we generated a number of reagents and further characterized the system (Figs 1 and 2). Expression of the hsp16 transcript was induced in response to heat shock treatment and this response was further elevated in a cdc22-M45 mutant background (Fig. 1A and B). Western blotting showed that the gene product, hsp16, is responsive to heat shock treatment (Fig. 1C, lanes 1 and 2) and is further upregulated in a cdc22-M45 mutant background both at 25 and 36°C (Fig. 1C, lanes 5 and 6). Hsp16 expression is clearly regulated by heat shock, in contrast to previously published results (6).
Figure 1.
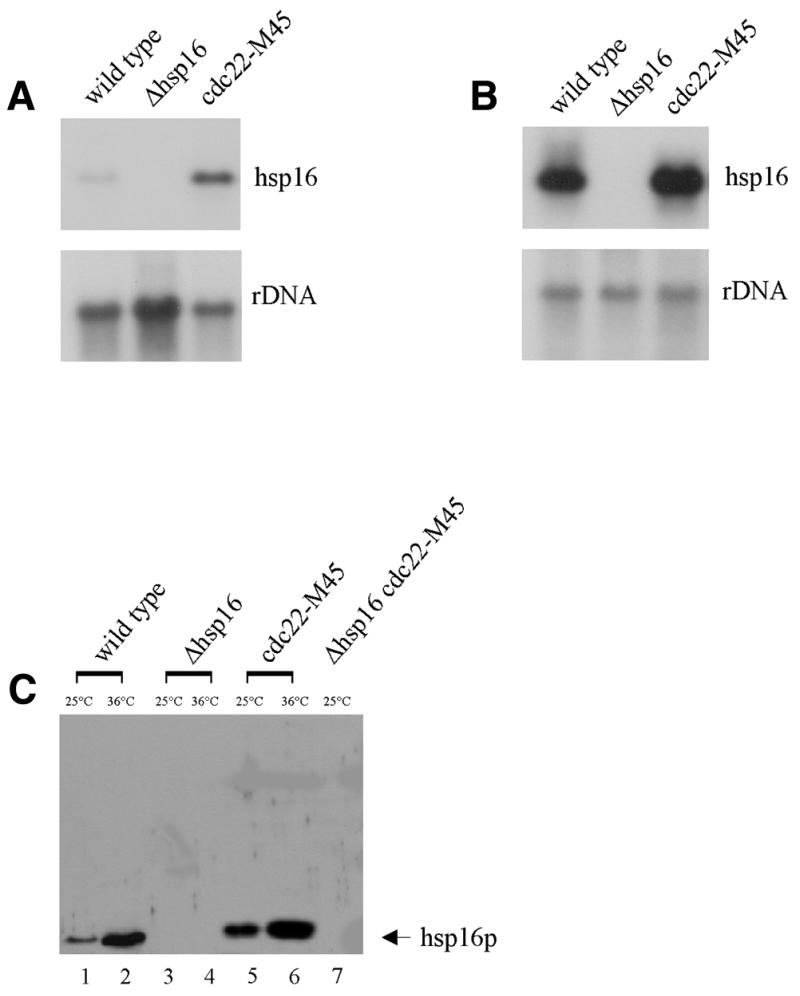
Expression of hsp16. (A and B) Northern blot analysis of hsp16 expression in a cdc22-M45 mutant background. (A) Total RNA was isolated from the strains indicated at 25°C, (B) total RNA was isolated following 4 h heat shock treatment at 36°C. RNA was analyzed by northern hybridization with a probe specific for hsp16 (A and B, upper panels). A ribosomal DNA probe was used as a loading control (A and B, lower panels). (C) hsp16 protein level correlates with RNA expression levels. Hsp16 protein levels are increased in a cdc22-M45 mutant background (lanes 1, 3, 5 and 7 at 25°C; lanes 2, 4 and 6 at 36°C). All strains were cultured at 25°C, then shifted to 36°C for 4 h in YEA medium.
Figure 2.
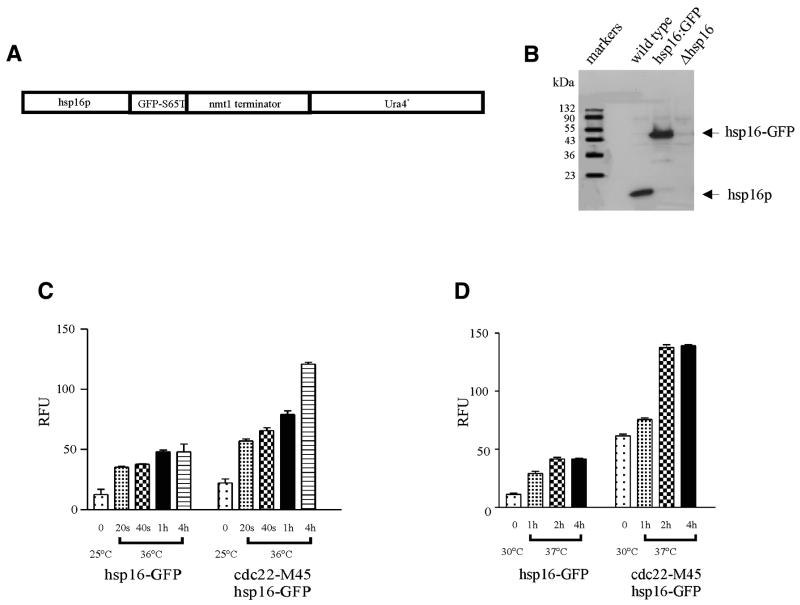
Hsp16–GFP expression. (A) Schematic of hsp16–GFP(S65T)-ura4+ chromosomal fusion placing expression under the control of the native promoter of hsp16. (B) Immunoblot showing that hsp16 and hsp16–GFP levels are similar following heat shock treatment at 36°C for 4 h. The strains were cultured at 25°C then shifted to 36°C for 4 h in YEA media. (C and D) Hsp16 is responsive to heat shock. (C) Expression of hsp16–GFP in wild-type and in a cdc22-M45 mutant background at 25°C and following heat shock treatment at 36°C for various times. (D) Expression of hsp16–GFP in wild-type and in a cdc22-M45 mutant background at 30°C and following heat shock treatment at 37°C for various times. Hsp16–GFP levels were quantitated by fluorimetry
A fusion construct, placing hsp16–GFP under the control of the native promoter of hsp16, was constructed by homologous recombination to generate a single copy chromosomal fusion (Fig. 2A). Expression of the hsp16–GFP behaved in a fashion similar to that of hsp16 alone in a wild-type background at 36°C (Fig. 2B). The presence of the GFP tag does not appear to affect expression or protein levels. The phenotype of wild-type cells expressing hsp16 tagged with GFP was indistinguishable from that of wild-type cells alone.
The hsp16–GFP fusion construct enabled us to easily and quantitatively study the normal regulation of hsp16 in a wide variety of contexts (Fig. 2C and D). The expression of hsp16–GFP was increased after heat shock treatment from 25 to 36°C at various times and by an additional 2–3-fold at 36°C in a cdc22-M45 background compared to wild-type. We also examined the heat shock response of hsp16–GFP in a shift from 30 to 37°C for various times. Expression under these conditions is similar to that in a treatment from 25 to 36°C (Fig. 2D).
hsp16 transcription is mediated in part by the spc1 MAPK pathway via atf1
Comparison to earlier work. Previous reports (6) have suggested that hsp16 does not respond to heat shock and that its expression is dependent on the activity of the ras1 pathway with expression being reduced 15-fold in a ras1 deletion background. However, in this investigation, northern blotting showed that heat shock alone is sufficient to elevate transcript levels ∼64-fold in a wild-type background (Table 2). Our data also clearly show that in both the ras1 and rasval17 backgrounds, hsp16 is transcribed at levels similar to that seen in wild-type.
Table 2. Quantitation of relative level of hsp16 transcript in various genetic backgrounds at 25 and 36°C.
| Strains |
Relative level at 25°C compared
to wild-type level |
Relative level at 36°C compared
to wild-type level at 25°C |
| wild-type | 1 | 64 ± 2.0 |
| Δhsp16 | na | na |
| Δras1 | 0.8 ± 0.4 | 60 ± 2.0 |
| ras1v17 | 1.4 ± 0.2 | 68 ± 2.0 |
| cdc22-M45 | 4.8 ± 0.2 | 141 ± 3.0 |
| Δspc1 | 1.0 ± 0.1 | 31 ± 1.0 |
| Δatf1 | 1.9 ± 0.3 | 52 ± 2.0 |
| Δpyp1 | 0.9 ± 0.0 | 44.5 ± 1.5 |
| rad1-1 | 2.2 ± 0.15 | 144 ± 4.0 |
| Δspc1 cdc22-M45 | 6.2 ± 0.5 | 78 ± 2.0 |
| Δatf1 cdc22-M45 | 2.8 ± 0.2 | 46 ± 2.0 |
| Δpyp1 cdc22-M45 | 6.5 ± 0.2 | 214 ± 4.0 |
| rad1-1 cdc22-M45 | 8.7 ± 0.2 | 158 ± 4.0 |
Relative level of hsp16 transcript was obtained by densiometric analysis of a more lightly exposed autoradiograph of Figure 1 and two other independent northern experiments. The values presented were first normalized to the ribosomal DNA for each RNA sample and then to the levels in the wild-type background at 25°C. Two independent values were averaged and the range is indicated. na, not available.
Effect of cdc22-M45. hsp16 expression was found to be elevated at both the restrictive and permissive temperatures in the cdc22-M45 mutant background (Fig. 1A and B; Table 2). The cdc22-M45 mutation inactivates deoxyribonucleotide production and is known to activate the DNA replication checkpoint. We therefore examined the dependence of induction on the checkpoint by analyzing hsp16 expression in the rad1-1 and rad1-1 cdc22-M45 mutant strains. Transcript levels in the rad1-1 cdc22-M45 mutant strain are similar to that seen in the cdc22-M45 mutant alone (Table 2). Curiously, at high temperature there is also increased expression in a rad1-1 mutant background alone. This suggests the possibility that heat shock alone elicits DNA damage. This damage rather than the cell cycle block per se appears to cause hsp16 activation since by removing the checkpoint the rad1-1 cdc22-M45 mutant strain proceeds through mitosis at 36°C.
Dependence on MAP kinase pathway. hsp27-mediated inhibition of actin polymerization is regulated in part by the p38 MAPK (46,47) corresponding to fission yeast spc1 (48–50). Therefore, we examined hsp16 expression in a Δspc1 mutant background to inactivate the pathway (48) and in a Δpyp1 mutant background, an inhibitor of spc1, to activate it (49). At 36°C, the transcript level for hsp16 was decreased by 2-fold in the Δspc1 mutant background compared to wild-type (Table 2). Interestingly, increased activity of spc1 MAPK as in the Δpyp1 mutant background has no additive effect on hsp16 expression in an otherwise wild-type strain (Table 2).
We next tested the effect of spc1 deletion on the cdc22-M45 response. At the reduced restrictive temperature of 30°C, cdc22-M45 Δspc1 arrested with an additive morphological phenotype yielding very elongated cells. However, hsp16 expression at 36°C in the cdc22-M45 Δspc1 mutant strain remained similar to that seen in wild-type (Table 2). This suggests that the spc1 pathway is necessary to respond to nucleotide depletion or to a DNA synthesis block as occurs in the cdc22-M45 mutant strain. This is reinforced by the observation that increased activity of spc1 (as occurs in Δpyp1 mutant strain) has an additive effect on hsp16 expression in the Δpyp1 cdc22-M45 mutant strain (Table 2).
Dependence on atf1. Conjugation, meiosis and osmotic stress response are affected by spc1 at least in part through atf1 whose expression and activity are stimulated by spc1 MAPK (51). hsp16 expression levels in a Δatf1 mutant background are similar to that in wild-type cells (Table 2). However, in a Δatf1 cdc22-M45 mutant background, hsp16 expression levels are greatly reduced compared to a cdc22-M45 mutant alone and comparable to or somewhat higher than in a Δspc1 mutant alone (Table 2). These results suggest that atf1 is an important part of the response pathway for this type of replicational stress and that it is sufficient to account for all spc1-dependent activation of hsp16 but not for its response to heat shock.
Search for synthetic interaction. We decided to generate double mutants with Δhsp16 and cdc22-M45, Δspc1, Δatf1 to see if a hsp16 deletion could interact genetically with these mutations. Δhsp16 cdc22-M45, Δhsp16 Δspc1, Δhsp16 atf1 double mutants were indistinguishable from the single mutants alone upon heat shock at 36°C (data not shown).
During the course of this study, we found another small HSP, hsp20 (accession no. AL02378I in the S.pombe Sanger database) that has significant homology within the C-terminal region of hsp16. Since this might provide a redundant function, we disrupted hsp20 and constructed a double mutant of Δhsp16 and Δhsp20. The Δhsp16 Δhsp20 double mutant did not exhibit a visible phenotype (data not shown).
Confirmation and extension of transcription results by monitoring hsp16 protein level
In most cases the level of hsp16–GFP fusion protein in the cells (Table 3) correlated well with the hsp16 transcriptional data (Table 2). One major difference is that the rad1-1 mutation suppressed the accumulation of hsp16–GFP which occurs in the cdc22-M45 mutant background. Some of the quantitative effects reflect the smaller average size of the rad1-1 cdc22-M45 mutant cells that proceed through cell division. However, this could not account for the 4-fold drop in rad1-1 cdc22-M45 mutant background relative to cdc22-M45 mutant background. This contrasts with the lack of a rad1 effect on transcript levels and it is clear that somehow there is an effect on translation or stability of the protein.
Table 3. Relative hsp16-GFP fusion protein levels in various genetic backgrounds as determined by GFP fluorescence.
| Strains |
25°C |
36°C (4 h) |
| hsp16–GFP | 12.5 ± 4.4 | 48 ± 6.6 |
| cdc22-M45 hsp16–GFP | 22.2 ± 3.3 | 122 ± 1.5 |
| rad1-1 | 19.4 ± 1.4 | 51 ± 1.5 |
| Δspc1 hsp16–GFP | 16.5 ± 5.2 | 42.0 ± 1.1 |
| Δatf1 hsp16–GFP | 5.3 ± 1.4 | 19.5 ± 1.3 |
| Δwis1 hsp16–GFP | 19.8 ± 1.9 | 71.2 ± 2.2 |
| Δpcr1 hsp16–GFP | 11.3 ± 1.9 | 34.5 ± 1.3 |
| Δras1 hsp16–GFP | 8.8 ± 0.4 | 53.8 ± 6.6 |
| rad1-1 cdc22-M45 hsp16–GFP | 15.0 ± 1.3 | 30 ± 2.5 |
| Δspc1 cdc22-M45 hsp16–GFP | 16.0 ± 5.5 | 43.5 ± 2.4 |
| Δatf1 cdc22-M45 hsp16–GFP | 9.5 ± 3.2 | 53.2 ± 3.0 |
| Δwis1 cdc22-M45 hsp16–GFP | 42.1 ± 2.3 | 124.2 ± 1.4 |
| Δpcr1 cdc22-M45 hsp16–GFP | 19.1 ± 3.8 | 121 ± 0.6 |
One unexpected finding is that a Δwis1 mutant background does not appear to affect expression (Table 3). It appears that the spc1 MAPK is essential for full response; however the wis1 MAPKK upstream of spc1 is not involved.
Since hsp16 expression in the Δatf1 mutant strain after heat shock was not completely abolished, we wanted to examine the possible role of the pcr1 transcription factor. A Δpcr1 mutant or Δpcr1 cdc22-M45 double mutant background had little effect on hsp16–GFP expression (Table 3). There was also no additive effect in the Δatf1 Δpcr1 double mutant background, the levels being comparable to Δatf1 alone (data not shown).
Hsp16 expression as a result of nucleotide depletion and DNA damage involves atf1
We have shown that hsp16–GFP is strongly induced in a cdc22-M45 mutant background at the restrictive temperature (Table 2). Since this blocks deoxyribonucleotide production and therefore DNA replication, we examined the effect of hydroxyurea and camptothecin in the absence of heat shock, two well characterized inhibitors of ribonucleotide reductase and of topoisomerase, respectively (52–55).
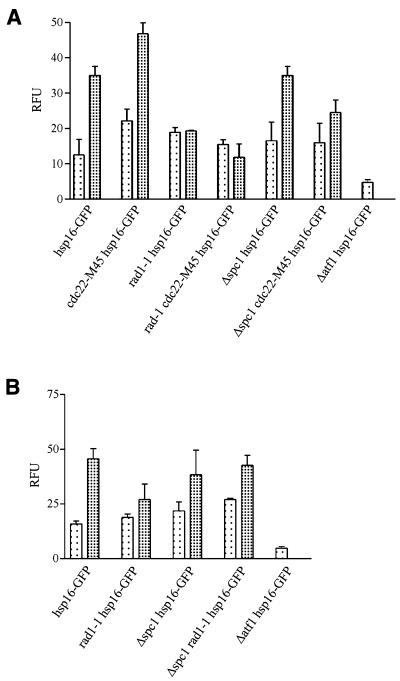
Hsp16–GFP expression was increased in the presence of hydroxyurea or camptothecin (Fig. 3). Both of these treatments have been shown to prevent DNA synthesis by their actions on a checkpoint system (56). The rad1-1 mutation suppressed the effects of hydroxyurea or camptothecin on hsp16–GFP expression (Fig. 3). This parallels the results seen in the cdc22-M45 mutant background. These results suggest that elevated accumulation of the protein is dependent on the cell cycle arrest, or direct signaling through the rad1 pathway. Neither occurs in the rad1-1 mutant background since the checkpoint is abolished and these cells continue to divide. The Δspc1 mutation had little effect on the level of hsp16–GFP following hydroxyurea or camptothecin treatment (Fig. 3). These experiments were all performed at 25°C and differences in cell size cannot account for these differences.
Figure 3.
Hsp16–GFP expression is activated following HU or CPT treatment. Analysis of hsp16–GFP levels in cdc22-M45 mutant. (A) Cells were cultured at 25°C (lightly shaded bars) in YEA to mid-exponential phase and then hydroxyurea (heavily shaded bars) was added at a final concentration of 11 mM to one-half of the culture and incubated for 4 h. (B) Cells were cultured at 25°C (lightly shaded bars) in YEA to mid-exponential and then camptothecin (heavily shaded bars) was added at a final concentration of 40 µM to one-half of the culture and incubated for 2 h. Hsp16–GFP levels were quantitated by fluorimetry.
Hsp16–GFP accumulation in response to heat shock is partly dependent upon the spc1 MAPK pathway (Tables 2 and 3). The question arises as to whether atf1 participates in the hydroxyurea or camptothecin responses. Hsp16–GFP expression was completely abolished in a Δatf1 mutant background in the presence of hydroxyurea or camptothecin (Fig. 3). In a Δatf1 mutant strain under these conditions the hsp16 gene appears to be negatively regulated.
Localization of hsp16 protein
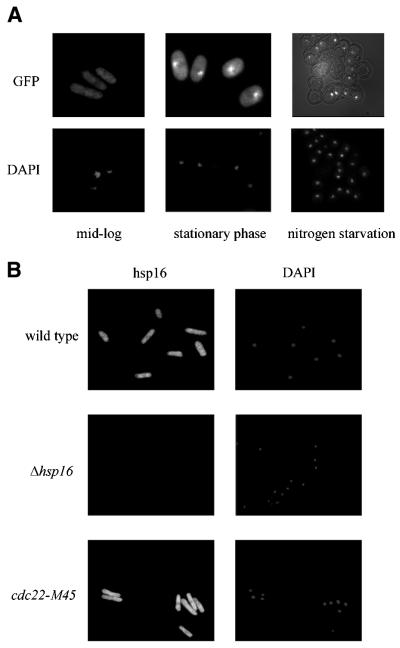
Hsp16–GFP protein was localized in the cytoplasm and the nucleus in rich media (Fig. 4A, left panel). In stationary phase, hsp16–GFP localization changed and was distributed to one or sometimes two sharply defined structures close to, but not within, the nucleus as judged by DAPI (Fig. 4A, middle panel). The same pattern of expression was seen in cells upon nitrogen starvation in spores (Fig. 4A, right panel) and in cells following heat shock treatment (data not shown). Spheroplasted cells lysed with 1% Triton X-100 showed that the hsp16–GFP protein was not membrane bound since it was not dispersed by detergent and therefore it is likely to be an inclusion body aggregate. Following hydroxyurea or camptothecin treatment the distribution was similar to that seen in stationary phase (data not shown).
Figure 4.
The cellular localization of hsp16 protein. (A) Hsp16–GFP localization in the absence of stress (left panel), upon glucose starvation (middle panel) and nitrogen starvation in spores (right panel). (B) Exponentially growing cells were immunostained with polyclonal anti-GST–hsp16 antibody and Alexa 488 anti-rabbit IgG (H + L) and stained with DAPI. Left panels show Alexa 488 images and the right panels show DAPI images (top panel, wild-type; middle panel, Δhsp16; bottom panel, cdc22-M45). Cells were cultured at 25°C in YEA to a density of 2–5× 106 cells/ml.
Immunolocalization using antibodies against hsp16 also showed hsp16 to localize to the cytoplasm and the nucleus in both wild-type and in the cdc22-M45 mutant background. This signal was absent in exponentially growing Δhsp16 mutant cells at 25°C (Fig. 4B). Cells were examined following the various treatments described earlier in the paper and in all cases the localization was the same.
Two-hybrid screen using hsp16
To identify potential hsp16 interacting proteins we performed a yeast two-hybrid screen using full-length hsp16 as bait. A total of 750 000 transformants were screened. The screen produced 41 reproducible interactions with eight different targets (Table 4). One of the clones repeatedly isolated in the screen was hsp16 indicating that the protein interacts with itself. We found that the hsp16–hsp16 interaction produces 224 U β-galactosidase activity. We did not pursue these targets further at this time.
Table 4. Results of cDNA protein interactions with hsp16 protein.
| Bait |
Target ORF name |
Target gene name |
Start-end domain amino acid |
Number of clones |
| LexA-hsp16 | SPAC19A8.10 | Hypothetical zfp | 5–254 | 12 |
| SPBC3E7.02C | Hsp16 | 5–143 | 11 | |
| SPCP31B10.06 | Eft1=Etf2 (elongation | 543–842 | 6 | |
| (SPAC513.01C) | factor 2) | |||
| SPBC365.06 | Pmt3 | 2–117 | 4 | |
| SPAC630.14C | Tup1 | 141–586 | 4 | |
| SPBC1734.06 | Putative DNA repair and | 16–387 | 2 | |
| recombination protein | ||||
| SPCC417.08 | Ef-3 (putative elongation | 836–1047 | 1 | |
| factor 3) | ||||
| SPBC8E4.07C | Hypothetical serine/threonine | 219–1283 | 1 | |
| repeat containing protein |
All the clones isolated included the C-termini and were deleted from the N-terminus to varying degrees, reflecting the nature of the cDNA library.
DISCUSSION
We found the hsp16 gene to be responsive to heat shock treatment with a 64-fold induction of the transcript at 36°C. We have also examined hsp16 protein levels using both a reporter fusion construct, hsp16–GFP, as well as a polyclonal antibody against hsp16. The protein levels are elevated at 36°C with ∼5-fold increase in the steady-state level of protein in a wild-type background. This contradicts a recent publication by Danjoh and Fujiyama (6) who reported that hsp16 is not heat shock responsive, based on northern blot analysis. Close inspection of these data (6; Fig. 4) shows a relatively constant, but highly expressed, level of hsp16 signal at the permissive temperature and upon heat shock conditions, hence their conclusion. However, the control lane (30°C) shows a substantial deficit of cdc2 transcript relative to the heat shocked samples. Since cdc2 is not heat shock responsive (57), if one normalized the data to cdc2 levels, this blot may suggest a very strong expression of hsp16 at 30°C and reduced levels at 37°C. There is no indication of the reproducibility of this result. The high level of expression before heat shock treatment shown in this publication is in direct contradiction to all of our findings. We have no simple explanation for this but are confident, based on our protein data as well, that hsp16 has a strong heat shock response. It is possible, depending on how cells were harvested and held prior to RNA isolation, that a MAPK-dependent stress response was induced and this accounts for the expression in all strains used in their northern blots.
In support of our conclusion two potential heat shock consensus sites are located at positions –157 to –170 (AGAAagaAAaaTTCt) and –529 to –543 (CGAAttTTCtcGtAa) from the hsp16 ORF (fission yeast cosmid accession no. AL023534) (58). These inverted nGAAn motifs presumably account for the heat shock-inducible transcription of hsp16 (59,60).
Danjoh and Fujiyama (6) identified their hsp16 clone using a differential display between wild-type and ras1– mutant cells. In our experiment we found no effect of ras1 on hsp16 transcription or protein accumulation, nor was there any effect of activating the ras pathway using the rasval17 allele. It is conceivable that their ras1– cell population was exhibiting a stress response.
In agreement with previously published results (6), we find that hsp16 is non-essential because we have independently disrupted this gene and found no phenotype affecting cell growth, viability or mating. It also has no discernible phenotype in the double mutant with a hsp20 deletion.
Role of the spc1 MAPK pathway
The spc1 MAPK pathway is known to play a role in adaptation to adverse external stimuli including heat stress through atf1 (61–64). We provide the first example of a heat shock gene being regulated in part by the spc1 MAPK pathway via atf1. The heat shock factor pathway presumably accounts for the remainder of hsp16 induction and the two pathways appear to be additive. It is interesting that both the spc1 and atf1 promoters have heat shock element consensus sites. In other systems it has been shown that the spc1 MAPK homolog, p38 in mammalian cells (65) and HOG1 in budding yeast (66) are involved in the regulation of a small HSP but for different environmental stresses such as oxidative stress and osmolarity.
The wis1 MAPKK is not involved in the regulation of hsp16 in response to heat shock. Earlier studies have shown that activation of wis1 is weak and transient after heat shock (67). It seems likely that another MAPKK dedicated to heat shock stimuli might interact with spc1 although no candidate gene suggests itself. This model would be similar to that found in vertebrates where p38 is phosphorylated by two different MAPKKs, SEK1 and MKK3/6 (68,69).
Relationship to DNA replication
Our data demonstrate a novel role for a small heat shock gene in response to nucleotide depletion or potential DNA damage. Ribonucleotide reductase is an essential enzyme for DNA precursor metabolism. Failure to regulate dNTP levels can lead to genetic abnormalities or cell death (70). Our findings strongly suggest that the spc1 pathway responds to this stimulus via atf1 and stimulates hsp16 induction under these conditions.
We were able to generalize the cdc22 response to other types of DNA synthesis block such as hydroxyurea or camptothecin treatment. The induction of hsp16 in response to these agents was not blocked by the Δspc1 mutation. However, a Δatf1 mutation appears to completely abolish hsp16 expression in the presence of these two drugs. This contrasts with the response to a cdc22 block.
Treatments that interfere with DNA synthesis arrest the cell cycle by activation of the checkpoint pathway dependent upon rad1. It is interesting that releasing the block by inactivating the checkpoint (rad1-1) does not affect the transcript level for hsp16. It does however cause a reduction in the accumulation of the hsp16 protein, presumably by affecting translation or stability. This contrasts with the response of the small subunit of ribonucleotide reductase encoded by suc22+, which requires the rad1+ gene for induction in response to DNA damage but not in response to heat shock (71). We found that the hsp16 transcript was also induced by heat shock in a rad1-1 mutant background. This is similar to levels as in a cdc22-M45 mutant background, which suggests the possibility that heat shock itself is damaging to DNA. This damage rather than cell cycle progress leads to hsp16 activation since a rad1-1 cdc22-M45 mutant proceeds through mitosis at 36°C, yet the transcript is still induced.
Two-hybrid screen
The two-hybrid screen isolated a number of targets that potentially interact with hsp16 and one of the targets was hsp16 itself. Another target that was isolated as frequently as hsp16 was a novel hypothetical zinc finger protein (accession no. SPAC19A8.10) that has no apparent homolog in S.cerevisiae. Zinc finger proteins are components of transcription factors involved in mediating protein–protein interactions.
In S.cerevisiae one of the genes that was found to negatively regulate the DNA damage response of ribonucleotide reductase was the tup1 transcription factor (72). We isolated tup12, the tup1 homolog of S.cerevisiae as a potential target of hsp16. The recent identification of tup11 and tup12 proteins in S.pombe have been linked to signaling through the spc1 MAPK pathway (73).
We also isolated the SUMO-1 (ubiquitin-like protein modifier) homolog in fission yeast, pmt3. Recently, pmt3 was isolated as a target of an accessory factor of DNA polymerase δ, PCNA (74). PCNA is required for DNA replication, repair or damage. An additional DNA related target was the fission yeast homolog of RAD18 of S.cerevisiae required for postreplicative repair (75,76).
Lastly the isolation of translation elongation factor-2 and translation elongation factor-3 suggests the possibility that hsp16 also stabilizes components of the translation machinery. Thus, the two-hybrid targets of hsp16 taken at face value suggest that the protein may be involved in a number of processes in the cell, but many of them can be related to replication and others to translational regulation. Both processes are logical targets of a heat and replicational stress inducible HSPs.
Model of pathways
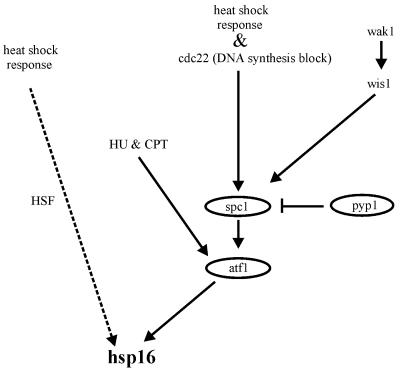
We propose a multiple pathway model (Fig. 5) for hsp16 regulation in response to heat shock treatment and nucleotide depletion or DNA damage. Our data suggests that hsp16 is downstream of atf1 and this is independent of the heat shock factor (HSF) pathway. In response to heat shock treatment both the spc1 MAPK pathway and the HSF pathway regulate hsp16 expression. However, when cells are treated with hydroxyurea or camptothecin, hsp16 expression is regulated by atf1 but not through the spc1 MAPK pathway suggesting the possibility of another pathway.
Figure 5.
A model of the pathway involved in the regulation of hsp16 expression in S.pombe. This model is based on results in this study.
To date no single small HSP has been shown to be indispensable under heat shock or other environmental stresses. This may not be surprising since most organisms contain two or more small HSPs and they may have overlapping functions. If hsp16 is important in protecting the cell against heat damage or nucleotide depletion, there must also be other stress proteins or HSPs that can compensate for its loss. In any event, the function of hsp16 in S.pombe is yet to be determined, like its homolog hsp26 in S.cerevisiae, which has been studied since 1986, with a specific function still to be determined.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by grants from the Natural Sciences and Engineering Research Council of Canada to P.G.Y.
References
- 1.DeJong W.W., Leunissan,J.A.M. and Voorter,C.E.M. (1993) Evolution of the α-crystallin/small heat shock protein family. Mol. Biol. Evol., 10, 103–126. [DOI] [PubMed] [Google Scholar]
- 2.Carver, J.A. (1999) Probing the structure and interactions of crystallin proteins by NMR spectroscopy. Prog. Retin. Eye Res., 18, 431–462. [DOI] [PubMed] [Google Scholar]
- 3.Boelens W.C., Croes,Y., deRuwe,M., deReu,L. and deJong,W.W. (1998) Negative charges in the C-terminal domain stabilize the αβ-crystallin complex. J. Biol. Chem., 273, 28085–28090. [DOI] [PubMed] [Google Scholar]
- 4.Kim K.K., Kim,R. and Kim,S.H. (1998) Crystal structure of a small heat shock protein. Nature, 394, 595–599. [DOI] [PubMed] [Google Scholar]
- 5.Arrigo A.-P. and Landry,J. (1994) Expression and function of the low-molecular weight heat shock proteins. In Morimoto,R., Tissieres,H. and Georgeopoulos,C. (eds), The Biology of Heat Shock Proteins and Molecular Chaperones. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. pp. 335–373.
- 6.Danjoh I and Fujiyama,A. (1999) Ras-mediated signalling pathway regulates the expression of a low molecular weight heat shock protein in fission yeast. Gene, 236, 347–352. [DOI] [PubMed] [Google Scholar]
- 7.Orlandi I., Cavadini,P., Popolo,L. and Vai,M. (1996) Cloning, sequencing and regulation of cDNA encoding a small heat shock protein from Schizosaccharomyces pombe. Biochim. Biophys. Acta, 1307, 129–131. [DOI] [PubMed] [Google Scholar]
- 8.Petko L. and Lindquist,S. (1986) Hsp26 is not required for growth at high temperatures, nor for thermotolerance, spore development, or germination. Cell, 45, 885–894. [DOI] [PubMed] [Google Scholar]
- 9.Praekelt U.M. and Meacock,P.A. (1990) HSP12, a new small heat shock gene of Saccharomyces cerevisiae: analysis of structure, regulation and function. Mol. Gen. Genet., 223, 97–106. [DOI] [PubMed] [Google Scholar]
- 10.Wotton D., Freeman,K. and Shore,D. (1996) Multimerization of Hsp42p, a novel heat shock protein of S. cerevisiae is dependent on a conserved-terminal sequence. J. Biol. Chem., 271, 2717–2723. [DOI] [PubMed] [Google Scholar]
- 11.Bateman A., Birney,A., Durbin,R., Eddy,S.R., Howe,K.L. and Sonnhammer,E.L.L. (2000) The Pfam protein families database. Nucleic Acids Res., 28, 263–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beissenger M. and Buchner,J. (1998) How chaperones fold proteins. Biol. Chem., 379, 245–259. [PubMed] [Google Scholar]
- 13.Chen C.F., Chen,Y., Dai,K., Chen,P.L., Riley,D.L. and Lee,W.H. (1996) A new member of the hsp90 family of molecular chaperone interacts with the retinoblastoma protein during mitosis and after heat shock. Mol. Cell Biol., 16, 4691–4699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alique R., Akhavan,N.H. and Russell,P. (1994) A role for Hsp90 in cell cycle control: Wee1 tyrosine kinase activity requires interaction with Hsp90. EMBO J., 13, 6099–6106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morimoto R.I., Tissières,A. and Georgopoulos,C. (1994) The Biology of Heat Shock Proteins and Molecular Chaperones. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 16.Ehrnsperger M., Graber,S., Gaestel,M. and Buchner,J. (1997) Binding of non-native protein to Hsp25 during heat shock creates a reservoir of folding intermediates for reactivation. EMBO J., 16, 221–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chang Z., Primm,T.P., Jakana,J., Lee,I.H., Seysheva,I., Chiu,W., Gilbert,H.F. and Quiocho,F.A. (1996) Mycobacterium tuberculosis 16kDa antigen (Hsp16.3) functions as a oligomeric structure in vitro to suppress thermal aggregation. J. Biol. Chem., 271, 7218–7223. [PubMed] [Google Scholar]
- 18.Hartl F.-U., Hlodan,R. and Langer,T. (1994) Molecular chaperones in protein folding: the art of avoiding sticky situations. Trends Biochem. Sci.,19, 20–25. [DOI] [PubMed] [Google Scholar]
- 19.Jakob U., Gaeste,M., Engel,K. and Buchner,J. (1993) Small heat shock proteins are molecular chaperones. J. Biol. Chem., 268, 1517–1520. [PubMed] [Google Scholar]
- 20.Horwitz J. (1992) α-crystallin can function as a molecular chaperone. Proc. Natl Acad. Sci. USA, 89, 10449–10453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haslbeck M., Walke,S., Stromer,T., Ehrnsperger,M., White,H.E., Chen,S., Saibil,H.R. and Buchner,J. (1999) Hsp26: a temperature-regulated chaperone. EMBO J., 18, 6744–6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leroux M., Melki,R., Gordon,B., Batelier,G. and Candido,E.P. (1997) Structure-function studies on small heat shock protein oligomeric assembly and interaction with unfolded polypeptides.J. Biol. Chem., 272, 24646–24656. [DOI] [PubMed] [Google Scholar]
- 23.Bentley N.J., Fitch,I.T. and Tuite,M.F. (1992) The small heat shock protein Hsp26 of Saccharomyces cerevisiae assembles into a high molecular weight aggregate. Yeast, 8, 95–106. [DOI] [PubMed] [Google Scholar]
- 24.Lavoie J.N., Gingras-Breton,G., Tanguay,R.M. and Landry,J. (1993) Induction of Chinese hamster HSP27 gene expression in mouse cells confers resistance to heat shock. Hsp27 stabilization of the microfilament organization. J. Biol. Chem., 268, 3420–3429. [PubMed] [Google Scholar]
- 25.Rollet E., Lavoie,J.N., Landry,J. and Tanguay,R.M. (1992) Expression of Drosophila’s 27 kDa heat shock protein into rodent cells confers thermal resistance. Biochem. Biophys. Res. Commun., 185, 116–120. [DOI] [PubMed] [Google Scholar]
- 26.Landry J., Chretien,P., Lambert,H., Hickey,E. and Weber,L.A. (1989) Heat shock resistance conferred by expression of the human HSP27 gene in rodent cells. J. Cell Biol., 109, 7–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Landry J. and Huot,J. (1995) Modulation of actin dynamics during stress and physiological stimulation by a signaling pathway involving p38 MAP kinase and heat shock protein 27. Biochem. Cell Biol., 73, 703–707. [DOI] [PubMed] [Google Scholar]
- 28.Nicholl I.D. and Quinlan,R.A. (1994) Chaperone activity of α-crystallins modulates intermediate filament assembly. EMBO J., 13, 945–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bennardini F., Wrzosek,A. and Chiesi,M. (1992) αβ-crystallin in cardiac tissue. Association with actin and desmin filaments. Circ. Res., 71, 288–294. [DOI] [PubMed] [Google Scholar]
- 30.Arrigo A.-P. (1998) Small stress proteins: chaperones that act as regulators of intracellular redox state and programmed cell death. Biol. Chem., 379, 19–26. [PubMed] [Google Scholar]
- 31.Mehlen P., Mehlen,A., Godet,J. and Arrigo,A.-P. (1997) hsp27 as a switch between differentiation and apoptosis in murine embryonic stem cells. J. Biol. Chem., 272, 31657–31665. [DOI] [PubMed] [Google Scholar]
- 32.Kurtz S., Rossi,J., Petko,L. and Lindquist,S. (1986) An ancient developmental induction: heat shock proteins induced sporulation and oogenesis. Science, 231, 1154–1157. [DOI] [PubMed] [Google Scholar]
- 33.Varela J.C., van Beekvelt,C., Planta,R.J. and Mager,W.H. (1992) Osmostress-induced changes in yeast gene expression. Mol. Microbiol., 6, 2183–2190. [DOI] [PubMed] [Google Scholar]
- 34.Susek R.E. and Lindquist,S.L. (1989) Hsp26 of Saccharomyces cerevisiae is related to the superfamily of small heat shock proteins but is without a demonstrable function. Mol. Cell. Biol., 9, 5265–5271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leupold U. (1970) Genetical methods for Schizosaccharomyces pombe. Methods Cell. Physiol., 4, 169–177. [Google Scholar]
- 36.Alfa C., Fantes,P., Hyams,J., McLeod,M. and Warbrick,E. (1993) Experiments With Fission Yeast. A Laboratory Course Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 37.Moreno S., Klar,A. and Nurse,P. (1991) Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol., 194, 795–823. [DOI] [PubMed] [Google Scholar]
- 38.Weaver C. (1986) Differences in messenger RNA populations in cell cycle mutant-arrested Schizosaccharomyces pombe. Master’s thesis. Queens University, Kingston, ON.
- 39.Feilotter H.E. (1986) An environmentally and cell cycle regulated gene of Schizosaccharomyces pombe. Master’s thesis. Queens University, Kingston, ON.
- 40.Albano C.R., Randers-Eichhorn,L., Bentley,W.E. and Govind,R. (1998) Green Fluorescent Protein as a real time quantitative reporter of heterologous protein production. Biotechnol. Prog., 14, 351–354. [DOI] [PubMed] [Google Scholar]
- 41.Li X., Zhao,X., Fang,Y., Jiang,X., Duong,T., Fan,C., Huang,C.C. and Kain,S.R. (1998) Generation of destabilized green fluorescent protein as a transcription reporter. J. Biol. Chem., 273, 34970–34975. [DOI] [PubMed] [Google Scholar]
- 42.Sawin K.E. and Nurse,P. (1998) Regulation of cell polarity of microtubules in fission yeast. J. Cell Sci., 97, 509–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gyuris J., Golemis,E., Chertkov,H. and Brent,R. (1993) Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell, 75, 791–803. [DOI] [PubMed] [Google Scholar]
- 44.Durfee T., Becherer,K., Chen,P.-L., Yeh,S.-H., Yang,Y., Kilburn,A.E., Lee,W.-H. and Elledge,S.J. (1993) The retinoblastoma protein associates with the protein phosphatase type I catalytic subunit. Genes Dev., 7, 555–569. [DOI] [PubMed] [Google Scholar]
- 45.Evangelista M., Blundell,K., Longtine,M.S., Chow,C.J., Adams,N., Pringle,J.R., Peter,M. and Boone,C. (1997) Bni1p, a yeast formin linking Cdc42p and the actin cytoskeleton during polarized morphogenesis. Science, 276, 118–122. [DOI] [PubMed] [Google Scholar]
- 46.Hedges J.C., Dechert,M.A., Yamboliev,I.A., Martin,J.L., Hickey,E., Weber,L.A. and Gerthoffer,W.T. (1999) A role for p38(MAPK)/HSP27 pathway in smooth muscle cell migration.migration. J. Biol Chem., 274, 24211–24219. [DOI] [PubMed] [Google Scholar]
- 47.Guay, J., Lambert,H., Gingras-Breton,G., Lavoie,J.N., Huot,J. and Landry,J. (1997) Regulation of actin filament dynamics by p38 map kinase-mediated phosphorylation of heat shock protein 27. J. Cell Sci., 110, 357–368. [DOI] [PubMed] [Google Scholar]
- 48.Miller J.B.A., Buck,V. and Wilkinson,M.G. (1995) Pyp1 and Pyp2 PTPases dephosphorylate an osmosensing MAPK controlling cell size at division in fission yeast. Genes Dev., 9, 2117–2130. [DOI] [PubMed] [Google Scholar]
- 49.Shiozaki K. and Russell,P. (1995) Cell-cycle control linked to the extracellular environment by MAP kinase pathway in fission yeast. Nature, 378, 739–743. [DOI] [PubMed] [Google Scholar]
- 50.Kato T.J., Okazaki,K., Murakami,H., Stettler,S., Fantes,P.A. and Okayama,H. (1996) Stress signal, mediated by a HOG1-like MAP kinase, controls sexual development in fission yeast. FEBS Lett., 378, 207–212. [DOI] [PubMed] [Google Scholar]
- 51.Shiozaki K. and Russell,P. (1996) Conjugation, meiosis and the osmotic stress response are regulated in spc1 kinase through atf1 transcription factor in fission yeast. Genes Dev., 10, 2276–2288. [DOI] [PubMed] [Google Scholar]
- 52.Mitchison J.M. and Creanor,J. (1971) Further measurements of DNA synthesis and enzyme potential during cell cycle of fission yeast Schizosaccharomyces pombe. Exp. Cell Res., 69, 244–247. [DOI] [PubMed] [Google Scholar]
- 53.Wan S., Capasso,H. and Walworth,N.C. (1999) The Topoisomerase I poison camptothecin generates a chk1-dependent DNA damage checkpoint signal in fission yeast. Yeast, 15, 821–828. [DOI] [PubMed] [Google Scholar]
- 54.D’Arpa P., Beardmore,C. and Liu,L.F. (1990) Involvement of nucleic acid synthesis in cell killing mechanisms of topoisomerase poisons. Cancer Res., 50, 6919–6924. [PubMed] [Google Scholar]
- 55.Hsiang Y.-H., Lihou,M.G. and Liu,L.F. (1989) Arrest of replication forks by drug-stabilized topoisomerase I-DNA cleavable complexes as a mechanism of cell killing by camptothecin. Cancer Res., 49, 5077–5082. [PubMed] [Google Scholar]
- 56.Carr A.M. (1995) DNA structure checkpoints in fission yeast. Semin. Cell Biol., 6, 65–72. [DOI] [PubMed] [Google Scholar]
- 57.Shibuya T., Tsuneyoshi,S., Azad,A.K., Urushiyama,S., Ohshima,Y. and Tani,T. (1999) Characterization of ptr6+ gene in fission yeast: a possible involvement of a transcriptional coactivator TAF in nucleocytoplasmic transport of mRNA. Genetics, 152, 869–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Heinemeyer T., Wingender,E., Reuter,I., Hermjakob,H., Kel,A.E., Kel,O.V., Ignatieva,E.V., Ananko,E.A., Podkolodnaya,O.A., Kolpakov,F.A. et al. (1998) Databases on transcriptional regulation: TRANSFAC, TRRD and COMPEL. Nucleic Acids Res., 26, 364–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Perisic O., Xiao,H. and Lis,J.T. (1989) Stable binding of Drosophila heat shock factor to head-to-head and tail-to-tail repeats of a conserved 5 bp recognition unit. Cell, 59, 797–806. [DOI] [PubMed] [Google Scholar]
- 60.Amin J., Ananthan,J. and Voellmy,R. (1988) Key features of heat shock regulatory elements. Mol. Cell. Biol., 8, 3761–3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shieh J.-C., Martin,H. and Millar,J.B.A. (1998) Evidence for a novel MAPKKK-independent pathway controlling the stress activated sty1/spc1 MAP kinase in fission yeast. J. Cell Sci., 111, 2799–2807. [DOI] [PubMed] [Google Scholar]
- 62.Degols G., Shiozaki,K. and Russell,P. (1996) Activation and regulation of the Spc1 stress-activated protein kinase in Schizosaccharomyces pombe. Mol. Cell Biol., 16, 2870–2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wilkinson M.G., Samuels,M., Takeda,T., Toone,W.M., Shieh,J.-C., Toda,T., Millar,J.B.A. and Jones,N. (1996) The atf1 transcription factor is a target for the sty1 stress-activated MAP kinase pathway in fission yeast. Genes Dev., 10, 2289–2301. [DOI] [PubMed] [Google Scholar]
- 64.Shiozaki K., Shiozaki,M. and Russell,P. (1998) Heat stress activates fission yeast spc1/sty1 MAPK by a MEKK-independent mechansim. Mol. Biol. Cell., 9, 1339–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Huot J., Houle,F., Marceau,F. and Landry,J. (1997) Oxidative stress-induced actin reorganization mediated by the p38 mitogen-activated protein kinase/heat shock protein 27 pathway in vascular endothelial cells. Circ. Res., 80, 383–392. [DOI] [PubMed] [Google Scholar]
- 66.Varela J.C., Praekelt,U.M., Meacock,P.A., Planta,R.J. and Mager,W.H. (1995) The Saccharomyces cerevisiae HSP12 gene is activated by the high-osmolarity glycerol pathway and negatively regulated by protein kinase A. Mol. Cell Biol., 15, 6232–6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nguyen A.N. and Shiozaki,K. (1999) Heat shock-induced activation of stress MAP kinase is regulated by threonine- and tyrosine-specific phosphatases. Genes Dev., 13, 1653–1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ganiatsas S., Kwee,L., Fujiwara,Y., Perkins,A., Ikeda,T., Labow,M.A. and Zon,L.I. (1998) SEK1 deficiency reveals mitogen-activated protein kinase cascade crossregulation and leads to abnormal hepatogenesis. Proc. Natl Acad. Sci. USA, 95, 6881–6886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Raingeaud J., Whitmarsh,A.J., Barrett,T., Derijard,B. and Davis,R.J. (1996) MKK3- and MKK6-regulated gene expression is mediated by the p38 mitogen-activated protein kinase signal transduction pathway. Mol. Cell Biol.., 16, 1247–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Reichard P. (1988) Interaction between deoxyribonucleotides and DNA synthesis. Annu. Rev. Biochem., 57, 349–374. [DOI] [PubMed] [Google Scholar]
- 71.Harris P., Kersey,P.J., McInerny,C.J. and Fantes,P.A. (1996) Cell cycle, DNA damage and heat shock regulate suc22+ expression in fission yeast. Mol. Gen. Genet., 252, 284–291. [DOI] [PubMed] [Google Scholar]
- 72.Elledge S.J., Zhou,Z., Allen,J.B. and Navas,T.A. (1993) DNA damage and cell cycle regulation of ribonucleotide reductase. Bioessays, 15, 333–339. [DOI] [PubMed] [Google Scholar]
- 73.Janoo R.T.K., Neeley,L.A., Braun,B.R., Whitehall,S.K. and Hoffman,C.S. (2001) Transcriptional regulators of the Schizosaccharomyces pombe fbp1 gene include two redundant Tup1-like corepressors and the CCAAT binding factor activation complex. Genetics, 157, 1205–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tanaka K., Nishide,J., Okazaki,K., Kato,H., Niwa,O., Nakagawa,T., Matsuda,H., Kawamukai,M. and Murakami,Y. (1999) Characterization of a fission yeast SUMO-1 homologue, Pmt3p, required for multiple nuclear events, including the control of telomere length and chromosome segregation. Mol. Cell. Biol., 19, 8660–8672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jones J.S., Weber,S. and Prakash,L. (1988) The Saccharomyces cerevisiae RAD18 gene encodes a protein that contains potential zinc finger domains for nucleic acid binding and a putative nucleotide binding sequence. Nucleic Acids Res., 16, 7119–7131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Xiao W., Chow,B.L., Broomfield,S. and Hanna,M. (2000) The Saccharomyces cerevisiae RAD6 group is composed of error-protein and two error-free postreplication repair pathways. Genetics, 155, 1633–1641. [DOI] [PMC free article] [PubMed] [Google Scholar]