Abstract
Increasing evidences suggest that circular RNAs (circRNAs) exert crucial functions in regulating gene expression. In this study, we perform RNA‐seq and identify 6,154 distinct circRNAs from human bladder cancer and normal bladder tissues. We find that hundreds of circRNAs are significantly dysregulated in human bladder cancer tissues. We further show that circHIPK3, also named bladder cancer‐related circular RNA‐2 (BCRC‐2), is significantly down‐regulated in bladder cancer tissues and cell lines, and negatively correlates with bladder cancer grade, invasion as well as lymph node metastasis, respectively. Over‐expression of circHIPK3 effectively inhibits migration, invasion, and angiogenesis of bladder cancer cells in vitro and suppresses bladder cancer growth and metastasis in vivo. Mechanistic studies reveal that circHIPK3 contains two critical binding sites for the microRNA miR‐558 and can abundantly sponge miR‐558 to suppress the expression of heparanase (HPSE). Taken together, our findings provide evidence that circRNAs act as “microRNA sponges”, and suggest a new therapeutic target for the treatment of bladder cancer.
Keywords: bladder cancer, circHIPK3, circular RNA, heparanase, miR‐558
Subject Categories: Cancer, RNA Biology
Introduction
Bladder cancer is the most commonly occurring tumor of the urinary system and the ninth most frequently diagnosed cancer in the world 1. In the USA, bladder cancer is the eighth leading cause of cancer death in 2015 2 and over 40,000 patients dying from disease each year in EU 3. Metastasis is the main reason of death elicited by bladder cancer, with a 5‐year survival rate of 8.1% 4. Even worse, it is estimated that one‐third of patients with muscle‐invasive bladder cancer already have undetected metastases at the time of treatment 5. Although considerable progress has been made in surgical techniques and adjuvant chemotherapies, the mortality of metastatic bladder cancer has not sharply decreased 2. Thus, a better knowledge of changes in gene expression during invasion and metastasis may lead to improvements for the treatment of bladder cancer.
The existence of covalently closed circular RNAs (circRNAs) in eukaryotic cells was observed by electron microscopy almost 40 years ago 6. Subsequently, circular RNAs were sporadically reported 7, 8 and misinterpreted as by‐products of splicing errors 9. With the advent of high‐throughput sequencing and bioinformatic analysis, thousands of circRNAs have been successfully identified in multiple cell lines and across various species 10, 11, 12, 13, 14. Unlike unidimensional RNAs, circRNAs usually originated from back splicing events of exons or introns. “Exon skipping” and “direct back splicing” 15, 16, 17 are the two mechanisms to format exonic or exon–intron circRNAs, in which 3′ and 5′ terminis of circRNAs are covalently joined together forming single‐stranded continuous loop structures, with no polyadenylated tail, and can be regulated by some splice factors 18, 19. Salient features of circRNAs include remarkable stability, high abundance, evolutionary conservation, and tissue‐ and development‐specific expression 10, 11, 12, 20. Besides, certain circRNAs are considerably more abundant than cognate mRNAs 11, 12, 13. All these findings strongly indicate that the formation of circRNAs is a regulated process rather than just the products of splicing errors, thus may shed new light on the potential functions of circRNAs in regulating gene expression 19, 21, 22.
miRNAs are 19–25 nucleotides‐long non‐coding RNAs that directly regulate the expression of most mRNAs in various ranges of biological functions. Increasing evidence suggests that miRNAs are aberrantly expressed in bladder cancer, and exert multiple influences on promoting or inhibiting carcinogenesis, development, apoptosis, invasion, and metastasis 23, 24. Recently, circRNAs were reported function as “miRNA sponges” and can make a negative regulation on miRNAs 10, 14, 25. The most well‐known circRNA is CDR1as or ciRS‐7 10, 14, which harbors up to 74 canonical binding sites for miR‐7, abundantly sequesters miR‐7 away from CDR1, and makes it become an effective “miR‐7 sponges”. However, the function of circRNAs act as “miRNA sponges” has not been clearly elucidated in bladder cancer yet.
In this study, we identified thousands of distinct circRNAs from human bladder cancer tissues and normal bladder tissues by using RNA‐seq. We further demonstrated that circHIPK3, which originated from exon 2 of HIPK3 gene 12, 26 and we also named it bladder cancer‐related circular RNA‐2 (BCRC‐2, GenBank: KU921433.1), was significantly down‐regulated in bladder cancer tissues and cell lines. Furthermore, we found that enforced expression of circHIPK3 obviously inhibited invasion, metastasis, and angiogenesis of bladder cancer cells, via sponging miR‐558 to suppress the expression of heparanase (HPSE).
Results
CircRNAs expression profiles in bladder cancer tissues and paired normal bladder tissues
Firstly, we characterized circRNA transcripts by performing RNA‐seq on ribosomal RNA‐depleted total RNA from three pairs of human bladder cancer tissues and paired normal bladder tissues. A computational pipeline based on the anchor alignment of unmapped reads was used to identify circRNAs 10. We mapped the RNA‐seq data to the human reference genome (GRCH38/hg38) by TopHat2. Counts of reads mapping (including TopHat mapping and TopHat‐fusion mapping reads) across an identified backsplice were normalized by SRPBM (spliced reads per billion mapping), which could be used to quantitatively compare the backsplices between different tissues (Fig EV2A). Detailed information about the summary for each sample and the computational analysis pipeline is provided in Dataset EV1 and Fig EV1. Collectively, 16,353 distinct circRNAs were found in these samples, in which 6,154 circRNAs contained at least two backspliced reads in at least two different samples (Dataset EV1). Furthermore, we found that there were 4,531 identical circRNAs and 1,623 novel circRNAs in our study (Dataset EV1) compared with the generally acknowledged circRNA database circBase 27.
Figure EV1. RNA‐seq pipeline.
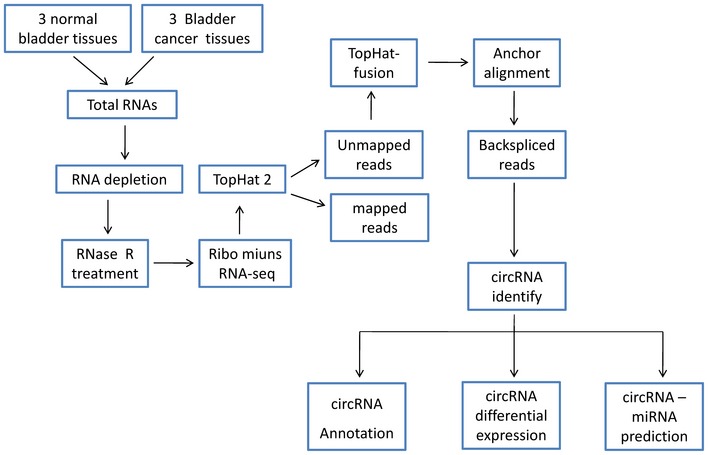
The RNA‐seq pipeline for identifying circRNAs in three pairs of bladder cancer tissues and paired adjacent normal bladder tissues is shown.
We annotated all these circRNAs using the ANNOVAR database 28; 88.35% of the circRNAs originated from exons (Fig 1A), and the others originated from introns, intergenic region, 3′UTR, and 5′UTR, etc. A total of 524 circRNAs were significantly down‐regulated (Dataset EV2), and 47 circRNAs were up‐regulated (Dataset EV2) in bladder cancer tissues (filtered by |FC(fold change)| ≥ 2 and P < 0.05) (Fig EV2B). The total differentially expressed circRNAs were displayed directly by hierarchical clustering analysis (Fig 1B). These differentially expressed circRNAs may have potential function in bladder cancer progression and need to be further explored.
Figure 1. Differential expression profiles of circRNAs in human bladder cancer and normal bladder tissues; circHIPK3 validation and expression in bladder cancer tissues and cells.
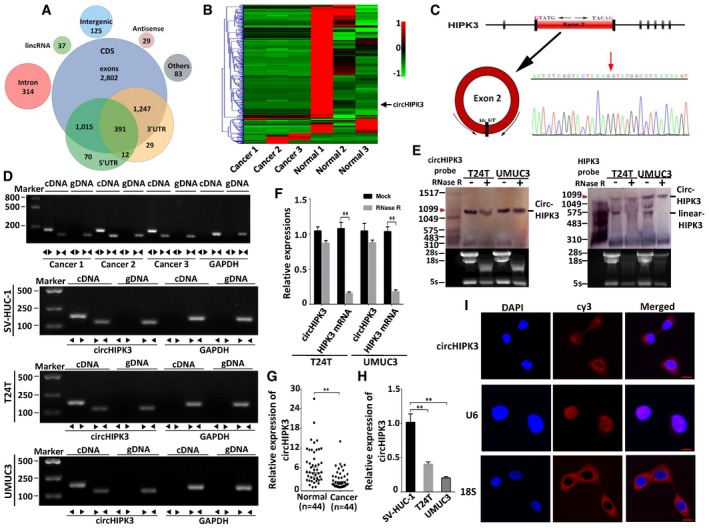
- Genomic origin of human bladder cancer circRNAs.
- Heat map and hierarchical clustering analysis of circRNAs which were differentially expressed between bladder cancer and normal bladder tissues. Each column represents the expression profile of a tissue sample (three bladder cancer and normal bladder samples, respectively), and each row corresponds to a circRNA (log fold change (FC) ≥ 2 and P < 0.05). Red color represents up‐regulated circRNAs, and green color represents down‐regulated circRNAs.
- Schematic illustration showing the circularization of HIPK3 exon 2 forming circHIPK3 (black arrow). The existence of circHIPK3 was validated by RT–PCR and followed by Sanger sequencing. Red arrow represents “head‐to‐tail” splicing sites of circHIPK3.
- The existence of circHIPK3 was validated in three bladder cancer tissues and SV‐HUC‐1, T24T, UMUC3 cell lines by RT–PCR. Divergent primers amplified circHIPK3 in cDNA but not genomic DNA (gDNA). GAPDH was used as negative control.
- Northern blots for detecting circHIPK3 and HIPK3 linear form RNA in T24T and UMUC3 cells treated with or without RNase R digestion. The upper panels show the probed blots of circHIPK3 and HIPK3 linear form, and the red triangle represents circHIPK3 band size (1,099 bp). The lower panels show the gel electrophoretic results of RNA with or without RNase R digestion.
- The expression of circHIPK3 and HIPK3 mRNA in T24T and UMUC3 cells treated with or without RNase R was detected by real‐time PCR. The relative levels of circHIPK3 and HIPK3 mRNA were normalized to the value measured in the mock treatment. Data are mean ± SEM, n = 3. **P < 0.01 versus mock (Student's t‐test).
- The expression of circHIPK3 was detected by real‐time PCR in 44 pairs of bladder cancer and normal bladder tissues. GAPDH was used as internal control. Data are mean ± SEM, n = 3. **P < 0.01 versus normal bladder tissues (Student's t‐test).
- The expression of circHIPK3 was tested by real‐time PCR in SV‐HUC‐1, T24T, and UMUC3 cells. GAPDH was used as internal control. Data are mean ± SEM, n = 3. **P < 0.01 versus SV‐HUC‐1 (Student's t‐test).
- RNA fluorescence in situ hybridization (FISH) showed that circHIPK3 was predominantly localized in cytoplasm. U6 was mainly localized in nucleus, used as negative control. 18S was mainly localized in cytoplasm, used as positive control. circHIPK3, U6, and 18S probes were labeled with Cy3, Nuclei were stained with DAPI. Scale bar, 10 μm.
Figure EV2. Mapping results of linear and circular RNA reads on human chromosomes and differentially expressed circRNAs.
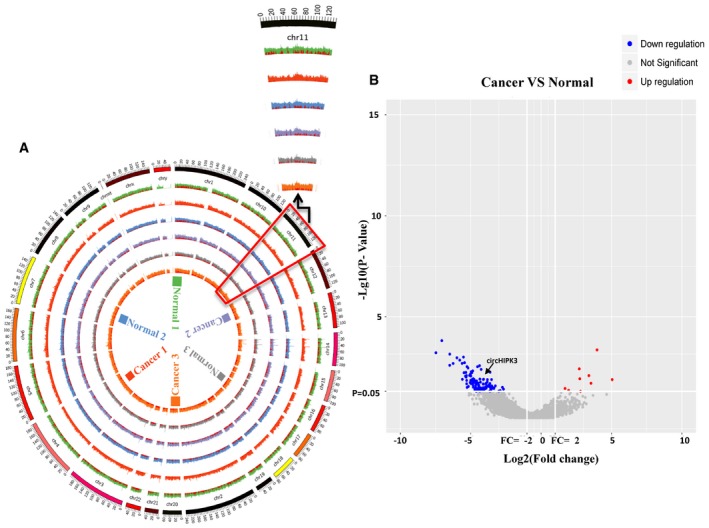
- The outside circle represents the genomic DNA, and the red color represents circRNAs reads junction of each sample. Different color represents different sample. The zoomed‐in part is chromosome 11, and the locus of circHIPK3 is chr11:33286413|33287511 (−). The scale of the axis is 106 bp.
- Volcano plots were constructed for visualizing differentially expressed circRNAs between bladder cancer and normal bladder samples. Differentially expressed circRNAs were filtered by |FC (fold change)| ≥ 2 (Log2 scaled) and P < 0.05 (−Log10 scaled). The red points represent significantly up‐regulated circRNAs, and blue points represent significantly down‐regulated circRNAs.
circHIPK3 is relatively low‐expressed in bladder cancer tissues and cell lines, and predominantly localized in cytoplasm
We then chose several significant differentially expressed circRNAs to verify their existence in bladder cancer cell lines. By using RT–PCR, we found that a single, distinct product of the expected size was amplified from T24T and UMUC3 cells (Appendix Table S1, Appendix Fig S1) with specially designed divergent primers to each selected circRNA, and circHIPK3 was the one that was stably expressed in both cell lines. circHIPK3 (hsa_circ_0000284) arose from the HIPK3 gene and consisted of the head‐to‐tail splicing of exon 2 (1,099 bp) (Fig 1C). According to the RNA‐seq results, circHIPK3 was down‐regulated in bladder cancer tissues (Log2FC = −4.57, Dataset EV2). Subsequently, we confirmed the head‐to‐tail splicing in the RT–PCR product of circHIPK3 with expected size by Sanger sequencing (Fig 1C). However, head‐to‐tail splicing could be produced by trans‐splicing or genomic rearrangements. Thus, we took several steps to rule out these possibilities according to previously described methodology 10, 17. Firstly, we designed convergent primers to amplify HIPK3 mRNA and divergent primers to amplify circHIPK3. Using cDNA and gDNA (genomic DNA) from three bladder cancer tissues and SV‐HUC‐1, T24T, UMUC3 cell lines as templates, circHIPK3 was only amplified by divergent primers in cDNA, and no amplification product was observed in gDNA (Fig 1D). Secondly, we performed Northern blot analysis for circHIPK3 with total RNA extracted from T24T and UMUC3 cells. As shown in Fig 1E, one band of expected size (1,099 bp) was detected by DIG‐labeled circHIPK3‐specific probe targeting the junction region in both cell lines with or without RNase R digestion. Meanwhile, we used the probe targeting exon 2 of HIPK3 mRNA to detect both the circular and linear form of HIPK3 transcripts, and it showed that the fragment of linear form of HIPK3 was digested by RNase R and circHIPK3 was retained after RNase R treatment (Fig 1E). By using real‐time PCR, we further confirmed that circHIPK3 was resistant to RNase R, while HIPK3 mRNA was significantly reduced after RNase R treatment (Fig 1F).
Next, we detected the expression level of circHIPK3 in 44 pairs of bladder cancer tissues and normal bladder tissues, as well as bladder cancer cell lines. Consistent with the RNA‐seq results, the expression of circHIPK3 was obviously decreased in 79.5% of the bladder cancer tissues compared with normal bladder tissues and was negatively correlated with bladder cancer grade, invasion, and the lymph nodes metastasis, respectively (Fig 1G, Table 1). Down‐regulation of circHIPK3 was also found in human muscle‐invasive bladder cancer cells T24T and UMUC3 compared with human immortalized uroepithelium cells SV‐HUC‐1 (Fig 1H). Using RNA fluorescence in situ hybridization (FISH) assay, we demonstrated that circHIPK3 predominately localized in the cytoplasm (Fig 1I).
Table 1.
Clinicopathological features of 44 bladder cancer patients and the expression of circHIPK3 and miR‐558
| Parameters | Group | Cases | circHIPK3 expression | P‐value | miR‐558 expression | P‐value | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Low | % | High | % | Low | % | High | % | |||||
| Gender | Male | 27 | 21 | 78 | 6 | 22 | 0.7141 | 5 | 19 | 22 | 81 | 0.9418 |
| Female | 17 | 14 | 82 | 3 | 18 | 3 | 18 | 14 | 82 | |||
| Age at surgery | < 55 | 21 | 17 | 81 | 4 | 19 | 0.6788 | 4 | 19 | 17 | 81 | 0.8869 |
| ≥ 55 | 23 | 18 | 78 | 5 | 22 | 4 | 17 | 19 | 83 | |||
| Pathological stage | pTa‐T1 | 11 | 4 | 36 | 7 | 64 | < 0.0001 | 5 | 45 | 6 | 55 | 0.0068 |
| pT2‐T4 | 33 | 31 | 94 | 2 | 6 | 3 | 9 | 30 | 91 | |||
| Grade | Low | 13 | 7 | 54 | 6 | 46 | 0.0062 | 6 | 46 | 7 | 54 | 0.0018 |
| High | 31 | 28 | 90 | 3 | 10 | 2 | 6 | 29 | 94 | |||
| Lymph node metastasis | Absent | 27 | 19 | 70 | 8 | 30 | 0.0479 | 7 | 26 | 20 | 74 | 0.0933 |
| Present | 17 | 16 | 94 | 1 | 6 | 1 | 6 | 16 | 94 | |||
| Vascular invasion | Absent | 29 | 20 | 69 | 9 | 31 | 0.0156 | 7 | 24 | 22 | 76 | 0.1544 |
| Present | 15 | 15 | 100 | 0 | 0 | 1 | 7 | 14 | 93 | |||
| Total | 44 | 35 | 80 | 9 | 20 | 8 | 18 | 36 | 82 | |||
P < 0.05 represents statistical significance (Chi‐square test).
Over‐expression of circHIPK3 inhibits migration and invasion of bladder cancer cell in vitro
To explore the function of circHIPK3 in bladder cancer cells, we transfected circHIPK3 expression construct into T24T and UMUC3 cells and established stable transfectants. The expression of circHIPK3 was significantly increased in these stable transfectants, and the over‐expressed circHIPK3 was resistant to RNase R digestion (Fig 2A). Meanwhile, the expression of HIPK3 mRNA, which could be degraded by RNase R, had no apparent change (Fig 2A). Wound healing assay demonstrated that over‐expression of circHIPK3 significantly inhibited cell migration in T24T and UMUC3 cells (Fig 2B). Consistently, over‐expression of circHIPK3 also suppressed migration and invasion of bladder cancer cell lines in transwell migration and matrigel invasion assays (Fig 2C and D). On the other hand, we transfected siRNAs targeting the junction sites of circHIPK3 into T24T and UMUC3 cells to evaluate the influence on the expression of circHIPK3 and HIPK3 mRNA. These siRNAs significantly decreased the expression of circHIPK3, but had no effect on HIPK3 mRNA (Fig 2E). Accordingly, the migration and invasion capabilities of bladder cancer cells increased upon transfection of the most effective circHIPK3 siRNA (si circHIPK3‐1) (Fig 2F–H). These findings indicate that over‐expression of circHIPK3 can inhibit migration and invasion of bladder cancer cells in vitro.
Figure 2. Over‐expression of circHIPK3 inhibits migration and invasion of bladder cancer cell lines in vitro .
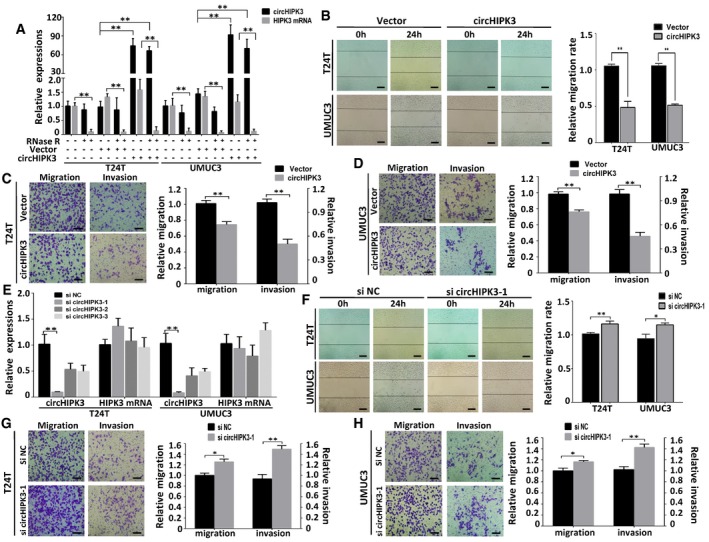
-
AThe expression levels of circHIPK3 and HIPK3 mRNA in T24T and UMUC3 cells after stable transfection of circHIPK3 or vector plasmids were detected by real‐time PCR in mock, vector, and circHIPK3 over‐expression groups treated with or without RNase R digestion. Data are mean ± SEM, n = 3. **P < 0.01 (Student's t‐test).
-
BThe effect of circHIPK3 on cell migration capability was evaluated by wound healing assay in T24T and UMUC3 cells, respectively. Data are mean ± SEM, n = 3. **P < 0.01 (Student's t‐test). Scale bar, 200 μm.
-
C, DCell migration and invasion abilities of T24T and UMUC3 cells transfected with circHIPK3 or vector were evaluated by transwell migration and matrigel invasion assay. Data are mean ± SEM, n = 3. **P < 0.01 (Student's t‐test). Scale bar, 100 μm.
-
EThree siRNAs specifically targeting circHIPK3 were transfected into T24T and UMUC3 cells. The interfering efficacy of each siRNA on circHIPK3 and HIPK3 mRNA was tested by real‐time PCR. Data are mean ± SEM, n = 3. **P < 0.01 versus si NC (Student's t‐test). NC represents negative control, same hereinafter.
-
FThe effect of si circHIPK3 on cell migration capability was evaluated by wound healing assay in T24T and UMUC3 cells, respectively. Data are mean ± SEM, n = 3. *P < 0.05, **P < 0.01 (Student's t‐test). Scale bar, 200 μm.
-
G, HCell migration and invasion abilities of T24T and UMUC3 cells transfected with si circHIPK3‐1 or si NC were evaluated by transwell migration and matrigel invasion assay. Data are mean ± SEM, n = 3. *P < 0.05, **P < 0.01 (Student's t‐test). Scale bar, 100 μm.
circHIPK3 abundantly sponges miR‐558 in bladder cancer cells
It has been reported that circHIPK3 functions as “miRNA sponge” in HEK‐293T cells 26. To address whether circHIPK3 could sponge miRNAs in bladder cancer cells, we selected 12 candidate miRNAs through overlapping the prediction results of miRNA recognition elements in circHIPK3 sequence by miRanda, PITA, and RNAhybrid (Fig 3A, Dataset EV3). Next, we investigated whether circHIPK3 could directly bind these candidate miRNAs. A biotin‐labeled circHIPK3 probe was designed and verified to pull down circHIPK3 in bladder cancer cell lines, and the pull‐down efficiency was significantly enhanced in circHIPK3 over‐expression stable transfectants (Fig 3B and C). The miRNAs were extracted after pull‐down assay, and the level of 12 candidate miRNAs was detected by real‐time PCR. As shown in Fig 3D, miR‐558 was the only one that was abundantly pulled down by circHIPK3 in both T24T and UMUC3 cells. CircHIPK3 contains six predictive binding sites of miR‐558 (Fig EV3). To confirm which binding site was functional, we mutated each site in circHIPK3 expression construct to test whether it could still pull down miR‐558 (Appendix Fig S2). The mutated circHIPK3 could also be pulled down by biotin‐circHIPK3 probe (Fig 3E), which was consistent with the fact that all the mutated binding sites were not located in the biotin‐circHIPK3 probe complementary region. Subsequently, we evaluated the relative level of miR‐558 pulled down by each mutated circHIPK3, respectively. The relative binding of miR‐558 was significantly decreased after mutating site1 or site2, while mutation of the other four sites did not show such decrease (Fig 3F). These results demonstrate that binding site1 and site2, but not the others four binding sites, are critical for circHIPK3 to sponge miR‐558.
Figure 3. circHIPK3 abundantly sponges miR‐558 in bladder cancer cells.
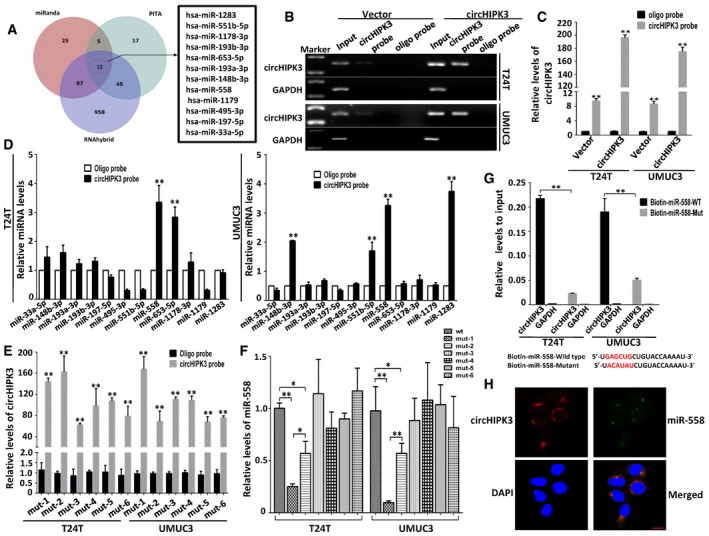
-
ASchematic illustration showing overlapping of the target miRNAs of circHIPK3 predicted by miRanda, PITA, and RNAhybrid.
-
B, CLysates prepared from T24T and UMUC3 cells stably transfected with circHIPK3 or vector were subject to RNA pull‐down assay and tested by RT–PCR (B) and real‐time PCR (C). Relative level of circHIPK3 was normalized to input. GAPDH was used as negative control. Data are mean ± SEM, n = 3. **P < 0.01 versus oligo probe (Student's t‐test).
-
DThe relative level of 12 miRNA candidates in the T24T and UMUC3 lysates was detected by real‐time PCR. Multiple miRNAs can be pulled down by circHIPK3, and miR‐558 was pulled down by circHIPK3 in two cell lines. Data are mean ± SEM, n = 3. **P < 0.01 versus vector (Student's t‐test).
-
ESix binding sites of miR‐558 on circHIPK3 were mutated and transfected into T24T and UMUC3 cells, respectively. The efficacies of circHIPK3 probe on the six mutants were tested by real‐time PCR. Relative level of circHIPK3 was normalized to input. Data are mean ± SEM, n = 3. **P < 0.01 versus oligo probe (Student's t‐test). mut‐1, mut‐2, etc. represent mutant of binding site1, mutant of binding site2, etc.
-
FThe pull‐down efficacies of six mutated circHIPK3 on miR‐558 were tested by real‐time PCR. Relative level of miR‐558 was normalized to input. Data are mean ± SEM, n = 3. **P < 0.01 versus wt group (Student's t‐test).
-
GThe biotinylated wild‐type miR‐558 (Bio‐miR‐558‐WT) or its mutant (Bio‐miR‐558‐Mut) was transfected into T24T and UMUC3 cells. After streptavidin capture, circHIPK3 levels were quantified by real‐time PCR, and the relative immunoprecipitate (IP)/input ratios are demonstrated. GAPDH was used as negative control. Biotin‐coupled miR‐558 wild‐type and mutant sequences are shown below. Data are mean ± SEM, n = 3. **P < 0.01 versus wild type (Student's t‐test).
-
HFluorescence in situ hybridization (FISH) showing the co‐localization between circHIPK3 and miR‐558 in T24T cells. CircHIPK3 probes were labeled with Cy3. Locked nucleic acid miR‐558 probes were labeled with Dig. Nuclei were stained with DAPI. Scale bar, 10 μm.
Figure EV3. Binding sites of miR‐558 on circHIPK3.
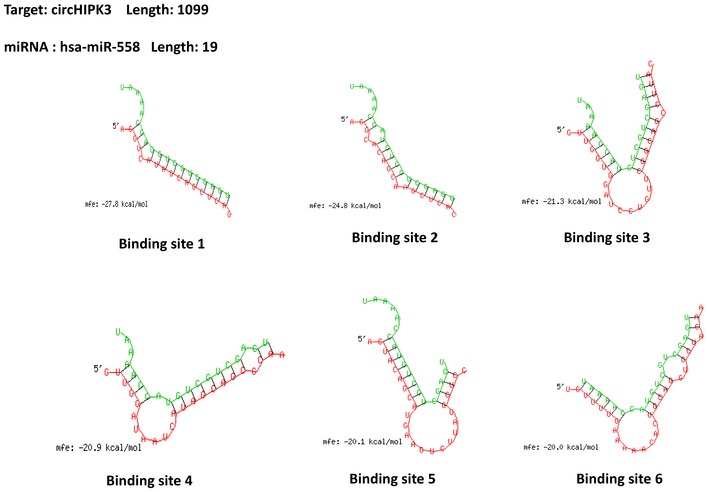
Detailed information of six binding sites of miR‐558 on circHIPK3 that were analyzed by the bioinformatics program RNAhybrid.
We next applied biotinylated miR‐558 mimics to further verify the direct binding of miR‐558 and circHIPK3. T24T and UMUC3 cells with stable over‐expression of circHIPK3 were transfected with biotinylated miR‐558 or its mutant. The binding of circHIPK3 with the miRNA mimics or mutant was tested by real‐time PCR. We found a higher enrichment of circHIPK3 in the captured fraction of wild‐type miR‐558 compared with the mutant that disrupted base pairing between circHIPK3 and miR‐558 (Fig 3G). Moreover, RNA FISH assay revealed that circHIPK3 and miR‐558 were co‐localized in cytoplasm (Fig 3H). The above results demonstrate that circHIPK3 can directly bind to miR‐558 in T24T and UMUC3 cells.
miR‐558 is up‐regulated in bladder cancer tissues and cell lines, and promotes cell migration, invasion, and angiogenesis through targeting HPSE in vitro
It has been reported that miR‐558 can promote migration, invasion, and angiogenesis in neuroblastoma via facilitating HPSE mRNA transcription 29. Previous studies of HPSE have confirmed its high expression in human bladder cancer 30, 31, and HPSE can increase the expression of MMP‐9 and VEGF 31, 32. Kaplan–Meier survival plots of 224 well‐defined bladder cancer cases derived from R2 Genomics Analysis and Visualization Platform (http://r2.amc.nl) showed that patients with high HPSE expression (P = 1.5 × 10−4) had worse survival probability (Fig 4A). Using real‐time PCR, we found that miR‐558 was up‐regulated in human bladder cancer tissues compared with normal bladder tissues (Fig 4B, Table 1). miR‐558 was also up‐regulated in T24T and UMUC3 cells compared with SV‐HUC‐1 cells (Fig 4C). To investigate the function of miR‐558 in bladder cancer cell lines, we performed the miRNA inhibition and over‐expression experiments. The wound healing (Fig EV4A) and transwell assays (Fig 4D) showed that over‐expression of miR‐558 significantly promoted migration and invasion of bladder cancer cells. The tube formation ability of HUVEC cells was also enhanced by treatment with the preconditioned medium of the T24T and UMUC3 cells transfected with miR‐558 mimics (Fig 4F). In contrast, transfection of anti‐miR‐558 inhibitor obviously suppressed cell migration, invasion of T24T and UMUC3 cells, and inhibited the tube formation of HUVEC cells (Figs 4C, E and F, and EV4A). Real‐time PCR and Western blot assays demonstrated that miR‐558 mimics could increase the expression of HPSE, VEGF, and MMP‐9. Accordingly, anti‐miR‐558 inhibitor decreased HPSE, VEGF, and MMP‐9 levels in bladder cancer cells (Figs 4G and H, and EV4B and C). These results reveal that miR‐558 can remarkably promote the migration, invasion, and angiogenesis of bladder cancer through targeting HPSE in vitro.
Figure 4. miR‐558 is up‐regulated in bladder cancer tissues and cell lines, and promotes cell migration, invasion, and angiogenesis through targeting HPSE in vitro .
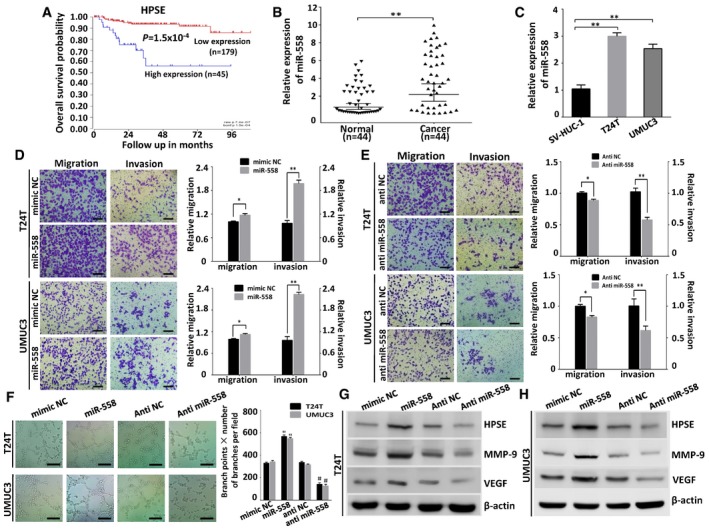
-
AKaplan–Meier survival plots of 224 well‐defined bladder cancer cases derived from R2 microarray analysis and visualization platform (http://r2.amc.nl) showing the survival probability of patients with high or low expression of HPSE.
-
B, CReal‐time PCR revealed that miR‐558 was up‐regulated in bladder cancer tissues (n = 44) and cell lines (T24T, UMUC3), compared with normal adjacent non‐tumorous bladder tissues (n = 44) or SV‐HUC‐1 cells, respectively. Data are mean ± SEM, n = 3. **P < 0.01 (Student's t‐test).
-
D, ETranswell migration and matrigel invasion assays indicating the increased migration and invasion capabilities in T24T and UMUC3 cells transfected with miR‐558 mimics. In contrast, the transfection of anti‐miR‐558 inhibited the migration and invasion of bladder cancer cells. Data are mean ± SEM, n = 3. *P < 0.05, **P < 0.01 (Student's t‐test). Scale bar, 100 μm.
-
FThe tube formation of HUVEC cells was promoted by treatment with the preconditioned medium of T24T and UMUC3 cells transfected with miR‐558 mimics; however, knockdown miR‐558 inhibited the tube formation ability of HUVEC cells. Data are mean ± SEM, n = 3. **P < 0.01 versus mimic NC; ## P < 0.01 versus anti‐NC (Student's t‐test). Scale bar, 100 μm.
-
G, HWestern blot indicating that miR‐558 up‐regulated the expression of HPSE, MMP‐9, and VEGF in T24T and UMUC3 cells, while anti‐miR‐558 down‐regulated the expression of HPSE, MMP‐9, and VEGF.
Figure EV4. miR‐558 promotes cell migration, invasion, and angiogenesis via up‐regulating the expression of HPSE in vitro .
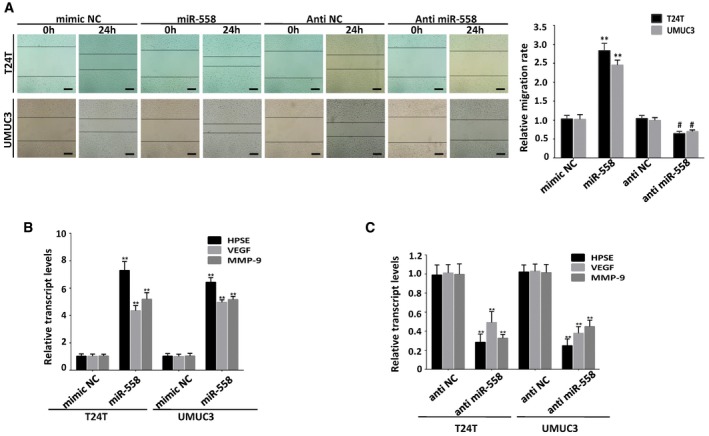
-
AWound healing assay indicating that transfection of miR‐558 mimics promoted cell migration, while transfection with anti‐miR‐558 inhibited cell migration. Data are mean ± SEM, n = 3. **P < 0.01 versus mimic NC; # P < 0.05 versus anti‐NC (Student's t‐test). Scale bar, 200 μm.
-
B, CReal‐time PCR assay indicating that transfection of miR‐558 mimics up‐regulated the expression of HPSE and its downstream targets MMP‐9 and VEGF in bladder cancer cells, and anti‐miR‐558 down‐regulated the expression of HPSE, MMP‐9, and VEGF. Data are mean ± SEM, n = 3. **P < 0.01 versus mimic NC or anti‐NC (Student's t‐test).
Over‐expression of circHIPK3 reverses miR‐558‐induced enhancement of cell migration, invasion, and angiogenesis in bladder cancer cells
In bladder cancer cells which with stable over‐expression of circHIPK3, the expression levels of HPSE, MMP‐9, and VEGF were obviously decreased (Fig EV5A, C and D). Accordingly, the expression of HPSE, MMP‐9, and VEGF was up‐regulated after knockdown of circHIPK3 (Fig EV5B–D). To address whether circHIPK3 inhibited migration and invasion of bladder cancer cells via interacting with miR‐558, we co‐transfected miR‐558 mimics and circHIPK3 expression construct into bladder cancer cells. It showed that the expression of HPSE, MMP‐9, and VEGF was significantly decreased in the bladder cancer cells co‐transfected with circHIPK3 plasmids and miR‐558 mimics, compared with the cells transfected with miR‐558 mimics alone (Fig 5A–D). Meanwhile, transwell matrigel invasion and wound healing assays indicated that the bladder cancer cells co‐transfected with miR‐558 mimics and circHIPK3 expression construct showed a reduction in invasion and migration capabilities compared with miR‐558 mimics‐transfected cells (Fig 5E and G). Besides, the tube formation ability of HUVEC cells was also down‐regulated by treatment of preconditioned medium of miR‐558 mimics and circHIPK3 co‐transfected cells (Fig 5F). Taken together, these data suggest that circHIPK3 suppresses cell migration, invasion, and angiogenesis through sponging miR‐558 and subsequent inhibition of HPSE expression in vitro.
Figure EV5. Over‐expression of circHIPK3 down‐regulates the expression of HPSE, MMP‐9, and VEGF in bladder cancer cells.
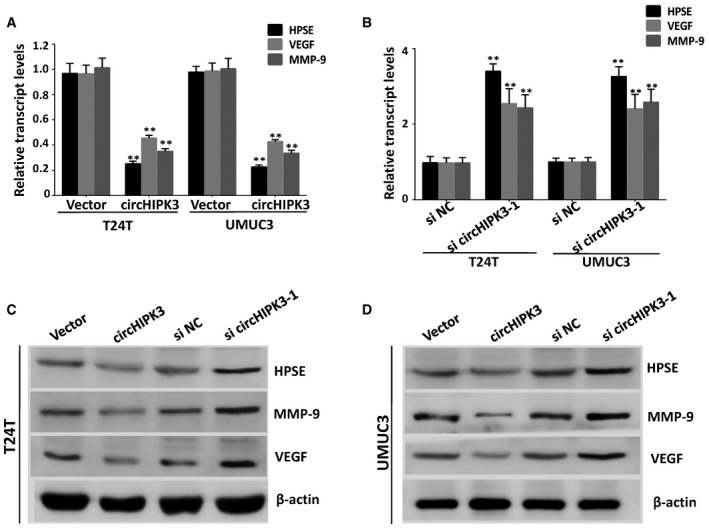
-
A–DReal‐time PCR and Western blot assays demonstrated that over‐expression of circHIPK3 in bladder cancer cells significantly down‐regulates the expression of HPSE, together with its downstream targets MMP‐9 and VEGF. In contrast, knockdown of circHIPK3 up‐regulates the expression of HPSE, MMP‐9, and VEGF. Data are mean ± SEM, n = 3. **P < 0.01 versus vector (A) or si NC (B) (Student's t‐test).
Figure 5. Over‐expression of circHIPK3 reverses miR‐558‐induced enhancement of cell migration, invasion, and angiogenesis in bladder cancer cells.
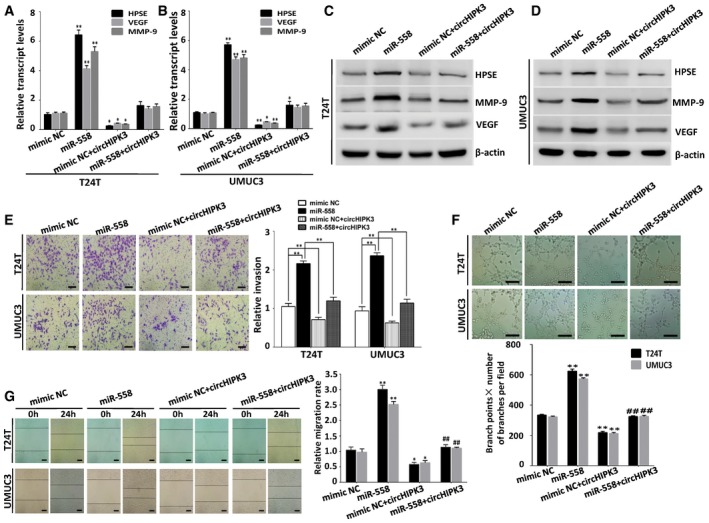
-
A–DReal‐time PCR and Western blot indicating that circHIPK3 counteracted the up‐regulation effect of miR‐558 on HPSE, MMP‐9, and VEGF. Data are mean ± SEM, n = 3. *P < 0.05, **P < 0.01 versus mimic NC (Student's t‐test).
-
ETranswell matrigel invasion assay demonstrating that miR‐558 increased the invasion ability of T24T and UMUC3 cells; however, when co‐transfected with circHIPK3, the function of miR‐558 was reduced. Data are mean ± SEM, n = 3. **P < 0.01 (Student's t‐test). Scale bar, 100 μm.
-
FThe tube formation of HUVEC cells was promoted by treatment with conditioned medium of bladder cancer cells transfected with miR‐558 mimics, when compared with mimic NC transfected cells. Co‐transfection with circHIPK3 inhibited the angiogenic capabilities of HUVEC cells that could be promoted by miR‐558. Data are mean ± SEM, n = 3. **P < 0.01 (Student's t‐test). Scale bar, 100 μm.
-
GIn wound healing assay, the migration of bladder cancer cells transfected with miR‐558 mimics was significantly increased. Co‐transfection with circHIPK3 reduced the migration of T24T and UMCU3 cells that could be increased by miR‐558. Data are mean ± SEM, n = 3. *P < 0.05 and **P < 0.01 versus mimic NC; ## P < 0.01 miR‐558 versus miR‐558 + circHIPK3 (Student's t‐test). Scale bar, 200 μm.
Enforced expression of circHIPK3 suppresses the growth, metastasis, and angiogenesis of bladder cancer cells in vivo
We further investigated the effects of enforced expression of circHIPK3 on regulating tumor growth, metastasis, and angiogenesis in vivo. T24T cells stably transfected with circHIPK3 expression construct or control vector were subcutaneously injected into BALB/c nude mice. The decreased growth rate and tumor weight of xenograft tumors were established in circHIPK3 transfectants compared with vector group (Fig 6A and B). Immunohistochemical staining showed that the expression of HPSE, MMP‐9, and VEGF was inhibited by over‐expressing circHIPK3 (Fig 6C). Moreover, over‐expression of circHIPK3 led to decrease in CD31+ microvessels and mean intra‐tumor vessel density (Fig 6C). We also injected T24T cells stably transfected with circHIPK3 expression construct or control vector via tail vein into BALB/c nude mice to establish metastasis model. The results showed that circHIPK3 transfectant‐treated nude mice formed fewer lung metastasis colonies than those treated with control vector transfectants (Fig 6D). Taken together, these results demonstrate that enforced expression of circHIPK3 efficiently inhibits the growth, angiogenesis, and metastasis of bladder cancer in vivo.
Figure 6. Enforced expression of circHIPK3 suppresses the growth, metastasis, and angiogenesis of bladder cancer cells in vivo .
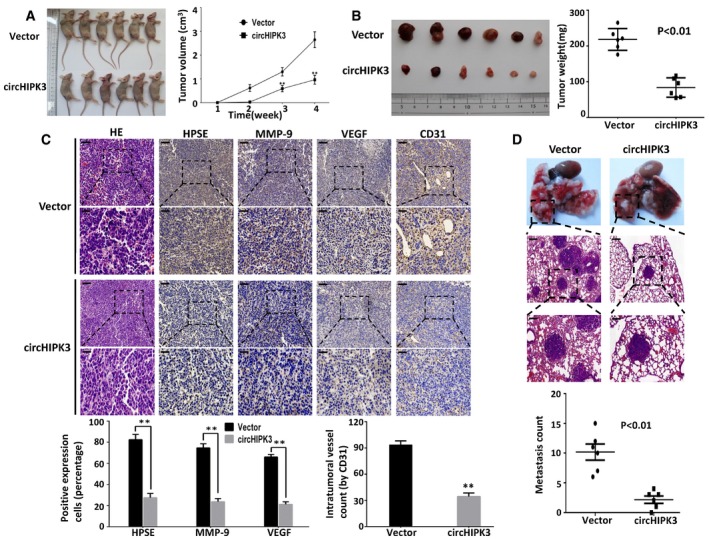
-
A, BHypodermic injection of T24T cells stably transfected with circHIPK3 or vector control into BALB/c nude mice established subcutaneous xenograft tumors (5 × 106 cells per mouse, n = 6 for each group). Compared with vector group, the tumor growth rate and weight significantly decreased in circHIPK3‐treated nude mice. Data are mean ± SEM, **P < 0.01 (Student's t‐test).
-
CImmunohistochemical staining revealed that over‐expression of circHIPK3 led to decreased expression of HPSE, MMP‐9, VEGF, and CD31 within tumors. Stable transfection of circHIPK3 reduced the mean vessel density within tumors. Data are mean ± SEM, **P < 0.01 (Student's t‐test). Scale bars, 50 and 100 μm.
-
DT24T cells stably transfected with circHIPK3 or vector control were injected into the tail vein of BALB/c nude mice (2 × 106 cells per mouse, n = 6 for each group). The nude mice treated with circHIPK3 transfected cells demonstrated significantly fewer lung metastatic colonies. Data are mean ± SEM (Student's t‐test). Scale bars, 100 and 200 μm.
Discussion
For more than 30 years, circular RNAs were reported sporadically and long considered to be molecular flukes 17. Recently, with the advent of next‐generation sequencing, numerous of circRNAs were identified from various animal genomes, and many of them were highly stable and abundantly expressed, thus largely reshaped the conventional perspective on circRNAs. circRNAs have been reported to be dysregulated in diverse cancer types, such as colorectal cancer 22, hepacellular carcinoma 26, esophageal squamous cell carcinoma 33, basal cell carcinoma 34, and laryngeal cancer 35. It is widely accepted that these differentially expressed circRNAs may have certain potential functions in regulation of gene expression 19, 22, 26, 36. RNA‐seq results provide useful information for revealing the general tendency of circRNAs expression and help us to select candidate circRNAs for further research. In the present study, we successfully identified thousands of circRNAs in human bladder cancer tissues and normal bladder tissues, and hundreds of them were differentially expressed. Nevertheless, the expression of each candidate circRNA still needs to be verified in a large cohort of clinical samples and cell lines.
CircRNAs arise from diverse genomic locations, and most of them are formed by circularization of exons 10. Consistent with previous studies, we found that the majority of circRNAs are originated from protein‐coding exons. Two main mechanisms both involving the canonical spliceosome have been proposed for the biogenesis of exonic circRNAs 37, 38. One is “direct backsplicing”, where the 3′‐end of downstream exon as splice donor pairs with the 5′‐end of unspliced upstream exon as splice acceptor, and then, intervening RNA is circularized. The other is “exon skipping”, for which a lariat containing an exon is created by exon skipping, and then, the intron is removed and circRNA is generated. Here, we demonstrated that circHIPK3, which was an exonic circRNA originated from exon 2 of HIPK3 gene, was down‐regulated in bladder cancer tissues and predominantly localized in cytoplasm. The expression of circHIPK3 has also been reported in previous studies through RNA deep sequencing of diverse cell lines and tissues 12, 21, 26. It is suggested that large exons are preferentially circularized because they are sterically easier for 3′ to 5′ splicing at canonical splice sites 17. The exon 2 of HIPK3 gene is large (1,099 bp) and flanked by long introns on both sides, which contain many complementary Alu repeats to further promote its circularization 21. All these features indicate that circHIPK3 is formed by “direct splicing” and stably expressed in different cell lines and tissues 17, 38.
Stable transcripts with a host of miRNA‐binding sites or miRNA response elements (MREs) can function as miRNA sponges, and exonic circRNAs also contain certain MREs. Moreover, with the decreasing polymorphisms of MREs, exonic circRNAs become more efficient miRNA sponges 10, 39. One extreme case is CDR1as 10, which contains over 70 binding sites of miR‐7 and significantly suppresses the activity of miR‐7. circHIPK3 was reported to promote proliferation of human liver cancer HuH‐7 cells, human colon cancer HCT‐116 cells, and human cervical cancer HeLa cells via sponging multiple miRNAs 26. In the present study, we found that circHIPK3 inhibited migration, invasion, and angiogenesis of human invasive bladder cancer T24T and UMUC3 cells by targeting miR‐558, which has not been reported by previous studies. Furthermore, we demonstrated that two binding sites were critical for circHIPK3 to sponge miR‐558. These findings suggest that circHIPK3 involves in complex regulatory networks, and confers cell‐type‐specific regulation of cell function in different cancers.
Some miRNAs are derived from miniature inverted‐repeat transposable elements (MITEs), such as miR‐548 family 40. It has been reported that miR‐558 is co‐expressed with its host gene baculoviral IAP repeat containing 6 (BIRC6), indicating that they are likely to be expressed from the same promoter 29. Canonically, with incomplete or complete base pairing to 3′UTR of mRNA, miRNAs can either retard translation or induce degradation of the target mRNA and then decrease the expression of the corresponding protein 23. Accumulating evidences suggest that miRNAs can also promote the transcription of mRNA via binding to the promoter of target genes 41. HPSE is an endoglycosidase that cleaves the heparan sulfate proteoglycans, which regulates tumor invasion and metastasis through releasing multiple types of cytokine, including VEGF and MMP‐9 31, 32. miR‐558 was reported to directly bind to the promoter of HPSE gene, and promote migration, invasion, angiogenesis, and tumor growth of neuroblastoma cells in vitro and in vivo via increasing the expression of HPSE mRNA 29, 42. In this study, we found that patients with higher HPSE expression have worse survival probability by using R2 genomics analysis. However, it remains unclear whether HPSE expression adds any additional prognostic value regarding grade and stage, or whether it might just correlate strongly with these. A multivariate analysis would be needed to determine whether high HPSE expression indeed holds independent prognostic value. Interestingly, our results showed that over‐expression of circHIPK3 efficiently interacted with miR‐558 and subsequently down‐regulated the expression of HPSE and its downstream targets MMP‐9 and VEGF to attenuate the promoting effect of miR‐558 on bladder cancer cell migration, invasion, and angiogenesis. Since circHIPK3 and miR‐558 were found to be predominantly co‐localized in cytoplasm, it is indicated that circHIPK3 could sponge miR‐558 and prevent miR‐558 from being transported into nucleus to bind the promoter of HPSE gene in bladder cancer cells.
Of note, not all circRNAs can act as “miRNA sponges” 17. Small sized circRNAs, which are apparently not suitable for miRNA sponges, can be absorbed into exosomes and function as promising biomarkers for cancer diagnosis 43. Intronic circRNAs and exon–intron RNAs, which mainly localize in nucleus with little enrichment for miRNA target sites, have been reported to regulate their parental genes expression via specific RNA–RNA interaction 44, 45. Moreover, some circRNAs, such as circMbl, cricFmn and circDMD, can strongly bind to cognate linear transcripts to sequester mRNA from translation and finally lead to the reduction in protein expression 17, 18. This process is also termed as “mRNA trap”. Thus, various functions of the differentially expressed circRNAs in bladder cancer cells still need to be explored beyond “miRNAs sponges”.
In conclusion, we show that circHIPK3 is down‐regulated in human bladder cancer, and it can efficiently sponge miR‐558 to inhibit heparanase expression. We also demonstrate that over‐expression of circHIPK3 can effectively inhibit aggressiveness and metastasis of bladder cancer cells through targeting miR‐558/heparanase axis. Our findings provide novel evidences that circRNAs act as “microRNA sponges” and also provide a new therapeutic target for the treatment of bladder cancer.
Materials and Methods
Human tissue specimens
Forty‐four pairs of bladder cancer tissues and paired adjacent normal bladder tissues were obtained from patients who underwent radical cystectomy at Department of Urology of the Union Hospital of Tong Medical College (Wuhan, China) between 2014 and 2016. With the instruction of a skillful pathologist, we collected the normal bladder urothelium samples (≥ 200 mg/sample) with a distance of ≥ 3 cm from the edge of cancer tissues in the resected bladder. All specimens were immediately snap‐frozen in liquid nitrogen after surgical removal. Histological and pathological diagnoses were confirmed, and the specimens were classified by at least two experienced clinical pathologist according to the 2004 World Health Organization Consensus Classification and Staging System for bladder neoplasms. All specimens were obtained with appropriate informed consent from the patients and approved by the Institutional Review Board of Tongji Medical College of Huazhong University of Science and Technology. Detailed information is presented in Table 1.
RNA sample treatment, library synthesis, and RNA sequencing
The total RNA samples of three pairs of bladder cancer and normal bladder tissues were treated with the RiboZero rRNA Removal Kit (Epicentre, WI, USA) for deleting rRNA, according to the manufacturer's instructions. Next, the rRNA depleted and RNase R digested RNA samples were fragmented and synthesized cDNA with random primer. Purified the PCR amplification products of cDNA, then the libraries were quality controlled and sequenced by HiSeq2000 (Illumina, San Diego, CA, USA).
Identification and annotation of human bladder cancer circRNAs
Reference genomes (GRCH38/hg38) were obtained from UCSC genome browser (http://genome.ucsc.edu/). In a first step, FASTQ reads that aligned contiguously and full length to the genomes by TopHat2 were discarded. Next, from the unmapped reads we extracted 20‐nt from both ends and aligned them independently to find unique anchor positions within spliced exons by TopHat2 again. Anchors that aligned in the reversed orientation (head‐to‐tail) indicated circRNA splicing. Using ANNOVAR database 28, we mapped the splicing ends of each circRNA to the genomic regions and then compared the results to the RefSeq and UCSC databases to annotate the functional element of circRNA. Detailed information is listed in Dataset EV1.
Differentially expressed circRNA selection
We adopted edgeR package to analyze the differences in expression of circRNAs between bladder cancer tissues and normal bladder tissues. Fold change was calculated by SRPBM (spliced reads per billion mapping). SRPBM = number of circular reads/total mapped reads (units in billion). Differentially expressed circRNAs were filtered by |FC (fold change)| ≥ 2 and P < 0.05. Detailed information is listed in Dataset EV2.
miRNA targets prediction of circHIPK3
We predicted the miRNA‐binding sites of circHIPK3 using the bioinformatic database miRanda (http://www.microrna.org/microrna/getMirnaForm.do), PITA (http://genie.weizmann.ac.il/pubs/mir07/mir07_data.html) and RNAhybrid (http://bibiserv.techfak.uni-bielefeld.de/rnahybrid/). Filtering restrictions were as follows: (i) Total Sore ≥ 140, Total Energy < −17kcal/mol; (ii) combined interaction energy ΔΔG < −10; (iii) minimum free energy (MFE) ≤ −20 kcal/mol. Detailed information is listed in Dataset EV3.
Cell culture and treatment
The human metastatic bladder cancer cell line T24T, which is a lineage‐related lung metastatic variant of invasive bladder cancer cell line T24, was kindly provided by Dr. Dan Theodorescu (Departments of Urology, University of Virginia, Charlottesville, VA) in 2010 and was subjected to DNA tests and authenticated in our previous studies 46. Human invasive bladder cancer cell line UMUC3(CRL‐1749), human immortalized uroepithelium cell line SV‐HUC‐1 (CRL‐9520), and endothelial cell line HUVEC (CRL‐1730) were purchased from American Type Culture Collection (ATCC, USA) in 2015. T24T and UMUC3 cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% FBS (Gibco, Australia origin), 1% penicillin/streptomycin, and 2 mM l‐glutamine (Life Technologies, Carlsbad, CA, USA). SV‐HUC‐1 cells were cultured in the F‐12K medium supplemented with 10% FBS, 1% penicillin/streptomycin. HUVEC cells were cultured in the RPMI1640 medium supplemented with 10% FBS, 1% penicillin/streptomycin. Cells were maintained at 37°C in a humidified atmosphere of 5% CO2.
RNA preparation, treatment with RNase R, and PCR
Total RNA was isolated with RNeasy Mini Kit (QIAGEN, Germany) according to the manufacturer's instructions. RNase R treatment was carried out for 15 min at 37°C using RNase R (Epicenter) 3 U/mg. For RT–PCR, the treated RNA was directly reverse transcribed using the Prime Script RT Master Mix (Takara, Japan) from 500 ng of RNA with random or oligo(dT) primer. The PCR reactions were performed using PCR Master Mix (2×) (Thermo Fisher Scientific, Waltham, MA, USA). To quantify the amount of circRNA, miRNA, and mRNA, the real‐time PCR analyses were performed using SYBR® Premix Ex Taq™ kit (Takara). GAPDH and U6 were used as internal control. All analyses were performed using the StepOnePlus Real‐Time PCR System (Applied Biosystems, Carlsbad, CA, USA). The ΔΔCT method was used to calculate the relative expression of different genes. The primers are listed in Appendix Table S2.
Northern blots
DIG‐labeled circHIPK3 or HIPK3 linear form probe was synthesized with DIG Labeling Kit (MyLab Corporation, Beijing, China) following the manufacturer's instructions. Total RNA with or without RNase R treatment and digoxigenin‐labeled RNA molecular weight marker (Roche) were loaded on a 1.2% agarose gel containing 1% formaldehyde and were run in 1× MOPS buffer. RNA was transferred onto Hybond‐N+ membranes (GE Healthcare) by capillary transfer. Washed the membrane and fixed the RNA onto the membrane by UV cross‐linker (UVP, CA, USA). Pre‐hybridization was performed at 65°C for 1–2 h and then hybridized with circHIPK3 probe or HIPK3 linear form probe at 65°C overnight. Membranes were stringently washed twice in 0.1× SSC and 0.1% SDS at 50°C for 30 min. The detection was performed using DIG colorimetric detection kit (MyLab) according to the manufacturer's instructions. The probe sequences are listed in Appendix Table S2.
CircRNA plasmids construction and stable transfection
To construct circHIPK3 over‐expression plasmids, human circHIPK3 cDNA was synthesized by TSINGKE (Wuhan, China) and cloned into pcD‐ciR vector (Geenseed Biotech Co, Guangzhou, China). The pcD‐ciR vector contained a front circular frame and a back circular frame. Transfection was carried out using Lipofectamine 2000 (Life Technologies) according to the manufacturer's instructions. The transfected cells were then, respectively, selected with G418 (Life Technologies) for 4–6 weeks. Surviving cells were used as stable mass transfectants.
Oligonucleotide transfection
SiRNA, miRNA mimics, and inhibitors were synthesized by GenePharma (Shanghai, China). The sequences used are listed in Appendix Table S2. Transfection was carried out using Lipofectamine RNAiMax (Life Technologies) according to the manufacturer's instructions.
RNA in situ hybridization
Cy3‐labeled circHIPK3 probes and Dig‐labeled locked nucleic acid miR‐558 probes were designed and synthesized by RiboBio (Guangzhou, China), and the probes sequences were available upon request. The signals of the probes were detected by Fluorescent In Situ Hybridization Kit (RiboBio, Guangzhou, China) according to the manufacturer's instructions. The images were acquired on Nikon A1Si Laser Scanning Confocal Microscope (Nikon Instruments Inc, Japan).
Pull‐down assay with biotinylated circHIPK3 probe
Pull‐down assay was performed as described 25. In brief, 1 × 107 bladder cancer cells were harvested, lysed, and sonicated. The circHIPK3 probe was incubated with C‐1 magnetic beads (Life Technologies) at 25°C for 2 h to generate probe‐coated beads. The cell lysates were incubated with circHIPK3 probe or oligo probe at 4°C overnight. After washing with the wash buffer, the RNA complexes bound to the beads were eluted and extracted with RNeasy Mini Kit (QIAGEN) for RT–PCR or real‐time PCR. Biotinylated‐circHIPK3 probe was designed and synthesized by RiboBio (Guangzhou, China) (Appendix Table S2).
Pull‐down assay with biotinylated miRNA
The pull‐down assay with biotinylated miRNA was performed as described 25. In brief, stably expressed circHIPK3 bladder cancer cells were transfected with biotinylated miRNA mimics or mutant (50 nM) using Lipofectamine RNAiMax (Life Technologies) and harvested 48 h after transfection. The cells were harvested and sonicated. 50 μl of the cell lysates were aliquot for input. The remaining cell lysates were incubated with C‐1 magnetic beads (Life Technologies) at 4°C for 3 h and then washed in wash buffer. The bound RNAs were purified using RNeasy Mini Kit (QIAGEN) for the analysis. The sequences are listed in Appendix Table S2.
Wound healing assay
T24T and UMUC3 cells were cultured in six‐well plates and scraped with the fine end of 200 μl pipette tips (time 0 h). Cell migration was photographed using 10 high‐power fields at 0 and 24 h after injury. Remodeling was measured as diminishing distance across the induced injury, normalized to the 0 h control, and expressed as relative migration.
Transwell migration and matrigel invasion assays
The migration and matrigel invasion assays were conducted by using transwell chamber (for migration assay) or transwell pre‐coated matrigel chamber (for invasion assay) according to the manufacturer's protocol (BD Science, Bedford, MA, USA). The homogeneous single cell suspensions (5 × 104 cells/well for migration, 1 × 105/well for invasion) were added to the upper chambers and incubated for 24 h. The migration and invasion rates were quantified by counting the migration and invaded cells at least three random fields.
Tube formation assay
Fifty microliters of growth factor‐reduced matrigel was polymerized on 96‐well plates. HUVEC cells were suspended in the RPMI1640 medium preconditioned with tumor cells, added to the matrigel‐coated wells at the density of 5 × 104 cells/well, and incubated at 37°C for 18 h. Quantification of angiogenic activity was calculated by measuring the length of tube walls formed between discrete endothelial cells in each well relative to the control.
Western blots
Cell lysates were prepared with RIPA buffer (Thermo Scientific). The concentration was determined using a bicinchoninic acid (BCA) protein assay kit (Pierce, Thermo Scientific). Immunoreactive bands were detected by using the Immobilon ECL substrate kit (Millipore, Merck KGaA, Germany). The images were acquired by using BioSpectrum 600 Imaging System (UVP, CA, USA). Antibodies used included primary antibodies against HPSE (Cat No:24529‐1‐AP, 1:800 dilution, Proteintech, USA), VEGF (Cat No:19003‐1‐AP, 1:1,000 dilution, Proteintech, USA), MMP‐9(Cat.No: 10375‐2‐AP, 1:1,000 dilution, Proteintech, USA), and β‐actin (Cat.No: 60008‐1‐Ig 1:1,000 dilution, Proteintech, USA); HRP‐conjugated secondary goat anti‐mouse(Cat.No: SA00001‐1) or goat anti‐rabbit (Cat.No: SA00001‐2) antibodies (1:4,000 dilution, Proteintech, USA).
Animal experiments
All animal experiments were approved by the Animal Care Committee of Tongji Medical College. For the in vivo tumor growth studies, 4‐week‐old female BALB/c nude mice were randomly divided into two groups (n = 6 for each group). T24T cells stably transfected with circHIPK3 plasmids or control vector were subcutaneously injected into upper back of the nude mice (5 × 106 cells per mouse), respectively. One month later, mice were sacrificed and examined for tumor weight, gene expression, and angiogenesis. For the in vivo tumor metastasis studies, BALB/c nude mice were divided into two groups as described before. T24T cells stably transfected with circHIPK3 plasmids or control vector were injected into the nude mice (2 × 106 cells per mouse) via tail vein, respectively. 50 days later, mice were sacrificed and examined for numbers of lungs metastatic colonies.
Immunohistochemistry
Immunohistochemical staining was performed as previously described 46, with antibodies specific for HPSE, VEGF, MMP‐9, and CD31 (1:200 dilution, Proteintech). The immunoreactivity in each tissue section was assessed by at least two pathologists. The degree of positivity was determined according to the percentage of positive tumor cells.
Statistical analysis
Unless otherwise stated, all data are shown as mean ± standard error of the mean (SEM). Statistical analyses were performed using SPSS 19.0 statistical software. Statistical significance (P < 0.05) was determined by Student's t‐test (two‐tailed), chi‐square test followed by assessment of difference.
Additional information
Accession codes: The RNA‐seq data have been deposited in the Gene Expression Omnibus database under accession code GSE97239.
Author contributions
YL performed all the RNA‐seq data analysis, primers design, FISH, circRNA pull‐down, miRNA pull‐down, and Northern/Western blots experiments. FZh and XX contributed transwell and animal experiments. FX collected and classified the human bladder cancer tissue samples. CH, DT, and DL contributed RT–PCR and real‐time PCR experiments. MW contributed tube formation experiments. LW contributed immunohistochemistry experiment. YL, GJ, and FZe analyzed the data and wrote the paper.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Dataset EV1
Dataset EV2
Dataset EV3
Review Process File
Acknowledgements
We thank Dr. Thomas B. Hansen from the Department of Molecular Biology and Genetics, Aarhus University. He patiently instructed us to design the divergent primers and the principles to construct over‐expression plasmids for circRNAs. We also thank Shanghai Biotechnology Ltd., Co and Guangzhou RiboBio Ltd., Co for the support of bioinformatics analysis. This research was supported by the National Natural Science Foundation of China (Nos. 81672529, 81472406, 81402113, 81202018, 81272816, and 81602234).
EMBO Reports (2017) 18: 1646–1659
Contributor Information
Fuqing Zeng, Email: zengfuqingpro@163.com.
Guosong Jiang, Email: jiangguosongdoc@hotmail.com.
References
- 1. Antoni S, Ferlay J, Soerjomataram I, Znaor A, Jemal A, Bray F (2016) Bladder cancer incidence and mortality: a global overview and recent trends. Eur Urol 71: 96–108 [DOI] [PubMed] [Google Scholar]
- 2. Siegel RL, Miller KD, Jemal A (2015) Cancer statistics, 2015. CA Cancer J Clin 65: 5–29 [DOI] [PubMed] [Google Scholar]
- 3. Leal J, Luengo‐Fernandez R, Sullivan R, Witjes JA (2016) Economic burden of bladder cancer across the european union. Eur Urol 69: 438–447 [DOI] [PubMed] [Google Scholar]
- 4. Abdollah F, Gandaglia G, Thuret R, Schmitges J, Tian Z, Jeldres C, Passoni NM, Briganti A, Shariat SF, Perrotte P et al (2013) Incidence, survival and mortality rates of stage‐specific bladder cancer in United States: a trend analysis. Cancer Epidemiol 37: 219–225 [DOI] [PubMed] [Google Scholar]
- 5. Alfred Witjes J, Lebret T, Comperat EM, Cowan NC, De Santis M, Bruins HM, Hernandez V, Espinos EL, Dunn J, Rouanne M et al (2016) Updated 2016 EAU guidelines on muscle‐invasive and metastatic bladder cancer. Eur Urol 71: 462–475 [DOI] [PubMed] [Google Scholar]
- 6. Hsu MT, Coca‐Prados M (1979) Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature 280: 339–340 [DOI] [PubMed] [Google Scholar]
- 7. Nigro JM, Cho KR, Fearon ER, Kern SE, Ruppert JM, Oliner JD, Kinzler KW, Vogelstein B (1991) Scrambled exons. Cell 64: 607–613 [DOI] [PubMed] [Google Scholar]
- 8. Capel B, Swain A, Nicolis S, Hacker A, Walter M, Koopman P, Goodfellow P, Lovell‐Badge R (1993) Circular transcripts of the testis‐determining gene Sry in adult mouse testis. Cell 73: 1019–1030 [DOI] [PubMed] [Google Scholar]
- 9. Cocquerelle C, Mascrez B, Hetuin D, Bailleul B (1993) Mis‐splicing yields circular RNA molecules. FASEB J 7: 155–160 [DOI] [PubMed] [Google Scholar]
- 10. Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M et al (2013) Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495: 333–338 [DOI] [PubMed] [Google Scholar]
- 11. Rybak‐Wolf A, Stottmeister C, Glazar P, Jens M, Pino N, Giusti S, Hanan M, Behm M, Bartok O, Ashwal‐Fluss R et al (2015) Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell 58: 870–885 [DOI] [PubMed] [Google Scholar]
- 12. Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO (2013) Cell‐type specific features of circular RNA expression. PLoS Genet 9: e1003777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO (2012) Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS ONE 7: e30733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J (2013) Natural RNA circles function as efficient microRNA sponges. Nature 495: 384–388 [DOI] [PubMed] [Google Scholar]
- 15. Barrett SP, Wang PL, Salzman J (2015) Circular RNA biogenesis can proceed through an exon‐containing lariat precursor. eLife 9: 18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chen LL, Yang L (2015) Regulation of circRNA biogenesis. RNA Biol 12: 381–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jeck WR, Sharpless NE (2014) Detecting and characterizing circular RNAs. Nat Biotechnol 32: 453–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ashwal‐Fluss R, Meyer M, Pamudurti NR (2014) circRNA biogenesis competes with Pre‐mRNA splicing. Mol Cell 56: 55–66 [DOI] [PubMed] [Google Scholar]
- 19. Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA, Roslan S, Schreiber AW, Gregory PA, Goodall GJ (2015) The RNA binding protein quaking regulates formation of circRNAs. Cell 160: 1125–1134 [DOI] [PubMed] [Google Scholar]
- 20. Barrett SP, Salzman J (2016) Circular RNAs: analysis, expression and potential functions. Development 143: 1838–1847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE (2013) Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 19: 141–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bachmayr‐Heyda A, Reiner AT, Auer K, Sukhbaatar N, Aust S, Bachleitner‐Hofmann T, Mesteri I, Grunt TW, Zeillinger R, Pils D (2015) Correlation of circular RNA abundance with proliferation–exemplified with colorectal and ovarian cancer, idiopathic lung fibrosis, and normal human tissues. Sci Rep 5: 8057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yoshino H, Seki N, Itesako T, Chiyomaru T, Nakagawa M, Enokida H (2013) Aberrant expression of microRNAs in bladder cancer. Nat Rev Urol 10: 396–404 [DOI] [PubMed] [Google Scholar]
- 24. Catto JW, Alcaraz A, Bjartell AS, De Vere White R, Evans CP, Fussel S, Hamdy FC, Kallioniemi O, Mengual L, Schlomm T et al (2011) MicroRNA in prostate, bladder, and kidney cancer: a systematic review. Eur Urol 59: 671–681 [DOI] [PubMed] [Google Scholar]
- 25. Wang K, Long B, Liu F, Wang JX, Liu CY, Zhao B, Zhou LY, Sun T, Wang M, Yu T et al (2016) A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR‐223. Eur Heart J 37: 2602–2611 [DOI] [PubMed] [Google Scholar]
- 26. Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, Luo Y, Lyu D, Li Y, Shi G et al (2016) Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun 7: 11215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Glazar P, Papavasileiou P, Rajewsky N (2014) circBase: a database for circular RNAs. RNA 20: 1666–1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang K, Li M, Hakonarson H (2010) ANNOVAR: functional annotation of genetic variants from high‐throughput sequencing data. Nucleic Acids Res 38: e164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Qu H, Zheng L, Pu J, Mei H, Xiang X, Zhao X, Li D, Li S, Mao L, Huang K et al (2015) miRNA‐558 promotes tumorigenesis and aggressiveness of neuroblastoma cells through activating the transcription of heparanase. Hum Mol Genet 24: 2539–2551 [DOI] [PubMed] [Google Scholar]
- 30. Gohji K (2001) Heparanase protein and gene expression in bladder cancer. J Urol 166: 1286–1290 [PubMed] [Google Scholar]
- 31. Jiang G, Zheng L, Pu J, Mei H, Zhao J, Huang K, Zeng F, Tong Q (2012) Small RNAs targeting transcription start site induce heparanase silencing through interference with transcription initiation in human cancer cells. PLoS ONE 7: e31379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Luan Q, Sun J, Li C, Zhang G, Lv Y, Wang G, Li C, Ma C, Gao T (2011) Mutual enhancement between heparanase and vascular endothelial growth factor: a novel mechanism for melanoma progression. Cancer Lett 308: 100–111 [DOI] [PubMed] [Google Scholar]
- 33. Li F, Zhang L (2015) Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/β‐catenin pathway. Oncotarget 6: 13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sand M, Bechara FG, Sand D, Gambichler T, Hahn SA, Bromba M, Stockfleth E, Hessam S (2016) Circular RNA expression in basal cell carcinoma. Epigenomics 8: 619–632 [DOI] [PubMed] [Google Scholar]
- 35. Xuan L (2016) Circular RNA: a novel biomarker for progressive laryngeal cancer. Am J Transl Res 8: 8 [PMC free article] [PubMed] [Google Scholar]
- 36. Zhong Z, Lv M, Chen J (2016) Screening differential circular RNA expression profiles reveals the regulatory role of circTCF25‐miR‐103a‐3p/miR‐107‐CDK6 pathway in bladder carcinoma. Sci Rep 6: 30919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Petkovic S, Muller S (2015) RNA circularization strategies in vivo and in vitro . Nucleic Acids Res 43: 2454–2465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Starke S, Jost I, Rossbach O, Schneider T, Schreiner S, Hung LH, Bindereif A (2015) Exon circularization requires canonical splice signals. Cell Rep 10: 103–111 [DOI] [PubMed] [Google Scholar]
- 39. Thomas LF, Saetrom P (2014) Circular RNAs are depleted of polymorphisms at microRNA binding sites. Bioinformatics 30: 2243–2246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Piriyapongsa J, Jordan IK (2007) A family of human microRNA genes from miniature inverted‐repeat transposable elements. PLoS ONE 2: e203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Younger ST, Corey DR (2011) Transcriptional gene silencing in mammalian cells by miRNA mimics that target gene promoters. Nucleic Acids Res 39: 5682–5691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Shohet JM, Ghosh R, Coarfa C, Ludwig A, Benham AL, Chen Z, Patterson DM, Barbieri E, Mestdagh P, Sikorski DN et al (2011) A genome‐wide search for promoters that respond to increased MYCN reveals both new oncogenic and tumor suppressor microRNAs associated with aggressive neuroblastoma. Cancer Res 71: 3841–3851 [DOI] [PubMed] [Google Scholar]
- 43. Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J, Chen D, Gu J, He X, Huang S (2015) Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res 25: 981–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, Zhu S, Yang L, Chen LL (2013) Circular intronic long noncoding RNAs. Mol Cell 51: 792–806 [DOI] [PubMed] [Google Scholar]
- 45. Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, Zhong G, Yu B, Hu W, Dai L et al (2015) Exon‐intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol 22: 256–264 [DOI] [PubMed] [Google Scholar]
- 46. Jiang G, Wu AD, Huang C, Gu J, Zhang L, Huang H, Liao X, Li J, Zhang D, Zeng X et al (2016) Isorhapontigenin (ISO) inhibits invasive bladder cancer formation in vivo and human bladder cancer invasion in vitro by targeting STAT1/FOXO1 axis. Cancer Prev Res 9: 567–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Dataset EV1
Dataset EV2
Dataset EV3
Review Process File


