Abstract
Hematopoiesis involves a series of lineage differentiation programs initiated in hematopoietic stem cells (HSCs) found in bone marrow (BM). To ensure lifelong hematopoiesis, various molecular mechanisms are needed to maintain the HSC pool. CCCTC-binding factor (CTCF) is a DNA-binding, zinc-finger protein that regulates the expression of its target gene by organizing higher order chromatin structures. Currently, the role of CTCF in controlling HSC homeostasis is unknown. Using a tamoxifen-inducible CTCF conditional knockout mouse system, we aimed to determine whether CTCF regulates the homeostatic maintenance of HSCs. In adult mice, acute systemic CTCF ablation led to severe BM failure and the rapid shrinkage of multiple c-Kithi progenitor populations, including Sca-1+ HSCs. Similarly, hematopoietic system-confined CTCF depletion caused an acute loss of HSCs and highly increased mortality. Mixed BM chimeras reconstituted with supporting BM demonstrated that CTCF deficiency-mediated HSC depletion has both cell-extrinsic and cell-intrinsic effects. Although c-Kithi myeloid progenitor cell populations were severely reduced after ablating Ctcf, c-Kitint common lymphoid progenitors and their progenies were less affected by the lack of CTCF. Whole-transcriptome microarray and cell cycle analyses indicated that CTCF deficiency results in the enhanced expression of the cell cycle-promoting program, and that CTCF-depleted HSCs express higher levels of reactive oxygen species (ROS). Importantly, in vivo treatment with an antioxidant partially rescued c-Kithi cell populations and their quiescence. Altogether, our results suggest that CTCF is indispensable for maintaining adult HSC pools, likely by regulating ROS-dependent HSC quiescence.
Introduction
Hematopoiesis in the human body is primarily maintained by a complex differentiation program initiated in hematopoietic stem cells (HSCs).1 These cells undergo a tightly coordinated regimen of self-renewal and differentiation that is finely regulated by several molecular mechanisms, including (1) a specific set of transcription factors, such as RUNX1, GATA2, GFI1, and TAL1;1, 2, 3 (2) signaling pathways, such as the Wnt/β-catenin and Notch pathways;4, 5 and (3) bone marrow (BM) niches.6 In addition, several reports emphasize the critical roles of epigenetic and chromatin modifications in maintaining HSC homeostasis.7, 8, 9 DNA methyltransferases have been found to be important to HSC homeostasis and differentiation by downregulating myeloid progenitor-related factors, including GATA1, ID2 and CEBPα.10, 11, 12 The components of polycomb-repressive complexes, including BMI-1,13 RAE2814 and RING1B,15 as well as the histone H2A deubiquitinase MYSM1,16 have been shown to be critical in the maintenance of HSC function. Another study has also demonstrated that HSC function is controlled by the mediator component MED12, which regulates H3K27Ac at enhancers of key HSC genes.17 Further understanding how HSC homeostasis and function are maintained by other epigenetic factors could be important for developing new therapeutic strategies. Indeed, epigenetic changes have been implicated in the pathogenesis of myelodysplastic syndrome and acute myeloid leukemia.18
CCCTC-binding factor (CTCF) is a highly conserved DNA-binding protein that contains an 11-zinc-finger domain. CTCF shows a genome-wide distribution of DNA occupancy, and 30–60% of its binding is cell type specific.19 Although CTCF was first described as a transcription factor,20 and subsequently as a chromatin insulator,21 recent studies have revealed that CTCF functions to mediate long-range DNA interactions and to identify the borders of topologically associated domains that contribute to three-dimensional chromatin interactions.22, 23, 24 Topological remodeling of the genome by CTCF can affect the expression of cell differentiation-associated and function-associated genes. Interestingly, CTCF has been shown to play multiple roles in hematopoietic cell lineages, both in lymphoid and in myeloid cells.25, 26 Recently, we discovered that CTCF is required for maintaining the systemic dendritic cell (DC) pools and the self-renewal of epidermal Langerhans cells in a conditional knockout (cKO) system.27 Nevertheless, the precise role of CTCF in controlling HSC homeostasis remains unknown.
Here, we aimed to identify the homeostatic role of CTCF in maintaining adult HSCs in mice. We generated inducible CTCF-cKO mice and analyzed the HSC populations in combination with the BM chimera approach. The CTCF-dependent gene expression was assessed by microarray-based transcriptome analysis.
Materials and methods
Mice
Mice carrying a conditional Ctcf allele (Ctcfflox) were described in our previous study.27 The Ctcfflox mice were crossed with Rosa26-CreER mice28 (CreER; kindly provided by Weonsang Simon Ro, Institute of Gastroenterology, Yonsei University College of Medicine) to generate a tamoxifen (TMX) -inducible systemic Ctcf-cKO strain (CTCF-cKO). For Cre-mediated recombination, mice were intraperitoneally injected with tamoxifen (Sigma-Aldrich; St Louis, MO, USA) dissolved in corn oil (Sigma-Aldrich) for 4–5 consecutive days (2 mg day−1). Wild-type (WT) C57BL/6 (CD45.2) and B6.SJL (CD45.1) mice were purchased from The Jackson Laboratory (Bar Harbor, ME, USA). All animals were bred in specific pathogen-free facilities at Yonsei University College of Medicine. Age- and sex-matched CreER littermate mice were used as WT controls throughout the study. All animal studies were approved by the Department of Laboratory Animal Resources Committee of the Yonsei University College of Medicine.
Blood cell count
Whole blood was collected from the superficial facial vein of the animals into EDTA-treated tubes at specified time points. The total blood cell and differential counts were evaluated using a hematology analyzer (HEMAVET 950FS, Drew Scientific).
Bone marrow transplantation
A total of 5 × 106 BM cells from CreER and CTCF-cKO mice (CD45.2) were transplanted into lethally irradiated (9.5 Gy) B6.SJL mice (CD45.1) or vice versa. To generate mixed BM chimeras, 2.5 × 106 BM cells from either CreER or CTCF-cKO mice (CD45.2) were mixed with an equal number of BM cells from B6.SJL (CD45.1) mice and transferred into the irradiated B6.SJL (CD45.1) mice. All resulting chimeric mice were provided with water that contained antibiotics (sulfamethoxazole/trimethoprim) for the first two weeks. Six to eight weeks after transplantation, Cre-mediated recombination was induced with daily injections of tamoxifen. Thereafter, the BM and spleen cells were harvested from the reconstituted mice and analyzed.
Antibodies and flow cytometry
Total BM cells were flushed and harvested from both hind legs. Single-cell suspensions from the spleen were obtained by enzymatic digestion as previously described.27 Fluorochrome-conjugated anti-mouse B220 (RA3-6B2), CD3 (145-2C11), CD4 (RM4-5), CD8 (53-6.7), CD11b (M1/70), CD11c (N418), CD19 (eBio1D3), CD16/32 (93), CD34 (RAM34), CD45.1 (A20), CD45.2 (104), CD48 (HM48-1), CD117/c-Kit (2B8), CD135/Flt3 (A2F10), CD150 (TC15-12F12.2), F4/80 (BM8), CD127/IL-7Rα (A7R34), Ki-67 (SolA15), Ly-6G (RB6-8C5), Sca-1 (D7) and Siglec-H (eBio440C) antibodies were used for surface immunophenotyping (eBioscience; San Diego, CA, USA). Lineage-positive cells in the BM were excluded using the following biotinylated antibodies, followed by treatment with APC-Cy7-conjugated streptavidin: B220, CD3, CD4, CD8, CD11b, CD11c, CD19, Gr-1, NK1.1 (PK136) and Ter119 (TER-119) (all from eBioscience). Cell death and apoptosis were analyzed using an Annexin V/propidium iodide (PI) staining kit (eBioscience). Stained cells were acquired using a FACSVerse or LSRFortessa flow cytometer (BD Biosciences, San Jose, CA, USA). All flow cytometry data were analyzed with the FlowJo software (Treestar, Ashland, OR, USA).
Cell cycle assays
Complete RPMI medium (HyClone, Logan, UT, USA) that was supplemented with 10% heat-inactivated fetal bovine serum (HyClone), 50 μM of 2-mercaptoethanol, 2 mM of L-glutamine, 100 units per ml of penicillin, and 100 μg ml−1 of streptomycin was used for the cell culture. The cells were incubated in complete RPMI containing 10 μg ml−1 of Hoechst 33342 (Molecular Probes, Eugene, OR, USA) for 45 min at 37 °C. After surface antigen staining, the cells were fixed and permeabilized, and subsequent nuclear staining of Ki-67 was performed. The cell cycle was analyzed by flow cytometer after gating out doublet cells.
Measurement of reactive oxygen species
Total BM cells were incubated with 1 μM of 5-(and-6)-carboxy-2′,7′-dichlorofluorescein diacetate (carboxy-DCFDA; Molecular Probes) diluted in phosphate-buffered saline for 30 min at 37 °C in the dark. After extensive washing with phosphate-buffered saline, the cells were stained for lineage and HSC markers at 4 °C and assayed by flow cytometry.
Colony-forming unit assay
Colony-forming units were assessed using a Methocult assay (M03434, Stem Cell Technologies; Vancouver, BC, Canada) according to the manufacturer’s instructions. The number of rising colonies was counted at specified time points. In brief, 2 × 104 total BM cells were plated in methylcellulose medium containing recombinant mouse stem cell factor (SCF), IL-3, IL-6 and erythropoietin and cultured at 37 °C in a 5% CO2 incubator for 10 to 14 days without changing the medium.
In vivo N-acetyl-L-cysteine treatment
Daily, N-acetyl-L-cysteine (NAC; Sigma) was intraperitoneally injected into WT and CTCF-cKO mice for 8 consecutive days at a dose of 100 mg per kg of body weight during and after tamoxifen-induced Ctcf genetic recombination.
Microarray
One day after the last tamoxifen treatment, BM single-cell suspensions were prepared, and the LSKs were sorted using a FACSAria II cell sorter (BD Biosciences) at the Flow Cytometry Core Lab in the Avison Biomedical Research Center (Yonsei University College of Medicine). Sorted LSKs were immediately collected in TRIzol (Invitrogen, Carlsbad, CA, USA), and the total RNA was extracted using the isopropanol precipitation method. Sample preparation and microarray data analyses were performed as described previously.27 The accession number for the data reported in this paper is GEO: GSE88995.
Real-time quantitative polymerase chain reaction
Total RNA from purified cells was isolated using the Hybrid-R Total RNA kit (GeneAll Biotechnology, Seoul, Korea) as described in our previous study.27 cDNA was synthesized using PrimeScript RT Master Mix (Takara Bio, Shiga, Japan). Quantitative real-time PCR was performed using the ABI StepOnePlus Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) by monitoring the synthesis of double-stranded DNA during various PCR cycles using SYBR Green (Takara Bio). For each sample, duplicate test reactions were analyzed for the expression of the gene of interest, and the results were normalized using the level of Gapdh mRNA. Primer sequences are listed in Supplementary Table 1.
Statistical analysis
The data were analyzed with unpaired Student’s two-tailed t-tests, unless otherwise stated, using Prism software (GraphPad Software Inc.; San Diego, CA, USA). Blood cell counts within the same mouse before and after tamoxifen treatment were compared using paired t-tests. All P-values <0.05 were considered statistically significant.
Results
Loss of CTCF leads to rapid BM failure
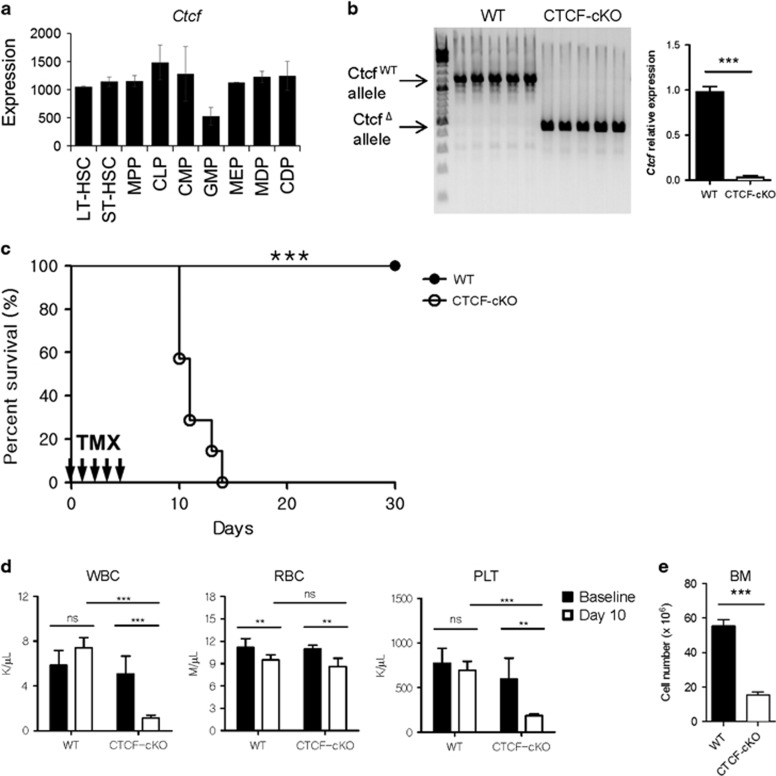
To examine the relative expression levels of Ctcf mRNA in primitive hematopoietic stem/progenitor cells, we analyzed the Immunological Genome microarray database, which includes data on a large number of immune cells and their progenitor cell populations.29 Ctcf mRNA was expressed in all the subsets of HSCs and progenitor cells examined (Figure 1a). To determine whether CTCF is required for general hematopoiesis in vivo, we used the Cre-loxP system to knockout the Ctcf gene with injections of TMX in adult mice. In CTCF-cKO mice, Ctcf ablation was achieved with intraperitoneal injections of TMX after 5 consecutive days (Figure 1b). Within 10 to 14 days after TMX treatment, all CTCF-cKO mice had died, while the survival rate of the WT mice was not affected (Figure 1c). Next, we analyzed the peripheral blood cell counts in these mice to evaluate the effect of CTCF loss on overall hematopoiesis. Although both mouse genotypes showed normal white blood cell and platelet counts before CTCF deletion, TMX treatment led to a significant reduction in these hematological parameters in CTCF-cKO mice (Figure 1d and Supplementary Figure 1). Interestingly, red blood cell (RBC) counts decreased slightly in both WT and CTCF-cKO groups upon treatment with TMX, although the difference therein was not statistically significant, suggesting that TMX alone could have a possible adverse effect on erythropoiesis. Importantly, CTCF-ablated mice exhibited markedly lower total BM cellularity than the WT control mice (Figure 1e). These data indicate that CTCF is critical to hematopoiesis and that the acute loss of CTCF results in rapid hematopoietic failure.
Figure 1.
Acute systemic CTCF ablation leads to hematopoietic failure. (a) The relative expression levels of Ctcf among various stem/progenitor cells (data from ImmGen). (b) WT and CTCF-cKO mice were treated with TMX for 5 consecutive days, and genomic DNA and total RNA from the BM were isolated at day 8. Efficient Ctcf deletion by TMX treatment in CTCF-cKO mice at the DNA (left) and the RNA (right) levels is shown. (c) Kaplan–Meier curves plotting the survival of WT and CTCF-cKO mice (n=10) are shown. (d) Peripheral blood counts of control and CTCF-cKO mice at baseline and 10 days after the first TMX injection (n=4−5). (e) Bone marrow counts (femurs and tibiae) of WT and CTCF-cKO mice 8 days after TMX treatment (n=5). Data are from at least three independent experiments with four or five mice per group. Error bars indicate the standard error of the mean (s.e.m.). **P<0.01; ***P<0.001; ns, not significant.
Acute depletion of CTCF results in the rapid shrinkage of c-Kithi HSC and progenitor cell pools
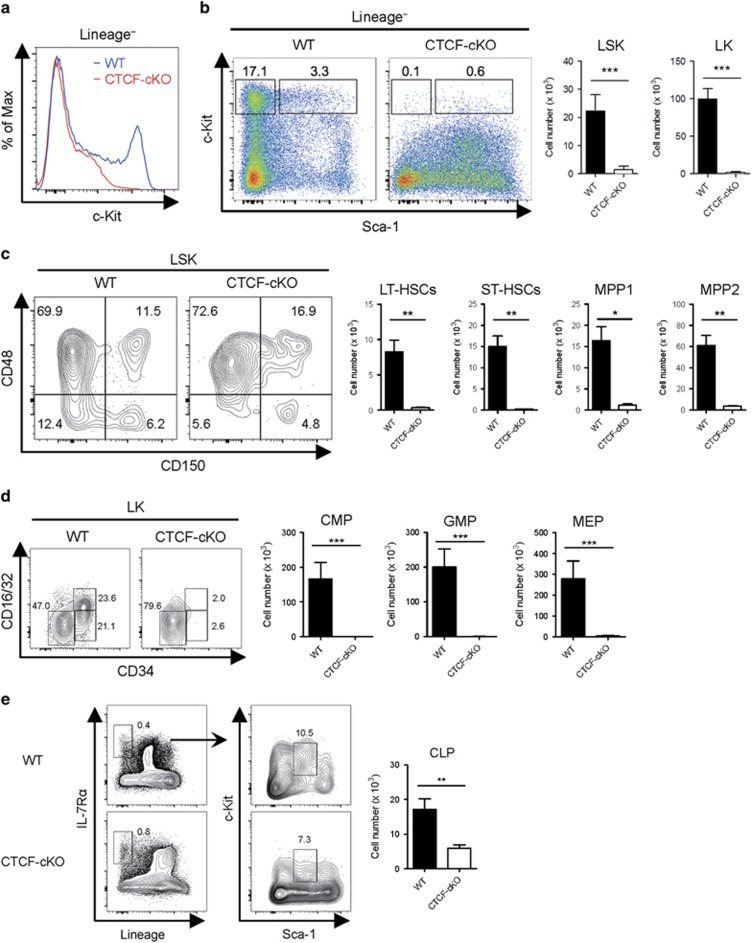
As systemic depletion of CTCF led to rapid BM failure, we next examined heterogeneous HSC populations by flow cytometry. CD117/c-Kit is a receptor for SCF that is highly expressed on most primitive stem/progenitor cells in the BM.30, 31 Surprisingly, within 8 days after CTCF ablation, we found a marked reduction in Lineage− c-Kithi cells from the BM (Figure 2a). Lineage− BM cells from CTCF-ablated mice also exhibited lower expression of CD135/Flt3 and CD34 and increased expression of Sca-1 and CD16/32 compared with cells from WT mice (Supplementary Figure 2). Lineage− c-Kithi cell populations can be divided into Lineage− Sca-1+c-Kithi (LSK) and Lineage− Sca-1−c-Kithi (LK) subsets, which are HSC-enriched and early myeloid progenitor-enriched compartments, respectively. In line with the observed acute hematopoietic failure, we noted a marked decrease in both the LSK and the LK populations after CTCF ablation (Figure 2b). As HSCs reflect a hierarchy of stem cell differentiation,1 we further assessed HSCs from the LSK compartment using different surface markers (Supplementary Figure 3). The most primitive, long-term reconstituting HSCs (LT-HSCs), which are distinguishable from other LSK cells by CD150+CD48− surface characteristics, were profoundly diminished in TMX-treated BM from CTCF-cKO mice (Figure 2c). In addition, we also found fewer short-term HSCs and other primitive multipotent progenitors upon CTCF deletion compared with WT controls. In the myeloid progenitor compartment, common-myeloid progenitors (CMPs), granulocyte-macrophage progenitors and megakaryocyte-erythroid progenitors (MEPs) were nearly absent from CTCF-cKO BM (Figure 2d). However, the number of common lymphoid progenitors (CLPs) was fairly preserved after CTCF depletion compared with those of HSCs and myeloid progenitors (Figure 2e). Accordingly, the population of splenic myeloid cells, which are directly derived from BM myeloid progenitors, was markedly reduced in CTCF-cKO mice; however, the numbers of lymphoid T and B cells were not affected (Supplementary Figure 4). These results suggest that CTCF is required for the homeostatic maintenance of c-Kithi HSCs and early progenitors in vivo.
Figure 2.
Acute systemic depletion of CTCF results in severe decreases in c-Kithi stem/progenitor cell numbers. (a, b) The surface levels of c-Kit in lineage− cells (a) and the frequencies and absolute numbers of LSK (lineage−Sca-1+c-Kithi) and LK (lineage−Sca-1−c-Kithi) cells (b) from WT and CTCF-cKO mice at 8 days after TMX injection. (c) The frequencies and absolute number of LT-HSCs (CD150+CD48−), ST-HSCs (CD150−CD48−), MPP1 (CD150+CD48+) and MPP2 (CD150−CD48+) populations among LSKs are shown. (d) FACS analysis with cell numbers of CMP (CD34+CD16/32−), GMP (CD34+CD16/32+) and MEP (CD34−CD16/32−) among the LK population are shown. (e) FACS analysis and quantitation of CLP. Data are from at least three independent experiments with three or four mice per group. Error bars indicate the s.e.m. *P<0.05; **P<0.01; ***P<0.001. FACS, fluorescence-activated cell sorting; GMP, granulocyte-macrophage progenitors; ST-HSC, short-term HSC.
Loss of CTCF in the hematopoietic compartment results in the rapid exhaustion of HSCs
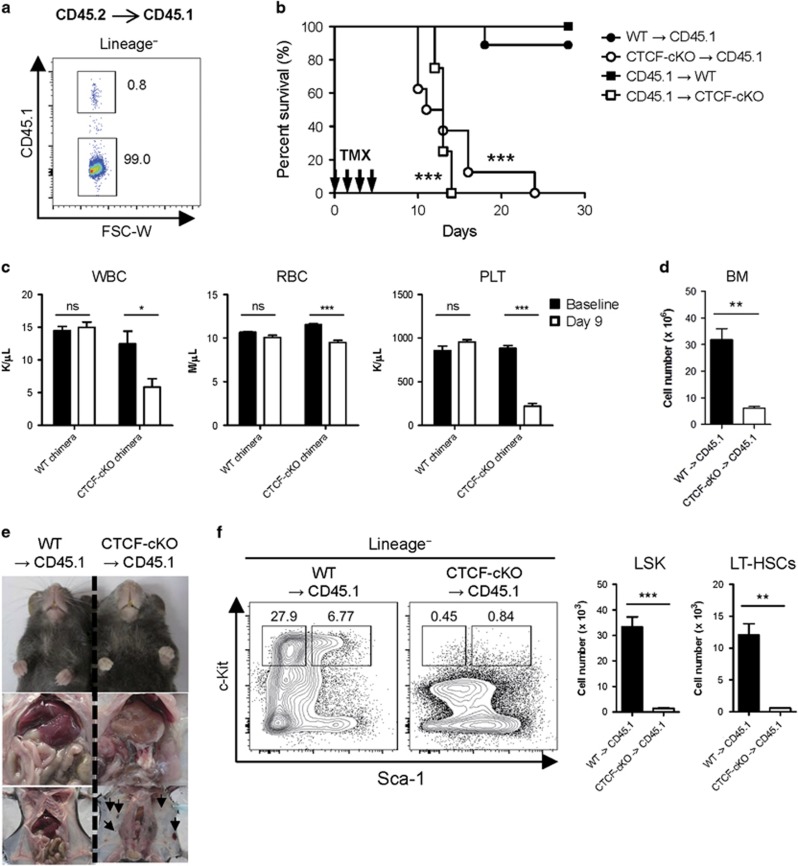
As TMX-inducible Cre recombinase is ubiquitously expressed in the tissues of ROSA26-CreER mice, the observed hematopoietic failure and rapid lethality after inducing CTCF depletion in the CTCF-cKO mice may have resulted from the dysfunction of cells from the non-hematopoietic compartment. To address this issue, we performed BM chimera experiments. BM chimerism was consistently achieved at a rate of ~98% (Figure 3a). TMX treatment of both CTCF-cKO BM-transplanted CD45.1 (CTCF-cKO→CD45.1) and CD45.1 BM-transplanted CTCF-cKO (CD45.1→CTCF-cKO) chimeras showed similar decreases in survival compared with other chimeras (WT→CD45.1 or CD45.1→WT), suggesting that CTCF ablation in both radio-sensitive and radio-resistant compartments led to rapid lethality (Figure 3b). Peripheral blood and BM analysis revealed similar hematopoietic failure in TMX-treated CTCF-cKO BM-transplanted CD45.1 chimeras, as seen in the systemic CTCF-deficient mice (Figures 3c and d). Consistent with the decreased numbers of RBCs and platelets in blood, CTCF-cKO BM chimeras exhibited severe anemic and hemorrhagic clinical features (Figure 3e). Furthermore, TMX treatment led to rapid losses of c-Kithi HSC/myeloid progenitor cells in BM-specific CTCF-null chimeras (Figure 3f). These results indicate that the ablation of CTCF in the hematopoietic compartment is sufficient to induce the rapid exhaustion of HSCs, resulting in hematopoietic failure and mortality.
Figure 3.
CTCF loss in the hematopoietic system leads to severe BM failure and consequent lethality. (a) The efficiency of chimerism was analyzed in lineage-negative BM cells from CD45.1+ recipients transplanted with CD45.2+ donor BM. (b) Kaplan–Meier curves plotting the survival of single chimeric mice (n=5–9). Statistical analyses were performed between (1) WT→CD45.1 and CTCF-cKO→CD45.1 and (2) CD45.1→WT and CD45.1→CTCF-cKO. (c) Peripheral blood counts of control and CTCF-cKO→CD45.1 chimeric mice at 9 days after the first TMX injection (n=5). (d) Total BM cellularity for each chimera is shown. (e) Clinical pictures of WT and CTCF-cKO chimeras showing anemic (upper and middle) and spontaneous hemorrhagic (lower) propensities. (f) The frequencies and absolute numbers of LSK and LT-HSCs from WT and CTCF-cKO chimeras at 8 days after TMX injection. Data are from at least two independent experiments with three or four mice per group. Error bars indicate the s.e.m. *P<0.05; **P<0.01; ***P<0.001; ns, not significant.
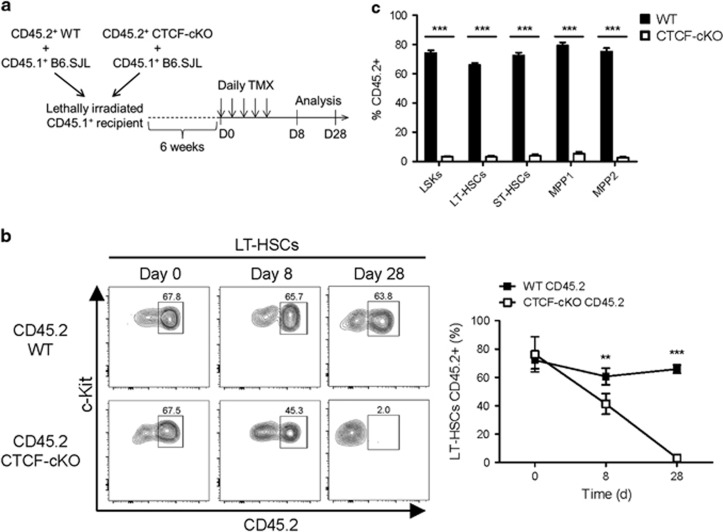
HSC homeostasis is finely regulated by heterogeneous cross-talk between HSCs and the surrounding stromal components within the BM niche.6 As CTCF ablation in CTCF-cKO mice also occurred in BM stromal cells, CTCF deficiency-mediated HSC exhaustion and the accompanied BM failure might have resulted from HSC-extrinsic effects. To test this possibility, we transplanted CD45.2+ WT or CD45.2+ CTCF-cKO BM cells together with CD45.1+-supporting BM cells at a 1:1 ratio into lethally irradiated CD45.1+ recipient mice. Six weeks after reconstitution, the mixed BM chimeras were injected with TMX to induce CTCF depletion, and the BM cells were analyzed up to 4 weeks after treatment (Figure 4a). In contrast to the systemic and BM-confined CTCF-depleting experiments, we found a moderate reduction in the number of LT-HSCs derived from CTCF-null donors during the acute phase (day 8), which eventually disappeared (day 28) (Figures 4b and c). These results imply that the genetic loss of CTCF results in HSC exhaustion in both cell-extrinsic and cell-intrinsic manners.
Figure 4.
CTCF deficiency-mediated HSC depletion has both cell-non-autonomous and cell-autonomous effects. (a) The experimental scheme of the generation and the use of mixed BM chimeras. (b) The frequencies of CD45.2+ BM-derived LT-HSCs from the TMX-treated mixed chimeric mice. (c) Multiple early progenitor cells from the TMX-treated mixed chimeric mice at day 28 are shown. Data are from two or three independent experiments with four or five mice per group. Error bars indicate the s.e.m. ***P<0.001.
CTCF-deficient HSCs show defective myeloid lineage development
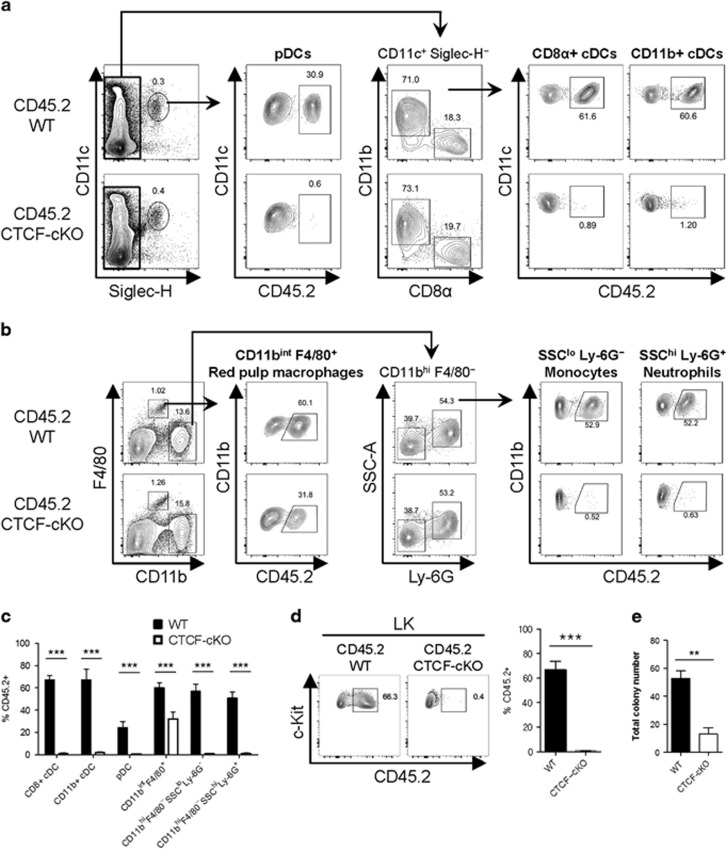
As CTCF depletion led to a marked reduction in the number of c-Kithi stem/progenitor cells in BM, we next aimed to investigate the development of HSC-driven early myeloid progenitors and the subsequent peripheral myeloid cells in the mixed chimera system. In the spleen, all subsets of DCs derived from CTCF-deficient BM, including plasmacytoid (Siglec-H+CD11c+), conventional CD8α+ (Siglec-H−CD11chi CD8α+CD11b−) and CD11b+ (Siglec-H−CD11chiCD8α−CD11b+) DCs, were markedly reduced compared with those from WT BM (Figures 5a and c). Splenic monocytes (CD11bhiF4/80−SSCloLy-6G−) and neutrophils (CD11bhiF4/80−SSChiLy-6G+), which develop from the HSC-myeloid progenitor axis, also showed a marked reduction in the absence of CTCF (Figures 5b and c). Meanwhile, however, F4/80+ red pulp macrophages in the spleen (CD11bintF4/80+), which are maintained by self-renewal without substantial monocyte input, only partially declined after inducing CTCF depletion, confirming their distinct homeostasis in the steady-state (Figures 5b and c).32, 33 Consistent with the reductions in the differentiated myeloid compartment after CTCF deletion, myeloid progenitor LK cells in the BM from CTCF-deficient donors were scarce (Figure 5d). To delineate the role of CTCF in the colony-forming capabilities of stem and myeloid progenitor cells, we performed in vitro colony-forming unit assays. Total untouched BM cells from either WT or CTCF-cKO mice were plated in methylcellulose medium supplemented with SCF, IL-3, IL-6, and erythropoietin, followed by ex vivo TMX treatment. CTCF-cKO BM cells treated with TMX showed marked decreases in colony-forming units compared with WT BM cells (Figure 5e). These data indicate that CTCF is critical in HSC-driven myeloid lineage development both in vivo and ex vivo.
Figure 5.
The loss of CTCF results in the exhaustion of myeloid progenitors and their differentiation. (a and b) Gating strategies for dendritic cells (a), red pulp macrophages, monocytes, and neutrophils (b) in the spleen at day 28 are shown. (c) The frequencies of CD45.2+ BM-derived myeloid lineage cells from the spleen of TMX-injected mixed chimeric mice are shown. (d) The proportion of CD45.2+ BM-derived LK cells is shown. (e) The number of total colonies from WT and CTCF-cKO mice cultured in methylcellulose medium supplemented with myeloid-driving factors over 9 days. Data are from at least three independent experiments with four or five mice per group. Error bars indicate the s.e.m. **P<0.01; ***P<0.001.
CLPs and lymphoid lineage cells are less sensitive to CTCF ablation
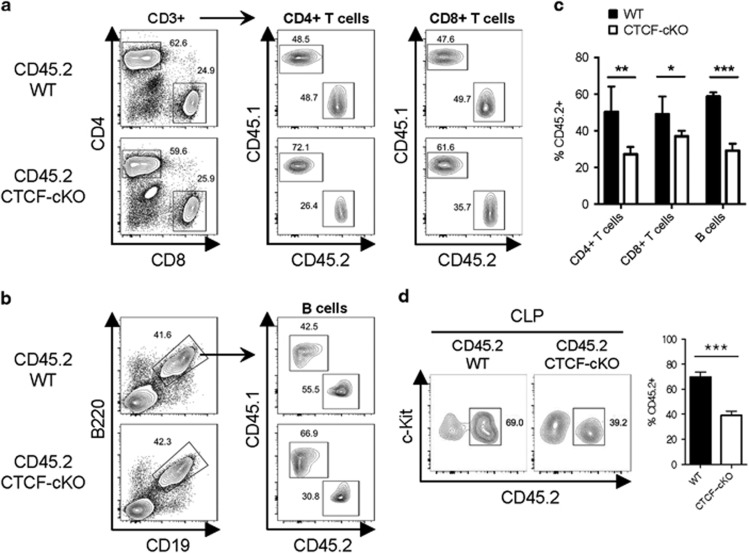
We next examined the role of CTCF in lymphoid lineage development. Similar to TMX-treated systemic CTCF-deficient mice, CTCF-ablated BM cells in the mixed chimeras gave rise to significant amounts of both CD4+ and CD8+ T cells in the spleen, unlike those in the myeloid lineage experiments (Figures 6a and c). B lymphocyte development was also relatively preserved, as the frequency of B cells from the CTCF-deficient BM was only reduced to approximately half that of the WT CD45.2+ BM-derived B cells (Figures 6b and c). Furthermore, we observed a discrete population of CTCF-deficient CLPs from CTCF-ablated BM in the mixed chimeras similar in number to T and B lymphocytes (Figure 6d). Therefore, these results suggest that lymphoid lineage development is only partially affected by CTCF depletion in vivo.
Figure 6.
CLP-derived lymphoid differentiation is less sensitive to CTCF depletion. (a and b) Gating strategies for T cells (a) and B cells (b) in the spleen at day 28 are shown. (c) The frequencies of CD45.2+ BM-derived lymphoid lineage cells from the spleens of TMX-treated mixed chimeric mice are shown. (d) The proportion of CD45.2+ BM-derived CLPs is shown. Data are from at least three independent experiments with four or five mice per group. Error bars indicate the s.e.m. *P<0.05; **P<0.01; ***P<0.001.
CTCF deficiency results in defective HSC quiescence
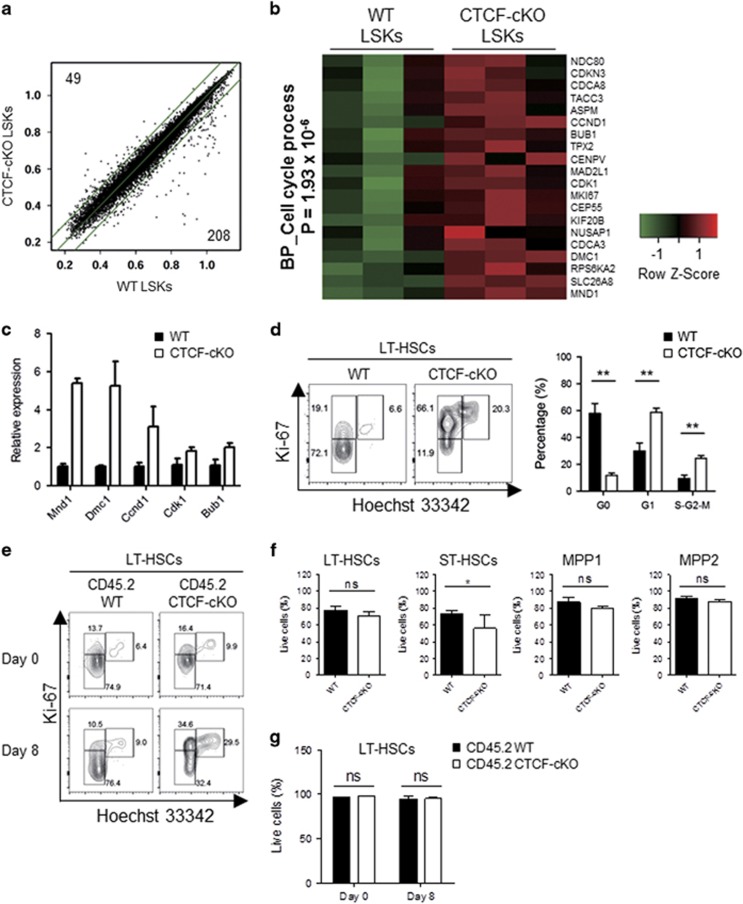
To identify genes regulated by CTCF in hematopoietic stem/progenitor cells, we performed microarray-based gene expression analyses. LSK cells were sorted by flow cytometry after three treatments with TMX and subjected to microarray experiments before the LSKs went missing (Supplementary Figure 5a). Acute loss of CTCF led to rapid changes in the gene expression profiles between WT and CTCF-deficient LSKs (Figure 7a). Subsequent systematic gene ontology analysis revealed that CTCF-depleted LSKs express high levels of cell cycle-promoting genes (Figure 7b), the results of which were confirmed by quantitative PCR (Figure 7c). In the steady state, the maintenance of HSC quiescence is critical for regulating stem cell pools. To determine whether HSC quiescence is affected by CTCF deletion, we performed a Ki-67/Hoechst-based cell cycle assay. Notably, CTCF-deficient LSKs showed significant increases in the proportion of cells in the G1 and the S-G2-M phases (Supplementary Figure 5b). Among LT-HSCs, CTCF deficiency led to a marked reduction in the number of cells in the G0 phase and increased the number of cells in the G1 and the S-G2-M phases, reflecting a quiescence defect (Figure 7d). Furthermore, cell cycle analyses performed in the mixed chimera system revealed a rapid induction of quiescence defects in LT-HSCs by CTCF ablation (Figure 7e). However, CTCF-depleted HSCs, multiple progenitors and differentiated lineage+ cells demonstrated similar levels of cell death compared with that of WT cells, regardless of supporting BM (Figures 7f and g and Supplementary Figure 6). Collectively, these data demonstrate that CTCF is critically involved in maintaining HSC quiescence in vivo.
Figure 7.
CTCF deficiency leads to an augmented cell cycle progression in HSCs. (a) LSKs were sorted 24 h after the last TMX treatment. Scatter plots show the raw expression levels (log2), and the number of genes that exhibited greater than two-fold increases in WT (lower) and CTCF-cKO (upper) LSKs with a P-value of less than 0.05 is indicated. Outer green lines indicate the two-fold boundary. (b) Heat map showing the genes that belong to the cell cycle biological process as revealed by the Gene Ontology analysis. (c) The expression levels of the selected genes from (b) were confirmed by real-time quantitative PCR. (d) Ki-67/Hoechst-based cell cycle analysis of LT-HSCs from WT and CTCF-cKO mice at 8 days after TMX treatment. (e) Cell cycle assay of LT-HSCs from the mixed BM chimeric mice at days 0 and 8 after initiating TMX treatment. (f and g) Apoptosis profiles of multiple HSCs in non-chimeric (f) and mixed BM chimeric mice (g) 8 days after TMX treatment. Data are from at least two or three independent experiments with four or five mice per group. Error bars indicate the s.e.m. *P<0.05; **P<0.01.
CTCF maintains quiescence of HSCs by downregulating ROS levels
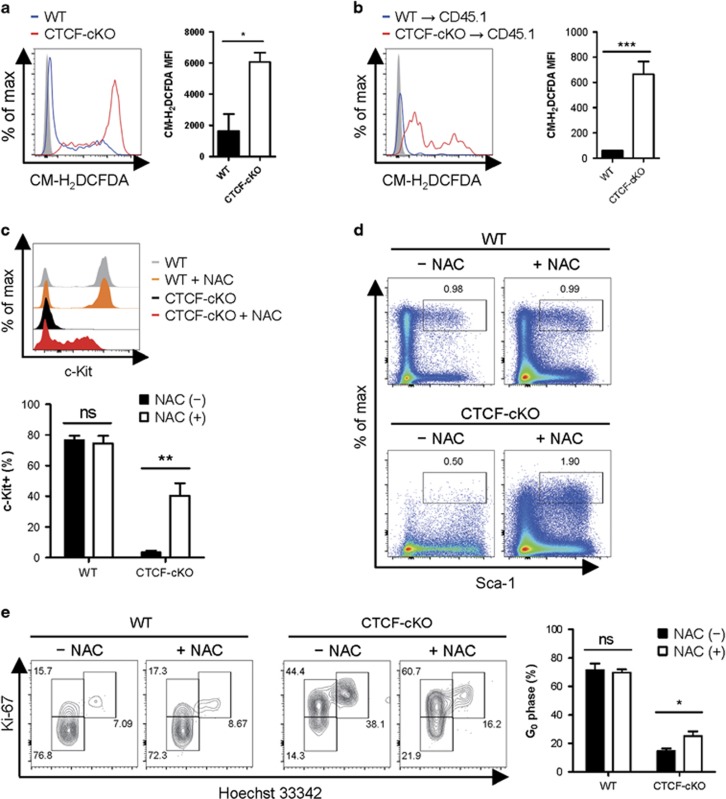
Maintaining a low level of reactive oxygen species (ROS) is essential to retaining HSC quiescence.34 HSCs maintain a low intracellular level of ROS by adapting metabolic shift to enhance glycolysis, which is associated with reduced mitochondrial oxidation and ROS generation.35, 36, 37 From our microarray results, we noted that the levels of several ROS-regulating genes, such as TNF, NCF1, AKT1 and SIRT5, were altered in CTCF-depleted LSKs (Supplementary Table 2). In addition, CTCF-deficient LSKs had a higher level of the gene expression of gluconeogenic enzyme fructose-1,6-bisphosphatase 1 (Fbp1), which may suppress glycolysis in HSCs (Supplementary Figure 7).38 Thus, we examined intracellular ROS production using the general oxidative stress indicator CM-H2DCFDA. CTCF-depleted HSCs displayed marked increases in ROS levels in both the acute systemic and the BM-confined CTCF ablation settings (Figures 8a and b). To test whether increased ROS levels confer a severe loss of c-Kithi stem/progenitor cells, we treated WT and CTCF-cKO mice with the antioxidant NAC. Importantly, in vivo NAC treatment significantly recovered the depleted c-Kit+ cell populations (Figure 8c) and the expression levels of c-Kit surface protein (Supplementary Figure 8a) on the Sca-1+CD150+CD48− cells from CTCF-cKO mice. In addition, treatment with NAC partially restored the c-Kithi LSK population and its subset divisions (Figure 8d and Supplementary Figure 8b). Further cell cycle analysis revealed that NAC treatment caused a slight increase in the number of cells in the quiescent G0 phase from the CTCF-deficient LT-HSC compartment, suggesting that the increases in ROS were responsible for the defect in CTCF ablation-mediated HSC quiescence (Figure 8e). Altogether, these results indicate that CTCF maintains the c-Kithi HSC pools and quiescence, partially in a ROS-dependent manner.
Figure 8.
CTCF maintains the c-Kithi HSC pool and quiescence in a ROS-dependent manner. (a and b) Intracellular ROS levels were measured using CM-H2DCFDA from TMX-treated straight cKO mice (a) and chimeric mice (b) at 8 days after starting TMX treatment. The MFI of the ROS levels was calculated. (c) The percentage of the c-Kit+ population in the Lin−Sca-1+CD150+CD48− cell population in the indicated experimental group is shown. (d) The representative flow cytometry plots of the lineage− population in WT and CTCF-cKO mice at 8 days after inducing CTCF depletion by TMX with or without NAC treatment. (e) Cell cycle analysis of LT-HSCs from WT and CTCF-cKO mice treated with the same conditions as in d. Data are from two independent experiments with three to five mice per group. Error bars indicate the s.e.m. *P<0.05; **P<0.01; ***P<0.001. MFI, mean fluorescence intensity; ns, not significant.
Discussion
Hematopoietic differentiation is a multistep process that is tightly controlled by complex molecular and cellular networks. Although CTCF has been shown to regulate the cellular development and differentiation of human erythroid cells,39 murine thymocytes,25 mature T cells,25, 40 B cells41 and myeloid lineage cells, including DCs,26, 27, 42 whether CTCF controls the homeostasis of adult HSCs has been unknown. In the present study, we generated a TMX-inducible CTCF-cKO mouse system to conditionally ablate CTCF from the adult hematopoietic system, as CTCF-null embryos experience prenatal lethality.43
The combined genetic and BM transplantation approaches used in our study demonstrated that acute systemic and hematopoietic-specific ablation of CTCF leads to rapid hematopoietic failure. Although peripheral blood analyses showed a minor effect on RBC concentration, we noted severe anemic clinical features in CTCF-cKO mice. As CTCF is required for erythroid differentiation44 and for the chromatin dynamics of globin genes,22 peripheral RBCs that are found in CTCF-cKO mice may represent functionally immature erythroid cells. Therefore, deregulated RBC differentiation in CTCF-cKO mice may cause lethality.
Notably, the ablation of CTCF in mice resulted in severe decreases in the heterogeneous c-Kithi stem/progenitor cell populations, including the most primitive LT-HSCs. Subsequent co-transplantation with supporting BM was able to delay the decreases in LT-HSCs, suggesting that the exhaustion of HSCs by CTCF depletion has both cell-extrinsic and cell-intrinsic effects. To the best of our knowledge, these results are the first to demonstrate the necessity of CTCF in the maintenance of HSC pools in vivo. c-Kit is the surface receptor of SCF, a cytokine that is essential for HSC self-renewal, growth, and survival.45 Interestingly, a recent study reported that MED12, a component of the Mediator complex, is required for the transcriptional regulation of c-Kit in HSCs.17 As our CTCF-ablated mice showed reductions in c-Kithi HSCs, assessing whether CTCF directly regulates c-Kit transcript expression in our model in association with mediator and/or chromatin modifiers could prove to be of value.
The delayed disappearance of CTCF-deficient HSCs in the mixed chimeras implicated a possible role for CTCF in BM niche-supporting cells for maintaining HSC homeostasis. HSC niches are comprised of heterogeneous cell populations, such as osteoblasts, CXCL12-abundant reticular cells, lepr+ perivascular cells, endothelial cells, and macrophages.6 However, how CTCF regulates the HSC niche has not yet been understood. Recently, a population of DARC+ macrophages has been shown to be crucial for maintaining LT-HSC quiescence.46 As multiple populations of tissue macrophages were moderately decreased by conditional CTCF deletion using the LysM-Cre system,47 it remains to be examined whether the pool and function of DARC+ macrophages in the BM are also regulated by CTCF, which may ultimately lead to non-autonomous HSC exhaustion.
Through intermediate multipotent progenitor cells, HSCs continuously give rise to both myeloid and lymphoid progenitor cells.1, 2 Interestingly, our mixed chimera experiments demonstrated that CTCF-deficient HSCs fail to give rise to myeloid lineage cells, while lymphoid lineage development was relatively preserved. A previous study using CTCF-silenced common-myeloid progenitors (CMPs) showed that CTCF knockdown increases the rate of CMP differentiation in vitro.26 As CMPs arise directly from HSCs and there is a lack of further CMP input from HSCs in the in vitro system, our data suggest that CTCF is required in order to sustain myeloid lineage development in vivo. For defective myeloid differentiation, we assume that the CTCF-mediated chromatin dynamics of certain loci, such as Irf8 and Sfpi1, may be pivotal for ensuring HSCs differentiate into early myeloid lineage cells.48 In lymphoid lineage development, CTCF has been shown to mediate the silencing of the B cell lymphoma oncogene BCL6, and CTCF haploinsufficiency leads to multiple lymphomas in vivo, which may be associated with the longer survival and maintenance of CTCF-deficient lymphoid lineage cells in our study.49, 50 Future work will be needed to clarify the possible role of CTCF in lineage commitment.
In the absence of specific mitogenic stimuli, HSCs remain in a quiescent or a dormant state.51 The microarray analyses conducted in the current study revealed that CTCF-deficient hematopoietic stem/progenitor cells show defective quiescence and striking elevations in cell cycle-promoting gene expression. However, CTCF has been shown to be associated with cell cycle progression and cell cycle arrest in a cell type-dependent manner. CTCF promotes cell cycle progression and the proliferation of αβ T cells in the thymus,25 epidermal Langerhans cells,27 and human embryonic stem cells,52 while it negatively regulates the cell cycle in several types of tumor cells.53, 54, 55 As our transcriptome analysis was conducted using LSK cells before a loss of surface c-Kit expression, any increase in the cell cycle program would have been, in part, directly regulated by CTCF deficiency at the transcription level.
Under the hypoxic conditions of the BM, low levels of intracellular ROS are critical to maintaining HSC quiescence.34, 56 According to the intracellular ROS level, HSCs can be divided into ROShigh and ROSlow fractions, and ROShigh HSCs have a higher myeloid differentiation capacity, indicating a positive correlation between ROS level and myeloid differentiation status.57, 58, 59, 60, 61 In the present study, CTCF-ablated HSCs showed marked elevations in their ROS levels that coincided with considerable decreases in the number of cells in the quiescence phase. The in vivo administration of the antioxidant NAC recovered the c-Kit+ population and defective quiescence in HSCs, implying that CTCF-mediated ROS regulation is crucial to HSC maintenance. Our microarray data supported the idea that CTCF transcriptionally regulates several ROS-associated genes in HSCs. An increase in the level of inflammatory products, including TNF and IFN-γ, has been implicated in inducing HSC proliferation and exhaustion.62, 63 Interestingly, we found that Fbp1 was highly elevated in CTCF-depleted LSKs. A previous study showed that cancer stem cells exhibited epigenetically silenced Fbp1 expression to maintain their active glycolysis and lower ROS generation status, which are metabolically stem cell-favorable environments.38, 64 Thus, we speculate that CTCF is somehow involved in the regulatory process of oxidative stress in HSCs by promoting glycolysis through silencing Fbp1 expression. Consistent with our findings, a recent study found that, in collaboration with Cockayne syndrome group B protein, CTCF protects cells from oxidative stress.65 Accordingly, we suggest that CTCF may have beneficial antioxidative activity in certain type of cells, including HSCs, by supporting metabolic glycolysis. Further studies will be needed to identify the CTCF-mediated metabolic characteristics of HSCs.
In summary, we have elucidated a role for CTCF in HSC homeostasis using genetic approaches. CTCF supports the homeostatic maintenance of HSC pools by sustaining HSC quiescence in a ROS-dependent manner. Further study is required to translate our findings into clinical applications to improve the outcomes of HSC transplantation or anti-leukemic therapies through modulating CTCF levels by RNA interference or genome editing of specific CTCF-binding sites in vivo.66, 67
Acknowledgments
This work was supported by grants from the National Research Foundation of Korea (NRF) funded by the Korean government (MSIP) (NRF-2011-0030086, 2012M3A9B4028272 and 2016R1A2B4014183 awarded to H-PK).
Footnotes
Supplementary Information accompanies the paper on Experimental & Molecular Medicine website (http://www.nature.com/emm)
The authors declare no conflict of interest.
Supplementary Material
References
- Orkin SH, Zon LI. Hematopoiesis: an evolving paradigm for stem cell biology. Cell 2008; 132: 631–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson A, Trumpp A. Bone-marrow haematopoietic-stem-cell niches. Nat Rev Immunol 2006; 6: 93–106. [DOI] [PubMed] [Google Scholar]
- Wilson NK, Foster SD, Wang X, Knezevic K, Schutte J, Kaimakis P et al. Combinatorial transcriptional control in blood stem/progenitor cells: genome-wide analysis of ten major transcriptional regulators. Cell Stem Cell 2010; 7: 532–544. [DOI] [PubMed] [Google Scholar]
- Bigas A, Espinosa L. Hematopoietic stem cells: to be or Notch to be. Blood 2012; 119: 3226–3235. [DOI] [PubMed] [Google Scholar]
- Bigas A, Guiu J, Gama-Norton L. Notch and Wnt signaling in the emergence of hematopoietic stem cells. Blood Cells Mol Dis 2013; 51: 264–270. [DOI] [PubMed] [Google Scholar]
- Mendelson A, Frenette PS. Hematopoietic stem cell niche maintenance during homeostasis and regeneration. Nat Med 2014; 20: 833–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebel VI, Kung AL, Tanner EA, Yang H, Bronson RT, Livingston DM. Distinct roles for CREB-binding protein and p300 in hematopoietic stem cell self-renewal. Proc Natl Acad Sci USA 2002; 99: 14789–14794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T, Hazan I, Zhang J, Ng SY, Naito T, Snippert HJ et al. The role of the chromatin remodeler Mi-2beta in hematopoietic stem cell self-renewal and multilineage differentiation. Genes Dev 2008; 22: 1174–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cedar H, Bergman Y. Epigenetics of haematopoietic cell development. Nat Rev Immunol 2011; 11: 478–488. [DOI] [PubMed] [Google Scholar]
- Broske AM, Vockentanz L, Kharazi S, Huska MR, Mancini E, Scheller M et al. DNA methylation protects hematopoietic stem cell multipotency from myeloerythroid restriction. Nat Genet 2009; 41: 1207–1215. [DOI] [PubMed] [Google Scholar]
- Trowbridge JJ, Snow JW, Kim J, Orkin SH. DNA methyltransferase 1 is essential for and uniquely regulates hematopoietic stem and progenitor cells. Cell Stem Cell 2009; 5: 442–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trowbridge JJ, Orkin SH. DNA methylation in adult stem cells: new insights into self-renewal. Epigenetics 2010; 5: 189–193. [DOI] [PubMed] [Google Scholar]
- Park IK, Qian D, Kiel M, Becker MW, Pihalja M, Weissman IL et al. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature 2003; 423: 302–305. [DOI] [PubMed] [Google Scholar]
- Ohta H, Sawada A, Kim JY, Tokimasa S, Nishiguchi S, Humphries RK et al. Polycomb group gene rae28 is required for sustaining activity of hematopoietic stem cells. J Exp Med 2002; 195: 759–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cales C, Roman-Trufero M, Pavon L, Serrano I, Melgar T, Endoh M et al. Inactivation of the polycomb group protein Ring1B unveils an antiproliferative role in hematopoietic cell expansion and cooperation with tumorigenesis associated with Ink4a deletion. Mol Cell Biol 2008; 28: 1018–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Nandakumar V, Jiang XX, Jones L, Yang AG, Huang XF et al. The control of hematopoietic stem cell maintenance, self-renewal, and differentiation by Mysm1-mediated epigenetic regulation. Blood 2013; 122: 2812–2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aranda-Orgilles B, Saldana-Meyer R, Wang E, Trompouki E, Fassl A, Lau S et al. MED12 regulates HSC-specific enhancers independently of mediator kinase activity to control hematopoiesis. Cell Stem Cell 2016; 19: 784–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintas-Cardama A, Santos FP, Garcia-Manero G. Histone deacetylase inhibitors for the treatment of myelodysplastic syndrome and acute myeloid leukemia. Leukemia 2011; 25: 226–235. [DOI] [PubMed] [Google Scholar]
- Wang H, Maurano MT, Qu H, Varley KE, Gertz J, Pauli F et al. Widespread plasticity in CTCF occupancy linked to DNA methylation. Genome Res 2012; 22: 1680–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobanenkov VV, Nicolas RH, Adler VV, Paterson H, Klenova EM, Polotskaja AV et al. A novel sequence-specific DNA binding protein which interacts with three regularly spaced direct repeats of the CCCTC-motif in the 5'-flanking sequence of the chicken c-myc gene. Oncogene 1990; 5: 1743–1753. [PubMed] [Google Scholar]
- Bell AC, West AG, Felsenfeld G. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell 1999; 98: 387–396. [DOI] [PubMed] [Google Scholar]
- Splinter E, Heath H, Kooren J, Palstra RJ, Klous P, Grosveld F et al. CTCF mediates long-range chromatin looping and local histone modification in the beta-globin locus. Genes Dev 2006; 20: 2349–2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong CT, Corces VG. CTCF: an architectural protein bridging genome topology and function. Nat Rev Genet 2014; 15: 234–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vietri Rudan M, Hadjur S. Genetic tailors: CTCF and cohesin shape the genome during evolution. Trends Genet 2015; 31: 651–660. [DOI] [PubMed] [Google Scholar]
- Heath H, Ribeiro de Almeida C, Sleutels F, Dingjan G, van de Nobelen S, Jonkers I et al. CTCF regulates cell cycle progression of alphabeta T cells in the thymus. EMBO J 2008; 27: 2839–2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouboussad L, Kreuz S, Lefevre PF. CTCF depletion alters chromatin structure and transcription of myeloid-specific factors. J Mol Cell Biol 2013; 5: 308–322. [DOI] [PubMed] [Google Scholar]
- Kim TG, Kim M, Lee JJ, Kim SH, Je JH, Lee Y et al. CCCTC-binding factor controls the homeostatic maintenance and migration of Langerhans cells. J Allergy Clin Immunol 2015; 136: 713–724. [DOI] [PubMed] [Google Scholar]
- Ventura A, Kirsch DG, McLaughlin ME, Tuveson DA, Grimm J, Lintault L et al. Restoration of p53 function leads to tumour regression in vivo. Nature 2007; 445: 661–665. [DOI] [PubMed] [Google Scholar]
- Heng TS, Painter MW, Immunological Genome Project C. The Immunological Genome Project: networks of gene expression in immune cells. Nat Immunol 2008; 9: 1091–1094. [DOI] [PubMed] [Google Scholar]
- Huang E, Nocka K, Beier DR, Chu TY, Buck J, Lahm HW et al. The hematopoietic growth factor KL is encoded by the Sl locus and is the ligand of the c-kit receptor, the gene product of the W locus. Cell 1990; 63: 225–233. [DOI] [PubMed] [Google Scholar]
- Edling CE, Hallberg B. c-Kit—a hematopoietic cell essential receptor tyrosine kinase. Int J Biochem Cell Biol 2007; 39: 1995–1998. [DOI] [PubMed] [Google Scholar]
- Yona S, Kim KW, Wolf Y, Mildner A, Varol D, Breker M et al. Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity 2013; 38: 79–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto D, Chow A, Noizat C, Teo P, Beasley MB, Leboeuf M et al. Tissue-resident macrophages self-maintain locally throughout adult life with minimal contribution from circulating monocytes. Immunity 2013; 38: 792–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suda T, Takubo K, Semenza GL. Metabolic regulation of hematopoietic stem cells in the hypoxic niche. Cell Stem Cell 2011; 9: 298–310. [DOI] [PubMed] [Google Scholar]
- Simsek T, Kocabas F, Zheng J, Deberardinis RJ, Mahmoud AI, Olson EN et al. The distinct metabolic profile of hematopoietic stem cells reflects their location in a hypoxic niche. Cell Stem Cell 2010; 7: 380–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takubo K, Goda N, Yamada W, Iriuchishima H, Ikeda E, Kubota Y et al. Regulation of the HIF-1alpha level is essential for hematopoietic stem cells. Cell Stem Cell 2010; 7: 391–402. [DOI] [PubMed] [Google Scholar]
- Takubo K, Nagamatsu G, Kobayashi CI, Nakamura-Ishizu A, Kobayashi H, Ikeda E et al. Regulation of glycolysis by Pdk functions as a metabolic checkpoint for cell cycle quiescence in hematopoietic stem cells. Cell Stem Cell 2013; 12: 49–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C, Yuan T, Wu Y, Wang Y, Fan TW, Miriyala S et al. Loss of FBP1 by Snail-mediated repression provides metabolic advantages in basal-like breast cancer. Cancer Cell 2013; 23: 316–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrano V, Chernukhin I, Docquier F, D'Arcy V, Leon J, Klenova E et al. CTCF regulates growth and erythroid differentiation of human myeloid leukemia cells. J Biol Chem 2005; 280: 28152–28161. [DOI] [PubMed] [Google Scholar]
- Sekimata M, Perez-Melgosa M, Miller SA, Weinmann AS, Sabo PJ, Sandstrom R et al. CCCTC-binding factor and the transcription factor T-bet orchestrate T helper 1 cell-specific structure and function at the interferon-gamma locus. Immunity 2009; 31: 551–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro de Almeida C, Stadhouders R, de Bruijn MJ, Bergen IM, Thongjuea S, Lenhard B et al. The DNA-binding protein CTCF limits proximal Vkappa recombination and restricts kappa enhancer interactions to the immunoglobulin kappa light chain locus. Immunity 2011; 35: 501–513. [DOI] [PubMed] [Google Scholar]
- Koesters C, Unger B, Bilic I, Schmidt U, Bluml S, Lichtenberger B et al. Regulation of dendritic cell differentiation and subset distribution by the zinc finger protein CTCF. Immunol Lett 2007; 109: 165–174. [DOI] [PubMed] [Google Scholar]
- Fedoriw AM, Stein P, Svoboda P, Schultz RM, Bartolomei MS. Transgenic RNAi reveals essential function for CTCF in H19 gene imprinting. Science 2004; 303: 238–240. [DOI] [PubMed] [Google Scholar]
- Stadhouders R, Thongjuea S, Andrieu-Soler C, Palstra RJ, Bryne JC, van den Heuvel A et al. Dynamic long-range chromatin interactions control Myb proto-oncogene transcription during erythroid development. EMBO J 2012; 31: 986–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domen J, Weissman IL. Hematopoietic stem cells need two signals to prevent apoptosis; BCL-2 can provide one of these, Kitl/c-Kit signaling the other. J Exp Med 2000; 192: 1707–1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hur J, Choi JI, Lee H, Nham P, Kim TW, Chae CW et al. CD82/KAI1 maintains the dormancy of long-term hematopoietic stem cells through interaction with DARC-expressing macrophages. Cell Stem Cell 2016; 18: 508–521. [DOI] [PubMed] [Google Scholar]
- Nikolic T, Movita D, Lambers ME, Ribeiro de Almeida C, Biesta P, Kreefft K et al. The DNA-binding factor Ctcf critically controls gene expression in macrophages. Cell Mol Immunol 2014; 11: 58–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schonheit J, Leutz A, Rosenbauer F. Chromatin dynamics during differentiation of myeloid cells. J Mol Biol 2015; 427: 670–687. [DOI] [PubMed] [Google Scholar]
- Lai AY, Fatemi M, Dhasarathy A, Malone C, Sobol SE, Geigerman C et al. DNA methylation prevents CTCF-mediated silencing of the oncogene BCL6 in B cell lymphomas. J Exp Med 2010; 207: 1939–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp CJ, Moore JM, Moser R, Bernard B, Teater M, Smith LE et al. CTCF haploinsufficiency destabilizes DNA methylation and predisposes to cancer. Cell Rep 2014; 7: 1020–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trumpp A, Essers M, Wilson A. Awakening dormant haematopoietic stem cells. Nat Rev Immunol 2010; 10: 201–209. [DOI] [PubMed] [Google Scholar]
- Balakrishnan SK, Witcher M, Berggren TW, Emerson BM. Functional and molecular characterization of the role of CTCF in human embryonic stem cell biology. PLoS One 2012; 7: e42424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasko JE, Klenova EM, Leon J, Filippova GN, Loukinov DI, Vatolin S et al. Cell growth inhibition by the multifunctional multivalent zinc-finger factor CTCF. Cancer Res 2001; 61: 6002–6007. [PubMed] [Google Scholar]
- Qi CF, Martensson A, Mattioli M, Dalla-Favera R, Lobanenkov VV, Morse HC 3rd. CTCF functions as a critical regulator of cell-cycle arrest and death after ligation of the B cell receptor on immature B cells. Proc Natl Acad Sci USA 2003; 100: 633–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Docquier F, Farrar D, D'Arcy V, Chernukhin I, Robinson AF, Loukinov D et al. Heightened expression of CTCF in breast cancer cells is associated with resistance to apoptosis. Cancer Res 2005; 65: 5112–5122. [DOI] [PubMed] [Google Scholar]
- Bigarella CL, Liang R, Ghaffari S. Stem cells and the impact of ROS signaling. Development 2014; 141: 4206–4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K, Hirao A, Arai F, Matsuoka S, Takubo K, Hamaguchi I et al. Regulation of oxidative stress by ATM is required for self-renewal of haematopoietic stem cells. Nature 2004; 431: 997–1002. [DOI] [PubMed] [Google Scholar]
- Ito K, Hirao A, Arai F, Takubo K, Matsuoka S, Miyamoto K et al. Reactive oxygen species act through p38 MAPK to limit the lifespan of hematopoietic stem cells. Nat Med 2006; 12: 446–451. [DOI] [PubMed] [Google Scholar]
- Tothova Z, Kollipara R, Huntly BJ, Lee BH, Castrillon DH, Cullen DE et al. FoxOs are critical mediators of hematopoietic stem cell resistance to physiologic oxidative stress. Cell 2007; 128: 325–339. [DOI] [PubMed] [Google Scholar]
- Kocabas F, Zheng J, Thet S, Copeland NG, Jenkins NA, DeBerardinis RJ et al. Meis1 regulates the metabolic phenotype and oxidant defense of hematopoietic stem cells. Blood 2012; 120: 4963–4972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J, Lu Z, Kocabas F, Bottcher RT, Costell M, Kang X et al. Profilin 1 is essential for retention and metabolism of mouse hematopoietic stem cells in bone marrow. Blood 2014; 123: 992–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King KY, Goodell MA. Inflammatory modulation of HSCs: viewing the HSC as a foundation for the immune response. Nat Rev Immunol 2011; 11: 685–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa MM, Thummar K, Mandelbaum J, Pasqualucci L, Rathinam CV. Lack of the ubiquitin-editing enzyme A20 results in loss of hematopoietic stem cell quiescence. J Exp Med 2015; 212: 203–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schieber MS, Chandel NS. ROS links glucose metabolism to breast cancer stem cell and EMT phenotype. Cancer Cell 2013; 23: 265–267. [DOI] [PubMed] [Google Scholar]
- Lake RJ, Boetefuer EL, Won KJ, Fan HY. The CSB chromatin remodeler and CTCF architectural protein cooperate in response to oxidative stress. Nucleic Acids Res 2016; 44: 2125–2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupianez DG, Kraft K, Heinrich V, Krawitz P, Brancati F, Klopocki E et al. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell 2015; 161: 1012–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Xu Q, Canzio D, Shou J, Li J, Gorkin DU et al. CRISPR inversion of CTCF sites alters genome topology and enhancer/promoter function. Cell 2015; 162: 900–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.