Abstract
Although aldehyde oxidase (AO) is an important hepatic drug-metabolizing enzyme, it remains understudied and is consequently often overlooked in preclinical studies, an oversight that has resulted in the failure of multiple clinical trials. AO’s preclusion to investigation stems from the following: (1) difficulties synthesizing metabolic standards due to the chemospecificity and regiospecificity of the enzyme and (2) significant inherent variability across existing in vitro systems including liver cytosol, S9 fractions, and primary hepatocytes, which lack specificity and generate discordant expression and activity profiles. Here, we describe a practical bacterial biotransformation system, ecoAO, addressing both issues simultaneously. ecoAO is a cell paste of MoCo-producing Escherichia coli strain TP1017 expressing human AO. It exhibits specific activity toward known substrates, zoniporide, 4-trans-(N,N-dimethylamino)cinnamaldehyde, O6-benzylguanine, and zaleplon; it also has utility as a biocatalyst, yielding milligram quantities of synthetically challenging metabolite standards such as 2-oxo-zoniporide. Moreover, ecoAO enables routine determination of kcat and V/K, which are essential parameters for accurate in vivo clearance predictions. Furthermore, ecoAO has potential as a preclinical in vitro screening tool for AO activity, as demonstrated by its metabolism of 3-aminoquinoline, a previously uncharacterized substrate. ecoAO promises to provide easy access to metabolites with the potential to improve pharmacokinetic clearance predictions and guide drug development.
Introduction
Aldehyde oxidase (AO) belongs to the molybdoflavo-containing family of enzymes that also includes xanthine oxidase. The AO monomer is a 148 kDa cytosolic protein and is active as a homodimer.1,2 Each monomer contains four cofactors, MoCo, FAD, and 2[2Fe–2S] clusters, that contribute to its redox-cycling catalytic mechanism.3 AO exhibits broad substrate specificities, performing oxidation and/or reduction reactions on an extensive range of endogenous and exogenous substrates.4 However, only limited studies have used human enzymes and most results summarized herein use nonhuman AO. AO oxidizes aliphatic and aromatic aldehydes, azaheterocycles.5 AO reduces nitro compounds,6−8 sulfoxides,9 azo-dyes,10 N-oxides,7 nitrite,11,12 and molecular oxygen. Although AO plays a significant role in drug metabolism, the physiological relevance of AO’s endogenous substrate transformations is not understood. AO constitutes 0.3% of human hepatocytes by weight (calculated from data published by Barr et al.),13 where it metabolizes many drugs and metabolic intermediates. It is also expressed in the intestine, kidney, lung, adrenal gland, and skin.14−17
The pharmaceutical industry’s trepidation toward AO has piqued in recent years as certain AO metabolites have achieved poor toxicological outcomes in clinical trials,18,19 AO has been implicated in potential drug–drug interactions (DDI),20,21 and because allometric scaling from AO preclinical species data consistently underpredicts human AO clearance.22−29 Consequently, promising drug candidates are discontinued every year, largely because AO metabolism has not been adequately characterized in existing preclinical in vitro systems. Predominant systems utilized to screen drug candidates for AO activity include liver cytosol, S9 fractions, and cryopreserved primary hepatocytes.30 All exhibit significant variability that has been attributed to interindividual phenotypic variability, hypervariable expression, low stability, and variation in biological fractionation methods.31 Additionally, these systems can suffer from lack of specificity due to the presence of other mechanistically similar metabolizing enzymes, including xanthine oxidase, xanthine dehydrogenase, and cytochromes P450.32 Furthermore, most human liver preparations are contaminated with the AO substrate allopurinol, which can affect the measured kinetic constants.31 In this publication, we combine MoCo-producing Escherichia coli strain TP1017 (Tracey Palmer Group, University of Dundee, unpublished) with pTHco-hAOX1, a codon-optimized, lactose-inducible human AO-containing plasmid33 to produce ecoAO, a standardized cell-paste-based in vitro AO assay system that is easily implemented by any lab.
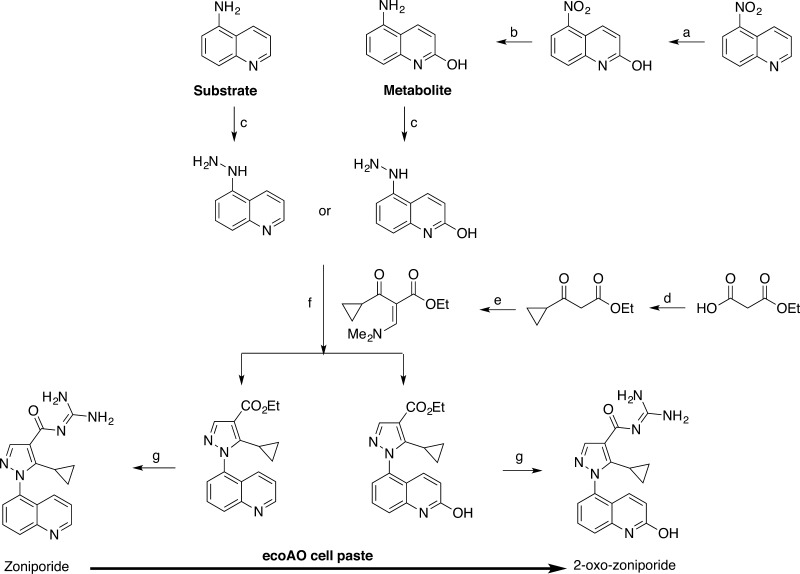
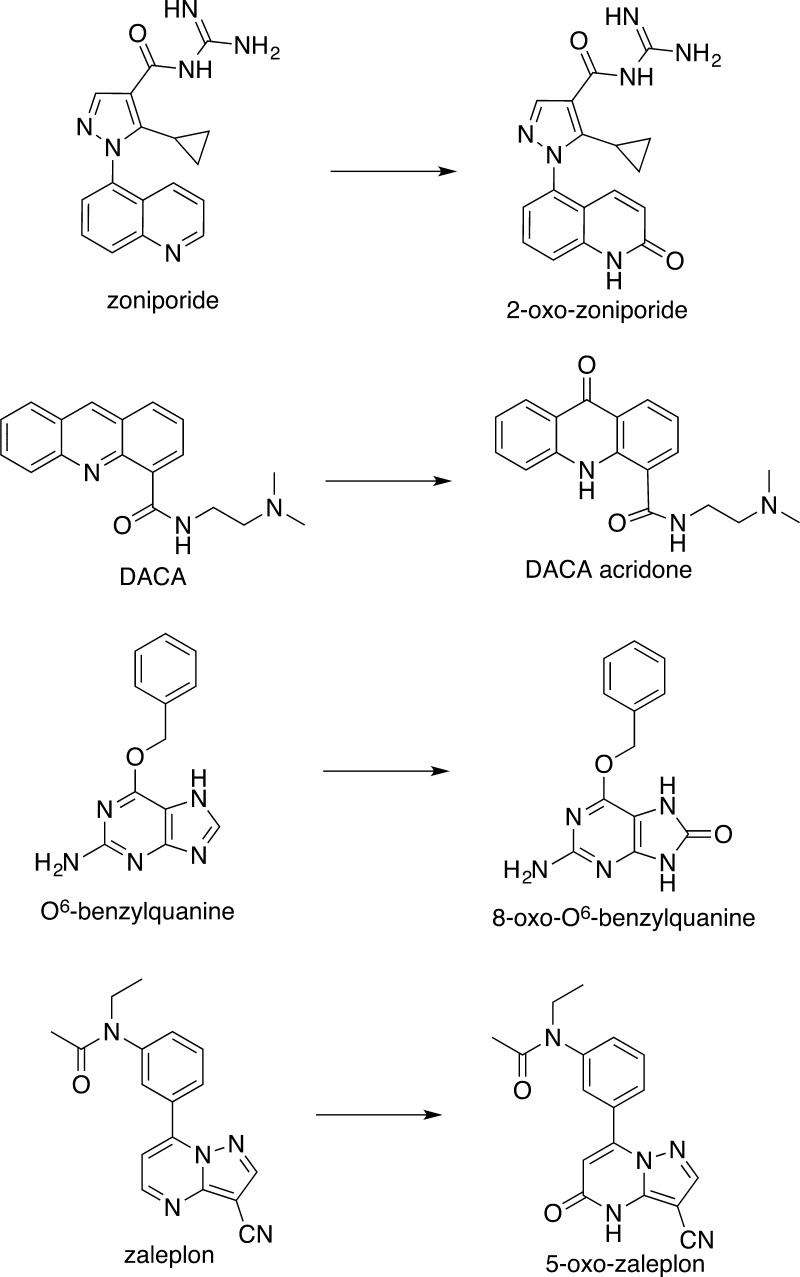
AO performs chemoselective and regioselective redox reactions that are difficult to reproduce via chemical synthesis. A good example is the synthesis of 2-oxo-zoniporide, a metabolite of zoniporide requiring seven synthetic steps (Scheme 1).34 Utilizing the ecoAO system, we demonstrate biotransformation of known AO-metabolized drugs (zoniporide to 2-oxo-zoniporide, 4-trans-(N,N-dimethylamino)cinnamaldehyde (DACA) to DACA-acridone, O6-benzylguanine (O6BG) to 8-oxo-O6BG, and zaleplon to 5-oxo-zaleplon) to produce milligram quantities of their respective metabolites in just 5 days, with minimal human intervention (Scheme 2). Furthermore, we provide proof of the concept of ecoAO’s utility to convert a previously uncharacterized AO substrate, 3-aminoquinoline (which has proven difficult to synthesize) to its regiospecifically oxidized metabolite, 2-oxo-3-aminoquinoline (Scheme 3). After solvent extraction and preparative high-performance liquid chromatography (HPLC), the system yields sufficient product for metabolite quantification, enabling determination of kinetic parameters, and providing starting materials to synthesize analogues.
Scheme 1. Synthesis of Zoniporide and 2-Oxo-zoniporide.
(a) 5.25% NaClO, overnight, room temperature (rt); (b) SnCl2 dihydrate, HCl (aq), 0–23 °C, 3 h; (c) (i) NaNO2, HCl (aq), 0 °C, 1 h; (ii) SNCl2 dihydrate, HCl (aq), 0–23 °C, 3 h, HCl (g), EtOAc, 0 °C, 15 min; (d) (i) trimethylsilyl chloride, pyridine, ether, 23 °C, 4 h; (ii) n-BuLi (2.5 M in hexanes), −78 °C, 75 min; (iii) cyclopropanecarbonyl chloride, 1,2-dimethoxyethane, (−78) to (+23) °C; (e) N,N-dimethylformamide dimethyl acetal, reflux, 1 h; (f) Et3N, EtOH, reflux, 1 h; (g) guanidine, EtOH, 80 °C, in vacuo, 45 min.
Scheme 2. Biotransformation of Known AO Substrates Using the ecoAO System.
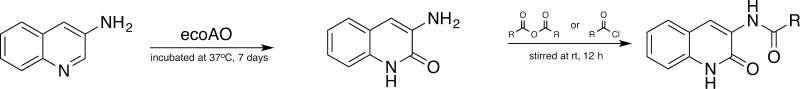
Scheme 3. Synthesis of 2-Oxo-3-aminoquinoline Analogs.
Results
Optimizing AO Activity Over Time by the Direct Expression Culture Assay
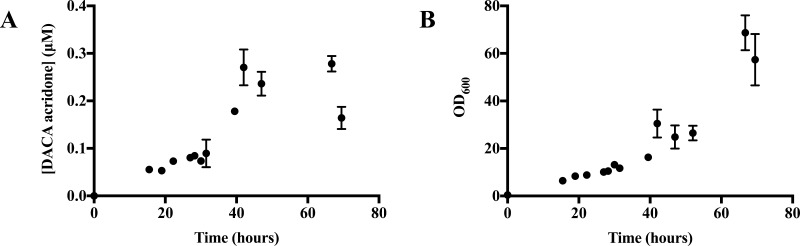
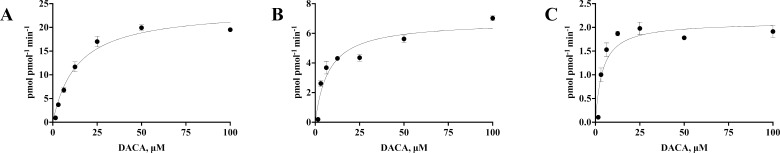
Figure 1 follows AO activity and growth over time in ecoAO expression cultures grown at 30 °C in aerated spinner flasks using DACA as a probe substrate. There was a correlation between the growth and activity for up to 30 h (Pearson r = 0.96 (R2 = 0.92, p < 0.0001), data not shown). However, after 30 h, the activity and growth no longer showed changes together. At around 42 h, AO activity, as quantified by DACA oxidation, peaked before it began to decrease; meanwhile, the E. coli concentration increased (OD600). On the basis of these results, it was determined that ecoAO cultures should be spun down to cell paste at 42 h and that optical density was not a reliable indicator of the optimal AO activity. Using the cell paste that resulted from the optimized expression, we determined the kinetic parameters, as shown in Figure 2, which depict the saturation kinetics for 0.15 pmol AO (based on the liquid chromatography–tandem mass spectrometry (LC–MS/MS) proteomic quantitation described below) in ecoAO cell paste. Furthermore, we were able to purify AO from the cell paste using the poly-His tag and determine the kinetic constants using the purified enzyme. The kinetic parameters obtained are presented in Table 1.
Figure 1.
ecoAO activity and growth curves for the culture grown at 30 °C with aeration. (A) The direct-culture activity assay of 1 mL of the culture with 37 μM DACA (5 min, 37 °C). (B) Optical density at 600 nm (OD600) after dilution correction (diluted to OD600 < 1). Error bars report the scanning electron microscopy (SEM) of experimental triplicates.
Figure 2.
Saturation kinetics of (A) ecoAO and (B) human liver cytosol (HLC) (C)-purified AO, oxidizing DACA to DACA-acridone. Error bars report the SD of experimental triplicates. The data for the purified enzyme is representative of results from three separate purifications and characterizations.
Table 1. Kinetic Parameters for ecoAO and Purified AO with DACAa.
| KM (μM) | kcat (min–1) | kcat/KM (min–1 μM–1) | |
|---|---|---|---|
| ecoAO | 14 ± 2 | 24 ± 1 | 1.7 ± 0.3 |
| HLC | 8 ± 2 | 6.8 ± 0.4 | 0.9 ± 0.3 |
| purified AO | 12.0 ± 2 | 3 ± 1 | 0.3 ± 0.1 |
Error is reported as the standard deviation (SD) for triplicate experiments. For purified AO, the value is the average of three purifications and separate measurements in triplicate.
Biocatalytic Production of AO Metabolites Using ecoAO
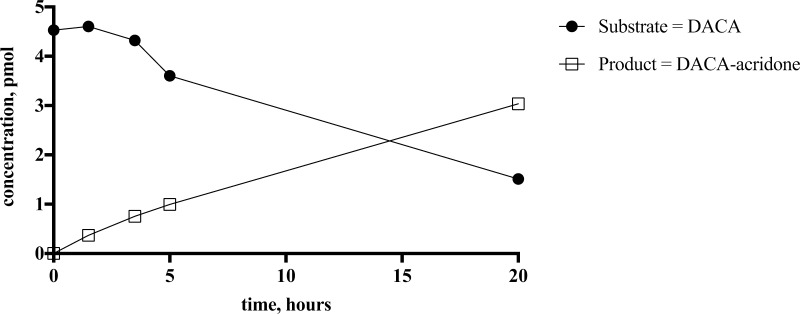
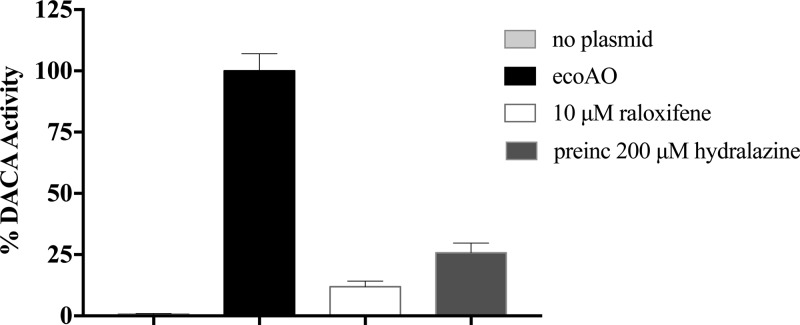
We used the substrate DACA to optimize the conditions for AO metabolite formation from cell paste and to test whether other enzymes native to E. coli would metabolize the substrate. Briefly, 50 mg/mL cell paste dissolved in AO buffer afforded significant substrate conversion over 20 h (Figure 3). It can be seen that for each pmol of substrate consumed, an equivalent amount of metabolite is formed, indicating that no other metabolite of DACA is being produced in significant amounts. Absolute AO quantification via LC–MS/MS revealed that 50 mg/mL cell paste (using the expression conditions outlined in the Methods section) contained 191 ± 7 nM AO. A control experiment monitoring TP1017 activity with DACA in the absence of the pTHco-hAOX1 expression plasmid revealed that no metabolite was formed (Figure 4). Furthermore, inhibitors of AO, raloxifene and hydralazine, significantly reduced ecoAO activity (Figure 4).
Figure 3.
Biotransformation of DACA by the ecoAO system. Substrate disappearance and product formation. S = DACA and P = DACA-acridone (n = 1).
Figure 4.
Percent of ecoAO DACA activity (light gray) in the absence of the AO-containing plasmid, pTHco-hAOX, in TP1017 (black), in the presence of 10 μM raloxifene (white), and after 30 min preincubation with 200 μM hydralazine (dark gray). Error bars report the SD of experimental triplicates.
The optimized ecoAO system was employed to biotransform preparative quantities of several common AO probe substrates to their metabolites. The substrates (see Scheme 1) were zoniporide (KM = 3.4 μM),23 DACA (KM = 6.3 μM),13 O6BG (KM = 120 μM),35 and zaleplon (KM = 124 μM).36 Furthermore, a previously uncharacterized AO substrate (3-aminoquinoline) was also evaluated (Scheme 3). Attempts by us to synthesize the metabolite by the method shown for 5-aminoquinoline in Scheme 1 failed. In general, substrates were incubated with cell paste and product formation monitored once daily by LC–MS/MS. When the product formation slowed, the cell paste was removed by centrifugation and more cell paste was added to the supernatant. The number of rounds of the addition of fresh ecoAO cell paste required for the reaction to reach completion was substrate dependent. After purification by solvent extraction and preparative HPLC, the metabolites were characterized by NMR (see Supporting Information). The extinction coefficients for each of the substrates and metabolites is given in Table 2.
Table 2. Preparative Scale Production of AO Metabolites Using ecoAOa.
| substrate | mM –1 cm –1 | metabolite | mM–1 cm –1 | % yield |
|---|---|---|---|---|
| zoniporide | ε315 6.48 ± 0.04 | 2-oxo-zoniporide | ε280 5.48 ± 0.15 | 10 |
| DACA | ε358 10.48 ± 0.09 | DACA-acridone | ε409 9.24 ± 0.07 | 25 |
| zaleplon | ε333 7.04 ± 0.03 | 5-oxo-zaleplon | ε330 7.70 ± 0.26 | 10 |
| O6-BG | ε282 9.10 ± 0.05 | 8-oxo-O6-BG | ε290 9.50 ± 0.22 | 12 |
All extinction coefficients were determined in buffer (pH 7.4) using an Agilent 8453 UV–vis spectrophotometer (Agilent Technologies, Santa Clara, CA). The error is reported as the SD for duplicate measurements.
Discussion
Previous expression systems used large volumes and specialized equipment. We have developed a system that we call ecoAO for production of human AO that uses small spinner flasks and stir plates, available in most labs. The ecoAO system can be utilized to synthesize metabolites, screen for AO activity, and carry out metabolic studies. In contrast to other in vitro systems, such as HLC, cryopreserved hepatocytes, S9 fractions, and purified enzymes, it offers the advantages of being simple, time-efficient, standardizable, widely applicable, accessible, economical, and reproducible. Importantly, ecoAO provides researchers with a tool to characterize AO activity against a nonmammalian background, ensuring specificity against xanthine oxidase, xanthine dehydrogenase, and P450 activities, as well as any still-uncharacterized contributions from other mammalian enzymes. Furthermore, HLC can contain different small molecules (drugs) depending on the donor, or on the preparation,31 which can affect kinetics.
ecoAO Biotransformation Yields Usable Amounts of Selectively Oxidized Azaheterocycles
We have provided evidence that ecoAO, a cell paste made from E. coli expressing human AO, is a promising new in vitro system for synthesis of AO metabolites and for the insertion of an oxygen atom adjacent to nitrogen in quinolines, acridines, and purines. We demonstrated that ecoAO cell paste can produce sufficient amounts of the metabolites of zoniporide, DACA, O6BG, and zaleplon for NMR characterization. At least for DACA, it appears that only the human AO metabolite is formed because substrate disappearance corresponds directly to the amount of the product formed (Figure 3). The yields range from 10 to 25% (see Table 2), which is reasonable given that chemical synthesis of these metabolites requires at least two steps and in the case of zoniporide replaces a 7-step reaction sequence.
Using an uncharacterized substrate (3-aminoquinoline), we demonstrate the utility of ecoAO to generate milligram quantities of AO metabolite from frozen cell paste reconstituted in buffer that can be used as starting material for the synthesis of more complex molecules. The biotranformation of 3-aminoquinoline, post-ecoAO 2-oxo-3-aminoquinoline analogue synthesis (Scheme 3), and ecoAO metabolism of the 3-aminoquinoline analogues, illustrate two important points: (1) these metabolites could not be synthesized by the same method as was used for zoniporide (see Scheme 1), although a 4-step synthesis has been reported37 and (2) even relatively sterically hindered compounds readily undergo oxidation adjacent to the nitrogen. From a broader perspective, the ecoAO system could also be employed in other settings where selective oxidation of azaheterocycles is desired in a synthesis.
ecoAO Enables Simple Optimization Expression Conditions for AO Purification
Although the human AO crystal structure was solved recently,32 the relationship between the AO structure and function remains poorly understood. Purified AO is necessary for its further biophysical characterization, and bacterial systems are a practical enzyme source; however, low yields and activities have been reported from bacterial expression systems in the past.38,39 Low yields may be attributed to complications associated with E. coli production of AO, for example, poor coexpression of molybdenum-containing enzymes, large AO (150 kDa) stressing the transcriptional and translational machinery, and incomplete or incorrect insertion of the four cofactors.40,41 Complete synthesis of the MoCo cofactor requires replacement of an oxygen atom with sulfur; however, even MoCo-modified E. coli (such as TP1017) is inefficient at this process.42,43 In addition, reports for molybdenum and iron incorporation for expressed proteins range from 20 to 70% and 45 to 90%, respectively.33,39,40 With many dynamic, complex, little-understood factors involved in heterologous AO expression, optical density or total protein quantification alone is not a good measure of AO activity. Our direct cell-culture DACA assay enables simple AO expression-monitoring to ensure protein is harvested at maximal activity and consequently when holoprotein quantity is highest. We have shown that we can purify human AO from the small batch ecoAO cultures that can be used to further characterize this enzyme.
How Does ecoAO Compare to Other AO In Vitro Systems?
The ecoAO cell paste metabolizes DACA with higher activity per pmol (kcat value of 24 min–1) of AO than human liver cytosol (HLC) and enzyme purified from the ecoAO system (Figure 2 and Table 1). The HLC-derived kcat value for DACA activity was found to be 6.8 min–1 in this study, with significant variations observed across batches reported in the literature.13 Enzymes purified from the ecoAO expression system had kcat of 3 ± 1 min–1 from three separate purifications. Thus, overall, the AO enzyme in cell paste is slightly more active than in either HLC or purified protein per nmol of AO.
It is possible that nonspecific binding in the cell paste could affect the kinetics (KM) of the reaction. If nonspecific binding was significant, the (KM) value would be predicted to be higher in cell paste than for purified enzymes. However, the ecoAO cell paste had KM of 13.7 ± 1.6 μM, which was not statistically different from KM values for AO activity with DACA that have been reported as 6.7–8.7 μM and 9.3 μM for HLC and purified enzyme, respectively (we achieved a very similar KM of 12 ± 2 μM for pure enzyme). However, it should be noted that at very high levels of cell paste, the reaction appeared to slow the reaction (data not shown).
ecoAO is a Comprehensive Platform for the Study AO Metabolism
From a single small-scale growth of ecoAO, one has the ability to synthesize significant amounts of metabolite and determine the in vitro kinetics. Even without extensive purification, the metabolite can be used to construct a standard curve that can be used to determine the amount of metabolite by using an NMR internal standard. If the desired metabolite is one of those produced herein, they can use the extinction coefficients in Table 2 (data and spectra are given in Supporting Information). However, as we have shown, you can produce and purify enough metabolites to directly measure the amount. This is significant in that even people without synthetic experience can make the metabolite. Without access to the metabolite, one is forced to use substrate disappearance to determine the in vitro kinetics. For substrate disappearance to give valid clearance estimates, the amount of substrate used must be below the KM. However, without the metabolite, it is difficult to know if these conditions are satisfied.44 Furthermore, metabolite formation data provides important information concerning mass balance, time-dependent inhibition, and drug–drug interactions (DDI). These important clearance-contributing processes are often not first-order, yet they have only been described to a limited extent in AO.20,21,35,45 Finally, inaccurate scaling between in vitro and in vivo clearance data has led to unanticipated rapid first-pass metabolism in several clinical trials, resulting in their failures.22,24,27 A plausible explanation for AO clearance underprediction is that AO activity is not constant over the time of the incubation due to enzyme inhibition and/or inactivation.13 Such kinetic phenomena cannot be identified by substrate depletion alone.
Conclusions
In future, we intend to expand the capabilities of the ecoAO system to address several issues facing the drug development community. For example, comparing AO activities across preclinical species has proven problematic due to issues with low activity or loss of activity of protein purified from heterologous expression systems, hypervariability of biological fractions, substrate competition with other mammalian enzymes, and resource-intensive quantification of holoenzymes. By generating ecoAO cell-paste strains containing active AO isoforms from various common preclinical species and making them readily available, we aim to provide a comprehensive, standardized platform for in vitro preclinical evaluation of AO. Furthermore, genotypic variability of AO has also been associated with altered drug metabolism phenotypes.39 Our human ecoAO strain is readily modifiable so that any AO genotype may be assessed for expression and activity.
In conclusion, the ecoAO system provides researchers with a new tool to characterize the role of AO in drug metabolism, addressing many of the established issues in other in vitro systems. Furthermore, it is well suited to high-throughput implementation for screening drug candidates for AO activity, producing AO metabolites, and performing metabolic studies.
Experimental Section
Materials
The lactose-inducible TP1017 E. coli strain (as JM101/Δ mobAB::Kan; unpublished) was a gift from Tracey Palmer of the University of Dundee. E. coli codon-optimized human AO-containing plasmid pTHco-hAOX1 was the expression vector.33 All E. coli expression media components were obtained from Sigma. Human liver cytosol (HLC), pooled from 50 individual donors (male and female), was purchased from XenoTech (Batch H06610.C Lot no. 141002, Lenexa, KS). DACA was synthesized using a method described previously13 with some revisions (described in Supporting Information). DACA-acridone hydrochloride was a gift from William Denny, University of Auckland. O6BG was purchased from Toronto Research Chemicals, Inc. (Toronto, Canada). Raloxifene hydrochloride was obtained from Enzo Life Sciences (Farmingdale, NY). Zoniporide hydrochloride hydrate, zaleplon, allopurinol, hydralazine, acetic anhydride, and internal standards 2-methyl-4(3H)-quinazolinone and phenacetin were purchased from Sigma-Aldrich (St. Louis, MO). 3-Aminoquinoline and 4-methylvaleric acid chloride were purchased from TCI America (Portland, OR).
Methods
AO Expression System
TP1017-competent cells were transformed with pTHco-hAOX1 and stored at −80 °C as 20% glycerol stocks. When required, the stock was cultured on an agar plate and a single ampicillin-resistant colony was used to inoculate Complete Luria Broth (LB supplemented with 100 μg/mL ampicillin, 0.25 μL/mL Trace Element solution, 0.2 mM sodium molybdate, and 1 μg/mL riboflavin).3 The starter culture was shaken overnight at 37 °C and diluted by 1:100 to inoculate Complete Terrific Broth,46 with identical supplementation to Complete LB (described above). The culture was grown at 30 °C, shaking at 220 rpm until OD600 = 0.7 (UV–vis), at which time AO expression was induced by addition of 5 mg/mL lactose dissolved in minimal double-distilled H2O. After induction, the culture was divided into 200 mL aliquots and transferred to 250 mL spinner flasks at 30 °C (spinning at 120 rpm), supplemented with filtered (50 mm, 0.2 μm, PFTE (EMD Millipore, Billerica, MA)) air bubbled in via a submerged glass tube (airflow was maintained at 4 scfh by a Dwyer RMB-51 flow-meter).
Direct Assay of TP1017(pTHco-hAOX1) Expression Culture Using DACA
Aliquots of 1 mL were removed from the expression culture and spun down at 1000g for 3 min. The supernatant was decanted off, and the pellet of intact E. coli cells was resuspended in 200 μL of prewarmed (37 °C) AO buffer (25 mM KPi (pH 7.4), 0.5 mM ethylenediaminetetraacetic acid). The assay was initiated by addition of 37 μM DACA in dimethyl sulfoxide (DMSO) (0.5% v/v) and allowed to run at 37 °C for 5 min. The reaction was quenched with 50 μL of 1 M formic acid containing a known amount of 2-methyl-4(3H)-quinazolinone as an internal standard (IS) and centrifuged at 16 100g for 10 min. DACA-acridone was quantified in the supernatant against IS using LC–MS/MS.
Purification, Quantitation, and DACA Saturation Kinetics of Pure AO and ecoAO
His-tagged AO was purified from the ecoAO expression culture according to a previously described method;38 additionally, the protein was further purified via anion exchange using the AKTA system with a Mono Q column (GE Healthcare, Pittsburgh, PA; buffer A: 25 mM KPi (pH 7.4); buffer B: 25 mM KPi (pH 7.4) + 2 M NaCl). Protein was eluted at 15% buffer B. Total AO concentration in the ecoAO cell paste, purified AO, and human liver cytosol was quantified via LC–MS/MS using a previously described method.13 DACA saturation kinetics were performed using 0.15 pmol AO (ecoAO), 1.9 pmol purified AO, and 0.51 pmol human liver cytosol.
Bioanalytics of AO Substrate Biotransformation Using ecoAO
Samples were analyzed using the LC-20AD series HPLC system (Shimadzu, Columbia, MD) fitted with an HTC PAL autosampler (LEAP Technologies, Carrboro, NC). Chromatography was performed on a Luna C18 column (50 x 2.0 mm2, 5 μm; Phenomenex, Torrance, CA). The mobile phase A consisted of 0.05% formic acid and 0.2% acetic acid in water and the mobile phase B comprised 90% acetonitrile, 9.9% water, and 0.1% formic acid. The column was equilibrated with 90% mobile phase A (600 μL/min). Column elution was achieved using a linear gradient to 25% mobile phase A. Quantitation was conducted using an API 4000 Q-Trap tandem mass spectrometry system (Applied Biosystems/MDS Sciex, Foster City, CA), with turbospray electrospray ionization operating in positive-ion mode. The tune parameters used were as follows: collision gas, medium; declustering potential, 70; entrance potential, 10; collision energy, 25; collision cell exit potential, 15; curtain gas, 20; ion spray voltage, 5500; ion source gas 1, 60; ion source gas 2, 40; and desolvation temperature, 400. Substrates and metabolites were quantified against IS using multiple reaction monitoring to monitor the following m/z transitions: zoniporide, 321 → 262; 2-oxo-zoniporide, 337 → 278;23 DACA, 295.2 → 250.2; DACA-acridone, 310.2 → 265.0;13 O6BG, 242.2 → 199.1; 8-oxo-O6BG, 258.2 → 91;35 zaleplon, 306.1 → 236.1; 5-oxo-zaleplon, 322.3 → 280.1;30 2-oxo-3-aminoquinoline 161.2 → 143.2; N-(2-oxo-1,2-dihydroquinolin-3-yl)acetamide 203.1 → 185.1, 4-methyl-N-(2-oxo-1,2-dihydroquinolin-3-yl)pentanamide 259.1 → 241, and the internal standards (IS) were either 2-methyl-4(3H)-quinazolinone 161.0 → 120.0 or phenacetin 180.2 → 110.1, as specified.
Substrate Disappearance, Product Formation, and Control Experiments
Biotransformation of 10 μM DACA was monitored over 24 h to provide kinetic data for substrate depletion and metabolite (DACA-acridone) formation. Non-AO expressing E. coli (no pTHco-hAOX1 plasmid) and inhibitors (10 μM raloxifene47 and 30 min preincubation of 200 μM hydralazine48) were used as negative controls. For each assay, the resuspended cell paste (50 mg/mL, wet weight) was prewarmed to 37 °C and 10 μM substrate was used to initiate the reaction (500 μL of final volume). Catalysis was quenched with 125 μL of 1 M formic acid containing a known amount of the internal standard and centrifuged at 16 100g for 10 min. Both compounds were quantified in the supernatant using LC–MS/MS.13 The activity was reported as percent conversion after 24 h.
Preparative Scale Biotransformation Using ecoAO Cell Paste
Frozen cell paste was thawed at room temperature (rt) and resuspended in 20 mL (50 mg/mL) of AO buffer (pH 7.4) in a 50 mL centrifuge tube. The cell paste suspension was prewarmed for 5 min (37 °C); then, 100 μL of the substrate (zoniporide, 2 mg; DACA, 4 mg; zaleplon, 2 mg; O6BG, 2 mg) dissolved in DMSO was added. The incubation was performed at 37 °C with replacement of cell paste when substrate conversion slowed, as monitored by LC–MS/MS. Once the reaction had advanced to completion (about 5 days), the cells were spun down at 30 000g for 20 min. The supernatant was extracted with ethyl acetate. The organic layer was collected and concentrated in vacuo. Further purification was performed on an Accela ultra high-performance liquid chromatography system (Thermo Scientific, Waltham, MA) with a 4.60 × 250 mm2 Luna 5 μm C18 column (Phenomenex, Torrance, CA). Mobile phases A and B were identical to those used for mass spectrometry. The column was equilibrated with 90% mobile phase A (4.0 min; 800 μL/min). Chromatographic separation was achieved using a linear gradient for over 13 min to 0% mobile phase A. The purified metabolites were identified using LC–MS/MS and proton NMR. NMR spectra (see Supporting Information) were recorded using a Varian 600 MHz spectrometer.
Synthesis of N-(2-Oxo-1,2-dihydroquinolin-3-yl)acetamide, 4-Methyl-N-(2-oxo-1,2-dihydroquinolin-3-yl)pentanamide, and 3-(Dimethylamino)-N-(2-oxo-1,2-dihydroquinolin-3-yl)propanamide
The crude 2-oxo-3-aminoquinoline incubation mixture from the preparative biotransformation (20 mg by LC–MS/MS) was added with excess amount of either the anhydride or acid chloride of the desired analogue (Scheme 3). It was stirred at rt for 12 h. The crude reaction mixture is added with Na2CO3 and extracted with ethyl acetate (50 × 2 mL). The ethyl acetate layer is washed with 50 mL of brine solution and concentrated in vacuo. The amount and purity of the compounds were characterized by qNMR.
Data Analysis
Experimental data were processed and analyzed using a GraphPad Prism (version 7.0a; GraphPad Software Inc., San Diego, CA).
Acknowledgments
The authors would like to thank Tracy Palmer’s group, University of Dundee, Scotland, for the gift of the TP1017 E. coli strain, and Slater Weinstock and Greg Crouch (Washington State University) for synthesizing 4-carboxyacridone. Funding for the NMR spectrometer used in this project included: NIH grants RR0631401 RR12948, NSF grants CHE-9115282, and DBI-9604689 and the Murdock Charitable Trust. This work was supported by grant GM100874.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsomega.7b01054.
Characterization data for all compounds (PDF)
The authors declare no competing financial interest.
Author Status
§ E.M.P. and S.C.H. are co-first-authors.
Supplementary Material
References
- Beedham C. Molybdenum hydroxylases: biological distribution and substrate-inhibitor specificity. Prog. Med. Chem. 1987, 24, 85–127. 10.1016/S0079-6468(08)70420-X. [DOI] [PubMed] [Google Scholar]
- Itoh K.; Maruyama H.; Adachi M.; Hoshino K.; Watanabe N.; Tanaka Y. Lack of dimer formation ability in rat strains with low aldehyde oxidase activity. Xenobiotica 2007, 37, 709–716. 10.1080/00498250701397713. [DOI] [PubMed] [Google Scholar]
- Alfaro J. F.; Jones J. P. Studies on the mechanism of aldehyde oxidase and xanthine oxidase. J. Org. Chem. 2008, 73, 9469–9472. 10.1021/jo801053u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garattini E.; Fratelli M.; Terao M. Mammalian aldehyde oxidases: genetics, evolution and biochemistry. Cell. Mol. Life Sci. 2008, 65, 1019–1048. 10.1007/s00018-007-7398-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krenitsky T. A.; Neil S. M.; Elion G. B.; Hitchings G. H. A comparison of the specificities of xanthine oxidase and aldehyde oxidase. Arch. Biochem. Biophys. 1972, 150, 585–599. 10.1016/0003-9861(72)90078-1. [DOI] [PubMed] [Google Scholar]
- Bauer S. L.; Howard P. C. The kinetics of 1-nitropyrene and 3-nitrofluoranthene metabolism using bovine liver xanthine oxidase. Cancer Lett. 1990, 54, 37–42. 10.1016/0304-3835(90)90088-F. [DOI] [PubMed] [Google Scholar]
- Kitamura S.; Tatsumi K. Involvement of liver aldehyde oxidase in the reduction of nicotinamide N-oxide. Biochem. Biophys. Res. Commun. 1984, 120, 602–606. 10.1016/0006-291X(84)91297-X. [DOI] [PubMed] [Google Scholar]
- Wolpert M. K.; Althaus J. R.; Johns D. G. <tep-common:author-query>AQ6: Please provide a DOI number for ref 8 or indicate if one doesn&#x2019;t exist.</tep-common:author-query>Nitroreductase activity of mammalian liver aldehyde oxidase. J. Pharmacol. Exp. Ther. 1973, 185, 202–213. [PubMed] [Google Scholar]
- Tatsumi K.; Kitamura S.; Yamada H. Sulfoxide reductase activity of liver aldehyde oxidase. Biochim. Biophys. Acta 1983, 747, 86–92. 10.1016/0167-4838(83)90125-5. [DOI] [PubMed] [Google Scholar]
- Stoddart A. M.; Levine W. G. Azoreductase activity by purified rabbit liver aldehyde oxidase. Biochem. Pharmacol. 1992, 43, 2227–2235. 10.1016/0006-2952(92)90182-I. [DOI] [PubMed] [Google Scholar]
- Li H.; Kundu T. K.; Zweier J. L. Characterization of the magnitude and mechanism of aldehyde oxidase-mediated nitric oxide production from nitrite. J. Biol. Chem. 2009, 284, 33850–33858. 10.1074/jbc.M109.019125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maia L. B.; Pereira V.; Mira L.; Moura J. J. G. Nitrite reductase activity of rat and human xanthine oxidase, xanthine dehydrogenase, and aldehyde oxidase: evaluation of their contribution to NO formation in vivo. Biochemistry 2015, 54, 685–710. 10.1021/bi500987w. [DOI] [PubMed] [Google Scholar]
- Barr J. T.; Jones J. P. Evidence for substrate-dependent inhibition profiles for human liver aldehyde oxidase. Drug Metab. Dispos. 2013, 41, 24–29. 10.1124/dmd.112.048546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempsey G. T.; Vaughan J. C.; Chen K. H.; Bates M.; Zhuang X. Evaluation of fluorophores for optimal performance in localization-based super-resolution imaging. Nat. Methods 2011, 8, 1027–1036. 10.1038/nmeth.1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manevski N.; Balavenkatraman K. K.; Bertschi B.; Swart P.; Walles M.; Camenisch G.; Schiller H.; Kretz O.; Ling B.; Wettstein R.; Schaefer D. J.; Pognan F.; Wolf A.; Litherland K. Aldehyde Oxidase Activity in Fresh Human Skin. Drug Metab. Dispos. 2014, 42, 2049. 10.1124/dmd.114.060368. [DOI] [PubMed] [Google Scholar]
- Moriwaki Y.; Yamamoto T.; Takahashi S.; Tsutsumi Z.; Hada T. Widespread cellular distribution of aldehyde oxidase in human tissues found by immunohistochemistry staining. Histol. Histopathol. 2001, 16, 745–753. [DOI] [PubMed] [Google Scholar]
- Nishimura M.; Naito S. Tissue-specific mRNA expression profiles of human phase I metabolizing enzymes except for cytochrome P450 and phase II metabolizing enzymes. Drug Metab. Pharmacokinet. 2006, 21, 357–374. 10.2133/dmpk.21.357. [DOI] [PubMed] [Google Scholar]
- Diamond S.; Boer J.; Maduskuie T. P.; Falahatpisheh N.; Li Y.; Yeleswaram S. Species-specific metabolism of SGX523 by aldehyde oxidase and the toxicological implications. Drug Metab. Dispos. 2010, 38, 1277–1285. 10.1124/dmd.110.032375. [DOI] [PubMed] [Google Scholar]
- Lolkema M. P.; Bohets H. H.; Arkenau H.; Lampo A.; Barale E.; de Jonge M. J. A.; van Doorn L.; Hellemans P.; de Bono J. S.; Eskens R. A. L. M. The c-Met tyrosine kinase inhibitor JNJ-38877605 causes renal toxicity through species specific insoluble metabolite formation. Clin. Cancer Res. 2015, 21, 2297–2304. 10.1158/1078-0432.CCR-14-3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr J. T.; Jones J. P. Inhibition of human liver aldehyde oxidase: implications for potential drug-drug interactions. Drug Metab. Dispos. 2011, 39, 2381–2386. 10.1124/dmd.111.041806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetterberg C.; Maltais F.; Laitinen L.; Liao S.; Tsao H.; Chakilam A.; Hariparsad N. VX-509 (Decernotinib)-Mediated CYP3A Time-Dependent Inhibition: An Aldehyde Oxidase Metabolite as a Perpetrator of Drug-Drug Interactions. Drug Metab. Dispos. 2016, 44, 1286–1295. 10.1124/dmd.116.071100. [DOI] [PubMed] [Google Scholar]
- Akabane T.; Tanaka K.; Irie M.; Terashita S.; Teramura T. Case report of extensive metabolism by aldehyde oxidase in humans: Pharmacokinetics and metabolite profile of FK3453 in rats, dogs, and humans. Xenobiotica 2011, 41, 372–384. 10.3109/00498254.2010.549970. [DOI] [PubMed] [Google Scholar]
- Dalvie D.; Zhang C.; Chen W.; Smolarek T.; Obach R. S.; Loi C.-M. Cross-species comparison of the metabolism and excretion of zoniporide: contribution of aldehyde oxidase to interspecies differences. Drug Metab. Dispos. 2010, 38, 641–654. 10.1124/dmd.109.030783. [DOI] [PubMed] [Google Scholar]
- Dittrich C.h.; Greim G.; Borner M.; Weigang-Köhler K.; Huisman H.; Amelsberg A.; Ehret A.; Wanders J.; Hanauske A.; Fumoleau P. Phase I and pharmacokinetic study of BIBX 1382 BS, an epidermal growth factor receptor (EGFR) inhibitor, given in a continuous daily oral administration. Eur. J. Cancer 2002, 38, 1072–1080. 10.1016/S0959-8049(02)00020-5. [DOI] [PubMed] [Google Scholar]
- Ishii Y.; Iwanaga M.; Nishimura Y.; Takeda S.; Ikushiro S.-I.; Nagata K.; Yamazoe Y.; Mackenzie P. I.; Yamada H. Protein-protein interactions between rat hepatic cytochromes P450 (P450s) and UDP-glucuronosyltransferases (UGTs): evidence for the functionally active UGT in P450-UGT complex. Drug Metab. Pharmacokinet. 2007, 22, 367–376. 10.2133/dmpk.22.367. [DOI] [PubMed] [Google Scholar]
- Jones J. P.; Korzekwa K. R. Predicting Intrinsic Clearance for Drugs and Drug Candidates Metabolized by Aldehyde Oxidase. Mol. Pharmaceutics 2013, 10, 1262–1268. 10.1021/mp300568r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaye B.; Offerman J. L.; Reid J. L.; Elliott H. L.; Hillis W. S. A species difference in the presystemic metabolism of carbazeran in dog and man. Xenobiotica 1984, 14, 935–945. 10.3109/00498258409151492. [DOI] [PubMed] [Google Scholar]
- Zhang X.; Liu H. H.; Weller P.; Zheng M.; Tao W.; Wang J.; Liao G.; Monshouwer M.; Peltz G. In silico and in vitro pharmacogenetics: aldehyde oxidase rapidly metabolizes a p38 kinase inhibitor. Pharmacogenomics J. 2011, 11, 15–24. 10.1038/tpj.2010.8. [DOI] [PubMed] [Google Scholar]
- Zientek M.; Jiang Y.; Youdim K.; Obach R. S. In vitro-in vivo correlation for intrinsic clearance for drugs metabolized by human aldehyde oxidase. Drug Metab. Dispos. 2010, 38, 1322–1327. 10.1124/dmd.110.033555. [DOI] [PubMed] [Google Scholar]
- Hutzler J. M.; Yang Y.; Albaugh D.; Fullenwider C. L.; Schmenk J.; Fisher M. B. Characterization of aldehyde oxidase enzyme activity in cryopreserved human hepatocytes. Drug Metab. Dispos. 2012, 40, 267–275. 10.1124/dmd.111.042861. [DOI] [PubMed] [Google Scholar]
- Barr J. T.; Choughule K. V.; Nepal S.; Wong T.; Chaudhry A. S.; Joswig-Jones C. A.; Zientek M.; Strom S. C.; Schuetz E. G.; Thummel K. E.; Jones J. P. Why do most human liver cytosol preparations lack xanthine oxidase activity?. Drug Metab. Dispos. 2014, 42, 695–699. 10.1124/dmd.113.056374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coelho C.; Foti A.; Hartmann T.; Santos-Silva T.; Leimkühler S.; Romão M. J. Structural insights into xenobiotic and inhibitor binding to human aldehyde oxidase. Nat. Chem. Biol. 2015, 11, 779–783. 10.1038/nchembio.1895. [DOI] [PubMed] [Google Scholar]
- Foti A.; Hartmann T.; Coelho C.; Santos-Silva T.; Romao M. J.; Leimkuhler S. Optimization of the expression of human aldehyde oxidase for investigations of single nucleotide polymorphisms. Drug Metab. Dispos. 2016, 44, 1277–1285. 10.1124/dmd.115.068395. [DOI] [PubMed] [Google Scholar]
- Dalvie D.; Sun H.; Xiang C.; Hu Q.; Jiang Y.; Kang P. Effect of structural variation on aldehyde oxidase-catalyzed oxidation of zoniporide. Drug Metab. Dispos. 2012, 40, 1575–1587. 10.1124/dmd.112.045823. [DOI] [PubMed] [Google Scholar]
- Barr J. T.; Jones J. P.; Oberlies N. H.; Paine M. F. Inhibition of human aldehyde oxidase activity by diet-derived constituents: structural influence, enzyme-ligand interactions, and clinical relevance. Drug Metab. Dispos. 2015, 43, 34–41. 10.1124/dmd.114.061192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renwick A. B.; Ball S. E.; Tredger J. M.; Price R. J.; Walters D. G.; Kao J.; Scatina J. A.; Lake B. G. Inhibition of zaleplon metabolism by cimetidine in the human liver: in vitro studies with subcellular fractions and precision-cut liver slices. Xenobiotica 2002, 32, 849–862. 10.1080/00498250210158221. [DOI] [PubMed] [Google Scholar]
- Juárez-Gordiano C.; Hernandez-Campos A.; Castillo R. An improved method for the synthesis of 3-amino-1H-quinolin-2-one. Synth. Commun. 2002, 32, 2959–2963. 10.1081/SCC-120012984. [DOI] [Google Scholar]
- Alfaro J. F.; Joswig-Jones C. A.; Ouyang W.; Nichols J.; Crouch G. J.; Jones J. P. Purification and mechanism of human aldehyde oxidase expressed in Escherichia coli. Drug Metab. Dispos. 2009, 37, 2393–2398. 10.1124/dmd.109.029520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann T.; Terao M.; Garattini E.; Teutloff C.; Alfaro J. F.; Jones J. P.; Leimkühler S. The Impact of Single Nucleotide Polymorphisms on Human Aldehyde Oxidase. Drug Metab. Dispos. 2012, 40, 856–864. 10.1124/dmd.111.043828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumann S.; Terao M.; Garattini E.; Saggu M.; Lendzian F.; Hildebrandt P.; Leimkuhler S. Site directed mutagenesis of amino acid residues at the active site of mouse aldehyde oxidase AOX1. PLoS One 2009, 4, e5348 10.1371/journal.pone.0005348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl R. C.; Rajagopalan K. V. Evidence for the inorganic nature of the cyanolyzable sulfur of molybdenum hydroxylases. J. Biol. Chem. 1982, 257, 1354–1359. [PubMed] [Google Scholar]
- Mendel R. R.; Leimkuhler S. The biosynthesis of the molybdenum cofactors. J. Biol. Inorg. Chem. 2015, 20, 337–347. 10.1007/s00775-014-1173-y. [DOI] [PubMed] [Google Scholar]
- Palmer T.; Santini C.; Iobbi-Nivol C.; Eaves D. J.; Boxer D. H.; Giordano G. Involvement of the narJ and mob gene products in distinct steps in the biosynthesis of the molybdoenzyme nitrate reductase in Escherichia coli. Mol. Microbiol. 1996, 20, 875–884. 10.1111/j.1365-2958.1996.tb02525.x. [DOI] [PubMed] [Google Scholar]
- Dahal U. P.; Jones J. P.; Davis J. A.; Rock D. A. Small molecule quantification by liquid chromatography-mass spectrometry for metabolites of drugs and drug candidates. Drug Metab. Dispos. 2011, 39, 2355–2360. 10.1124/dmd.111.040865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strelevitz T. J.; Orozco C. C.; Obach R. S. Hydralazine as a selective probe inactivator of aldehyde oxidase in human hepatocytes: estimation of the contribution of aldehyde oxidase to metabolic clearance. Drug Metab. Dispos. 2012, 40, 1441–1448. 10.1124/dmd.112.045195. [DOI] [PubMed] [Google Scholar]
- Sambrook J.; Russell D. W.. Molecular Cloning: A Laboratory Manual, 3rd ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, 2001; p 3 volumes. [Google Scholar]
- Obach R. S. Potent inhibition of human liver aldehyde oxidase by raloxifene. Drug Metab. Dispos. 2004, 32, 89–97. 10.1124/dmd.32.1.89. [DOI] [PubMed] [Google Scholar]
- Johnson C.; Stubley-Beedham C.; Godfrey J.; Stell P. Hydralazine: A potent inhibitor of aldehyde oxidase activity in vitro and in vivo. Biochem. Pharmacol. 1985, 34, 4251–4256. 10.1016/0006-2952(85)90280-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.