Abstract
Efficient extraction of genomic DNA (gDNA) from biological materials found in harsh environments is the first step for successful forensic DNA profiling. This study aimed to evaluate two methods for DNA recovery from animal tissues (livers, muscles), focusing on the best storage temperature for DNA yield in term of quality, quantity, and integrity for use in several downstream molecular techniques. Six male Swiss albino mice were sacrificed, liver and muscle tissues (n=32) were then harvested and stored for one week in different temperatures, -20°C, 4°C, 25°C and 40°C. The conditioned animal tissues were used for DNA extraction by Chelex-100 method or NucleoSpinC Blood and Tissue kit. The extracted gDNA was visualized on 1.5% agarose gel electrophoresis to determine the quality of gDNA and analysed spectrophotometrically to determine the DNA concentration and the purity. Both methods, Chelex-100 and NucleoSpin Blood and Tissue kit found to be appropriate for yielding high quantity of gDNA, with the Chelex 100 method yielding a greater quantity (P < 0.045) than the kit. At -20°C, 4°C, and 25°C temperatures, the concentration of DNA yield was numerically lower than at 40°C. The NucleoSpinC Blood and Tissue kit produced a higher (P=0.031) purity product than the Chelex-100 method, particularly for muscle tissues. The Chelex-100 method is cheap, fast, effective, and is a crucial tool for yielding DNA from animal tissues (livers, muscles) exposed to harsh environment with little limitations.
Keywords: DNA degradation, DNA extraction, DNA profiling, Purity, Temperature
Introduction
In forensic science, purification of high quality and suitable quantity of DNA from challenged biological samples is a key tool for subsequent DNA profiling. Several organic and inorganic protocols are available for DNA extraction. They vary in their nucleic acids yield, processing time, and the ability of removing the Polymerase Chain Reaction inhibitors (PCR inhibitors) (Phillips et al., 2012). DNA can be successfully extracted from a wide range of biological samples, these include, blood residues, urine, semen, saliva, soft and hard tissues. These samples presented in several environmental conditions such as temperature, humidity changes, chemical, physical and microbial contamination, all of which require different extraction strategies to remove the inhibitors and ensure efficient DNA yields (Willard et al., 1998).
“Upon the death of an organism, internal nucleases contained within the cells cause autolysis, cellular organelles and nuclear DNA degradation over time” (El-Harouny et al., 2008). Determining the quantity and quality of DNA may provide precise way to estimate the post mortem interval (Liu et al., 2001). Therefore, it is important to know which organ is most reliable for DNA extraction, and also to know the effect of post mortem interval on DNA degradation (El-Harouny et al., 2008).
Silica membranes and chelating resin are widely used for DNA extraction in forensic laboratory (Bogas et al., 2011). Silica membranes based technology provides reliable and reproducible DNA recovery. Its strategy follows four main steps: lysis of cellular membranes using a combination of enzymatic and mechanical approaches; selective binding of DNA on the designed silica membrane; washing away of contaminants and DNA elution (Phillips et al., 2012; Dhaliwal, 2013). Chelating resin such as Chelex.100 is simple and rapid DNA extraction method utilise inorganic solvent and do not require multiple tube transfer steps. It involves disruption of the cell membrane under boiling temperature while preventing DNA degradation using chelex suspension. Chelex has been described as efficient method for removing of PCR inhibitors such as hem in porphyrin compounds from blood (Walsh et al., 1991).
The aim of this work was to compare the efficiency of two extraction methods; Chelex-100 and NucleoSpinC Blood and Tissue kit for extraction of genomic DNA (gDNA) from mice tissue (liver and muscle) exposed to different temperature conditions, focusing on the best storage temperature for DNA yield in terms of quantity, purity, and integrity for forensic biology use.
Materials and Methods
Animals and tissue recovery
All experiments were performed in accordance with the regulation of the Animal Experimentation Committees of Faculty of Science, University of Tripoli (Tripoli, Libya). All efforts were made to fulfil the ethical experimentation standards such as minimizing the pain during animal handling and experiments as well as reducing the number of animals used. Six male Swiss albino mice, with an age range of four to five weeks and weight range of 11 g to 14 g, were used in this study. They were bred in the animal house of the Zoology Department, Faculty of Science, University of Tripoli, (Tripoli, Libya), and housed under natural conditions of light (12-hour cycle), temperature (24 ± 2°C) and 55 ± 5% relative humidity. During this period food and water were available ad libitum.
At the age of six weeks, mice were sacrificed by cervical dislocation and the intended tissue samples were removed (livers and muscles, n=32, 16 per each extraction method). The tissue samples were cut into small pieces (~ 1gm) and stored immediately in sterile Eppendorf tubes at -20°C (n=4), 4°C (n=4), 25°C (n=4) and 40°C (n=4) without using any preservative for one week.
gDNA extraction
Chelex-100 method
DNA was extracted using the Chelex-100 method as described previously. A homogenized tissue samples was added to 500 μL 5% Chelex-100 resin (Bio-Rad, US), with the subsequent addition of 10 μL proteinase K (QIAGEN, Venlo, Limburg, Netherlands) and 10µl of dithiothreitol (DTT; 1M). The mixture was vortexed, incubated at 56°C for 45 min, and boiled in a water bath for 8 min to inactivate proteinase K. After vigorous vortexing for 10 secs, the samples were centrifuged at 11,000 rpm for 5 min, and the supernatant was collected and stored in a new tube at -20°C until use.
NucleoSpinN Blood and Tissue kit
DNA was isolated from animal tissues (livers, muscles) using the NucleoSpinD Blood and Tissue kit (QIAGEN, Venlo, Limburg, Netherlands), according to manufacturer instructions, and the extracted gDNA were stored at -20°C until use.
Quantity and purity of gDNA
As described previously, the gDNA concentration and purity were assessed by optical density measurements using the NanoDrop™ 2000 Lite Spectrophotometer (Thermo Scientific, Wilmington. USA). For this purpose, DNA absorbance was measured at 260 nm to determine the quantity of DNA, and DNA purity was estimated by determining the A260/A280 ratio and comparing it to the reference value 1.8 (Desjardins and Conklin, 2010).
DNA visualization on agarose gel
The presence and quality/integrity of gDNA extracted by the two methods were analysed on 1% agarose gel. Ten animal tissue DNA aliquots were stained with 1 μL RedGel fluorescence and subjected to electrophoresis on the agarose gel.
The gDNA was visualized under an UV transilluminator, and the image was digitalized. The degraded DNA of the samples was observed by the visibility of the bands compared against a known molecular weight marker.
Statistical analysis
The Statistical Package for the Social Sciences (IBM SPSS Statistics for Windows, Version 20.0. Armonk, NY: IBM Corp.) was used to perform the statistical analysis.
The gDNA quantity and purity values were statistically determined by one-way analysis of variance (ANOVA) test, and differences were compared with a two-way analysis of variance with post-hoc Tukey test. P < 0.05 was considered significant. The results are shown as means ± SD.
Results
Assessment of gDNA quality extracted from animal tissues
The spectrometric assay demonstrated that the quantity of DNA extracted from animal tissue samples was higher (P < 0.05) for the Chelex-100 method than for the NucleoSpin Blood and Tissue kit, particularly for liver tissues.
Using Chelex-100 method, it was found that the quantities of the DNA extracted from liver samples stored at -20°C, 4°C, 25°C and 40°C were 1068.9±62.5, 507.8±118.51, 613.2±127.98 and 637.1±123.24, respectively. The greatest amount of DNA was obtained from tissue samples stored under -20°C (P<0.05).
However, there was no a significant difference between other conditions in terms of quantity of the gDNA extracted (P > 0.05) (Fig. 1). Furthermore, the results showed limited quantity of the DNA extracted from these samples using NucleoSpin® Blood and Tissue kit. It was found that the quantities of DNA extracted from liver tissues stored at -20°C, 4°C, 25°C and 40°C were 294.3±38.8, 18.05±4.31, 12.1±0.98 and 17.2±9.1, respectively (Fig. 1).
Fig. 1.
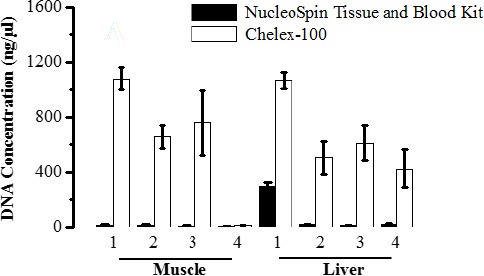
Average concentration of DNA measured in ng/µl. DNA was extracted from animal tissues (muscles, livers) stored in different temperature conditions using NucleoSpinl Blood & Tissue kit (black bar) and Chelex-100 method (white bar). Data are expressed as mean ± S.D, P < 0.05 significant.
Similarly, to the results obtained by Chelex-100 method, the samples processed using NucleoSpin® Blood and Tissue kit immediately after one week storage at -20°C appear to have greatest amount of DNA (294.3 ± 38.8) although far less than result obtained by Chelex (1068.9 ± 62.5) (Fig. 1).
For muscle tissue samples, the total yields of extracted DNA using Chelex method for samples stored at -20°C, 4°C and 25°C was 1081.5±80.8, 659.6±82.7, and 759.1±236.3 respectively, that was comparable to the amount of DNA extracted from liver tissues using the same extraction technique.
However, the muscle samples stored under 40°C for a week gave low DNA copies (11.85±2.3) indicative degradation effect. All muscle tissue samples processed with NucleoSpin® Blood and Tissue kit recovered lower DNA concentration compared to Chelex-100 method (Fig. 1).
Assessment of gDNA purity extracted from animal tissues
The NucleoSpini Blood and Tissue kit gave a higher (P < 0.05) DNA purity than did Chelex-100. Although there was a variation among the samples extracted, the kit yielded purities of nearly the reference value 1.8 (Desjardins and Conklin, 2010). The purity ratio (A260/A280) of DNA obtained by NucleoSpin/Blood and Tissue kit, particularly from liver samples, although it was not optimum, was higher than that extracted using Chelex-100 (0.9-1.9). DNA extracted from muscle tissues by Chelex-100 shown to have lower purity (0.7- 0.98) indicates contamination with protein (Fig. 2).
Fig. 2.
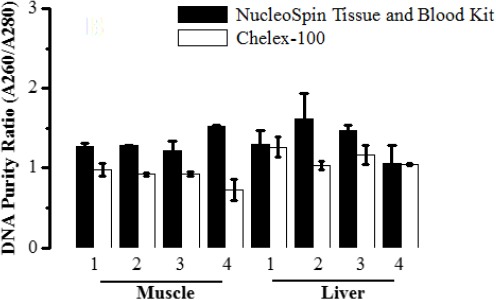
Average purity of extracted gDNA. DNA was spectrophotometrically analysed to determine the purity (A260/A280) of extracted DNA from animal tissues (muscles, livers) stored in different temperature conditions using NucleoSpinl Blood & Tissue kit (black bar) and Chelex-100 method (white bar). Data are expressed as mean ± S.D, P < 0.05 significant.
The integrity and quality of gDNA extracted by the two methods was also analysed on 1.5% agarose gel (Fig. 3). The quality of DNA observed by the visibility of the bands against a known molecular weight marker. Chelex produced DNA of minimum degradation from liver tissues stored at -20°C, 4°C, and 25°C as indicated by clear bands on the gel (Fig. 3).
Fig. 3.
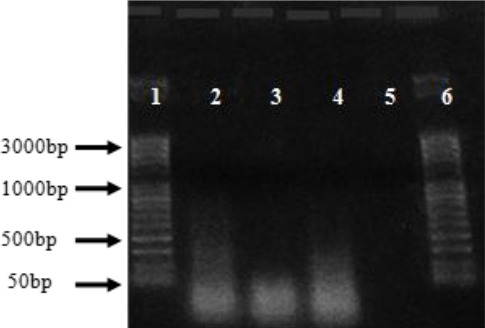
Agarose gel electrophoresis results on 1% agarose gel with DNA extracted from four animal tissue samples (2-5) by using the Chelex-100 method. Lanes: 1 and 6 ladders; lanes 2-5 liver DNA samples stored in different temperatures, L4°C, L 25°C, L-20°C and L 40°C respectively.
However, Liver tissues kept under 40°C and all DNA samples extracted by Chelex from muscle tissues did not show bands on the gel indicates degradation of the DNA produced even though the quantity of DNA recovered were relatively high. The entire DNA samples extracted by the NucleoSpine Blood and Tissue kit showed very shallow smearing, due to low copies of recovered DNA that could not have picked by the low-resolution camera used for documentation of the gel (data not shown).
Discussion
It has been well known that the best and efficient DNA extraction requires fresh tissue samples as a source material (Salman, 2000). Under some situations, particularly in case of criminal offences, fresh tissue cannot be obtained directly upon the crime occurrence. It is likely that the tissues in the field are present in several states of decomposition when exposed to different temperatures, humidity conditions, chemical, physical, and microbial contaminants (Willard et al., 1998; Vass et al., 2002).
This study was designed to keep the samples in different temperature settings, thus simulating different tissue states on the time of discovery at the crime scene. Such temperature settings (-20°C, 4°C, 25°C (room temperature), and 40°C) were maintained for one week, the amount of time assuming is adequate for transportation of the samples from the criminal scene into the forensic diagnostic laboratories where samples can then be store at -20°c for short-term manipulation or under -80°C for long-term storage.
Two protocols have been compared in this study for DNA extraction from the conditioned animal tissues in respect to the quantity, quality and time consumed for the extraction. The NucleoSpinv Blood and Tissue kit required less time (30 to 40min), compared to the Chelex protocol, which required 3.5 to 4 hrs, mostly for incubation time. However, the former protocol required multiple tubes transfer that made it laborious for manipulation of a large number of samples and subjecting to cross contamination and pipetting error.
Our preliminary results suggested that the amount of DNA extracted by Chelex from both muscle and liver tissues were far higher than the ones extracted by the NucleoSpinu Blood and Tissue kit for the same samples under the same conditions. However, the purity of the samples extracted by the later protocol was better than the ones extracted by Chelex.
Previously, it has been shown that typical DNA yield ranges from 1000–5000 ng·mg−1 in animal tissue (Pereira et al., 2011). Our results showed that the quantity of DNA extracted from liver tissues that have been stored under 40°C using Chelex protocol was far higher (637.1 ± 123.24 ng/µl) comparing with the amount of DNA recovered from the muscle tissue stored under the same condition and extracted with the same technique (11.85±2.3 ng/µl). Such results could be explained by the ability of the liver to hold intact DNA longer than the muscle cells. In a comparative study by Ebuehi et al. (2015) for study of the effects of post mortem interval (PMI) on degradation of the DNA presented in the brain, liver, kidney, and heart tissues of male mice.
The results revealed that the degradation of gDNA was a time dependent process. While the brain showed slowest DNA, degradation compared to the other organs, the liver and brain tissues were similar rigidity when viewing the profile of random segments of gDNA on the agarose gel electrophoresis. Furthermore, the study suggested that at a later PMI, the brain, followed by the liver were preferred organs for forensic studies than the heart and kidney (Ebuehi et al., 2015).
Our results agreed with Ebuehi and colleagues in term of quantity and the integrity of gDNA extracted from liver tissues which was prominent than the ones extracted from muscle tissues although we use muscle tissues rather than heart tissues, both which are multinucleated muscle tissues. Similarly, a study by Pooniya et al. (2014) suggested that muscle tissues were found to be the worst for gDNA extraction while the brain tissue preserved at -80°C and 4°C was the best among other soft tissue studied (Brain, Muscle, Kidney and heart) (Pooniya et al., 2014).
One important reason regarding the needs for high purity of extracted gDNA is the suitability of the extract for long term banking for subsequence genotyping analysis, the time needed for collections and manipulation of all samples. It is well known that the Secondary compounds and heavy metals ions can result in gDNA damage (Psifidi et al., 2015). The ability of Chelex to remove inhibitors remains controversial. Phillips et al. (2012) reported failure of the PCR to produce profile when Chelex was used for DNA extraction from blood samples suggesting that either the Chelex resin or haem were left within the samples, whereas a study by Walsh et al. (1991) suggested that gDNA extracted from bloodstain samples prepared by Chelex were less likely to have PCR inhibitors (Walsh et al., 1991).
Our results showed that the gDNA recovered by Chelex was below the acceptable level of purity as indicated by the ratio of absorbance at 260nm and 280nm that should be ~ 1.8 (Desjardins and Conklin, 2010). These results suggested the presence of protein residue which can inhibit the subsequent PCR applications and hindered gDNA-banking, particularly for gDNA extracted from muscle tissue that have shown very low purity (0.7- 0.98). DNA extracted by NucleoSpino Blood and Tissue kit for all samples was far purer than the ones extracted by Chelex. However, the DNA concentration recovered by NucleoSpinN Blood and Tissue kit was very poor and below the adequate amount of DNA concentration required for genotyping studies (~ 50ng/µl) (Psifidi et al., 2015). Furthermore, they were unable to produce visible bands on gel electrophoresis indicated the lower molecular weight of the yielded gDNA (Salman, 2000). The reason for poor quantity of gDNA produced from liver and muscle tissue when the NucleoSpin. Blood and Tissue kit was used for gDNA extraction could be explained by the fact that the technique failed to incorporate a vital step that takes into account the special feature of animal cells.
When using animal tissue, enzymatic lysis step or mechanical disruption of the tissue should be proceeding the separation of DNA from other cell components (Hofstetter et al., 1997; Dhaliwal, 2013). However, we did not examine the performance of DNA extracts by the two methods for PCR applications to see whether a small amount of gDNA can be used for PCR profiling. Walsh et al. (1991) suggested, that native high molecular weight DNA is not required to amplify the target sequence and only the target sequences are required to be intact, thus partially degraded or denatured DNA could be successfully used for PCR applications (Walsh et al., 1991).
In conclusion, Chelex protocol appeared to be acceptable regarding the quantity of the DNA product whereas NucleoSpinI Blood and Tissue kit recovered purer gDNA. Furthermore, a lysis step should be incorporated before processing the tissue samples by NucleoSpin. Blood and Tissue kit to enhance the productivity of gDNA. Although the difference between the two extraction methods was clear, a small data set was used for this study. More samples are needed to be tested to make a solid conclusion.
Acknowledgments
Authors would like to thank the staff of the National Center of Disease Control Laboratory for their practical assistance. The authors also would like to thank Dr. Taher Shaibi, a staff member at the Faculty of Science, University of Tripoli, Tripoli-Libya, for his assistance.
Conflict of interest
The authors declare that there is no conflict of interests.
References
- Bogas V, Balsa F, Carvalho M, Anjos M. J, Pinheiro M. F, Corte-Real F. Comparison of four DNA extraction methods for forensic application. Forensic Science International: Genetics Supplement Series. 2011;3(1):e194–e195. [Google Scholar]
- Desjardins P, Conklin D. NanoDrop microvolume quantitation of nucleic acids. J Vis Exp. 2010;45:2565. doi: 10.3791/2565. doi:10.3791/2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhaliwal A. DNA Extraction and Purification. MATER METHODS [Online] 2013. Available at: https://www.labome.com/method/DNA-Extraction-and-Purification.html .
- Ebuehi O.A.T, Amode M, Balogun A, Fowora A. Postmortem Time Affects Brain, Liver, Kidney and Heart DNA in Male Rat. Am. J. Biochem. 2015;5(1):1–5. [Google Scholar]
- El-Harouny M, El-Dakroory S, Attalla S, Hasan N, Hassab El-Nabi S.E. The relationship between postmortem interval and DNA degradation in different tissues of drowned rats. Mansoura J. Forensic Med. Clin. Toxicol. 2008;16(2):45–61. [Google Scholar]
- Hofstetter J.R, Zhang A, Mayeda A.R, Guscar T, Nurnberger J.I, Jr, Lahiri D.K. Genomic DNA from Mice: A Comparison of Recovery Methods and Tissue Sources. Biochem. Mol. Med. 1997;62(2):197–202. doi: 10.1006/bmme.1997.2637. [DOI] [PubMed] [Google Scholar]
- Liu L, Peng D.B, Liu Y, Deng W.N, Liu Y.L, Li J.J. A study on the relationship between postmortem interval and the changes of DNA content in the kidney cellule of rat. Fa. Yi. Xue. Za. Zhi. 2001;17(2):65–68. [PubMed] [Google Scholar]
- Pereira J.C, Chaves R, Bastos E, Leitão A, Guedes-Pinto H. An Efficient Method for Genomic DNA Extraction from Different Molluscs Species. Int. J. Mol. Sci. 2011;12:8086–8095. doi: 10.3390/ijms12118086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips K, McCallum N, Welch L. A comparison of methods for forensic DNA extraction: Chelex-100(R) and the QIAGEN DNA Investigator Kit (manual and automated) Forensic Sci. Int. Genet. 2012;6(2):282–285. doi: 10.1016/j.fsigen.2011.04.018. [DOI] [PubMed] [Google Scholar]
- Pooniya S, Lalwani S, Raina A, Millo T, Dogra T.D. Quality and quantity of extracted deoxyribonucleic Acid (DNA) from preserved soft tissues of putrefied unidentifiable human corpse. J. Lab. Physicians. 2014;6(1):31–35. doi: 10.4103/0974-2727.129088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Psifidi A, Dovas C.I, Bramis G, Lazou T, Russel C.L, Arsenos G, Banos G. Comparison of Eleven Methods for Genomic DNA Extraction Suitable for Large-Scale Whole-Genome Genotyping and Long-Term DNA Banking Using Blood Samples. PLoS One. 2015;10(1):e0115960. doi: 10.1371/journal.pone.0115960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salman A.H. Effect of Storage Temperature on the Quality and Quantity of DNA Extracted from Blood. Pakistan J. Biol. Sci. 2000;3:392–394. [Google Scholar]
- Vass A.A, Barshick S.A, Sega G, Caton J, Skeen J.T, Love J.C, Synstelien J.A. Decomposition chemistry of human remains: a new methodology for determining the postmortem interval. J. Forensic Sci. 2002;47:542–553. [PubMed] [Google Scholar]
- Walsh P.S, Metzger D.A, Higuchi R. chelex 100 as a medium for simple extraction of DNA for PCR-based typing from forensic material. Biotechniques. 1991;10(4):506–513. [PubMed] [Google Scholar]
- Willard J.M, Lee D.A, Holland M.M. Recovery of DNA for PCR amplification from blood and forensic samples using a chelating resin. Methods Mol. Biol. 1998;98:9–18. doi: 10.1385/0-89603-443-7:9. [DOI] [PubMed] [Google Scholar]


