Abstract
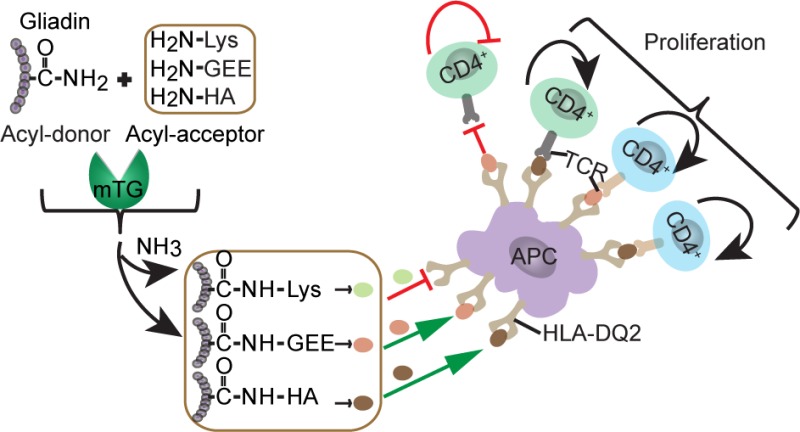
Wheat gluten confers superior baking quality to wheat based products but elicits a pro-inflammatory immune response in patients with celiac disease. Transamidation of gluten by microbial transglutaminase (mTG) and tissue transglutaminase (tTG) reduces the immunogenicity of gluten; however, little information is available on the minimal modification sufficient to eliminate gliadin immunogenicity nor has the effectiveness of transamidation been studied with T-cell clones from patients. Here we demonstrate that mTG can efficiently couple three different acyl-acceptor molecules, l-lysine, glycine ethyl ester, and hydroxylamine, to gliadin peptides and protein. While all three acyl-acceptor molecules were cross-linked to the same Q-residues, not all modifications were equally effective in silencing T-cell reactivity. Finally, we observed that tTG can partially reverse the mTG-catalyzed transamidation by its isopeptidase activity. These results set the stage to determine the impact of these modifications on the baking quality of gluten proteins and in vivo immunogenicity of such food products.
Keywords: acyl-acceptor molecule, gliadin, immunogenic properties, microbial transglutaminase, transamidation
Introduction
Celiac disease (CD) is a T-cell-mediated autoimmune enteropathy, induced by the ingestion of prolamins from wheat, rye, or barley, with an estimated worldwide prevalence of approximately 1%.1 CD has a strong genetic component as approximately 95% of patients carry HLA (human leukocyte antigen)-DQ2 (A1*0501/B1*0201), while the remainder is usually HLA-DQ8 (A1*0301/B1*0302) positive.2 This association is explained by the observation that T-cell specific for gluten peptides bound to either HLA-DQ2 or HLA-DQ8 are found in patients with CD.3,4 At present, there is no effective treatment for CD patients except a permanent life-long gluten free diet (GFD).
A number of gluten T-cell epitopes associated with HLA-DQ receptors of CD have been identified.5 A highly antigenic 33-mer peptide from α-gliadin is resistant to gastric and intestinal proteolysis and contains six partially overlapping T-cell epitopes: PFPQPQLPY (DQ2.5-glia-α1a), PYPQPQLPY (DQ2.5-glia-α1b, two copies), and PQPQLPYPQ (DQ2.5-glia-α2, three copies).6,7 It is well established that tissue transglutaminase (tTG) plays a critical role in CD pathogenesis. Specifically, the tTG mediated deamidation of Q → E strongly enhances the binding of gluten epitopes to HLA-DQ2 or HLA-DQ8 and such peptide-HLA-DQ complexes strongly activate gluten-specific T-cell clones.8−10
Microbial transglutaminase (mTG) from Streptomyces mobaraensis is a food-grade transamidase and widely used in the food industry. Similar to tTG, mTG can deamidate glutamine residues to glutamic acid residues, resulting in gluten peptides with T-cell stimulatory properties.11−14 In contrast mTG does not affect the reactivity of gliadin-specific antibodies.15 Here, the in vitro transamidation of Q by either tTG or mTG can prevent the deamidation process induced by tTG present in the human intestine. Indeed, previous studies indicated that selective modification of glutamine residues present in toxic epitopes by mTG transamidation using l-lysine or l-lysine methyl ester prevented the deamidation process.16,17 Moreover, Elli et al. found that the modification of gluten by tTG with l-lysine inhibited the duodenal immunological effects on cultured biopsies from CD patients.18 Additionally, gluten transamidated by tTG and l-lysine methyl ester reduced the number of clinical relapses in challenged patients but did not eradicate the antigenicity of gluten.19 Transamidated gluten can be used to produce bread with less immunoreactive gluten20 and recently, Ribeiro et al. exploited n-butylamine and mTG to obtain wheat flour with decreased CD toxic epitopes.21 Most importantly, mTG has much lower deamidation activity as compared with tTG22 and mTG is active with a variety of acyl-acceptor molecules so that several additional acyl-acceptor molecules can be used instead of lysine and link them to peptides or proteins of interest.23,24
Gluten has unique properties which make it highly suitable for the preparation of high quality dough, which are tightly linked to its characteristic amino acid composition dominated by high glutamine content. Therefore, modification of such glutamines by transamidation could have negative consequences.25 Minimal modification is thus desirable; however, little information on the minimal modification sufficient to eliminate gluten immunogenicity is available. Therefore, we investigated whether apart from lysine other acyl-acceptor molecules can also be used with mTG to eliminate the T-cell stimulatory properties of gliadin peptides and proteins. For this purpose we compared the effect of transamidation with l-lysine (Lys), glycine ethyl ester (GEE), and hydroxylamine (HA) as acyl-acceptor molecules.
Materials and Methods
Synthetic Peptides and Chemicals
DQ2.5-glia-α1a (native form LQPFPQPQLPYPQ and deamidated form LQPFPQPELPYAA), DQ2.5-glia-α2 (native form PFPQPQLPYPQPQ and deamidated form AAPQPELPYPQPQ), and deamidated 33-mer (LQLQPF (PQPELPY)3 PQPQPF) peptides were synthesized by standard Fmoc chemistry on a multiple peptide synthesizer (Syroll, MultiSynTech GmbH, Witten, Germany). The native 33-mer peptide (LQLQPF (PQPQLPY)3 PQPQPF) was synthesized by GL Biochem Ltd. (Shanghai, China). Details on the HLA-DQ2 binding properties of the gliadin peptides can be found in the reference Sollid et al.5 The purity and integrity of the peptides was confirmed by reversed-phase high-performance liquid chromatography (RP-HPLC) and mass spectrometry. mTG from Streptomyces mobaraensis was donated by Jiangsu Yiming Biological Products Co., Ltd. (Jiangsu, China) with a declared activity of 1 000 units (U)/ g and has an amino sequence that is identical to Ajinomoto transglutaminase. l-Lysine (Lys, >98%) was bought from Sangon Biotech (Shanghai, China). Glycine ethyl ester hydrochloride (GEE, >98%), hydroxylamine hydrochloride (HA, 98%), lysine methyl ester (LME), and n-butylamine (BL, >99.5%) were from Sigma-Aldrich. Gliadin, pepsin (3800 units/mg solid), trypsin (activity 10 000 BAEE units/mg), and tTG from guinea pig liver (4 units/mg solid) were purchased from Sigma-Aldrich. IMDM used in cell culture was obtained from Lonza (BioWhittaker, Belgium). Other chemicals were of analytical grade.
Modification of Peptides by mTG and tTG
In order to study the effect of pH on the transamidation and deamidation reaction, the experiments were performed according to a previous study with some modifications.26 In preliminary experiments, we observed that the mTG enzyme activity at pH 8.0 is lower than at pH 6.0, in agreement with previous work.22 On the basis of these results and with the aim of achieving optimal transmidation at both low and high pH, we choose to use 0.13 U mTG at pH 8.0 and 0.50 U mTG at pH 6.0. All mTG experiments were carried out in 200 mM HEPES buffer. For determination of the effect of pH on transamidation 0.19 μmol of 33-mer, 0.50 U mTG with either 40 μmol of Lys or 40 μmol of HA was incubated in 1 mL of buffer at either pH 6.0, 7.0, or 8.0 for 1 h. To determine the effect of the acyl-acceptor concentration on the transamidation reaction 0.19 μmol of 33-mer, 0.13 U mTG and a concentration range of GEE (0.16 μmol, 0.63 μmol, 2.50 μmol, 10 μmol, or 40 μmol) was incubated in 1 mL of buffer at pH 6.0 for 1 h.
On the basis of previous work27 and the effect of the acyl-acceptor concentration on the transamidation reaction, it was deduced that the 33-mer has three modification sites, whereas the DQ2.5-glia-α1a and DQ2.5-glia-α2 have one modification site. Therefore, a 1:210 molar ratio between the 33-mer peptide and acyl-acceptor molecule and a 1:70 molar ratio between DQ2.5-glia-α1a/DQ2.5-glia-α2 and acyl-acceptor molecule was selected. As Lys has a pKa value that is substantially higher as that of both GEE and HA,23 we choose to use pH 8.0 in the case of Lys, pH 6.0 in the case of HA, and GEE for all further experiments. In detail, 0.19 μmol of 33-mer, 0.50 U mTG, and 40 μmol of Lys was incubated in 1 mL of reaction mixture at pH 8.0, while 0.19 μmol of 33-mer, 0.13 U mTG, and 40 μmol of GEE/HA was incubated in 1 mL of reaction mixture at pH 6.0. For DQ2.5-glia-α1a and DQ2.5-glia-α2, 0.57 μmol of peptide, 0.50 U mTG, and 40 μmol of Lys was incubated in a final volume of 1 mL at pH 8.0, and 0.57 μmol of peptide, 0.13 U mTG, and 40 μmol of GEE, HA, LME, or BL was incubated in a final volume of 1 mL at pH 6.0. For deamidation, 0.19 μmol of 33-mer peptide or 0.57 μmol of DQ2.5-glia-α1a/α2 and 0.50 U mTG were mixed in 1 mL at pH 6.0, 7.0, or 8.0.
The reaction mixtures were preincubated for 5 min at 50 °C before adding mTG. mTG treatment was carried out at 50 °C for 1 h, followed by termination of the enzymatic reaction by heating at 85–90 °C for 10 min. Subsequently, in selected experiments, tTG treatment was performed by incubating 50 μL of mTG-transamidated product with 15 μL of tTG (1 mg/mL) in a final volume of 65 μL at 37 °C, pH 6.5 for 16 h, in 200 mM HEPES buffer with 8 mM CaCl2.
Mass Spectrometry
Mass spectrometric analysis of the synthetic gluten peptides before and after mTG and tTG-mTG treatment was performed on a Bruker Microflex (matrix-assisted laser desorption ionization-time-of-flight (MALDI TOF)) and a Thermo Fisher LTQ-FT Ultra. For the MALDI TOF part, matrix solution (10 mg/mL α-cyano-4-hydroxycinnamic acid in 50:50 acetonitrile/water with 0.2% TFA) was prepared. Next, peptide solutions were mixed with 1 μL of matrix solution on a 96 well target plate. Measurements were detected in reflectron mode with acquisition mass range of 500–5000 Da. For DQ2.5-glia-α1a, DQ2.5-glia-α2 peptides, and their modified forms, measured average masses were corrected based on the mass of the reference peptide (VNTPEHVVPYGLGSPSRS, monoisotopic mass 1895.96). Peptide Calibration Standard from Bruker was used to calibrate the data of the 33-mer peptide and its modified forms.
For Fourier transform-ion cyclotron resonance (FT-ICR) mass spectrometry, peptides were measured by tandem MS, equipped with a nanoflow liquid chromatography 1100 HPLC system (Agilent Technologies), as previously described.28 Peptides were trapped at 10 μL/min on a 1.5 cm column (100-μm i.d. ReproSil-Pur C18-AQ, 3 μm, Dr. Maisch GmbH, Germany) and eluted to a 20 cm column (50-μm ID; ReproSil-Pur C18-AQ, 3 μm) at 150 nL/min. The column was developed with a 20 min gradient from 0 to 30% acetonitrile in 0.1% formic acid. The end of the nanocolumn was drawn to a tip (i.d. about 5 μm), from which the eluent was sprayed into a LTQ-FT Ultra mass spectrometer (Thermo Electron). The mass spectrometer was operated in data-dependent mode, automatically switching between MS and MS/MS acquisition. Full scan MS spectra were acquired in the FT-ICR with a resolution of 25 000 at a target value of 3 000 000. The two most intense ions were then isolated for accurate mass measurements by a selected ion monitoring scan in FT-ICR with a resolution of 50 000 at a target accumulation value of 50 000. The selected ions were then fragmented in the linear ion trap using collision-induced dissociation at a target value of 10 000. The tandem mass spectra of modified and unmodified gliadin peptides were compared and manually interpreted to determine the site of modification.
Gliadin Preparation and Modification
For the purpose of modification of gliadin in solution, a pepsin/trypsin digest of gliadin was prepared as described previously with some modifications.29 First, 1 g of gliadin was solubilized in 10 mL of 1 M acetic acid and boiled for 10 min. Subsequently, 10 mg of pepsin was added and the mixture was incubated for 4 h at 37 °C followed by the adjustment of the pH to 7.8 with NaOH and the addition of 10 mg of trypsin. Next, the mixture was incubated for another 4 h at 25 °C. Finally, the mixture was dialyzed with water (molecular weight cutoff, 14 kDa), followed by concentrating using 10 kDa centrifugal filter units. The protein concentration was determined by a bicinchoninic acid (BCA) assay.
mTG treatment of the gliadin preparation was performed as follows: 1 mg of gliadin was mixed with 0.50 U mTG and 10 μmol, 40 μmol, or 160 μmol of Lys in a final volume of 1 mL at pH 8.0. For transamidation with GEE and HA, 1 mg of gliadin, 0.13 U mTG and 10 μmol, 40 μmol or 160 μmol acyl-acceptor molecules were mixed in a final volume of 1 mL at pH 6.0. For the mTG mediated deamidation, the reaction solution was prepared using 1 mg of gliadin and 0.50 U mTG.
The reaction mixtures were preincubated for 5 min at 50 °C before adding mTG. mTG treatment was carried out at 50 °C for 1 h, followed by termination of the enzymatic reaction by heating at 85–90 °C for 10 min. Subsequently, tTG treatment was performed by incubating 50 μL of mTG-transamidated product with 15 μL of tTG (1 mg/mL) in a final volume of 65 μL at 37 °C, pH 6.5 for 16 h, in 200 mM HEPES buffer with 8 mM CaCl2.
T-Cell Proliferation Assays
Proliferation assays were performed in 150 μL of IMDM medium supplemented with 2 mM glutamine and 10% human serum in 96-well flat-bottom plates as previously described.30 In brief, irradiated (3000 rad) HLA-DQ2-matched antigen-presenting cells (105) were incubated with 50 μL of antigen for 2 h, followed by the addition of 15 000 DQ2.5-glia-α1 or DQ2.5-glia-α2 specific T-cell clones. All conditions were carried out in triplicate. mTG- and tTG-mTG-treated 33-mer were used at 9.48 nmol/mL, 3.16 nmol/mL, 1.05 nmol/mL, 0.35 nmol/mL, and 0.12 nmol/mL. mTG- and tTG-mTG-treated DQ2.5-glia-α1a peptide or DQ2.5-glia-α2 peptide were used at 28.43 nmol/mL, 9.48 nmol/mL, 3.16 nmol/mL, 1.05 nmol/mL, or 0.35 nmol/mL. mTG- and tTG-mTG-treated gliadin was used at 20 μg/mL. Both native and deamidated versions of DQ2.5-glia-α1a, DQ2.5-glia-α2, and 33-mer peptides were used as controls. After 48 h at 37 °C, 0.5 μCi of 3H-thymidine/well was added to the cultures and the cells were harvested 18 h later. 3H-thymidine incorporation in the T-cell DNA was determined with a liquid scintillation counter (1205 Betaplate Liquid Scintillation Counter) and results are expressed as the mean counts per minute (c.p.m.).
Statistical Analysis
Statistical analyses were performed with GraphPad Prism 7 software. Values are presented as the mean ± standard deviation (SD) (n ≥ 3).
Results
Effect of pH and Concentration of Acyl-Acceptor Molecule on Modification of Gliadin Peptides by mTG
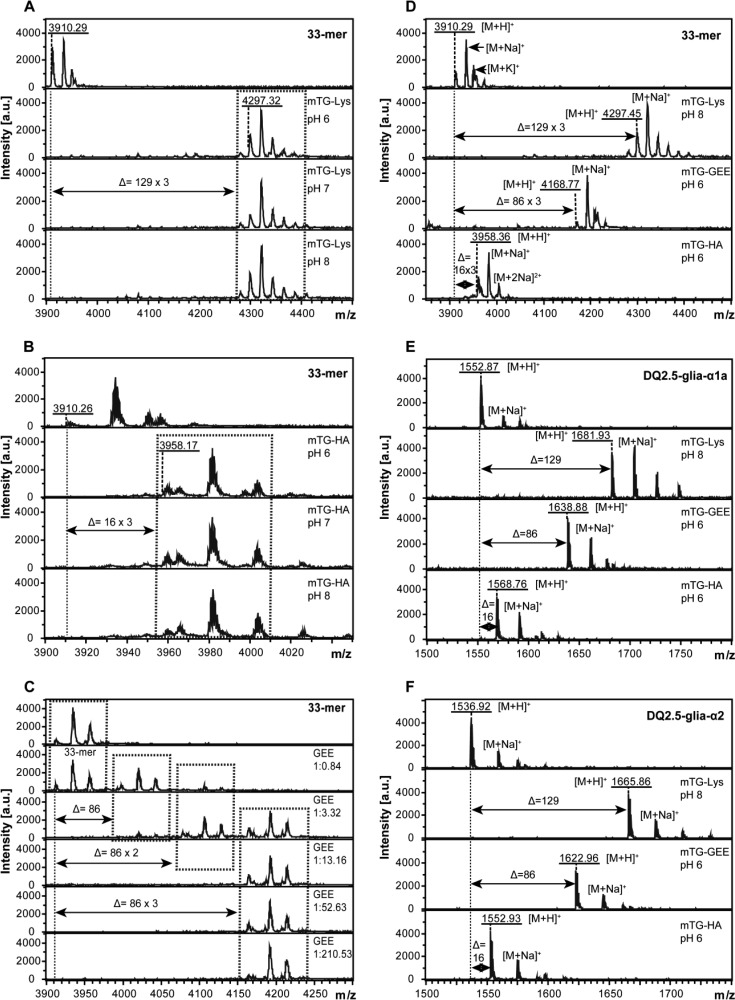
To achieve optimal modification of gliadin peptides by mTG, we first determined the effect of pH and the concentration of the acyl-acceptor molecule in two separate experiments. To determine the effect of pH we incubated the 33-mer α-gliadin peptide, known to contain three glutamine residues that can be targeted by transglutaminases, with mTG and a molar excess of Lys and HA at pH 6.0, pH 7.0, or pH 8.0. The MALDI-TOF analysis of the end product demonstrate that at all three pH values this resulted in shifts in molecular mass of 387 Da in the case of Lys (Figure 1A) and 48 Da in the case of HA (Figure 1B), indicative of the addition of three Lys and three HA molecules, respectively. Moreover, the modification was highly efficient as more than 99% of the 33-mer peptide was modified. Similarly, incubation of the 33-mer peptide with mTG and the acyl acceptor GEE also resulted in near complete modification at every pH tested as demonstrated by a shift of 258 Da in the mass of the peptide, corresponding to the addition of three GEE molecules (data not shown). Also, similar results were obtained with both the DQ2.5-glia-α1a (Supplementary Figure 1A–C) and DQ2.5-glia-α2 peptides (Supplementary Figure 1E–G). Together these results demonstrate that the pH did not have a major impact on the transamidation reaction with any of the three acyl-acceptor molecules.
Figure 1.
Transamidation pattern of DQ2.5-glia-α1a, DQ2.5-glia-α2, and 33-mer peptide after modification using mTG and three different acyl-acceptor molecules. (A) Effect of pH on 33-mer reaction induced by mTG in the presence of Lys. (B) Effect of pH on 33-mer reaction induced by mTG in the presence of HA. (C) Effect of GEE concentration on 33-mer transamidation induced by mTG at pH 6.0. Molar ratio between the 33-mer peptide and GEE is 1:0.84, 1:3.32, 1:13.16, 1:52.63, 1:210.53 as shown. (D) Transamidation pattern of 33-mer by mTG. As compared with native peptide, 33-mer cross-linked to Lys, GEE, and HA resulted in a shift of 129 × 3 Da, 86 × 3 Da and 16 × 3 Da, corresponding to the addition of three Lys, GEE, and HA groups, respectively. (E) Transamidation pattern of DQ2.5-glia-α1a by mTG. DQ2.5-glia-α1a presented 129 Da, 86 Da, and 16 Da shift responding to the addition of a single Lys, GEE, and HA group. (F) Transamidation pattern of DQ2.5-glia-α2 by mTG. DQ2.5-glia-α2 presented 129 Da, 86 Da, and 16 Da shift responding to the addition of a single Lys, GEE, and HA group. Monoisotopic mass are shown throughout.
To determine the effect of the concentration of the acyl-acceptor molecule on the transamidation reaction, the 33-mer α-gliadin peptide was incubated with mTG in the presence of 5 different concentrations of GEE. The MALDI-TOF analysis of the end products demonstrate that at low concentrations of GEE (0.16 and 0.63 μmol/mL) the modification was incomplete with either one, two, or three additions of the acyl-acceptor molecules to the gliadin peptide (Figure 1C). In contrast, at higher concentrations (2.50, 10, and 40 μmol/mL) more than 99% of the 33-mer was modified at three sites (Figure 1C). To ensure (near) complete modification of the gliadin peptides, we chose to use 40 μmol/mL acyl-acceptor concentrations for all further experiments.
In contrast, the pH did influence mTG-mediated deamidation (Supplementary Figure 1D,H). In line with previous results,22,31 the deamidation activity of mTG was maximal at pH 6.0 and lower at neutral and basic pH.
Next we determined the mTG mediated modification of the DQ2.5-glia-α1a, DQ2.5-glia-α2, and 33-mer peptides with three distinct acyl-acceptor molecules, Lys, GEE, and HA under the optimal conditions determined above. The MALDI-TOF analysis indicated that the 33-mer peptide shifted 387, 258, and 48 Da upon modification with Lys, GEE, and HA, respectively, corresponding to the addition of three Lys, GEE, and HA, respectively (Figure 1D). Similarly, the modification of the DQ2.5-glia-α1a (Figure 1E) and DQ2.5-glia-α2 (Figure 1F) peptides resulted in shifts of 129, 86, and 16 Da, corresponding to the addition of a single Lys, GEE, and HA group. In addition, we observed that both lysine methyl ester and n-butylamine could be similarly coupled to gliadin by mTG (Supplementary Figures 4 and 6). In all cases, the modification was nearly complete as demonstrated by the disappearance of the unmodified peptide and the sodium and potassium adducts thereof upon treatment with mTG. Together this demonstrates that mTG can effectively catalyze the transamidation reaction between all tested acyl-acceptor molecules and glutamine residues in the three tested gliadin peptides.
Identification of mTG Modified Glutamine Residues in Gliadin Peptides
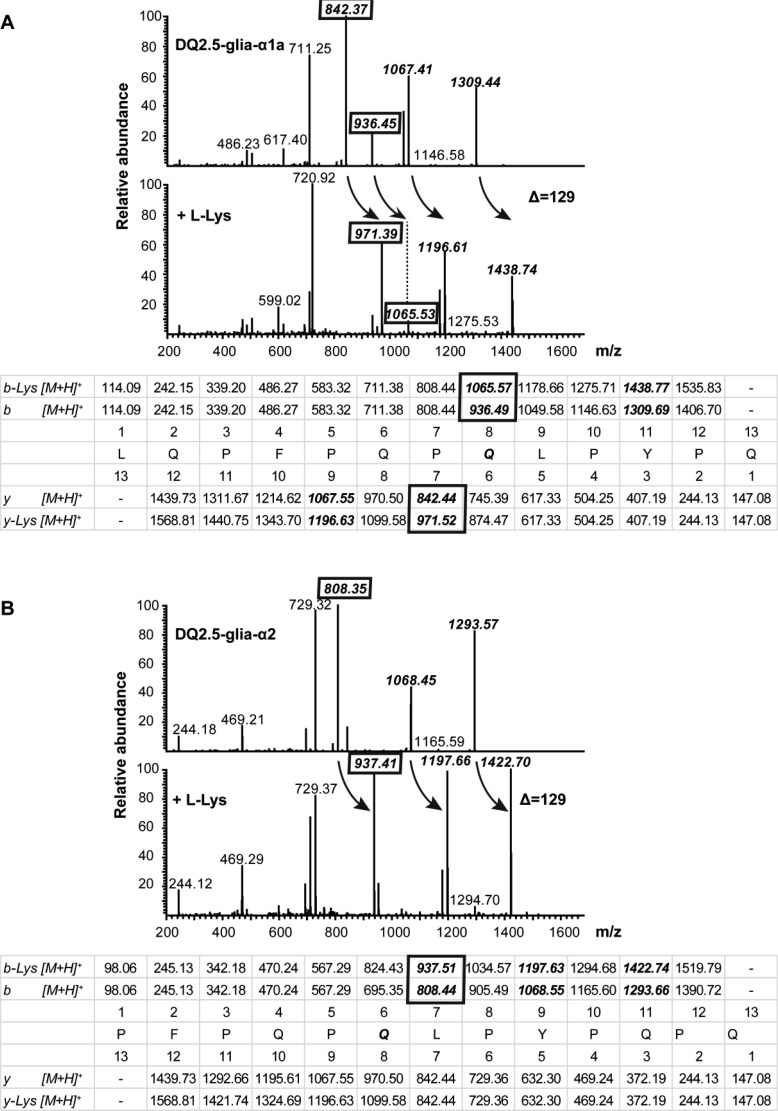
In the above experiments, we observed that a single acyl-acceptor molecule was coupled to the DQ2.5-glia-α1a and DQ2.5-glia-α2 peptides while three groups were added to the 33-mer peptide, which indicates that the three tTG target glutamines in these peptides are also modified by mTG. To verify that this is indeed the case, the DQ2.5-glia-α1a and DQ2.5-glia-α2 peptide were treated with mTG and acyl-acceptor molecules and the end products were analyzed by FT-ICR. The resulting MS/MS spectra of the peptides and fragments thereof indicated that over 99% of the peptides were transamidated with all three acyl-acceptor molecules (DQ2.5-glia-α1a, 99.7% (Lys), 99.6% (GEE), and 99.5% (HA); DQ2.5-glia-α2:99.8% (Lys), 99.6% (GEE), and 99.8% (HA)), confirming the results obtained with the MALDI-TOF analysis.
For DQ2.5-glia-α1a (Figure 2A), the MS/MS spectra obtained from the fragmentation of the native and Lys-modified peptides reveal a series of b- and y-ions where the shift in m/z of 936 for the b8-ion from the native peptide to 1065 in the modified peptide indicates that the Lys modification occurred at the glutamine residue of p8 in the peptide. This is confirmed by the observed shift in m/z from 842 to 971 for the y7-ion (Figure 2A). Similar results were obtained when GEE and HA were used as acyl-acceptor molecules (Supplementary Figure 2A).
Figure 2.
Identification of transamidated sites induced by mTG using LTQ-FT Ultra. (A) Observed fragment of DQ2.5-glia-α1a before treatment (upper spectrum) and after treatment with mTG in the presence of Lys (lower spectrum); predicted fragment ion masses are given in the table with identified ions illustrating transamidation shown in boxes. (B) Observed fragment of DQ2.5-glia-α2 before treatment (upper spectrum) and after treatment with mTG in the presence of Lys (lower spectrum); predicted fragment ion masses are given in the table with identified ions illustrating transamidation shown in boxes. Arrows indicate the shift in Da due to transamidation.
Similarly, for the DQ2.5-glia-α2 peptide the MS/MS spectra indicate a shift in m/z from 808 to 937 for the b7-ion, demonstrating that the Lys modification was at the glutamine at p6 in the peptide (Figure 2B). Similar results were obtained with the GEE and HA modifications (Supplementary Figure 2B). Thus, mTG selectively modifies the gliadin peptides at a single position which corresponds to Q8 in DQ2.5-glia-α1a and Q6 in DQ2.5-glia-α2. Moreover, the transamidation was nearly complete in all cases.
Abrogation of Immunogenic Properties of Gliadin Peptides Is Both Acyl-Acceptor Molecule and T-Cell Clone Dependent
To evaluate whether the treatment of peptides with mTG and the three acyl-acceptor molecules abrogated their immune stimulatory properties, four T-cell clones specific for DQ2.5-glia-α1a and four T-cell clones specific for DQ2.5-glia-α2 that have been previously isolated from intestinal biopsies of patients were used. The TCR usage of all T-cell clones was determined previously,4 and each T-cell clone expressed a unique TCR (Supplementary Table 1).
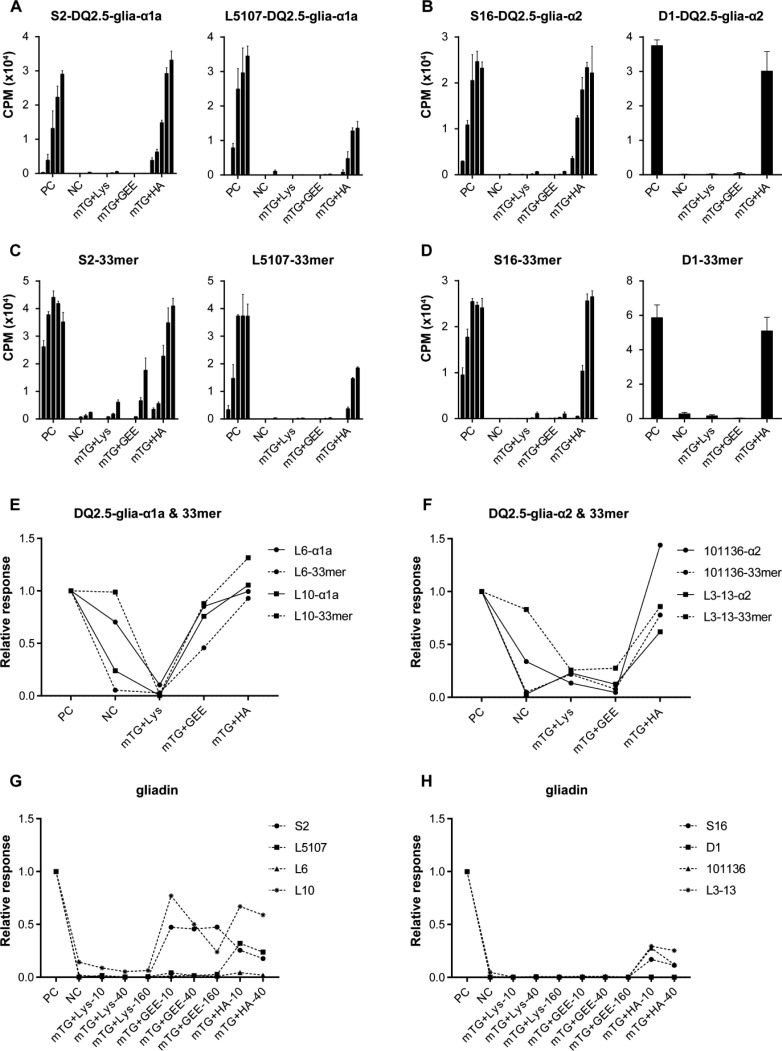
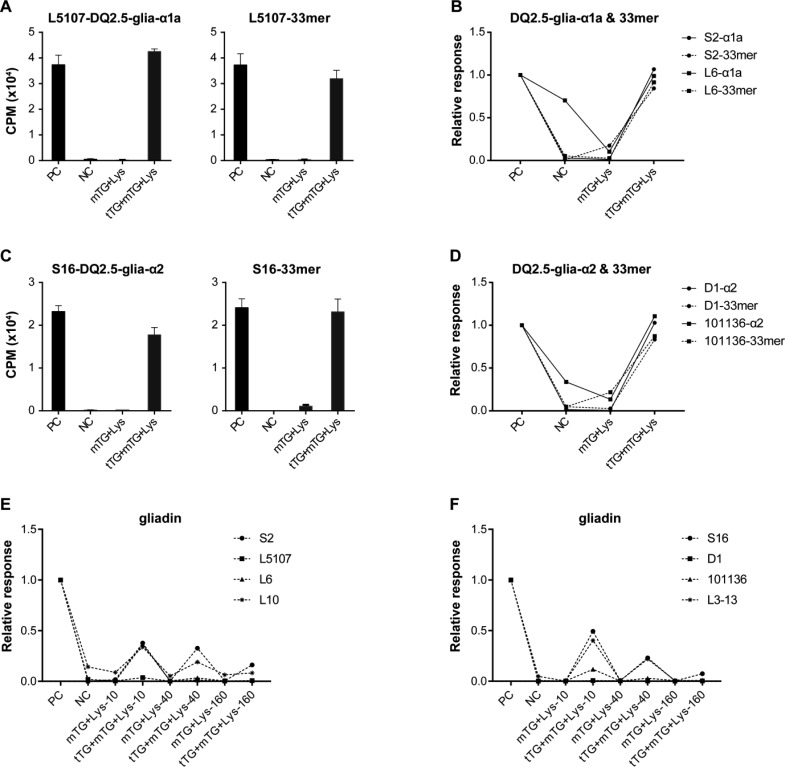
For both the DQ2.5-glia-α1a and DQ2.5-glia-α2-specific T-cell clones, the Lys and GEE modification of the DQ2.5-glia-α1a and DQ2.5-glia-α2 peptides virtually abrogated the T-cell response while modification with HA (Figure 3A–F,) and BL (Supplementary Figure 6B) was hardly effective. Similarly, Lys but not HA modification of the 33-mer peptide strongly reduced the response of the T-cell clones while GEE modification was less effective for T-cell clone S2 (Figure 3C). To further investigate the effectiveness of the modification with the acyl-acceptor molecules, we tested two additional DQ2.5-glia-α1a and two DQ2.5-glia-α2 specific T-cell clones. In line with the above observations, the Lys modification was found to be highly effective for the gliadin epitopes (Figure 3A,B and 3E,F), the 33-mer (Figure 3C,D and 3E,F) and the gliadin protein (Figure 3G,H) while the HA modification was hardly effective. In contrast, the GEE modification was found to inhibit the response of all DQ2.5-glia-α2 specific T-cell clones but proved ineffective in the case of two of the DQ2.5-glia-α1a specific T-cell clones tested. Additionally, the three T-cell clones, L6, L10, and L3-13 also responded significantly to the nondeamidated native gliadin peptides.
Figure 3.
Reactivity pattern of glia-α1and glia-α2-specific-T-cell clones to DQ2.5-glia-α1a, DQ2.5-glia-α2, 33-mer, gliadin, and their mTG-treated forms. (A) Reactivity pattern of glia-α1-specific T-cell clone S2 and L5107 to DQ2.5-glia-α1a and its modified products (5 concentrations tested in each group). (B) Reactivity pattern of glia-α2-specific T-cell clones S16 and D1 to DQ2.5-glia-α2 and its modified products (5 concentrations tested with S16 and only the highest concentration with D1). (C) Reactivity pattern of glia-α1-specific T-cell clones S2, and L5107 to 33-mer and its modified products (5 concentrations tested in each group). (D) Reactivity pattern of glia-α2-specific T-cell clones S16 and D1 to 33-mer and its modified products (5 concentrations tested with S16 and only the highest concentration with D1). (E) Reactivity pattern of glia-α1-specific T-cell clones L6 and L10 to DQ2.5-glia-α1a, 33-mer and their modified products (only the highest concentration tested). (F) Reactivity pattern of glia-α2-specific T-cell clones 101136 and L3-13 to DQ2.5-glia-α2, 33-mer and their modified products (only the highest concentration tested). (G) Reactivity pattern of glia-α1-specific T-cell clones to gliadin protein and its modified products (only the highest concentration tested). (H) Reactivity pattern of glia-α2-specific T-cell clones to gliadin protein and its modified products (only the highest concentration tested). PC, deamidated peptides as positive control; NC, native peptides as negative control. Relative response: CPM of T cell clone caused by peptide/that of caused by deamidated peptide (positive control).
tTG-Catalyzed Hydrolysis of Transamidated Products Can Generate Deamidated T-Cell Epitopes
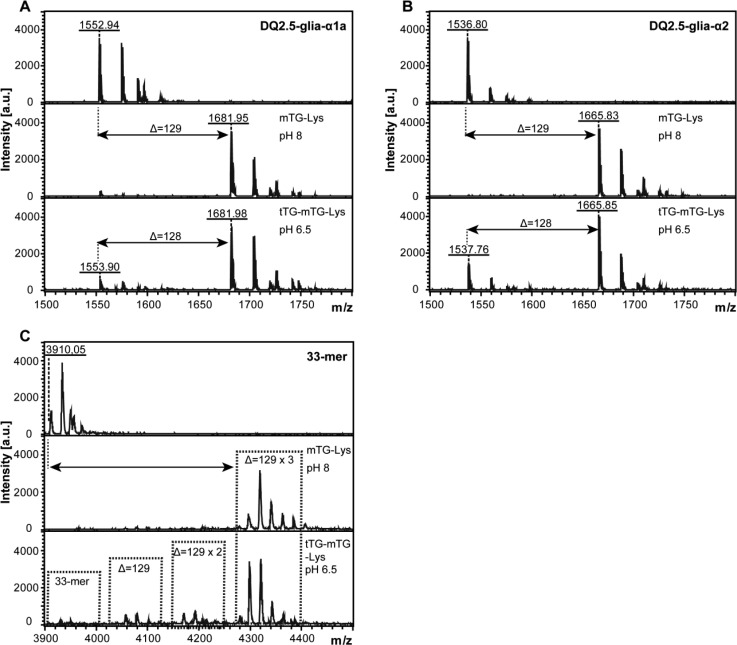
Upon ingestion of mTG transamidated gliadin, it could come into contact with intestinal tTG. To determine the potential effect of this on the immune stimulatory properties of the mTG treated gliadin, we treated mTG transamidated DQ2.5-glia-α1a, DQ2.5-glia-α2, and 33-mer with tTG and determined the effect through mass spectrometry. After incubation with tTG, a proportion of the Lys-modified peptide was found to be delysinated, resulting in deamidated gliadin (Figure 4A–C). Simultaneously, LME- and BL-modified peptide were also partially reversed by tTG (Supplementary Figures 4 and 6). Moreover, T-cell clones specific for the DQ2.5-glia-α1a (Figure 5A,B) and DQ2.5-glia-α2 (Figure 5C,D and Supplementary Figure 6B) epitopes responded to the tTG treated samples compared to the control and mTG treated samples, indicating that tTG had indeed reversed the transamidation of a proportion of the mTG treated epitopes. Similarly, we observed that tTG can partially reverse the mTG-mediated transamidation of gliadin protein (Figure 5E,F).
Figure 4.
Analysis of DQ2.5-glia-α1a and DQ2.5-glia-α2 after mTG and tTG treatment by MALDI-TOF. (A) Mass spectrum of DQ2.5-glia-α1a (upper panel), DQ2.5-glia-α1a after mTG transamidation with Lys (mTG-Lys, middle panel), and after tTG treatment of Lys transamidated DQ2.5-glia-α1a (tTG-mTG-Lys, lower panel). (B) Mass spectrum of DQ2.5-glia-α2 (upper panel), DQ2.5-glia-α2 after mTG transamidation with Lys (mTG-Lys, middle panel), and after tTG treatment of Lys transamidated DQ2.5-glia-α2 (tTG-mTG-Lys, lower panel). (C) Mass spectrum of 33-mer (upper panel), 33-mer after mTG transamidation with Lys (mTG-Lys, middle panel), and after tTG treatment of Lys transamidated 33-mer (tTG-mTG-Lys, lower panel). The deamidation of Q → E would result in a 1 Da shift. Monoisotopic mass are shown throughout. Arrows indicate the shift in Da after modification.
Figure 5.
tTG can revert mTG-mediated transamidation. (A) Reactivity pattern of glia-α1-specific T-cell clone L5107 to DQ2.5-glia-α1a (left panel), the 33-mer (right panel) before (NC) and after mTG transamidation with Lys (mTG-Lys) and after tTG treatment of mTG-Lys (tTG-mTG-Lys). (B) Reactivity pattern of glia-α1-specific T-cell clones S2 and L6 to DQ2.5-glia-α1a, 33-mer, and their modified products. (C) Reactivity pattern of glia-α2-specific T-cell clone S16 to DQ2.5-glia-α2 (left panel) and the 33-mer (right panel) before (NC) and after mTG transamidation with Lys (mTG-Lys) and after tTG treatment of mTG-Lys (tTG-mTG-Lys). (D) Reactivity pattern of glia-α2-specific T-cell clones D1 and 101136 to DQ2.5-glia-α2, 33-mer, and their modified products. (E) Reactivity pattern of glia-α1-specific T-cell clones to gliadin protein before (NC) and after mTG transamidation with Lys (mTG-Lys) and after tTG treatment of mTG-Lys (tTG-mTG-Lys). Three different concentrations of lysine were used, 10, 40, and 160 mM as indicated. (F) Reactivity pattern of glia-α2-specific T-cell clones to gliadin protein and its modified products.
Discussion
Wheat-based products are one of the most commonly consumed foods worldwide. As wheat gluten is the causative agent in celiac disease, this implies that a large number of food products are off limit for patients suffering from this condition. While the availability of bona fide gluten-free products has increased significantly in recent years, these are not always a good replacement for gluten-containing foods due to the special properties of gluten proteins that confer superior baking properties to wheat based-products.32 Also, an inappropriate gluten-free diet can cause nutritional shortcomings, often lacks sufficient fiber,33 and persists reduced digestibility.34 Detoxification of gluten proteins would be one approach to overcome such shortcomings. The molecular basis for the toxic properties of gluten is well established as T-cells specific for modified gluten fragments bound to the disease-predisposing HLA-DQ2 and -DQ8 molecules reside in the intestine of patients.1 Upon activation, such T-cell secrete pro-inflammatory cytokines leading to inflammation and remodelling of the intestinal morphology. The modification of gluten is an enzymatic conversion of particular glutamine residues in gluten fragments into glutamic acid, introducing a negative charge that allows high-affinity binding to either HLA-DQ2 or -DQ8. The enzyme involved, tTG, specifically modifies glutamine residues in QXP sequences (where X can be any amino acid except proline) but not in QP sequences, which results in highly selective modification of particular glutamine residues only due to the proline-rich nature of gluten proteins.11,27 Enzymatic modification of these glutamine residues has therefore been proposed as an approach to prohibit the conversion of glutamine to glutamic acid by tTG. Indeed, enzymatic coupling of Lys to the tTG target glutamines in gluten has been shown to diminish the immunogenicity of gluten.16 In addition, mTG has also been shown to effectively mediate this transamidation.16 However, a systematic analysis of the effectiveness of such an approach with a panel of well characterized gluten-specific T-cell clones from patients is currently lacking. Moreover, the coupling of the relatively large amino acid Lys to several glutamine residues in gluten proteins may affect their unique properties. Therefore, in the present study we have determined if smaller acyl-acceptor molecule would be equally capable of reducing the immunogenic properties of gluten proteins and peptides. Our results provide detailed insight into the transamidation of gliadin peptides and proteins by mTG and three distinct acyl-acceptor molecules. We have used mass spectrometry to determine the extent and exact location of the mTG mediated modifications and we have exploited the availability of gliadin-specific T-cell clones to monitor the impact of these modifications in detail. Finally, we have investigated if tTG could revert the effect of mTG-mediated transamidation.
Our results demonstrate that the DQ2.5-glia-α1a, DQ2.5-glia-α2, and 33-mer peptides were almost completely transamidated by mTG with all five acyl-acceptor molecules used, which confirms and extends the previous observation that mTG has a strong transamidation activity.22 Similarly, we observed that the pH did not have a major impact on the transamidation reaction, indicating that transamidation of gluten proteins by mTG would be robust under a variety of conditions. However, in agreement with previous observations22 mTG displayed pH-dependent deamidation activity which was maximal at pH 6.0 and minimal at pH 8.0. Thus, mTG mediated modification of gluten should preferably be carried out at higher pH values to avoid the deamidation of gluten which is known to increase immunogenicity. Importantly, our results indicated that mTG targeted the same glutamine residues that are modified by tTG in both the DQ2.5-glia-α1a, DQ2.5-glia-α2, and in the 33-mer gliadin peptide (Figure 2 and Supplementary Figure 2). Thus, the transamidation of gluten by mTG can be used to prevent the deamidation of gluten which enhances the binding of gluten epitopes to HLA-DQ molecules. Our results are in contrast to a recent study22 where two glutamine residues were transamidated in the peptide QPFPQPQLPYPQPQ, encompassing both the DQ2.5-glia-α1a and DQ2.5-glia-α2 epitopes. This may be due to different reaction conditions such as the concentration of acyl-acceptor molecules and reaction time employed.
We have used a panel of well characterized T-cell clones to evaluate the effect of the transamidation of gliadin peptides and proteins. While it is likely that abrogation of T-cell reactivity against transamidated gliadin peptides results from impaired binding to HLA-DQ, transamidation may also affect the conformation of the gliadin peptide in HLA-DQ, a possibility that cannot be inferred from the results with the T-cells. Nonetheless, whatever the mechanism, abrogation of T-cell stimulatory properties would render gluten safe for consumption by celiac disease patients.
While the transamidation of the Q residues in the DQ2.5-glia-α1a and DQ2.5-glia-α2 epitopes was nearly complete with all three acyl-acceptor molecules, significant differences were found with respect to the response of T-cell clones specific for these epitopes. In agreement with previous results,16 the modification with Lys drastically reduced the proliferation of all T-cell clones. In contrast, the modification with GEE was similarly effective in the case of DQ2.5-glia-α2-specific T-cell clones but less effective for DQ2.5-glia-α1-specific T-cell clones. This may relate to the featureless central region of the DQ2.5-glia-α1a peptide while bound to HLA-DQ2.5,4 where the relatively small addition of the GEE group may not affect the binding interface with the T-cell receptor. Strikingly, the modification with HA was largely ineffective even though this acyl-acceptor molecule was coupled as efficiently as the Lys and GEE groups. Presumably, the HA-modification had little effect on the binding of the modified peptides while that with GEE and Lys does. In this respect it is important to note that several of the T-cell clones also responded significantly to the nondeamidated native gliadin peptides, in line with previous results indicating that deamidation of gluten peptides is not always a prerequisite for T-cell recognition.29,30 As such, transamidation of gliadin would not only prevent deamidation but would also eliminate T-cell responses to native gliadin peptides. Importantly, mTG-transamidation of gliadin also abolished the T-cell stimulatory properties. However, as not all T-cell clones tested were equally affected by the mTG mediated transamidation, this may imply that the effect of mTG mediated transamidation can vary from patient to patient depending on the expressed T-cell receptor repertoire.
Finally, we demonstrate that guinea pig derived tTG can partially undo the mTG mediated transamidation reaction as we observed T-cell responses to mTG-transamidated gliadin peptides that were subsequently treated with tTG, a result consistent with a previous observation that tTG can hydrolyze iso-peptide bonds.35 While evidence for hydrolyzation of the iso-peptide bond was already apparent after 1 h, it is of note that most of the transamidated peptide was still intact even after 16 h incubation. Moreover, it is unclear whether human tTG can hydrolyze iso-peptide bonds as this is distinct from guinea pig tTG which could affect catalytic activity and substrate specificity.36,37 Finally, in future studies it may be worthwhile to combine mTG mediated transamidation with other approaches to reduce the immunogenicity of gluten.38,39
In conclusion, our results confirm previous results that mTG can effectively transamidate gliadin peptides and gluten proteins. We demonstrate that transamidation with Lys is most effective to abrogate the immune stimulatory properties of gluten while the coupling of smaller acyl-acceptor molecule is less (GEE) effective or mostly ineffective (HA). At present transamidation with lysine is thus the only available option to eliminate gluten toxicity. In this respect, it is important to note that food grade lysine is readily available on the market, potentially enabling the large scale production of transamidated gluten for incorpotation in food products. However, our results on the reversibility of the mTG mediated transamidation indicate that this may also occur in vivo. Thus, additional studies are required to test the impact of these modifications on gluten baking properties and in vivo antigenicity to evaluate the general applicability of this approach in practice.
Acknowledgments
We thank Cees Franken for help with the gliadin preparation.
Glossary
Abbreviations Used
- APC
antigen-presenting cell
- CD
celiac disease
- FT-ICR
Fourier transform ion cyclotron resonance
- GEE
glycine ethyl ester
- glia
gliadin
- HA
hydroxylamine
- Lys
l-lysine
- MALDI-TOF
matrix-assisted laser desorption/ionization time-of-flight
- mTG
microbial transglutaminase
- TCR
T-cell receptor
- tTG
tissue transglutaminase
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jafc.7b02557.
HLA-DQ2.5-glia-α1/α2-specificT-cell clones and their TCR gene usage, effect of pH on transamidation and deamidation of DQ 2.5-glia-α1a and DQ 2.5-glia-α2 peptides, identification of transamidated sites induced by mTG using LTQ-FT Ultra, identification of tTG-mediated deamidation of transamidated DQ2.5-glia-α2 by MALDI-TOF, transamidation of DQ2.5-glia-α2 with LME and effect of tTG treatment of transamidated peptide, time-tracking of mTG transamidation by MALDI-TOF, and transamidation of DQ2.5-glia-α2 with BL and reactivity pattern of glia-α2-specific-T-cell clone to mTG-treated and tTG-mTG treated peptide (PDF)
L.Z. was supported by International Science & Technology Cooperation Program of China, NO. 2013DFG31380.
The authors declare no competing financial interest.
Supplementary Material
References
- Di Sabatino A.; Corazza G. R. Coeliac disease. Lancet 2009, 373, 1480–1493. 10.1016/S0140-6736(09)60254-3. [DOI] [PubMed] [Google Scholar]
- Karell K.; Louka A. S.; Moodie S. J.; Ascher H.; Clot F.; Greco L.; Ciclitira P. J.; Sollid L. M.; Partanen J.; Disease E. G. C. o. C. HLA types in celiac disease patients not carrying the DQA1* 05-DQB1* 02 (DQ2) heterodimer: results from the European Genetics Cluster on Celiac Disease. Hum. Immunol. 2003, 64, 469–477. 10.1016/S0198-8859(03)00027-2. [DOI] [PubMed] [Google Scholar]
- Petersen J.; Kooy-Winkelaar Y.; Loh K. L.; Tran M.; Van Bergen J.; Koning F.; Rossjohn J.; Reid H. H. Diverse T Cell Receptor Gene Usage in HLA-DQ8-Associated Celiac Disease Converges into a Consensus Binding Solution. Structure 2016, 24, 1643–1657. 10.1016/j.str.2016.07.010. [DOI] [PubMed] [Google Scholar]
- Petersen J.; Montserrat V.; Mujico J. R.; Loh K. L.; Beringer D. X.; Van Lummel M.; Thompson A.; Mearin M. L.; Schweizer J.; Kooy-Winkelaar Y. T-cell receptor recognition of HLA-DQ2-gliadin complexes associated with celiac disease. Nat. Struct. Mol. Biol. 2014, 21, 480–488. 10.1038/nsmb.2817. [DOI] [PubMed] [Google Scholar]
- Sollid L. M.; Qiao S.-W.; Anderson R. P.; Gianfrani C.; Koning F. Nomenclature and listing of celiac disease relevant gluten T-cell epitopes restricted by HLA-DQ molecules. Immunogenetics 2012, 64, 455–460. 10.1007/s00251-012-0599-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao S.-W.; Bergseng E.; Molberg Ø.; Xia J.; Fleckenstein B.; Khosla C.; Sollid L. M. Antigen presentation to celiac lesion-derived T cells of a 33-mer gliadin peptide naturally formed by gastrointestinal digestion. J. Immunol. 2004, 173, 1757–1762. 10.4049/jimmunol.173.3.1757. [DOI] [PubMed] [Google Scholar]
- Shan L.; Molberg Ø.; Parrot I.; Hausch F.; Filiz F.; Gray G. M.; Sollid L. M.; Khosla C. Structural basis for gluten intolerance in celiac sprue. Science 2002, 297, 2275–2279. 10.1126/science.1074129. [DOI] [PubMed] [Google Scholar]
- Anderson R. P.; Degano P.; Godkin A. J.; Jewell D. P.; Hill A. V. In vivo antigen challenge in celiac disease identifies a single transglutaminase-modified peptide as the dominant A-gliadin T-cell epitope. Nat. Med. 2000, 6, 337–342. 10.1038/73200. [DOI] [PubMed] [Google Scholar]
- Arentz-Hansen H.; Körner R.; Molberg Ø.; Quarsten H.; Vader W.; Kooy Y. M.; Lundin K. E.; Koning F.; Roepstorff P.; Sollid L. M. The intestinal T cell response to α-gliadin in adult celiac disease is focused on a single deamidated glutamine targeted by tissue transglutaminase. J. Exp. Med. 2000, 191, 603–612. 10.1084/jem.191.4.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molberg Ø.; McAdam S.; Lundin K. E.; Kristiansen C.; Arentz-Hansen H.; Kett K.; Sollid L. M. T cells from celiac disease lesions recognize gliadin epitopes deamidated in situ by endogenous tissue transglutaminase. Eur. J. Immunol. 2001, 31, 1317–1323. . [DOI] [PubMed] [Google Scholar]
- Dekking E.; Van Veelen P.; De Ru A.; Kooy-Winkelaar E.; Gröneveld T.; Nieuwenhuizen W.; Koning F. Microbial transglutaminases generate T cell stimulatory epitopes involved in celiac disease. J. Cereal Sci. 2008, 47, 339–346. 10.1016/j.jcs.2007.05.004. [DOI] [Google Scholar]
- Kristiansen C.; Madsen L.; Fugger L.; Scott H.; Norén O.; Roepstorff P.; Lundin K. E.; Sjöström H.; Sollid L. M. Tissue transglutaminase selectively modifies gliadin peptides that are recognized by gut-derived T cells in celiac disease. Nat. Med. 1998, 4, 713–717. 10.1038/nm0698-713. [DOI] [PubMed] [Google Scholar]
- van de Wal Y.; Kooy Y.; van Veelen P.; Peña S.; Mearin L.; Papadopoulos G.; Koning F. Cutting edge: selective deamidation by tissue transglutaminase strongly enhances gliadin-specific T cell reactivity. J. Immunol. 1998, 161, 1585–1588. [PubMed] [Google Scholar]
- Cabrera-Chávez F.; Rouzaud-Sández O.; Sotelo-Cruz N.; Calderón de la Barca A. M. Transglutaminase treatment of wheat and maize prolamins of bread increases the serum IgA reactivity of celiac disease patients. J. Agric. Food Chem. 2008, 56, 1387–1391. 10.1021/jf0724163. [DOI] [PubMed] [Google Scholar]
- Ruh T.; Ohsam J. r.; Pasternack R.; Yokoyama K.; Kumazawa Y.; Hils M. Microbial transglutaminase treatment in pasta-production does not affect the immunoreactivity of gliadin with celiac disease patients’ sera. J. Agric. Food Chem. 2014, 62, 7604–7611. 10.1021/jf501275c. [DOI] [PubMed] [Google Scholar]
- Gianfrani C.; Siciliano R. A.; Facchiano A. M.; Camarca A.; Mazzeo M. F.; Costantini S.; Salvati V. M.; Maurano F.; Mazzarella G.; Iaquinto G. Transamidation of wheat flour inhibits the response to gliadin of intestinal T cells in celiac disease. Gastroenterology 2007, 133, 780–789. 10.1053/j.gastro.2007.06.023. [DOI] [PubMed] [Google Scholar]
- Lombardi E.; Bergamo P.; Maurano F.; Bozzella G.; Luongo D.; Mazzarella G.; Aufiero V. R.; Iaquinto G.; Rossi M. Selective inhibition of the gliadin-specific, cell-mediated immune response by transamidation with microbial transglutaminase. J. Leukocyte Biol. 2013, 93, 479–488. 10.1189/jlb.0412182. [DOI] [PubMed] [Google Scholar]
- Elli L.; Roncoroni L.; Hils M.; Pasternack R.; Barisani D.; Terrani C.; Vaira V.; Ferrero S.; Bardella M. T. Immunological effects of transglutaminase-treated gluten in coeliac disease. Hum. Immunol. 2012, 73, 992–997. 10.1016/j.humimm.2012.07.318. [DOI] [PubMed] [Google Scholar]
- Mazzarella G.; Salvati V. M.; Iaquinto G.; Stefanile R.; Capobianco F.; Luongo D.; Bergamo P.; Maurano F.; Giardullo N.; Malamisura B. Reintroduction of gluten following flour transamidation in adult celiac patients: a randomized, controlled clinical study. Clin. Dev. Immunol. 2012, 2012, 329150. 10.1155/2012/329150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heredia-Sandoval N. G.; Islas-Rubio A. R.; Cabrera-Chávez F.; de la Barca A. M. C. Transamidation of gluten proteins during the bread-making process of wheat flour to produce breads with less immunoreactive gluten. Food Funct. 2014, 5, 1813–1818. 10.1039/C4FO00118D. [DOI] [PubMed] [Google Scholar]
- Ribeiro M.; Nunes F. M.; Guedes S.; Domingues P.; Silva A. M.; Carrillo J. M.; Rodriguez-Quijano M.; Branlard G.; Igrejas G. Efficient chemo-enzymatic gluten detoxification: reducing toxic epitopes for celiac patients improving functional properties. Sci. Rep. 2016, 5, 18041. 10.1038/srep18041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heil A.; Ohsam J.; Büchold C.; Pasternack R.; Yokoyama K.; Kumazawa Y.; Hils M. Microbial transglutaminase has a lower deamidation preference than human tissue transglutaminase on a celiac disease relevant wheat gliadin T-cell epitope. J. Cereal Sci. 2016, 70, 47–56. 10.1016/j.jcs.2016.05.022. [DOI] [Google Scholar]
- Gundersen M. T.; Keillor J. W.; Pelletier J. N. Microbial transglutaminase displays broad acyl-acceptor substrate specificity. Appl. Microbiol. Biotechnol. 2014, 98, 219–230. 10.1007/s00253-013-4886-x. [DOI] [PubMed] [Google Scholar]
- Ohtsuka T.; Sawa A.; Kawabata R.; Nio N.; Motoki M. Substrate specificities of microbial transglutaminase for primary amines. J. Agric. Food Chem. 2000, 48, 6230–6233. 10.1021/jf000302k. [DOI] [PubMed] [Google Scholar]
- Grossmann I.; Döring C.; Jekle M.; Becker T.; Koehler P. Compositional changes and baking performance of rye dough as affected by microbial transglutaminase and xylanase. J. Agric. Food Chem. 2016, 64, 5751–5758. 10.1021/acs.jafc.6b01545. [DOI] [PubMed] [Google Scholar]
- Zhou L.; Wu Y.; Cheng Y.; Wang J.; Lu J.; Gao J.; Yuan J.; Chen H. Blocking celiac antigenicity of the glutamine-rich gliadin 33-mer peptide by microbial transglutaminase. RSC Adv. 2017, 7, 14438–14447. 10.1039/C6RA27893K. [DOI] [Google Scholar]
- Vader L. W.; de Ru A.; van der Wal Y.; Kooy Y. M.; Benckhuijsen W.; Mearin M. L.; Drijfhout J. W.; van Veelen P.; Koning F. Specificity of tissue transglutaminase explains cereal toxicity in celiac disease. J. Exp. Med. 2002, 195, 643–649. 10.1084/jem.20012028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiring H.; Van der Heeft E.; Ten Hove G.; De Jong A. Nanoscale LC-MS (n): technical design and applications to peptide and protein analysis. J. Sep. Sci. 2002, 25, 557–568. . [DOI] [Google Scholar]
- Van De Wal Y.; Kooy Y. M.; Van Veelen P. A.; Peña S. A.; Mearin L. M.; Molberg Ø.; Lundin K. E.; Sollid L. M.; Mutis T.; Benckhuijsen W. E. Small intestinal T cells of celiac disease patients recognize a natural pepsin fragment of gliadin. Proc. Natl. Acad. Sci. U. S. A. 1998, 95, 10050–10054. 10.1073/pnas.95.17.10050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vader W.; Kooy Y.; van Veelen P.; de Ru A.; Harris D.; Benckhuijsen W.; Peña S.; Mearin L.; Drijfhout J. W.; Koning F. The gluten response in children with celiac disease is directed toward multiple gliadin and glutenin peptides. Gastroenterology 2002, 122, 1729–1737. 10.1053/gast.2002.33606. [DOI] [PubMed] [Google Scholar]
- Ho M. L.; Leu S. Z.; Hsieh J. F.; Jiang S. T. Technical approach to simplify the purification method and characterization of microbial transglutaminase produced from Streptoverticillium ladakanum. J. Food Sci. 2000, 65, 76–80. 10.1111/j.1365-2621.2000.tb15959.x. [DOI] [Google Scholar]
- Pellegrini N.; Agostoni C. Nutritional aspects of gluten-free products. J. Sci. Food Agric. 2015, 95, 2380–2385. 10.1002/jsfa.7101. [DOI] [PubMed] [Google Scholar]
- Theethira T. G.; Dennis M.; Leffler D. A. Nutritional consequences of celiac disease and the gluten-free diet. Expert Rev. Gastroenterol. Hepatol. 2014, 8, 123–129. 10.1586/17474124.2014.876360. [DOI] [PubMed] [Google Scholar]
- Gulati P.; Li A.; Holding D.; Santra D.; Zhang Y.; Rose D. J. Heating Reduces Proso Millet Protein Digestibility via Formation of Hydrophobic Aggregates. J. Agric. Food Chem. 2017, 65, 1952–1959. 10.1021/acs.jafc.6b05574. [DOI] [PubMed] [Google Scholar]
- Stamnaes J.; Fleckenstein B.; Sollid L. M. The propensity for deamidation and transamidation of peptides by transglutaminase 2 is dependent on substrate affinity and reaction conditions. Biochim. Biophys. Acta, Proteins Proteomics 2008, 1784, 1804–1811. 10.1016/j.bbapap.2008.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy I.; Smith O.; Clouthier C. M.; Keillor J. W. Expression, purification and kinetic characterisation of human tissue transglutaminase. Protein Expression Purif. 2013, 87, 41–46. 10.1016/j.pep.2012.10.002. [DOI] [PubMed] [Google Scholar]
- Heil A.; Weber J.; Büchold C.; Pasternack R.; Hils M. Differences in the inhibition of coagulation factor XIII-A from animal species revealed by Michael Acceptor–and thioimidazol based blockers. Thromb. Res. 2013, 131, e214–e222. 10.1016/j.thromres.2013.02.008. [DOI] [PubMed] [Google Scholar]
- Pérot M.; Lupi R.; Guyot S.; Delayre-Orthez C.; Gadonna-Widehem P.; Thébaudin J.-Y.; Bodinier M.; Larré C. Polyphenol Interactions Mitigate the Immunogenicity and Allergenicity of Gliadins. J. Agric. Food Chem. 2017, 65, 6442–6451. 10.1021/acs.jafc.6b05371. [DOI] [PubMed] [Google Scholar]
- Berti C.; Roncoroni L.; Falini M. L.; Caramanico R.; Dolfini E.; Bardella M. T.; Elli L.; Terrani C.; Forlani F. Celiac-related properties of chemically and enzymatically modified gluten proteins. J. Agric. Food Chem. 2007, 55, 2482–2488. 10.1021/jf062623n. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.