Abstract
Using molecular genetic techniques, we have generated and characterized six temperature sensitive (ts) alleles of nop2. All failed to support growth at 37°C and one was also formamide sensitive (fs) and failed to grow on media containing 3% formamide. Conditional lethality is not due to rapid turnover of mutant Nop2p proteins at 37°C. Each allele contains between seven and 14 amino acid substitutions and one possesses a nonsense mutation near the C-terminus. Mapping experiments with one allele, nop2-4, revealed that a subset of the amino acid substitutions conferred the ts phenotype and that these mutations have an additive effect. All six mutants exhibited dramatic reductions in levels of 60S ribosome subunits under non-permissive conditions as well as some reduction at permissive temperature. Processing of 27S pre-rRNA to mature 25S rRNA was defective in all six mutants grown under non-permissive conditions. Levels of the 40S ribosomal subunit and 18S rRNA were not significantly affected. Amino acid substitutions in nop2 conditional alleles are discussed in the context of the hypothesis that Nop2p functions both as an RNA methyltransferase and a trans-acting factor in rRNA processing and large ribosomal subunit biogenesis.
INTRODUCTION
An essential aspect of ribosome synthesis in eukaryotic cells is the processing and modification of rRNA. In yeast and other eukaryotes in general, processing and modification reactions occur post-transciptionally, but begin prior to the completion of 35S pre-rRNA synthesis, which takes place in the nucleolus. In yeast, co- and post-transcriptional processing of 35S pre-rRNA yield mature 18S, 5.8S and 25S rRNAs. Modification of rRNA by small nucleolar ribonucleoprotein complexes (snoRNPs) and other trans-acting factors takes place co-transcriptionally as initial rRNA processing and subunit assembly steps are executed. In yeast cytoplasmic small and large subunit rRNAs, 55 ribose sugars are methylated at the 2′-hydroxyl position, 44 uridines are converted to pseudouracil (Ψ) and approximately nine covalent modifications are made to nucleotide bases (1–3). Pseudouridylation and 2′-O-methylation are carried out by snoRNPs by means of a targeting mechanism entailing antisense complementarity between a small nucleolar RNA (snoRNA) and a modification site in rRNA (4). Base modifications on the other hand are not known to involve snoRNA-based targeting mechanisms and may be carried out by enzymes that directly recognize modification sites in rRNA, by analogy with tRNA base modifying gene products from various species, including Escherichia coli and yeast (5). A number of factors required for modification have also been shown to be required for pre-rRNA processing and subunit assembly (reviewed in 6). Thus, rRNA modification is linked functionally and temporally with processing and assembly steps required for ribosome synthesis.
The yeast nucleolar protein Nop2p is required for processing of 27S pre-rRNA to 5.8S and 25S rRNAs (7), both of which are incorporated into the large 60S subunit. Nop2p and the human proliferation marker P120 are likely orthologs (8). Primary structure analysis in the form of motif and database comparisons have suggested that Nop2p and P120 may function as methyltransferases (7,9), although NOP2 was not identified in a recent motif-based search of the Saccharomyces cerevisiae genome for methyltransferases that possess an S-adenosyl-methionine (SAM)-binding domain (10). We previously addressed the question as to whether or not Nop2p functions in the 2′-O-methylation of riboses in 25S rRNA at positions UmGm2922 [nucleotide numbering according to the Saccharomyces Genome Database (SGD); ribose methylation is denoted ‘Nm’, whereas base methylation is denoted ‘mN’] (7). Methylation at these nucleotides was chosen for study because they are evolutionarily conserved from yeast to human (11) and methylation at these nucleotides is known to occur late in pre-rRNA processing, at the step of conversion of 27S to 25S rRNA (12). Our previous work demonstrated that during depletion of Nop2p, at which time 27S pre-rRNA accumulates, methylation remains low at UmGm2922 (7). We concluded that methylation and processing are tightly coupled at this processing step and we were not able to determine if low methylation was a cause or a consequence of the failure to process 27S pre-rRNA (7). Considering our current understanding of the role of C/D box snoRNPs in 2′-O-methylation and the antisense complementarity between UmGm2922 and snR52 (2), it is unlikely that Nop2p methylates these sites. Thus, the function of Nop2p remains an open question. Moreover, the mechanism by which processing of 27S pre-rRNA is coordinated with ‘late’ methylation of UmGm2922 by C/D snoRNPs remains a compelling mystery.
Recent progress in characterizing an rRNA methyltransferase from E.coli and a tRNA methyltransferase from yeast suggests that Nop2p may be a 5-methyl-cytosine (m5C) methyltransferase (13). Two of the most closely related proteins by sequence alignment with Nop2p are E.coli Fmu/RsmB/RrmB and yeast Trm4p/Ncl1p (7). Fmu and Trm4p have recently been shown to be m5C-methyltransferases for bacterial large subunit rRNA and yeast tRNA, respectively (14–16). In addition, King et al. have found a limited, but intriguing region of similarity, ‘motif II’, between Nop2p and m5C DNA methyltransferases and have shown that a conserved cysteine residue in this region is required for Nop2p function (17). Consistent with the notion that Nop2p and human P120 may function as rRNA m5C methyltransferases, the only known m5C modification sites in rRNA are located in 25S rRNA at a position conserved in higher eukaryotic 28S rRNA.
For the purpose of investigating Nop2p function, we report herein the generation and characterization of six conditional nop2 alleles, certain ones of which sustained amino acid substitutions in motifs found in SAM-dependent methyltransferases. To our knowledge, this is the first demonstration that point mutations in NOP2 can adversely affect processing of pre-25S rRNAs and production of 60S ribosome subunits, and the second report of an amino acid substitution in a putative SAM-binding motif (‘motif I’) of a nucleolar protein that induces a growth phenotype. A previously described temperature sensitive (ts) allele of NOP1, nop1-3, encodes four amino acid replacements (18), one of which lies within a motif recently identified as a putative SAM-binding domain (10). One of the nop2 alleles described here, nop2-10, appears to have a more pronounced effect on large ribosome subunit assembly than on pre-rRNA processing, suggesting a role for Nop2p in ribosome assembly per se. Also, nop2-4 has been useful in the identification of novel nucleolar proteins Nop12p and Nop13p, which have been described (19).
MATERIALS AND METHODS
Microbiology and molecular biology
Saccharomyces cerevisiae strains used in this study are listed in Table 1. L4717 is an S288c derivative (C. Styles, Whitehead Institute, personal communication). Yeast was grown on complex media (YPD or YPGal), or synthetic minimal media (SD or SGal), which were supplemented with appropriate nutrients, as described (19). For plasmid shuffling, 1 mg/ml 5-fluoroorotic acid (5-FOA) was added to SD medium (21). Formamide (molecular biology grade) was added to SD medium at 3% (v/v) (22). Ethanol was added to 50°C agar media at 6% (v/v) just prior to pouring plates, and plates were used immediately after cooling to room temperature (22).
Table 1. Yeast strains and plasmids.
|
Strain |
Description |
Plasmida |
Allele |
Source |
| L4717 | MATa ade2 can1-100 his3-11,15 leu2-3, 112 trp1-1 ura3-1 | (7) | ||
| YBH3 | L4717 nop2::LEU2 | pJPA40 | NOP2 | (7) |
| YBH15 | L4717 nop2::LEU2 | pBH60 | nop2-4 | This study |
| YBH16 | L4717 nop2::LEU2 | pBH67 | nop2-5 | This study |
| YBH17 | L4717 nop2::LEU2 | pBH72 | nop2-6 | This study |
| YBH20 | L4717 nop2::LEU2 | pBH55 | nop2-3 | This study |
| YBH21 | L4717 nop2::LEU2 | pBH74 | nop2-9 | This study |
| YBH23 | L4717 nop2::LEU2 | pBH77 | nop2-10 | This study |
| YBH31 | L4717 nop2::LEU2 | pBH49 | NOP2 | This study |
| YBH36 | L4717 nop2::LEU2 | pBH61 | nop2-41 | This study |
| YBH37 | L4717 nop2::LEU2 | pBH62 | nop2-42 | This study |
| YBH38 | L4717 nop2::LEU2 | pBH63 | nop2-43 | This study |
| YBH39 | L4717 nop2::LEU2 | pBH64 | nop2-44 | This study |
| YBH40 | L4717 nop2::LEU2 | pBH65 | nop2-45 | This study |
| YBH41 | L4717 nop2::LEU2 | pBH66 | nop2-46 | This study |
| YKW7 | L4717 nop2::LEU2 | pKW7 | nop2-442 | This study |
| YKW9 | L4717 nop2::LEU2 | pKW9 | nop2-441 | This study |
aAll plasmids except pJPA40 are derivatives of pRS313 (HIS3, CEN6) (28).
Plasmids used in this study are listed in Table 1 and were amplified in E.coli strain DH5α and prepared using standard methods (23). pJPA40 consists of NOP2 in pRS316 (URA3, CEN6) (7). DNA was transformed into yeast cells using a lithium acetate method (24). Recovery of plasmid DNAs from yeast was done according to Strathern and Higgins (25). Standard methods were used for molecular cloning (23,26). Restriction enzymes were from New England Biolabs. Manual DNA sequencing was done using dideoxy chain terminators (United States Biochemical). Automated sequencing was done using the Big Dye method (Perkin-Elmer). A total of 23 oligonucleotide primers (sequences available on request) were used to sequence the entire NOP2 ORF on both strands plus variable amounts of 5′- and 3′-flanking regions.
Isolation and construction of mutant alleles
In vitro mutagenesis of NOP2 was done by low fidelity PCR using reaction conditions modified from Muhlrad et al. (27). NOP2 on a linear KpnI fragment from pJPA30 (8) was used as template for PCR with primers 16, 5′-TTCCTTCTACCGGTACC-3′ and 19A, 5′-GCCGGACGATAACGTGC-3′. Four PCRs were carried out in parallel under nominally optimal conditions (1.5 mM MgCl2, 0.25 mM each dNTP) using Taq polymerase (Promega) with the exception that each reaction contained one dNTP at a final concentration of 0.05 mM. PCR products generated under identical conditions but in the presence of 50 µM MnCl2 (27) yielded a low frequency of transformants that were resistant to 5-FOA at 25°C (data not shown), suggesting that addition of MnCl2 to PCR mixtures containing low levels of dNTPs elevated polymerase misincorporation rates to an excessively high mutagenic level. PCR products were pooled and gel-purified. Gel-purified, gapped plasmid was prepared from pBH49 digested with AatII and SnaBI. pBH49 consisted of a ClaI–BglII fragment (coordinates 509544–513717 on chromosome XIV) inserted between the ClaI and BamHI sites of pRS313 (HIS3) (28). All plasmids listed in Table 1 are derivatives of pBH49, except pJPA40, which has been described (7). Sequence homology between gapped pBH49 and PCR products was ∼380 bp at the 5′ end and ∼715 bp at the 3′ end. Multiple transformations of YBH3 were done in parallel and plated onto selective medium containing uracil, but lacking histidine, followed by replica plating with velvets onto medium containing 5-FOA. Each transformation mixture contained ∼140 ng PCR product and ∼60 ng gapped plasmid DNA. Colonies that formed in the presence of 5-FOA were replica plated onto selective medium and incubated at 14, 37 or 25°C with or without 3% formamide or 6% ethanol. Plasmid DNAs recovered from yeast cells were analyzed by restriction digestion and the NOP2 ORF was excised with EcoRI, gel-isolated, recloned into pBH49 at the EcoRI site and reintroduced into YBH3 by plasmid shuffling.
The six nop2 conditional alleles generated by PCR mutagenesis contained a total of 78 nucleotide changes. For the six alleles considered together, there was good agreement between the frequency with which a nucleotide was mutated and the frequency with which a nucleotide occurs in the NOP2 ORF (data not shown). Thus, although there are well-documented biases in base incorporation rates by Taq polymerase used under various conditions (29), the strategy of pooling four PCRs, each of which contained a reduced concentration of a single dNTP, did not introduce bias into the generation of nop2 mutants.
Site-directed mutagenesis was done using the QuickChange kit (Stratagene), with the following primer pairs: JA48, 5′-GAAAATGAGCGTACGTTGGATATGGC-3′ and JA49, 5′-GCCATATCCAACGTACGCTCATTTTC-3′ (I349T); JA47, 5′-GATATGGCAGCGGATCCAGGTGG-3′ and JA46, 5′-CCACCTGGATCCGCTGCCATATC-3′ (A355D); and JA45, 5′-GTTTTCGCTAACGACACCAATAAATCTAGAAC and JA50, 5′-GTTCTAGATTTATTGGTGTCGTTAGCGAAAAC-3′ (A378T). The entire NOP2 ORF was sequenced in plasmids containing site-directed mutations.
Immunoblotting and immunofluorescence localization
Total cellular proteins were extracted in the presence of TCA, precipitated and separated on 10.5% polyacrylamide–SDS gels, transferred to nitrocellulose and analyzed by immunoblotting using chemiluminescent detection as previously described (30). Affinity-purified polyclonal antibody #3 (APpAb3) is directed against synthetic peptide A (DEELKDEFDLEQEYD) and has been previously described (8). Monoclonal antibody D68 emerged from a hybridoma screen described previously (31). Blots to be stripped and reprobed were kept moist following chemiluminescent detection, incubated for 30 min in 50 mM NaH2PO4, pH 6.5, 2% SDS, 0.1 M β-mercaptoethanol at 50°C, washed extensively with 0.1% Tween-20 in PBS and exposed to film prior to reprobing, which was done using standard procedures.
For indirect immunofluorescence, L4717 and YBH5 cells were grown in YPD or YPG medium, fixed in 3% freshly hydrolyzed paraformaldehyde, placed on slides and probed with antibodies as described (30). Application of Cy2-conjugated anti-rabbit and Cy3-conjugated anti-mouse secondary antibodies (Jackson ImmunoResearch) and counterstaining with 4′,6-diamidino-2-phenylindole (DAPI) were done as described (30). Data were collected on a Zeiss Axiophot microscope equipped with a 100× Neofuar objective lens and a CCD-based digital camera using the single color channel mode.
Polysome and pulse–chase labeling analysis
Sucrose density gradient analysis was done as described previously (7). Pulse–chase analysis of precursor–product relationships in rRNA synthesis was done as described (30). Pulse labeling and chase incubations were done under non-permissive growth conditions. Glyoxal gels were treated with En3Hance according to the manufacturer (DuPont NEN), dried and exposed to Kodak MR film.
RESULTS
Isolation and characterization of six ts nop2 alleles
In this study, random point mutations were generated in vitro in plasmid-borne NOP2 using low fidelity PCR and an estimated 32 000 colonies were screened to determine if they were sensitive to temperature, cold (cs), formamide (fs) or ethanol (es). In the first round, 26 ts, one cs, one fs and no es candidates were isolated. Of these, seven ts and one fs candidates were demonstrated to be plasmid-borne and not attributable to chromosomal mutations. Subcloning experiments demonstrated that in five cases the ts phenotype mapped to the NOP2 ORF and that both ts and fs phenotypes manifested by one isolate mapped to the NOP2 ORF. The six nop2 mutant alleles were present on pRS313-based plasmids in strains YBH15, 16, 17, 20, 21 and 23 (Table 1).
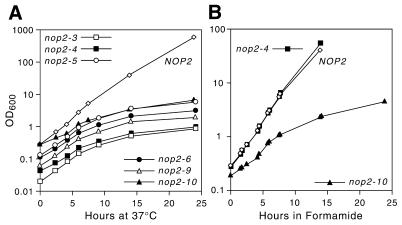
All six mutant alleles failed to grow at 37°C (Fig. 1A). Four of the six mutant alleles (nop2-3, nop2-5, nop2-6 and nop2-9) supported rates of growth at 25 and 30°C that were similar to wild-type NOP2 (Fig. 1A). Two alleles, nop2-4 and nop2-10, grew noticeably slower, even at permissive temperatures (Fig. 1A). The slowest growing allele, nop2-10, conferred sensitivity to both 37°C and 3% formamide (Fig. 1B). All six alleles could be rescued by NOP2 provided in trans (Fig. 1B and data not shown). None of the ts alleles exhibited cs growth at 14°C (data not shown). After shift to non-permissive temperature, the six alleles did not exhibit a pronounced growth defect for 2–4 doublings (4–6 h) (Fig. 2A). After ∼10 h growth under non-permissive conditions, however, growth rates were substantially diminished. Parental (L4717) and NOP2 strains (YBH3 and YBH31) did not exhibit reduced growth rates at 37°C (Fig. 2; data not shown). Shift to formamide-containing medium yielded similar growth dynamics for the strain bearing nop2-10 (Fig. 2B). After ∼10 h in medium containing formamide the growth rate slowed considerably (Fig. 2B). Prior to this, however, the growth rate was only slightly reduced, similar to the behavior of ts mutants during the initial hours after shift to 37°C (Fig. 2A).
Figure 1.
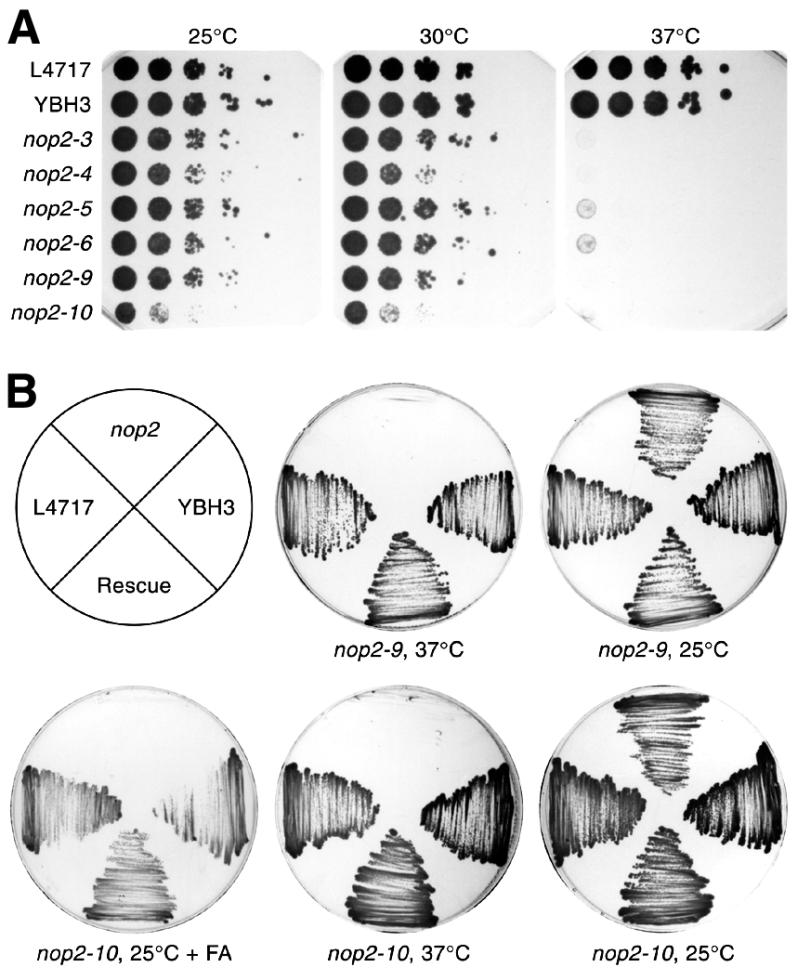
Six ts nop2 alleles. (A) Replica platings of serial dilutions of L4717 (NOP2), YBH3 (nop2::LEU2, pJPA40) and six strains bearing plasmid-borne nop2 ts alleles. Strains were grown for 3 days at the indicated temperatures on YPD medium. (B) Rescue of conditional phenotypes with pJPA40 (NOP2). YBH23 (nop2-10) is sensitive to growth on YPD containing 3% formamide (+ FA) at 25°C, as well as to 37°C. YBH21 (nop2-9) and YBH23 transformed with pJPA40 grow as well as L4717 and YBH3 under non-permissive conditions.
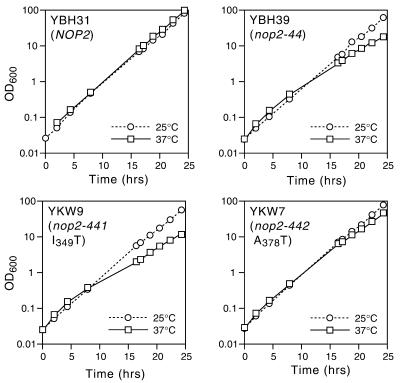
Figure 2.
(A) Growth of nop2 ts strains and strain YBH3 (NOP2) in YPD medium after shift from 25 to 37°C. (B) Growth of YBH23 (nop2-10), YBH15 (nop2-4) and YBH3 at 25°C after shift from YPD to YPD containing 3% formamide. Liquid cultures were diluted over the time course to maintain an OD600 <0.5.
In a number of experiments, we found evidence for influence of allele copy number and strain genetic background on conditional lethality. In the case of nop2-6, the ts phenotype seems to be dependent on both strain background and allele copy number. Several haploids that were derived from a cross of YBH17 (L4717 nop2::LEU2 pBH72 [nop2-6]; S288c background) with YCH128 (32; W303 background) contained nop2-6 in a ‘blended’ genetic background and failed to exhibit ts growth at 37°C (data not shown). In addition, nop2-6 present on a plasmid containing the 2 micron circle origin of replication in the original L4717 background did not exhibit ts growth (data not shown).
Temperature sensitivity is not caused by rapid turnover of mutant Nop2p proteins
To exclude the possibility that conditional phenotypes arise from rapid turnover of mutant proteins under non-permissive conditions, immunoblotting analysis of mutant versions of Nop2p was done. After 6 h at 37°C, all mutated versions of Nop2p were readily detected in whole cell extracts (Fig. 3). The affinity-purified antipeptide polyclonal antibody (APpAb3) crossreacts non-specifically with certain cellular proteins on the blot, which provides an internal control for protein loading in the immunoblotting experiment (Fig. 3 and data not shown). Although Nop2p levels were reduced compared with controls, no evidence of dramatically reduced protein half-life was observed, suggesting that mutated versions of Nop2p were relatively stable. Even after 24 h at 37°C, mutant versions of Nop2p were detected by western blotting of whole cell extracts (data not shown). It is noteworthy that immunoblotting results did not correlate with the severity of the ts phenotype. In particular, levels of Nop2-10p appeared similar to levels in control strains, despite the fact that nop2-10 confers the most severe growth phenotype. This supports the interpretation that conditional lethality at 37°C is due to alterations in protein function rather than reductions in protein stability.
Figure 3.
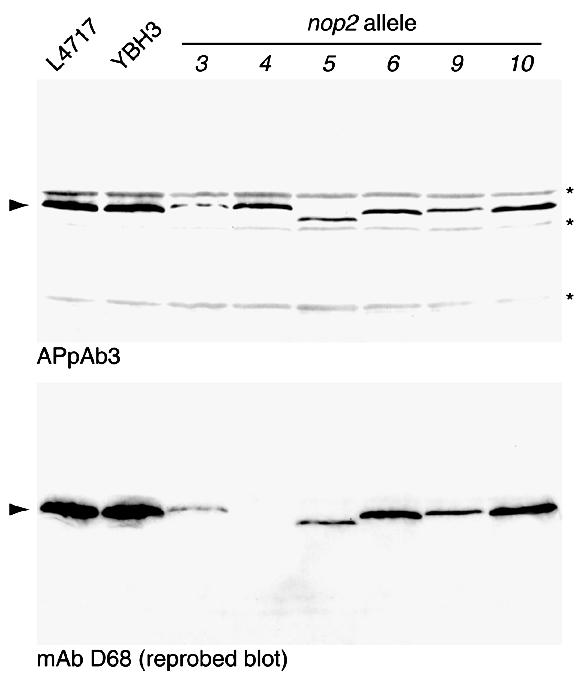
Western blot analysis. L4717, YBH3 and strains bearing nop2 ts alleles were shifted from 25 to 37°C and grown at 37°C for 6 h. Total cell protein extracts were prepared and analyzed by western blotting with an affinity-purified antipeptide polyclonal antibody directed against Nop2p (APpAb3). After stripping immunoglobulins from the membrane, it was reprobed with a monoclonal antibody (mAb D68). Nop2p (arrowhead) and non-specific protein bands (asterisks) are indicated.
Significantly, mutant versions of Nop2p migrated anomalously on SDS–polyacrylamide gels. Nop2-5p migrated faster than other mutants and controls, which is consistent with a reduction in apparent molecular mass of 1–2 kDa (Fig. 3). Sequencing of this allele revealed the existence of an amber mutation near the 3′ end of the ORF, causing a truncation of 14 C-terminal amino acids (1552 Da). Altered electrophoretic mobilities were also observed for Nop2-6p, Nop2-9p and Nop2-10p and are most likely due to physiochemical changes, such as changes in charge and/or post-translational modification of mutant proteins. In general, physiochemical alterations in mutant proteins are consistent with alterations in function and support the findings that mutations carried in the NOP2 ORF confer the mutant phenotypes.
Monoclonal antibody D68 is specific for Nop2p
During the course of these studies, we examined the effects of mutations in NOP2, under permissive and non-permissive conditions, on the distribution of various nucleolar proteins, nuclear pore complex proteins and nucleoplasmic proteins using indirect immunofluorescence (data not shown). We noticed that a previously uncharacterized monoclonal antibody (mAb) (D68) gave a nucleolar immunofluorescence localization pattern with all mutant Nop2p proteins except Nop2-4p (data not shown), suggesting that it may be directed against Nop2p. This finding was verified by immunoblotting extracts from nop2 mutant strains with mAb D68 (Fig. 3). mAb D68 specifically detects wild-type and mutant forms of Nop2p with the exception of Nop2-4p (Fig. 3). Close inspection of Figure 3 reveals that the Nop2-6p band is darker in the blot reprobed with mAb D68. This may result from the fact that nop2-6 encodes an amino acid substitution (E108G) within the peptide sequence (D102–D116) used to prepare the affinity-purified antipeptide polyclonal antibody 3 (APpAb3). Thus, the amount of Nop2-6p (and Nop2-4p, which contains the change E108V) may be underestimated by immunoblotting with APpAb3. In addition, the immunofluorescence signal generated by mAb D68 precisely colocalizes with Nop2p, as detected by APpAb3 (Fig. 4a–d). Furthermore, depletion of Nop2p in YBH5 (7) grown on glucose-containing medium for up to 12 h is accompanied by a time-dependent reduction of D68-specific nucleolar immunofluorescence signal (Fig. 4e–h). This confirms that D68 is specifically directed against the nucleolar protein Nop2p.
Figure 4.
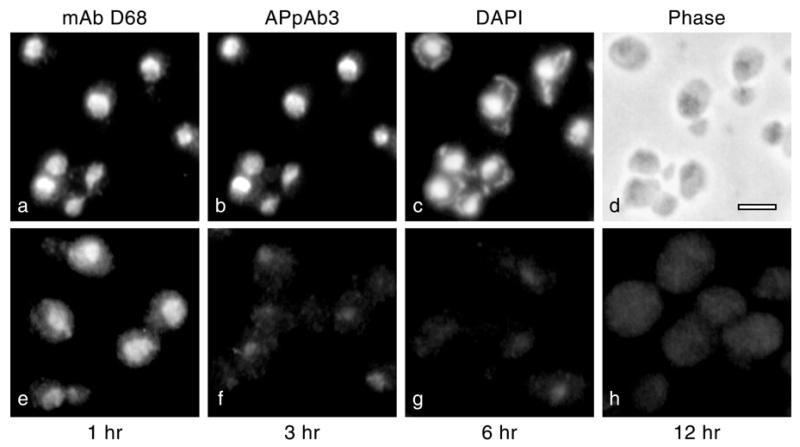
mAb D68 recognizes Nop2p. (a–d) Immunofluorescence localization in strain L4717. mAb D68 co-localizes with anti-Nop2p affinity-purified antipeptide polyclonal antibody 3 (APpAb3; 8). Counterstaining of chromatin with DAPI and a phase contrast image of the same field are shown. (e–h) Detection of Nop2p with mAb D68 in strain YBH5 (GAL::NOP2; 14). YBH5 was shifted from YPG to YPD, grown for 1, 3, 6 or 12 h and analyzed by immunofluorescence localization with mAb D68. Panels are overexposed (by 2-fold compared with a) to reveal a low level of non-specific background staining in yeast cell cytoplasm. Bar, 5 µm.
nop2 alleles are defective in ribosome synthesis
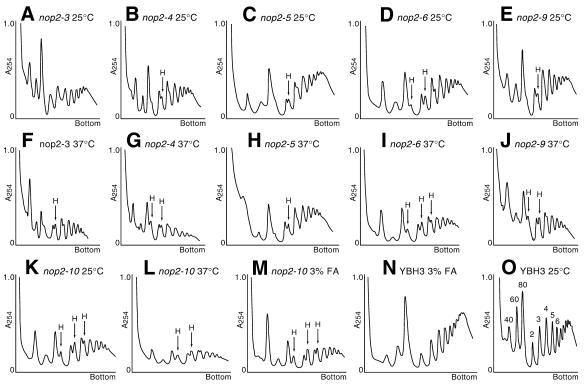
Each allele was analyzed for its effect on ribosome synthesis. The six ts strains and the control strain YBH3 (carrying plasmid-borne NOP2) were grown to mid-log phase at 25°C and half of the culture was shifted to 37°C for 2 h. Levels of ribosomal subunits, monosomes and polysomes were measured by sucrose density gradient analysis. Extracts from YBH3 grown at 25°C reveal a normal polysome profile (Fig. 5O), which does not change significantly if YBH3 is grown at 37°C (data not shown). In contrast, all of the ts strains exhibited significant changes in polysome profiles (Fig. 5). In all mutants, levels of 60S subunits, 80S monosomes and polysomes were reduced after 2 h growth at 37°C (Fig. 5F–J and L). The degree of severity of the defect varied among the six strains grown at 37°C, with the nop2-10 strain being most affected. Even at the permissive temperature (25°C), reductions in 60S subunit and 80S monosome peaks were clearly evident in all mutants, although to different extents (Fig. 5A–E and K). Concomitant with these reductions, an accumulation of 40S subunits and the appearance of ‘halfmer’ polysomes were also observed (Fig. 5A–E and K). Halfmer peaks correspond to 43S preinitiation complexes that accumulate under conditions in which the intracellular 40S:60S subunit ratio is increased. When YBH3 was incubated in medium containing 3% formamide at 25°C for 2 h, 40S, 60S, 80S and polysome peaks were reduced proportionally to a limited extent, but no halfmer polysome peaks were seen (Fig. 5N). When the nop2-10 strain was incubated in medium containing 3% formamide at 25°C, 60S subunit levels declined below levels detected in the absence of formamide. In addition, the polysome profile revealed more pronounced accumulation of 40S subunits and higher relative levels of halfmer polysomes, compared with growth at 25°C without formamide (Fig. 5L and M).
Figure 5.
Polysome analysis. Strains bearing nop2 ts alleles and YBH3 were grown at 25°C, shifted to 37°C or maintained at 25°C and grown for 2 h. YBH23 (nop2-10) was also shifted to medium containing 3% formamide (+ FA) and grown for 2 h at 25°C. Cell extracts were layered onto 15–50% sucrose gradients, centrifuged and analyzed using absorbance at 254 nm. See panel O for positions of ribosome subunits (40S and 60S), monosomes (80S) and polysomes (2–6). H, halfmer 43S preinitiation complexes are labeled.
We conclude from the polysome analyses that exposure to non-permissive conditions leads to a specific and pronounced impairment of 60S subunit synthesis in all nop2 mutant strains. This is consistent with previous findings that NOP2 is required for pre-25S rRNA processing and 60S ribosome subunit synthesis (7). The failure to produce normal levels of 60S subunits is most likely the basis for the growth defect in nop2 mutant strains under non-permissive conditions. Moreover, failure to produce normal levels of 60S subunits is most likely a direct result of altered function in mutant Nop2p isoforms given the fact that deficiencies in subunit levels are observed after only 2 h growth under non-permissive conditions, at which point growth rate and Nop2p protein levels have not changed significantly.
nop2 alleles are defective in 25S rRNA production
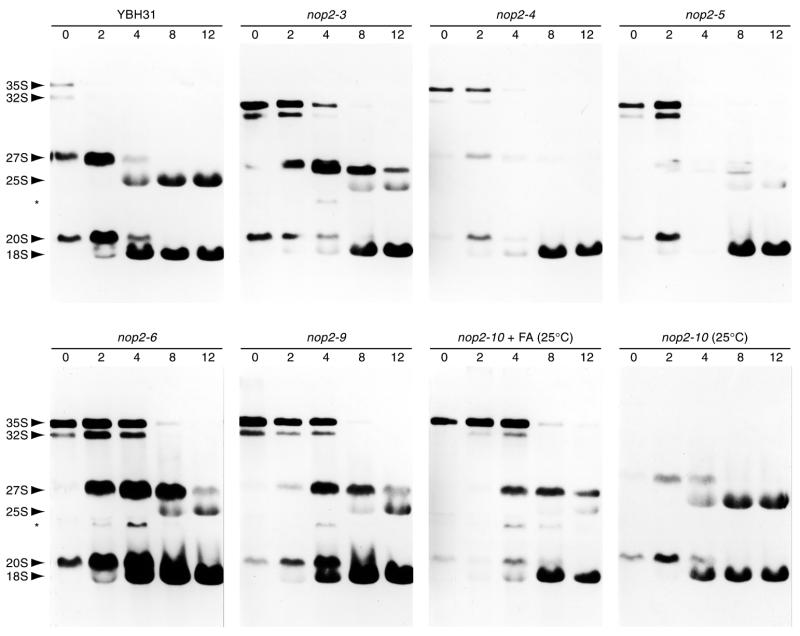
To further delineate the molecular cause underlying defects in 60S subunit synthesis, pre-rRNA processing in the nop2 mutants was evaluated by pulse–chase labeling with [3H-methyl]methionine. After 2 h growth at 37°C, all six ts strains exhibited significant accumulations of 35S and 32S precursors compared with YBH31 grown under identical conditions (Fig. 6; data not shown for nop2-10 at 37°C). At the same time, formation of 25S rRNA was significantly retarded, which is evident in low levels of 25S rRNA after 12 min of chase (Fig. 6). The pre-rRNA processing defect was severe in nop2-4 and nop2-5 strains. When grown in 3% formamide medium at 25°C, the nop2-10 strain exhibited a similar pre-rRNA processing defect (Fig. 6). However, when grown at 25°C in medium without 3% formamide, pre-rRNA processing in the nop2-10 strain appeared comparable to the control YBH31 (Fig. 6). Pulse–chase labeling of the nop2-10 strain after 2 h growth at 37°C yielded results virtually identical to those obtained with the nop2-4 strain: the 27S pre-rRNA being almost undetectable (data not shown). No accumulation of 20S pre-rRNA was observed and levels of 18S rRNA were not significantly affected in the six ts strains grown at 37°C (Fig. 6). The severe processing defect observed with nop2-4 was relieved if NOP2 was provided in trans in a ‘rescued’ strain (data not shown). To rule out an influence of the nop2-4 and nop2-5 alleles on incorporation of [3H-methyl]-label into the majority of methylated sites in 27S pre-rRNAs, pulse–chase labeling experiments were also done with [3H]uracil and virtually identical results were obtained (data not shown). Taken together, these results prove that Nop2p is required for the processing pathway leading to 25S rRNA and are consistent with previously published results from Nop2p depletion analysis (7).
Figure 6.
Pulse–chase analysis of pre-rRNA processing. Strains bearing nop2 ts alleles and YBH31 (NOP2) were grown at 25°C and shifted to 37°C for 2 h. YBH23 (nop2-10) was shifted instead to medium containing 3% formamide (+ FA) and grown for 2 h at 25°C. A 2 min labeling pulse with [3H-methyl]-methionine was followed by chase times of 2, 4, 8 or 12 min. Labeled RNAs were separated on 1% glyoxal-agarose gels and visualized by fluorography. Precursor and mature rRNAs are indicated on the left. The 23S intermediate is indicated with an asterisk.
Although all six nop2 mutants demonstrate a characteristic reduction in 25S rRNA production, subtle differences in pulse–chase labeling results are apparent. nop2-3, nop2-6, nop2-9 and nop2-10 (in formamide) strains result in a more pronounced accumulation of 27S, 32S and 35S precursors (Fig. 6). In particular, 27S pre-rRNA remains visible at 8 and 12 min of chase in these four alleles (Fig. 6). In contrast, 27S rRNAs are barely detectable in nop2-4 and nop2-5 strains after 8 and 12 min of chase (Fig. 6). These results suggest that Nop2p may function in both the stabilization and processing of 27S pre-rRNAs and that different mutants may differentially affect these two functions. The fact that the 27S processing intermediate did not accumulate in nop2-4 and nop2-5 strains suggests that one or more region(s) mutated in nop2-4 and nop2-5 may be required for the stability of 27S intermediates. In nop2-3, nop2-6, nop2-9 and nop2-10 strains, a relatively small amount of an aberrant 23S intermediate, which typically results from direct processing of the 35S primary transcript at sites A0 and A3 or B1, is also visible. The 23S intermediate is indicative of abnormal processing of pre-rRNAs and has been observed in connection with alterations in pre-25S rRNA processing in strains defective in the function of Nop4p/Nop77p and Nop8p (32–34). Finally, it is intriguing that YBH23 (nop2-10) exhibits virtually normal pre-rRNA processing in pulse–chase experiments at 25°C (Fig. 6), yet displays the slowest growth (Figs 1 and 2) and most abnormal polysome profile at 25°C (Fig. 5K). This suggests that although Nop2-10p function at 25°C may be sufficient to permit relatively normal rates of pre-rRNA processing, it is not sufficient to promote normal synthesis of the 60S large ribosomal subunit.
Sequence analysis of nop2 mutants
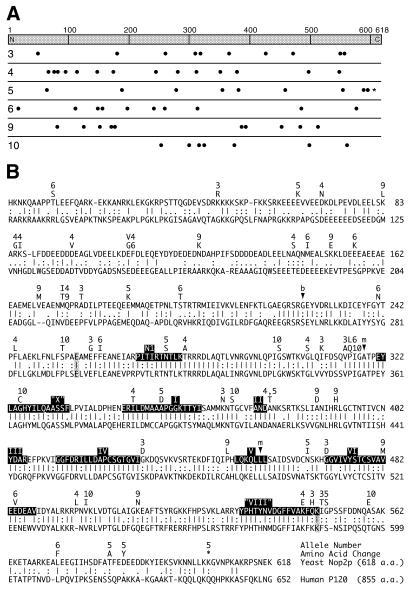
To identify changes in the primary structure of mutant Nop2p proteins, the six nop2 alleles were sequenced to identify missense and nonsense substitutions at the nucleotide level (Table 2). The number of nucleotide changes in each allele ranged from 10 in nop2-10 to 16 in nop2-4. The number of missense mutations in nop2 alleles ranged from seven (nop2-10) to 15 (nop2-4) (Tables 2 and 3). Only one nonsense mutation emerged (K605 to amber in nop2-5). In nop2-10, all mutations mapped to the C-terminal two-thirds of Nop2-10p, whereas in all other alleles missense mutations mapped across the entire protein span (Fig. 7A). Amino acid substitutions are summarized in Table 3 and Figure 7A and shown in Figure 7B in the context of conservation of sequence between Nop2p and the human ortholog P120 (Entrez Protein Record 1171747). Not shown are alignments with likely Nop2p orthologs from other species, including Mus musculus (477430), Arabidopsis thaliana (7487043), Drosophila melanogaster (7303448), Caenorhabditis elegans (7509110) and Schizosaccharomyces pombe (7492771). In general, the region of greatest conservation between members of the Nop2p/P120 protein family, based on degree of uninterrupted agreement with the consensus from a CLUSTALW alignment (data not shown), extends from E257 to K548 in Nop2p (Fig. 7B). Thus, the pairwise alignment between Nop2p and P120 shown in Figure 7B is representative of regions of conservation of amino acid sequence in the Nop2p/P120 protein family. Motif analysis and database mining (9,13) and recent progress in understanding the functions of Trm4p/Ncl1p (15) and Fmu/RsmB/RrmB (14,16) point to the likelihood that Nop2p may be an m5C-methyltransferase. We have previously noted the similarity between Nop2p, Ncl1p and Fmu (7). Ten motifs conserved among known and predicted SAM-dependent m5C-methyltransferases (13) are highlighted in Figure 7B.
Table 2. Nucleotide substitutions in nop2 ts alleles.
|
Allele |
Nucleotide changesa |
Total |
Amino acid changes |
| nop2-3 | A146G, T198A(s), G532A, A779G, T903A(s), T926A, | 13 | 10 |
| T941C, A1104T, C1203T(s), G1292A, G1409A, A1641C, T1646C | |||
| nop2-4 | G208A, A253G, A257T, A287T, A324T, G428T, G522A, | 16 | 14 |
| T730C, A838G, C840T, G916A, C1021T(s), T1046C, G1132A, C1490T, A1633G | |||
| nop2-5 | G193A, G562A, A827G, C1064A, G1132A, T1194C(s), A1397T, | 11 | 9 |
| G1648A, A1750C, G1759T, A1813T(amber) | |||
| nop2-6 | A43T, A323G, G438A, G466A, T595A, A618G(s), T721A, | 14 | 10 |
| T784A, CA945TT, C1006T(s), T1320A(s), A1451T, G1719T | |||
| nop2-9 | A87G(s), AA248TT, C351T(s), T369G, A451G, | 14 | 10 |
| T474A(s), G505A, T521C, C1163A, G1175A, C1346T, G1444A, A1530T | |||
| nop2-10 | T693C(s), A726T(s), T760A, G898A, C944A, | 10 | 7 |
| G973T, A1108T, T1293C(s), G1495A, C1676A |
aNumbering: 1 = ATG. Silent mutations are denoted (s).
Table 3. Predicted amino acid replacements.
|
Allele |
Amino acid and positiona |
|
|
|
|
|
|
|
|
|
|
|
|
|
| 3 | R | T | G | K | A | N | D | D | H | T | ||||
| WT | K49 | A178 | E260 | I309 | V314 | K368 | G431 | G470 | Q547 | I549 | ||||
| 4 | N | G | I | V | V | S | I | L | A | S | T | T | L | E |
| WT | D70 | R85 | K86 | D96 | E108 | A143 | M174 | F244 | T280 | G306 | I349 | A378 | P497 | K545 |
| 5 | K | K | S | D | T | I | S | A | Y | ter | ||||
| WT | E65 | E188 | N276 | A355 | A378 | N466 | G550 | T584 | D587 | K605 | ||||
| 6 | S | G | I | K | T | N | I | L | V | F | ||||
| WT | T15 | E108 | M146 | E156 | S199 | Y241 | F263 | P315 | E484 | L573 | ||||
| 9 | L | K | E | M | T | D | H | L | M | N | ||||
| WT | K83 | N123 | K151 | V169 | M174 | A388 | R392 | P449 | V482 | K510 | ||||
| 10 | T | S | Q | C | S | I | E | |||||||
| WT | S254 | G300 | P315 | G325 | T370 | V499 | A559 |
aAmino acid substitutions (single letter code) and a premature stop codon (ter) present in mutant alleles are shown above amino acids present in wild-type (WT) Nop2p.
Figure 7.
Amino acid substitutions in nop2 ts alleles. (A) Distribution of amino acid changes in six nop2 alleles. Nop2p is 618 amino acids in length. Amino acid substitutions are designated with a dot. A nonsense mutation in nop2-5 is denoted with an asterisk. Positions of changes are approximate. Silent mutations are not shown. (B) Allele numbers are listed above amino acid substitutions at the corresponding position in the Nop2p sequence, which is aligned with the sequence of human P120. Human P120 is 855 amino acids in length. Amino acids are numbered on the right. Four amino acids were mutated in two different alleles: E108V in nop2-4 and E108G in nop2-6; M174T in nop2-9 and M174I in nop2-4; P315L in nop2-6 and P315Q in nop2-10; and A378T in both nop2-4 and nop2-5. The nonsense mutation in nop2-5 introduced a stop codon at K605. Methyltransferase motifs N1, I-VI, ‘VIII’ and ‘X’ are shaded (31). The region of the alignment between E257 and K548 (endpoints highlighted in gray) is highly conserved among members of the Nop2p/P120 protein family. Approximate positions of BsgI and MfeI sites in the corresponding nucleotide sequence are indicated by arrowheads.
Because of the relatively high number of amino acid substitutions identified in each allele, we wished to determine which amino acids were responsible for the conditional growth phenotype and, by inference, the defect in ribosome synthesis. We considered it likely that not all amino acid replacements in a given nop2 allele would be required to elicit the conditional phenotype. To examine this issue, the nop2-4 allele was selected because it conferred a tight growth phenotype and had sustained the highest number of missense mutations, including substitutions in motifs conserved in SAM-dependent methyltransferases. To analyze nop2-4, BsgI and MfeI restriction sites were used to create alleles nop2-41 to nop2-46, which carried different combinations of mutations found in nop2-4 (Fig. 8 and Table 1). Growth of nop2-41 at 37°C makes clear that the seven mutations upstream of the BsgI site are not responsible for the ts phenotype (Fig. 8). Conversely, the seven point mutations downstream of the BsgI site in nop2-42 are responsible for the ts phenotype. These seven mutations were separated into different overlapping groups. Surprisingly, none of the groups tested conferred a complete ts phenotype. Only nop2-44 and nop2-45 conferred a partial phenotype (Fig. 8). Mutations I349T and A378T alone (nop2-44), or in combination with P497L and K545E (nop2-45), had an effect on growth at 37°C. In contrast, P497L and K545E did not cause impaired growth in combination with F244L, T280A and G306S (nop2-43). Likewise, F244L, T280A and G306S did not cause ts growth in combination with I349T and A378T (nop2-46). Clearly, I349T and A378T result in impaired growth at 37°C. But, whereas P497L and K545E exacerbate this growth defect, they have no discernible effect on growth in the context of F244L, T280A and G306S. These findings suggest that amino acid substitutions act in a combinatorial fashion to generate the ts phenotype. Clearly, however, the two substitutions in Nop2-44p (I349T and A378T) resulted in slow growth at 37°C. One of these (I349T) falls within methyltransferase motif I, which is the SAM binding domain (Fig. 7B). To determine if one or both of these mutations resulted in impaired growth at 37°C, site-directed mutagenesis was used to create individual changes I349T and A378T in alleles termed nop2-441 and nop2-442, respectively. Growth in liquid culture revealed that A378T had only minimal consequences for growth at 37°C, whereas the effect of I349T was more dramatic and primarily responsible for the growth defect in nop2-44 (Fig. 9).
Figure 8.
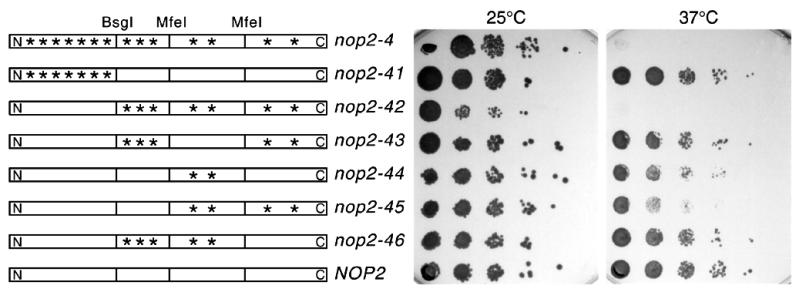
Mapping mutations in nop2-4. Restriction sites BsgI and MfeI were used to subclone regions of nop2-4 into a NOP2 plasmid to create alleles nop2-41 through nop2-46, which were introduced into a nop2::LEU2 strain (YBH3) by plasmid shuffling (Table 1). Serial dilutions of the resulting strains were replica plated onto YPD and grown at 25 or 37°C for 3 days. Asterisks represent approximate positions of amino acid substitutions in mutant alleles.
Figure 9.
Site-directed mutagenesis of NOP2. Two point mutant alleles, nop2-441 (I349T) and nop2-442 (A378T), were constructed to contain the individual amino acid substitutions found in nop2-44 (Fig. 8). Mutant and control NOP2 strains were grown in YPD at 25°C and either maintained at 25°C or shifted to 37°C. Cell densities (OD600) were measured at different times (in h) and liquid cultures were diluted during the time course to maintain an OD600 <0.5.
DISCUSSION
PCR-based mutagenesis of NOP2 yielded six mutants exhibiting ts growth at 37°C, one of which grew slowly and was fs, at 25°C. These mutants contained between 10 and 16 point mutations, only one of which was a nonsense mutation. The conditional lethal phenotypes observed were not due to rapid turnover of mutant proteins, which was shown through the use of a newly-described monoclonal antibody directed against Nop2p. Thus, impairment of a required function for Nop2p underlies the conditional lethality in nop2 mutant strains grown under non-permissive conditions. This is the first demonstration that point mutations in NOP2 can adversely affect processing of pre-25S rRNAs and production of 60S ribosome subunits.
All six mutant alleles showed very similar molecular defects after only 2 h growth at non-permissive conditions, when growth rate and Nop2p protein levels were not significantly diminished. This indicates that Nop2p plays a direct, rather than indirect, role in pre-25S rRNA processing and 60S subunit production. Mutations in nop2-4, the allele with the greatest number of amino acid substitutions (14), were subdivided by subcloning experiments in order to identify those mutations responsible for the growth phenotype. The seven N-terminal amino acid substitutions in Nop2-4p were neither necessary nor sufficient for ts growth. It may be generally true of the six nop2 alleles that many of the missense mutations do not contribute to the conditional lethal phenotype. Frameshift mutations were not detected in our screen. In contrast, PCR mutagenesis of SNM1, which encodes an essential 22.5 kDa subunit of RNase MRP, yielded a number of frameshift and nonsense mutations, in addition to missense mutations (36).
One of the most intriguing findings that emerged from our studies concerns results obtained with ts strains grown under permissive conditions (25°C). In all six mutant strains, production of 60S subunits was impaired at 25°C and became more so at 37°C. However, pulse–chase analysis plainly showed that production of 25S rRNA at 25°C was similar to wild type. This was most evident with YBH23 (nop2-10), which displayed the slowest growth and most abnormal polysome profile at 25°C, yet exhibited virtually normal pre-rRNA processing in pulse–chase experiments conducted at 25°C. One interpretation of these results is that 60S subunit production is exquisitely sensitive to changes in pre-25S rRNA processing. This may be due to transcriptional repression of ribosomal protein genes during heat shock (37,38), which may exacerbate the effects of a large subunit rRNA processing defect by interrupting the supply of ribosomal proteins during early subunit assembly steps that take place during rRNA processing reactions. An alternative interpretation is that mutations that affect a function of Nop2p required for 60S synthesis per se may be better tolerated than mutations that affect a Nop2p function required for 27S pre-rRNA processing per se. This suggests that Nop2p may perform more than one function required for pre-rRNA processing and large subunit assembly and that sensitivity to formamide may be a useful experimental tool for uncoupling these functions.
No evidence for essential functions unrelated to production of large ribosomal subunits emerged from our studies. In this sense, Nop2p is unlike Dim1p and Snm1p, which are also nucleolar proteins involved in pre-rRNA processing. Analysis of different ts alleles of DIM1 revealed that Dim1p carries out an essential quality control function in addition to its activity as an 18S rRNA diadenosine dimethylase (39). Certain conditional alleles of SNM1 result in plasmid missegregation, suggesting a role for Snm1p in cell cycle regulation during exit from mitosis (36).
We also tested for suppression of the nop2 ts phenotype by expression of pre-rRNA from a Pol II promoter. The rationale for this was two-fold: (i) in the course of conducting genetic screens for high copy suppressors of different nop2 ts alleles, we identified a plasmid carrying a portion of the rDNA repeat (data not shown); and (ii) LaFontaine et al. (39) reported that expression of rDNA from the PGK promoter (but not GAL10 promoter) suppressed pre-rRNA processing defects in dim1-1 and dim1-9 mutants. We found that expression from a GAL7::rDNA plasmid (40) did not suppress any of the nop2 ts alleles (data not shown). In contrast, allele-specific, transcription-dependent synthetic lethal interactions were observed between the GAL7::rDNA plasmid and the nop2-4 and nop2-6 alleles at 25°C (data not shown). This indicates the existence of a functional difference between Pol I-transcribed pre-rRNA and Pol II-transcribed pre-rRNA insofar as the function of Nop2p is concerned.
Interestingly, two mutations in nop2-4 (I349T and A378T) were found to yield a partial ts phenotype. This partial ts phenotype was augmented by combination with P497L and K545E, but ameliorated by combination with F244L, T280A and G306S. Thus, at least in nop2-4, a single amino acid change does not appear to block Nop2p function at 37°C. Changes encoded by nop2-4 (I349T and A378T) mapped to a region of Nop2p that is highly conserved in the human ortholog P120. This conserved region contains signature motifs found in a number of different classes of SAM-dependent methyltransferase enzymes (9,13). I349T lies in motif I (SAM-binding motif) and A378T is adjacent to motif II. Site-directed mutagenesis demonstrated that I349T was responsible for the partial ts phenotype. Another change, A355D in nop2-5, also fell within motif I. Site-directed mutagenesis demonstrated that this mutation in nop2-5 was responsible for a partial ts phenotype as well (data not shown). To our knowledge, this is the second report of a substitution in the SAM-binding motif of a nucleolar protein that induces a growth phenotype. The previously described ts allele nop1-3 results in four amino acid replacements (18), one of which (A175V) lies within a motif (VLYLGAASG178) recently identified as a putative SAM-binding domain in Nop1p (10). Random PCR mutagenesis of DIM1 yielded 10 conditional alleles with between one and six amino acid replacements, none of which was present in the SAM-binding motif (41). Significantly, the nop1-3 allele results in strong inhibition of nucleolar methylation of pre-rRNA (18). Alignment of putative SAM-binding motifs from Nop1p and Nop2p indicates that the position in motif I of A355D encoded by nop2-5 lies immediately adjacent to the position in motif I of A175V encoded by nop1-3. We note, however, that putative SAM-binding motif I in Nop2p (ILDMAAAPGGKTTYIS) and Nop1p (VLYLGAASGTTVSHVS) are not highly similar, which reflects the fact that a single consensus sequence has not been defined for the SAM-binding domain, which is found in methyltransferases exhibiting a broad range of substrate specificities.
A number of other amino acid substitutions in different nop2 ts alleles mapped to motifs present in methyltransferases. Figure 7 shows motifs conserved among members of the methyltransferase family that are most similar to Nop2p (13). Of these, motif IV is the second longest, yet did not sustain an amino acid replacement in any of the six nop2 ts alleles. This may be due to the fact that substitutions in this motif are tolerated bimodally; either relatively well or not at all. Six site-directed mutations made by King et al. (17) at conserved positions within this motif were tolerated well or not at all. Substitution of C424 for alanine or serine eliminated Nop2p function and the change D416A resulted in slower growth at 38°C, but only by a small extent. Two other site-directed mutations within seven residues of motif IV on its C-terminal side did not affect growth (17).
SAM-dependent methyltransferases modify numerous types of molecules in biological systems, including nucleic acids, proteins, lipids and small molecules, and can be classified on the basis of substrate type and common primary structure motifs. In this context, it is striking that Nop2p exhibits a high degree of similarity with a specific class of RNA methyltransferase from S.cerevisiae (Trm1p) and E.coli (Fmu) that methylate specific cytosines at ring position five. This suggests that Nop2p may function as an m5C methyltransferase. Consistent with this notion for Nop2p function, only one or two m5C modification(s) are known to be present in mature rRNA. Small subunit rRNA, in either yeast or human, does not contain m5C (12,42,43). On the other hand, large subunit rRNAs from yeast and human contain 1–2 m5C modifications. Quantitation and mapping of m5C using radiolabeling and RNA fingerprint analysis by Planta and colleagues (12,42,43) is consistent with either one or two moles of m5C per 25S rRNA, as has been reviewed by Maden (3,11). Only one m5C has been unequivocally mapped in 25S rRNA (43). This m5C is found in 25S rRNA at position 2278 in yeast (nucleotide numbering according to SGD), which is equivalent to position 3751 in human (3,11). The fact that this site is conserved in yeast and human is consistent with the notion that functional counterparts exist in yeast (Nop2p) and human (P120). The conditional alleles of nop2 described herein should be extremely useful in testing the hypothesis that Nop2p is an RNA methyltransferase and in revealing its role in rRNA processing and large ribosomal subunit biogenesis.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Ms Dana Sacco and Ms Catherine Avery-Jones for conducting screens for high copy suppressors of nop2 ts alleles and attending to the analysis of suppressor candidates. Dr Andrea Hofig assisted with clonings at an early stage of this work. This work was supported in part by NIH grant GM48586 to J.P.A.
References
- 1.Ofengand J. and Fournier,M.J. (1998) The pseudouridine residues of rRNA: number, location, biosynthesis and function. In Grosjean,H. and Benne,R. (eds), Modification and Editing of RNA. ASM Press, Washington, DC, pp. 229–253.
- 2.Lowe T.M. and Eddy,S.R. (1999) A computational screen for methylation guide snoRNAs in yeast. Science, 283, 1168–1171. [DOI] [PubMed] [Google Scholar]
- 3.Maden B.E.H. (1998) Intracellular locations of RNA-modifying enzymes. In Grosjean,H. and Benne,R. (eds), Modification and Editing of RNA. ASM Press, Washington, DC, pp. 421–440.
- 4.Tollervey D. and Kiss,T. (1997) Function and synthesis of small nucleolar RNAs. Curr. Opin. Cell Biol., 9, 337–342. [DOI] [PubMed] [Google Scholar]
- 5.Grosjean H., Motorin,Y. and Morin,A. (1998) RNA-modifying and RNA-editing enzymes: methods for their identification. In Grosjean,H. and Benne,R. (eds), Modification and Editing of RNA. ASM Press, Washington, DC, pp. 21–46.
- 6.Kressler D., Linder,P. and de La Cruz,J. (1999) Protein trans-acting factors involved in ribosome biogenesis in Saccharomyces cerevisiae. Mol. Cell. Biol., 19, 7897–7912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hong B., Brockenbrough,J.S., Wu,P. and Aris,J.P. (1997) Nop2p is required for pre-rRNA processing and 60S ribosome subunit synthesis in yeast. Mol. Cell. Biol., 17, 378–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Beus E., Brockenbrough,J.S., Hong,B. and Aris,J.P. (1994) Yeast NOP2 encodes an essential nucleolar protein with homology to a human proliferation marker. J. Cell Biol., 127, 1799–1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koonin E.V. (1994) Prediction of an rRNA methyltransferase domain in human tumor-specific nucleolar protein p120. Nucleic Acids Res., 22, 2476–2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Niewmierzycka A. and Clarke,S. (1999) S-Adenosylmethionine-dependent methylation in Saccharomyces cerevisiae. Identification of a novel protein arginine methyltransferase. J. Biol. Chem., 274, 814–824. [DOI] [PubMed] [Google Scholar]
- 11.Maden B.E.H. (1990) The numerous modified nucleosides in eukaryotic ribosomal RNA. Prog. Nucleic Acid Res. Mol. Biol., 39, 241–303. [DOI] [PubMed] [Google Scholar]
- 12.Brand R.C., Klootwijk,J., Van Steenbergen,T.J., De Kok,A.J. and Planta,R.J. (1977) Secondary methylation of yeast ribosomal precursor RNA. Eur. J. Biochem., 75, 311–318. [DOI] [PubMed] [Google Scholar]
- 13.Reid R., Greene,P.J. and Santi,D.V. (1999) Exposition of a family of RNA m(5)C methyltransferases from searching genomic and proteomic sequences. Nucleic Acids Res., 27, 3138–3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tscherne J.S., Nurse,K., Popienick,P., Michel,H., Sochacki,M. and Ofengand,J. (1999) Purification, cloning and characterization of the 16S RNA m5C967 methyltransferase from Escherichia coli. Biochemistry, 38, 1884–1892. [DOI] [PubMed] [Google Scholar]
- 15.Motorin Y. and Grosjean,H. (1999) Multisite-specific tRNA:m5C-methyltransferase (Trm4) in yeast Saccharomyces cerevisiae: identification of the gene and substrate specificity of the enzyme. RNA, 5, 1105–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gu X.R., Gustafsson,C., Ku,J., Yu,M. and Santi,D.V. (1999) Identification of the 16S rRNA m5C967 methyltransferase from Escherichia coli. Biochemistry, 38, 4053–4057. [DOI] [PubMed] [Google Scholar]
- 17.King M., Ton,D. and Redman,K.L. (1999) A conserved motif in the yeast nucleolar protein Nop2p contains an essential cysteine residue. Biochem. J., 337, 29–35. [PMC free article] [PubMed] [Google Scholar]
- 18.Tollervey D., Lehtonen,H., Jansen,R., Kern,H. and Hurt,E.C. (1993) Temperature-sensitive mutations demonstrate roles for yeast fibrillarin in pre-rRNA processing, pre-rRNA methylation and ribosome assembly. Cell, 72, 443–457. [DOI] [PubMed] [Google Scholar]
- 19.Wu, K.,Wu,P. and Aris,J.P. (2001) Nucleolar protein Nop12p participates in synthesis of rRNA in Saccharomyces cerevisiae.Nucleic Acids Res., 29, 2938–2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sherman F. (1991) Getting started with yeast. Methods Enzymol., 194, 3–21. [DOI] [PubMed] [Google Scholar]
- 21.Boeke J.D., Truehart,J., Natsoulis,G. and Fink,G.R. (1987) 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol., 154, 164–175. [DOI] [PubMed] [Google Scholar]
- 22.Aguilera A. (1994) Formamide sensitivity: a novel conditional phenotype in yeast. Genetics, 136, 87–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ausubel F.A., Brent,R., Kingston,R.E., Moore,D.D., Seidman,J.G., Smith,J.A. and Struhl,K. (eds) (2000) Current Protocols in Molecular Biology. Greene Publishing and Wiley-Interscience, New York, NY.
- 24.Gietz R.D., Schiestl,R.H., Willems,A.R. and Woods,R.A. (1995) Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast, 11, 355–360. [DOI] [PubMed] [Google Scholar]
- 25.Strathern J.N. and Higgins,D.R. (1991) Recovery of plasmids from yeast into Escherichia coli: shuttle vectors. Methods Enzymol., 194, 319–329. [DOI] [PubMed] [Google Scholar]
- 26.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 27.Muhlrad D., Hunter,R. and Parker,R. (1992) A rapid method for localized mutagenesis of yeast genes. Yeast, 8, 79–82. [DOI] [PubMed] [Google Scholar]
- 28.Sikorski R.S. and Hieter,P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics, 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Y., Mitaxov,V. and Waksman,G. (1999) Structure-based design of Taq DNA polymerases with improved properties of dideoxynucleotide incorporation. Proc. Natl Acad. Sci. USA, 96, 9491–9496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu P., Brockenbrough,J.S., Metcalfe,A.C., Chen,S. and Aris,J.P. (1998) Nop5p is a small nucleolar ribonucleoprotein component required for pre-18 S rRNA processing in yeast. J. Biol. Chem., 273, 16453–16463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aris J.P. and Blobel,G. (1988) Identification and characterization of a yeast nucleolar protein that is similar to a rat liver nucleolar protein. J. Cell Biol., 107, 17–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hardy C.F. (1996) Characterization of an essential Orc2p-associated factor that plays a role in DNA replication. Mol. Cell. Biol., 16, 1832–1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zanchin N.I. and Goldfarb,D.S. (1999) Nip7p interacts with Nop8p, an essential nucleolar protein required for 60S ribosome biogenesis and the exosome subunit Rrp43p. Mol. Cell. Biol., 19, 1518–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun C. and Woolford,J.L. (1994) The yeast NOP4 gene product is an essential nucleolar protein required for pre-rRNA processing and accumulation of 60S ribosomal subunits. EMBO J., 13, 3127–3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Berges T., Petfalski,E., Tollervey,D. and Hurt,E.C. (1994) Synthetic lethality with fibrillarin identifies NOP77p, a nucleolar protein required for pre-rRNA processing and modification. EMBO J., 13, 3136–3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cai T., Reilly,T.R., Cerio,M. and Schmitt,M.E. (1999) Mutagenesis of SNM1, which encodes a protein component of the yeast RNase MRP, reveals a role for this ribonucleoprotein endoribonuclease in plasmid segregation. Mol. Cell. Biol., 19, 7857–7869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Herruer M.H., Mager,W.H., Raue,H.A., Vreken,P., Wilms,E. and Planta,R.J. (1988) Mild temperature shock affects transcription of yeast ribosomal protein genes as well as the stability of their mRNAs. Nucleic Acids Res., 16, 7917–7929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim C.H. and Warner,J.R. (1983) Mild temperature shock alters the transcription of a discrete class of Saccharomyces cerevisiae genes. Mol. Cell. Biol., 3, 457–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lafontaine D.L., Preiss,T. and Tollervey,D. (1998) Yeast 18S rRNA dimethylase Dim1p: a quality control mechanism in ribosome synthesis? Mol. Cell. Biol., 18, 2360–2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oakes M., Aris,J.P., Brockenbrough,J.S., Wai,H., Vu,L. and Nomura,M. (1998) Mutational analysis of the structure and localization of the nucleolus in the yeast Saccharomyces cerevisiae. J. Cell Biol., 143, 23–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lafontaine D., Vandenhaute,J. and Tollervey,D. (1995) The 18S rRNA dimethylase Dim1p is required for pre-ribosomal RNA processing in yeast. Genes Dev., 9, 2470–2481. [DOI] [PubMed] [Google Scholar]
- 42.Klootwijk J. and Planta,R.J. (1973) Analysis of the methylation sites in yeast ribosomal RNA. Eur. J. Biochem., 39, 325–333. [DOI] [PubMed] [Google Scholar]
- 43.Veldman G.M., Klootwijk,J., de Regt,V.C., Planta,R.J., Branlant,C., Krol,A. and Ebel,J.P. (1981) The primary and secondary structure of yeast 26S rRNA. Nucleic Acids Res., 9, 6935–6952. [DOI] [PMC free article] [PubMed] [Google Scholar]